Abstract
Objective
To measure the transcript levels of Aurora kinases and compare them to their immunoreactivity patterns in prostate tumors.
Materials and Methods
A total of 26 cases of prostate cancer (PCa) and 38 cases of benign nodular hyperplasia (BPH) were sampled from archived formalin-fixed paraffin-embedded tissues. Tissue sections were lysed, total RNA extracted and cDNA made by random hexamer priming while slide sections were immunostained for the kinases. Normalized relative quantitation was performed for all the kinases using real-time PCR on TaqMan chemistry.
Results
The immunoreactivity profile showed 15.4, 53.8 and 30.7% positivity for Aurora kinases A, B and C in PCa cases, respectively, while the positivity was 76.3, 73.7 and 84.2% for the same kinases in BPH cases. The immunoreactivity was preponderant on epithelial tissue compared to stromal component.
Conclusion
Aurora kinases were significantly overexpressed in BPH cases compared to PCa cases. At the transcript level, there was no significant differential expression in the kinases between PCa and BPH cases.
Key Words: Aurora kinases, Aurora kinase A, Aurora kinase B, Aurora kinase C, Gene expression, Prostate cancer
Introduction
Three Aurora family members (A, B and C) of highly conserved serine/threonine kinases regulate cell entry into mitosis, centrosome maturation and the mitotic spindle checkpoint [1]. Cell cycle checkpoints have evolved to ensure the inheritance of undamaged DNA, and that each daughter cell receives the correct complement of chromosomes during mitosis [2]. Aberrant expression of the Aurora kinases can induce abnormal mitosis, centrosome dysfunction and chromosomal instability, which may lead to aneuploidy and neoplastic transformation [3]. Increased expression of Aurora kinase A (AURKA) and Aurora kinase B (AURKB) has been reported in prostatic intraepithelial neoplasms, a precursor to most prostate cancers (PCa), compared to normal prostate cells [4]. Novel therapeutic agents that target Aurora kinases may therefore be used to augment mitotic inhibitors like the taxanes (docetaxel and paclitaxel) in the management of PCa.
AURKA is localized on chromosome 20q13.2, and has 14 transcript variants with exons ranging from 5 to 10, transcript lengths of 584–2,238 bp, and encoding protein isoforms of 9–46 kDa (79–403 amino acid residues; http://genome.ucsc.edu/ and http://www.ensembl.org/index.html). Pseudogenes of AURKA have also been found on chromosomes 1 and 10 (www.pseudogenes.com). AURKA is involved in centrosome separation, microtubule formation and stabilization at the spindle during mitosis [5]. Overexpression of AURKA is reported to confer resistance to the taxane paclitaxel, a mitotic inhibitor, which interferes with normal breakdown of microtubules during mitosis [2]. AURKA has therefore been suggested as a predictive marker for taxane resistance [2]. It is generally believed that AURKA may play a role in tumor development and progression because of its critical role in mitotic entry as well as mitotic spindle checkpoint [6]. Two proteins known to bind and initiate activation of AURKA are TPX2 and Ajuba [2]. Quet al. [7] had reported that DNAzyme (DNA molecules with enzymatic cleavage activity) could be used to target AURKA mRNA in PCa cells as a potential therapeutic agent.
AURKB is localized on chromosome 17p13.1 and has one transcript variant with 9 exons, and a transcript length of 1,245 bp encoding a 39-kDa protein of 344 amino acid residues (http://www.ensembl.org/index.html). AURKB is a chromosomal passenger protein that begins mitosis localized to the centromeres but at the onset of anaphase relocates to the spindle equator [8]. AURKB localizes to microtubules near kinetochores, specifically to the specialized microtubules called K-fibers, and it is known to regulate processes such as kinetochore and microtubule interactions, segregation of sister chromatids and cytokinesis [2]. Immunohistochemical studies on prostate tissues and cell lines have demonstrated increased nuclear expression of AURKB in high Gleason score PCa compared to intermediate and low grades, as well as in all cancers compared to benign and normal glands [4].
AURKC is localized on chromosome 19q13.43 and has 3 transcript variants of 7 exons each, and variable transcript lengths (1,108–1,249 bp) encoding protein isoforms of variable molecular masses – 32–36 kDa containing 275–309 amino acid residues. AURKC is expressed specifically in the male testis and has meiotic functions [2].
