Abstract
Objective
This study was undertaken to determine the prevalence of congenital cytomegalovirus (CMV) infection in pregnant women at the end of pregnancy in Kuwait using cord blood and maternal urine.
Subjects and Methods
Urine samples were collected prior to childbirth, and cord blood was collected immediately after delivery from 983 women. Anti-CMV IgG and IgM antibodies were determined using ELISA; CMV DNA was detected using nested PCR, and viral load was calculated using real-time PCR. CMV concentration in samples was categorized as low when the viral load ≤103 copies/µl, intermediate when the viral load = 103−104 copies/µl, and high when the viral load >104 copies/µl. The cord blood serology outcome was compared to cord blood PCR, cord blood viral load, maternal urine PCR and viral load analyses.
Results
Serology showed that of the 983 cord blood samples, 89 (9%) were positive for anti-CMV IgM antibodies; PCR test showed 44 (4.5%) contained CMV DNA, and there was a high viral load in all. Maternal urine PCR showed that 9 (10.11%) women had CMV DNA, and there was a high viral load in 7 (78%). The kappa test for measures of agreement showed a reasonable agreement (0.45) between cord blood PCR and urine PCR.
Conclusion
This study showed that CMV infection in the cord blood sera of pregnant women is common in Kuwait and highlights the need for more clinically based studies to follow up newborns with congenital CMV infection.
Key Words: Active cytomegalovirus infection, Cord blood, Maternal urine, Polymerase chain reaction, Viral load
Introduction
Congenital cytomegalovirus (CMV) infection is one of the most common intrauterine infections in newborn infants with reported incident rates varying from 0.15 to 2% worldwide [1]. Congenital CMV infection is transmitted from mother to fetus as a consequence of primary or recurrent infection of the mother during pregnancy [2,3]. About 85–90% of infants with congenital primary infection have no signs or symptoms but around 5–15% of them may show sequelae later in life [3].
Many studies have been conducted to identify CMV DNA during pregnancy which related the outcome to the congenital infection of fetuses [4,5,6,7]. Revello et al. [4] screened amniotic fluids. Fabrri et al. [5] and Kaneko et al. [6] screened cord blood sera. Munro et al. [7] used maternal urine and Revello et al. [8] compared the presence of CMV DNA in the peripheral blood of normal blood donors with that of pregnant women. Khare et al. [9] used serial PCR assays of maternal urine to screen for primary CMV infection during pregnancy, compared the urine PCR results with those from cord blood, and found that 1 out of 609 (0.16%) women had viral DNA in the urine and subsequently developed seroconversion. CMV DNA was also detected in matching cord blood.
Noninvasive diagnostic tests for congenital CMV detection may be carried out when women are at the end of their pregnancies so that early diagnosis and early intervention of asymptomatic neonates who may be at risk of developing conditions such as sensorineural hearing loss can be treated [10]. This study was undertaken to investigate the prevalence of CMV infection in pregnant women in Kuwait. Cord blood serology, cord blood PCR and maternal urine PCR from women at the end of their pregnancy were analyzed and compared.
Subjects and Methods
Subjects
Nine hundred and eighty-three women attending the labor ward at the Maternity Hospital took part in the study. Informed consent was obtained from each woman. The study was approved by the Ministry of Health and the Faculty of Medicine Ethics Committee. The age range of the subjects was 16–48 years (median 29 years) and 392 of the women had a gestational age of 40 weeks or more at delivery. The majority, 740 women, had a vaginal delivery and 243 had caesarean delivery. A lesser proportion, 299 women, were primigravidas and 765 had no previous abortion or miscarriage. Other information was obtained from the hospital records of the women, including: (a) the number of pregnancies, (b) whether the woman was having term or preterm delivery and (c) the number of previous abortions or miscarriages.
Voided urine was collected from the women as they presented to the labor wards with no prior selection, and cord blood was taken immediately after childbirth. Cord blood serum from each woman was initially tested for anti-CMV IgG and IgM antibodies and then tested with urine samples for the presence of the CMV genome using the nested PCR technique. The viral load of CMV was detected in serum and urine samples that were positive for the CMV genome.
Serological Assays
Anti-CMV IgG and IgM antibodies were determined using a CMV IgG enzyme immunoassay test kit and a CMV IgM enzyme immunoassay test kit (ClinPro International Co., Union City, Calif., USA). The serology was performed according to the manufacturer's instructions and by reading the optical density for negative control, positive control and cut-off calibrators. Samples with optical density <0.99 IU/ml were considered negative for the presence of anti-CMV antibodies, and samples with optical density ≥0.99 IU/ml were considered positive for the presence of anti-CMV antibodies.
DNA Extraction, Primers and PCR Assays
DNA was extracted from 200 µl of cord blood serum using NucleoSpin® Genomic DNA from a blood kit (Macherey-Nagel GmbH & Co., Düren, Germany), according to the manufacturer's instructions. Genomic DNA was extracted from urine samples stored at −80°C using NucleoSpin genomic DNA from a tissue kit (Macherey-Nagel), according to the manufacturer's instructions. DNA from CMV reference strain AD169 (American Type Culture Collection, Rockville, Md., USA) was also extracted by the same procedure using the tissue kit (Macherey-Nagel). The extracted DNA was eluted with 80 µl of elution buffer.
PCR assays were carried out as follows: 3 µl of the extracted DNA was mixed with 12.5 µl of PCR mix composed of AmpliTaq Gold DNA polymerase (0.625 U/µl), GeneAmp PCR Gold Buffer containing 15 mM Tris/HCl at pH 8.05, 50 mM KCl, dNTP (200 µM each), 2.5 mM MgCl2 (Applied Biosystems, Foster City, Calif., USA) and 0.4 µM of each primer. PCR amplification was carried out in a DNA thermal cycler (Mastercycler® ep, Eppendorf AG, Hamburg, Germany). The amplified DNA was visualized on ethidium bromide-stained 1% agarose gel after electrophoresis.
β-Globin primers were used to determine the integrity of the target DNA by β-globin PCR assay, amplifying a target of 268 bp fragment, as described previously by Lum and Le Marchand [11]. The PCR amplification was initiated at 95°C for 10 min and completed in 35 amplification cycles (denaturation at 94°C for 1 min, annealing at 55 °C for 1 min and extension at 72°C for 1 min). The amplified DNA (268 bp) was visualized on ethidium bromide-stained 1% agarose gel after electrophoresis. The primers used in the nested CMV PCR assay were derived from the CMV major immediate-early exon 4 gene, as described previously by Dzieciatkowski et al. [12], and were custom synthesized (Syngyn, Inc., Chester, Pa., USA).
As for the CMV PCR assay, the first reaction mixture of the nested PCR assay was carried out using the outer primers. The PCR assay included 20 amplification cycles, each run at 94°C for 1 min, 58°C for 1 min and 72°C for 1 min. For the second reaction, 1 µl of the PCR product was transferred to a second reaction mixture containing 24 µl of the PCR mix and each of the inner primers. A total of 30 amplification cycles were carried out and the amplified DNA (146 bp) was visualized on ethidium bromide-stained 1% agarose gel after electrophoresis. DNA from AD169 strain was used as a positive control and water as a negative control.
Real-Time PCR
Real-time PCR was carried out on cord blood serum and urine samples using an Abbott CMV ASRs kit (Abbott Molecular, Des Plaines, Ill., USA), as previously described by Caliendo et al. [13] in combination with ABI PRISM® 7000 Sequence Detection System (Applied Biosystems). The kit included CMV Master Mix, CMV Mg-Sol (labeled with FAM), 4 CMV quantitation standards (QS) containing CMV target sequence (CMV QS1 at 1 × 10 copies/µl, CMV QS2 at 1 × 102 copies/µl, CMV QS3 at 1 × 103 copies/µl, and CMV QS4 at 1 × 104 copies/µl), water (PCR grade) and CMV internal control. This was used to assess the DNA extraction procedure and verify possible PCR inhibition.