Our study was designed to measure the quantity of transcripts (mRNA) of AURKA, AURKB and AURKC normalized to ABL1 housekeeping gene and to compare the transcript level with the immunoreactivity profile of formalin-fixed paraffin-embedded (FFPE) prostate tissues from PCa and benign cases. The study was undertaken because understanding gene expression of Aurora kinases in prostate tissues at both transcript and protein levels may help to define overexpression for a better therapeutic approach.
Materials and Methods
Ethical approval was obtained from the Meena Histopathology and Cytology Laboratory, Jos, and the University of Port Harcourt Teaching Hospital, Port Harcourt, Rivers State, Nigeria.
Samples
A total of 26 previously diagnosed PCa cases and 38 benign nodular hyperplasia (BPH) cases were sampled from archived FFPE prostate tissues in 2010 records of the University of Port Harcourt Teaching Hospital, Rivers State (21 PCa and 31 BPH cases) and the Meena Histopathology and Cytology Laboratory (5 PCa and 7 BPH cases), Jos, Nigeria. Each FFPE tissue material was sectioned and macrodissected to harvest tissue areas with the highest lesion. Three pieces of 5-µm-thick sections were aseptically collected into 2-ml Eppendorf tubes for total RNA extraction, and 3-µm-thick sections were used for immunohistochemistry (IHC). The 2-ml tubes were briefly centrifuged, deparaffinized in two washes of 1 ml xylene for 10 min each, the xylene decanted and the tissue rehydrated by two washes in 1 ml of 100% ethanol before allowing the pellets to dry for 5 min on a heat block kept at 37°C. The tissue pellets were digested overnight in 540 µl of all tissue lysis buffer (ATL) and 60 µl of proteinase K (Qiagen, UK). The digest was centrifuged for 5 min and the supernatant (containing nucleic acids) was collected.
RNA Extraction and cDNA Synthesis
For all samples, 350 µl of the tissue lysate (supernatant) was used for RNA extraction with the spin column RNeasy Mini kit (Qiagen, UK). The elution volume was 60 µl (50 µl of which was used for cDNA synthesis). RNA quality was checked using the BioRad automated electrophoresis system for 18S/28S ratio and the Eppendorf Biophotometer Plus for A260/280 ratio. The amount of RNA used for cDNA synthesis ranged from 0.5 to 2.0 µg. An equal volume of RNA elution was used in cDNA synthesis. Random primers (at a final concentration of 300 ng per reaction) from Invitrogen, UK, were used for cDNA synthesis with the Moloney murine leukemia virus reverse transcriptase and RNasin (RNase inhibitor; Promega, UK). The cDNA synthesis was done immediately after RNA extraction.
Antibodies and IHC
Monoclonal antibodies for AURKA and AURKB were purchased from ABCAM, UK, and rabbit polyclonal anti-AURKC. The mouse- and rabbit-specific HRP/DAB detection IHC kit was purchased from ABCAM, UK for immunostaining. Briefly, the slides were deparaffinized, rehydrated and microwaved for 10 min with citrate buffer pH 6.0 for antigen retrieval. They were immersed in hydrogen peroxide for 10 min before protein block for the same duration and then incubated in primary antibodies for 40 min in a humid chamber. Detection of immunoreactivity was performed according to the manufacturer's instruction for the mouse- and rabbit-specific HRP/DAB system. Slides were immersed in xylene and mounted in DPX mountant and viewed at ×400 magnification. The immunohistochemical staining was semi-quantitatively scored based on percentage of cells (area) that stained positive and the intensity of the staining (strong, moderate, weak). A score of 5+ was assigned to 80% or more of epithelial and/or stromal cells that stained positive with strong intensity, 4+ was assigned to 50% or more (but less than 80%) of epithelial cells and/or stromal cells with strong intensity or 80% of cells or more with moderate to weak intensity; 3+ was assigned to 30% or more of epithelial and/or stromal cells with strong intensity or 50% or more (but less than 80%) of positive cells with moderate to weak intensity; 2+ was assigned to 10% or more cells that stained positive with strong intensity or 30% or more (but less than 50%) that stained moderate to weak; 1+ was assigned to 10% or more cells (less than 30% of positive cells) that stained positive with moderate to weak intensity; 0 was assigned to less than 10% of positivity irrespective of the intensity of staining.