The real-time PCR assay was carried out in a 96-well plate at a reaction volume of 50 µl containing 20 µl of extracted DNA and 30 µl of Master Mix. The real-time PCR assay included 45 amplification cycles and, in each run, CMV QS at the 4 different concentrations; internal control, DNA extracts and a negative control were included. The outcome of real time was analyzed using Sequence Detection System software version 1.1 (Applied Biosystems) with a cut-off line of 0.2 for analytic PCR and internal control. A standard curve was generated by the serially 10-fold concentrated CMV QS, and the value of the CMV DNA copies in each sample was plotted on a CMV QS curve and determined as the value of copies per µl. The CMV concentration in the samples was categorized as follows: low, when the sample contained ≤103 copies/µl; intermediate, when the sample contained 103–104 copies/µl, and high, when the sample contained >104 copies/µl. In every real-time PCR run, the samples were analyzed in duplicate; the average values of the CMV copy number per µl from the samples, the 4 CMV QS and 15 negative controls were taken. A cycle threshold of 29 or less was used as a cut-off point for QS and samples.
Data Analysis
The t test for two independent samples was used to test whether there was a statistical difference in the values of viral load in cord blood or in urine between anti-CMV IgG and IgM antibodies. The measurement of agreement (kappa), which ranged from −1 to +1, was calculated for cord blood PCR and urine PCR outcomes and was considered good when >0.070.
Results
Serology Tests
The outcome of the enzyme immunoassay tests for the detection of anti-CMV IgG and IgM antibodies in 983 cord blood samples is shown in table 1. Of the 983 samples, 89 (9%) were positive for anti-CMV IgM antibodies. Of these, about 90% were positive for anti-CMV IgG antibodies. The results also show that 894 (91%) of the samples were negative for anti-CMV IgM antibodies, and 869 (88%) of these had IgG antibodies, indicating that these cord blood samples belonged to women who had CMV infection in the past.
Table 1.
Cord blood serology outcome of 983 samples
| Serology | Anti-CMV IgM antibodies n (%) |
Total | |
|---|---|---|---|
| positive | negative | ||
| Anti-CMV IgG antibodies | |||
| Positive | 80 (8) | 789 (80) | 869 (88) |
| Negative | 9 (1) | 105 (11) | 114 (12) |
| Total | 89 (9) | 894 (91) | 983 |
PCR and Viral Load of Cord Blood
All the 983 specimens were suitable for PCR assay, as judged by the successful amplification of the β-globin sequences. The results of nested PCR showed that CMV DNA was detected in 4 (0.4%) out of 983 cord blood samples.
Statistical tests of gestational age at delivery showed that women with CMV DNA-positive cord blood had a mean gestational age of 37.7 weeks at delivery, and women with CMV DNA-negative cord blood had a mean gestational age of 38.6 weeks (p = 0.028). Statistical analysis of the gestational age for women with anti-CMV IgM antibodies and women without anti-CMV IgM antibodies did not show any significant difference (p = 0.75).
Analysis of viral load in the cord blood samples showed that the median CMV viral load in 4 cord blood samples was 1.52 × 106 (copies/µl), and the range was 3.30 × 104 to 2.51 × 106 (copies/µl). The comparative result of the PCR and serology is shown in table 2. Of the 80 samples, 3 (3.75%) with anti-CMV IgM and IgG antibodies had CMV DNA, and quantitative PCR showed that the viral load was high in all of them (median viral load = 1.96 × 106 copies/µl). One sample (11.11%), which was positive for anti-CMV IgM antibodies and negative for anti-CMV IgG antibodies, had CMV DNA, and the viral load was at a high concentration (1.13 × 106 copies/µl).
Table 2.