Real-Time PCR Set-Up
TaqMan chemistries were used in the Applied Biosystem's One Step Plus Real Time PCR system (Applied Biosystems, UK). Commercially predesigned ×20 TaqMan primer mixes for AURKA, AURKB and AURKC based on TaqMan minor grove binding chemistry and TaqMan ×2 universal master mix were purchased from Applied Biosystems, UK. The assay IDs for AURKA, AURKB and AURKC were Hs0182073_m1, Hs00177782_m1 and Hs00152930_m1, respectively. The sequences of primers and probes for ABL1 endogenous control gene have previously been described [9] and are available on request. For absolute quantification of the ABL1 gene, in-house plasmid calibrators were used to construct standard curves for reading samples. The transcripts of AURKA, AURKB, and AURKC were measured by normalized relative quantitation using the formula NRQ = ECt AURKA/ECt ABL1 where E is the PCR efficiency for each target, and Ct is the cycle of threshold, also known as Cq (cycle of quantitation), and refers to the PCR cycle at which the amplification of the target gene crosses a threshold line [9,10]. The Ct value is inversely proportional to the log of the starting quantity of the target gene. PCR efficiency is related to the slope of the calibration curve. For all the Aurora kinases, in-house cDNA of a PNTIA prostate cell line was used in creating the calibration curve for the purposes of determining the slope and PCR efficiency. The predesigned primer mixes for AURKA, AURKB and AURKC were used at a final concentration of ×1 in a 20-µl reaction volume while the ABL1 primers were used at a concentration of 300 nm and the probe at a concentration of 200 nm. The cDNA was used at 3 µl for each reaction and all samples were measured in triplicates. The thermal profile for all the reactions was as follows: 50°C for 2 min, initial denaturation at 95°C for 10 min; cycle denaturation of 95°C for 15 s, annealing and extension at 60°C for 1 min for 40 cycles. Data were acquired at all stages.
Statistical Analysis
All statistical analyses were done on GraphPad Prism version 5.02 (www.graphpad.com).
Results
For the IHC staining, only 4 (15.4%) and 14 (53.8%) cases stained positive for AURKA and AURKB, respectively, in PCa tissues. AURKA and AURKB were preponderant in the epithelial not stromal cells. In the same pattern of staining, only 8 (30.7%) cases were positive for AURKC in PCa tissues (fig. 1). The median IHC scores for AURKA, AURKB and AURKC were 3+, 4+ and 3+ in the PCa cases.
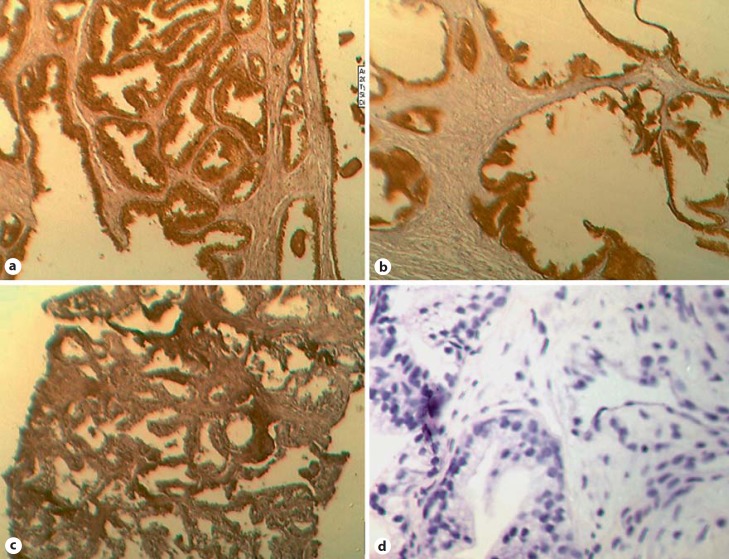
Fig. 1.
Photomicrographs of immunoreactivity of Aurora kinases and frequency. a AURKA-positive BPH (dark-brown stain), ×400 magnification. Epithelial cells are predominantly stained. The IHC score was 5+. b AURKB-positive BPH (dark-brown stain), ×400 magnification. The IHC score was 4+. c AURKC-positive PCa (dark-brown stain), ×400 magnification. The IHC score was 3+. d Negative control slide (blue nucleic stain), ×400 magnification. The IHC score was 0. Scale: 1 cm = 18 µM in ×400 magnification.