Serology and CMV DNA results in cord blood samples
| Serology | Anti-CMV IgM antibodies n (%) |
Total | |
|---|---|---|---|
| positive | negative | ||
| CMV DNA positive | 4 (0.4) | 0 (0) | 4 (0.4) |
| CMV DNA negative | 85 (8.6) | 894 (91) | 979 (99.6) |
| Total | 89 (9) | 894 (91) | 983 |
Correlation of Cord Blood and Maternal Urine PCR
The results of maternal urine PCR showed that CMV DNA was detected in 9 (0.9%) of the 983 women. CMV DNA was detected in 3 (0.3%) maternal urine samples that were obtained from women with cord blood positive for CMV DNA. The kappa test for agreement between cord blood PCR and urine PCR shows that there was a reasonable agreement between the two tests (value = 0.45).
The comparative result of urine PCR and serology outcome is shown in table 3 and demonstrates that 9 out of 89 (10.11%) women with anti-CMV IgM antibodies had CMV DNA in the urine.
Table 3.
Serology and CMV DNA results in urine samples of pregnant women
| Serology | Anti-CMV IgM antibodies n (%) |
Total | |
|---|---|---|---|
| positive | negative | ||
| CMV DNA positive | 9 (0.9) | 0 (0) | 9 (0.9) |
| CMV DNA negative | 80 (8.1) | 894 (91) | 974 (99.1) |
| Total | 89 (9) | 894 (91) | 983 |
The viral load quantification of CMV DNA in the 9 urine samples showed that the median CMV viral load was 0.9 × 106 (copies/µl) and the range was 2.1 × 101 to 1.89 × 107 (copies/µl). Quantitative PCR of the urine samples showed that the viral load was high in 7 samples (median viral load = 5.56 × 107 copies/µl) and low in 2 (median viral load = 1.12 × 102 copies/µl).
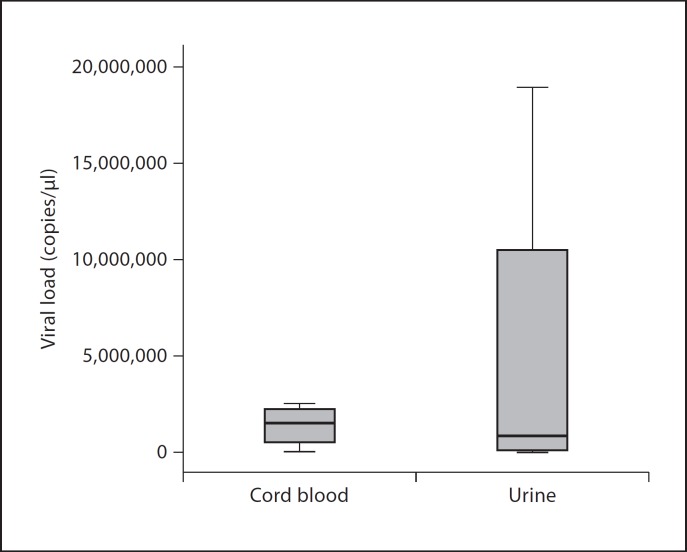
Box plot analysis of logarithmic-transformed viral load values of 4 cord blood samples with CMV DNA and anti-CMV IgM antibodies and 9 maternal urine samples with CMV DNA obtained from women with cord blood positive for anti-CMV IgM antibodies is shown in figure 1. Analysis of the figure showed that the CMV copy number in the urine samples was higher than in cord blood. The quantitative results of CMV also showed that the viral concentration in 3 cord blood samples and the corresponding maternal urine was high in both sets.
Fig. 1.
Box plot of logarithmic-transformed viral load values of 9 urine and 4 cord blood samples with anti-CMV IgM antibodies.