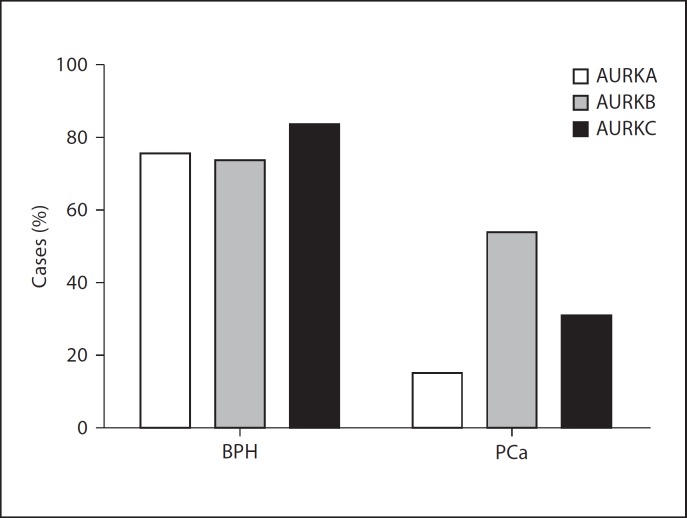
In contrast, 29 (76.3%) cases stained positive for AURKA in BPH tissues and were predominantly stained in epithelial compared to stromal cells. In the same pattern of staining, 28 (73.7%) cases and 32 (84.2%) cases of BPH tissues were immunopositive for AURKB and AURKC, respectively (fig. 1). The median IHC scores for AURKA, AURKB and AURKC were 4+, 5+ and 3+ in BPH cases. The χ2 analysis showed that Aurora kinases were expressed more in BPH than PCa cases (fig. 2).
Fig. 2.
Frequency distribution of immunoreactivity pattern of Aurora kinases. The Aurora kinases were significantly overexpressed in BPH compared to PCa, p value = 0.002 (χ2 test).
For the mRNA analysis, the average 28S/18S RNA ratio was 1.75 (range 1.60–1.95) similar to A260/280 readings. For all qPCR results, the slope was within −3.30 to −3.60 corresponding to 100–80% PCR efficiency. The y-intercept of the calibration curves was less than 40.
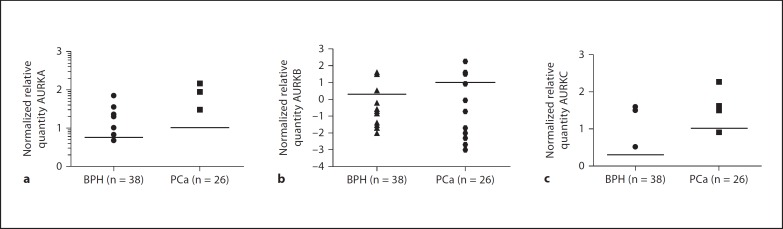
Data from normalized relative quantities of AURKA, AURKAB and AURKC were tested for gaussian approximation using the D'Agostino-Pearson omnibus normality test. The data were not normally distributed. Therefore, median values were calculated for normalized relative quantity and the results were 0.01, 0.02 and 0.02 for AURKA, AURKB and AURKC, respectively, in PCa cases. Same median values were obtained for the respective kinases in BPH cases. Since the normalized relative quantity values were not normally distributed, the most appropriate statistic test was the Mann-Whitney test which does not make any assumption for gaussian distribution. The results of the Mann-Whitney tests are shown in figure 3. Similar results were obtained for AURKA, AURKB and AURKC when their normalized relative quantities were compared between BPH and PCa cases.
Fig. 3.
Normalized relative quantity of Aurora kinases A, B and C. a Normalized relative quantity of AURKA. No significant difference between BPH and PCa cases, p value = 0.593 (Mann-Whitney test). b Normalized relative quantity of AURKB. There was no significant difference in AURKB expression between PCa and BPH case, p value = 0.7571 (Mann-Whitney test). c Relative quantity of AURKC. There was no significant difference.
Discussion
The immunoreactivity profile of AURKA, AURKB and AURKC in this study showed a significantly reduced expression in PCa cases compared to BPH cases. This finding is contrary to some reports of overexpression of AURKA and AURKB in PCa [4,11,12]. Changing antigen retrieval method did not change the immunoreactivity results. It is also very unlikely that the primary antibodies may not have targeted common epitopes in the cellular antigens. It is probable that the difference in the expression of Aurora kinases in PCa of a Nigerian population is influenced by ethnicity and race. Ethnicity is known to influence many biomarkers in PCa [13]. Although our sample size was relatively small, the results of immunoreactivity (IHC scores) were comparable to those of normalized relative quantities of AURKA, AURKB and AURKC mRNA transcripts. However, at transcript level there was no differential expression between BPH and PCa cases. It is possible that mutant isoforms of Aurora kinases are produced copiously in prostate cancer but more studies will be required to establish overexpression. It is also known that RAF kinase inhibitory protein, which is lost in about 20–50% of PCa, regulates NEK6, a protein that downregulates G2/M checkpoint molecules such as AURKB, cyclin G1 and Sertuin [14]. It is therefore plausible to think that the expression of Aurora kinases may be diminished in PCa due to upstream perturbations in G2/M regulatory molecules. Several other factors could influence transcript quantity and IHC score, such as formalin fixation and the age of the sample. These will contribute to differences observed in FFPE tissues compared to fresh-frozen samples.
Conclusion
AURKA, AURKB and AURKC were, at protein level, underexpressed in PCa cases compared to BPH cases in the study group. At transcript level, no differential expression was observed. Ethnic origin may influence expression of Aurora kinases. More studies on larger sample sizes and fresh-frozen tissues are still required to determine aberrant expression of Aurora kinases in PCa before they can be relied upon as therapeutic targets.
Acknowledgements
We express our gratitude to Chimdi Achi and Sowechi Ebi, both staff at the Safety Molecular Pathology Laboratory, for assisting in some laboratory preparations.
References
- 1.Marumoto T, Zhang D, Saya H. Aurora-A – A guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 2.Scharer CD, Laycock N, Osunkoya AO, Logani S, McDonald JF, Benigno BB, Moreno CS. Aurora kinase inhibitors synergize with paclitaxel to induce apoptosis in ovarian cancer cells. J Transl Med. 2008;6:79. doi: 10.1186/1479-5876-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee EC, Frolov A, Li R, Ayala G, Greenberg NM. Targeting Aurora kinases for the treatment of prostate cancer. Cancer Res. 2006;66:4996–5002. doi: 10.1158/0008-5472.CAN-05-2796. [DOI] [PubMed] [Google Scholar]
- 4.Chieffi P, Cozzolino L, Kisslinger A, Libertini S, Staibano S, Mansueto G, De Rosa G, Villacci A, Vitale M, Linardopoulos S, et al. Aurora B expression directly correlates with prostate cancer malignancy and influence prostate cell proliferation. Prostate. 2006;66:326–333. doi: 10.1002/pros.20345. [DOI] [PubMed] [Google Scholar]
- 5.Giet R, Glover DM. Drosophila Aurora B kinase is required for histone h3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–681. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu Y, Zhang L, Mao M, Zhao F, Huang X, Yang C, Xiong Y, Mu D. Effects of DNAzymes targeting Aurora kinase A on the growth of human prostate cancer. Cancer Gene Ther. 2008;15:517–525. doi: 10.1038/cgt.2008.22. [DOI] [PubMed] [Google Scholar]
- 8.Keen N, Taylor S. Aurora kinase inhibitors as anticancer agents. Nat Rev Cancer. 2004;4:927–936. doi: 10.1038/nrc1502. [DOI] [PubMed] [Google Scholar]
- 9.Nna E, Tothill IE, Ludeman L, Bailey T. Endogenous control genes in prostate cells: evaluation of gene expression using ‘real-time’ quantitative polymerase chain reaction. Med Princ Pract. 2010;19:433–439. doi: 10.1159/000320300. [DOI] [PubMed] [Google Scholar]
- 10.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katayama H, Sen S. Aurora kinase inhibitors as anticancer molecules. Biochim Biophys Acta. 2010;1799:829–839. doi: 10.1016/j.bbagrm.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haqq C, Li R, Khodabakhsh D, Frolov A, Ginzinger D, Thompson T, Wheeler T, Carroll P, Ayala G. Ethnic and racial differences in prostate stromal estrogen receptor α. Prostate. 2005;65:101–109. doi: 10.1002/pros.20272. [DOI] [PubMed] [Google Scholar]
- 13.Libertini S, Abagnale A, Passaro C, Botta G, Portella G. Aurora A and B kinases – Targets of novel anticancer drugs. Recent Pat Anticancer Drug Discov. 2010;5:219–241. doi: 10.2174/157489210791760517. [DOI] [PubMed] [Google Scholar]
- 14.Al-Mulla F, Bitar MS, Taqi Z, Rath O, Kolch W. RAF kinase inhibitory protein (RKIP) modulates cell cycle kinetics and motility. Mol Biosyst. 2011;7:928–941. doi: 10.1039/c0mb00208a. [DOI] [PubMed] [Google Scholar]