Discussion
This study showed that the seroprevalence of CMV was 88% in cord blood, consistent with other findings showing that CMV seroprevalence varies between countries and social status in normal populations and ranges from 50 to 90% [14]. This study also demonstrated that 9% of cord blood had anti-CMV IgM antibodies, consistent with the findings of Roback et al. [15], who screened umbilical cord blood units and found anti-CMV IgM antibodies were present in 8.3% of the units. Fabri et al. [5] have also shown that the prevalence of anti-CMV IgM in fetal cord blood was 7%. However, our findings are different from those reported by el-Mekki et al. [16], in which anti-CMV IgM antibodies were found in 2.6% of the 575 umbilical cord samples. The difference between the study of el-Mekki et al. [16] and this study could be due to the increase in the CMV infection trend in Kuwait [17], or it could be explained by the differences in the commercial tests and laboratory preparation procedures used in the two studies.
New approaches have been suggested for the diagnosis of congenital CMV infection, based on CMV DNA quantification in amniotic fluid or the combined assessment of ultrasound findings and low platelet count in fetal blood samples [18,19]. Recent studies have shown that the determination of specific virological, hematological and biochemical markers in fetal blood samples greatly help to predict a fetal neonatal CMV infection, once a positive viral detection has been established in amniotic fluid samples. Kaneko et al. [6] screened for CMV DNA in the cord blood of neonates with congenital CMV infection and found CMV DNA was present in 10% of cord blood sera. Khare et al. [9] screened seronegative pregnant women for the presence of CMV infection by using serial maternal urine PCR assays and found that 1 out of 609 (0.16%) women had viral DNA in the urine and subsequently developed seroconversion; CMV DNA was also detected in matching cord blood. Munro et al. [7] also screened the maternal urine of pregnant women with anti-CMV IgM antibodies by using nested PCR and found that CMV DNA was present in 1.1%. Analysis of real-time PCR results in the current study showed that the CMV load in cord blood was more than that in urine. This could be explained by the fact that cord blood PCR was based on the determination of cell-free CMV copies in the serum, whereas in urine the PCR was based on the determination of cell-free and intracellular CMV copies. This also explains why CMV DNA is more detectable in maternal urine than in cord blood.
The findings of this study revealed that maternal urine PCR was in agreement with 3 cord blood samples with CMV DNA did not confirm the presence of CMV DNA in a maternal urine sample corresponding to 1 cord blood with CMV DNA. This could be due to the fact that the CMV infection in this case was recent and viral shedding in the urine had not yet taken place.
A wide range of CMV DNA concentration in urine samples was encountered in the study. This could be because of the low number of urine samples with CMV infection. Another explanation is that some of the women had chronic CMV infection, CMV reactivation or viral clearance, resulting in the low level of CMV DNA in the urine samples. In cases with active infection a high level of CMV DNA was detected.
The current study showed that pregnant women with CMV DNA in their cord blood and urine had an earlier gestational age than women without CMV DNA, a finding that is in agreement with Yamamoto et al. [20].
Clinical management of CMV infection during pregnancy is based mainly on the detection of CMV in the amniotic fluid and there is a general opinion that fetal infection acquired late in pregnancy does not result in symptomatic newborns, whereas infection acquired in the first half of pregnancy carries a higher risk of symptomatic fetuses [21,22]. In this study, CMV infection was detected in the cord blood samples at the end of pregnancy and it was not possible to anticipate at which trimester the primary or recurrent infection took place.
The main limitation of this study is that it lacks a follow-up of newborns to correlate the virological markers with the development of congenital CMV infection and the appearance of long-term sequelae. Nevertheless, this study has laid the foundation for future studies to ascertain the diagnosis of congenital CMV infection by the association of serology and DNA results with any clinical diseases.
Conclusion
This study showed that the number of pregnant women with anti-CMV IgM antibodies in their cord blood sera has increased over the years in Kuwait, and their proportion was relatively high. This study also showed that CMV DNA was detected more in the urine than in cord blood sera of pregnant women with anti-CMV IgM.
Acknowledgements
The authors wish to thank Dr. Wassim Chehadeh for his help and assistance. They also wish to thank Ms. Gnana Priya Raju, Ms. Veeda Abu Arji, Ms. Bashayer Al-Zayer, Ms. Marina James, Ms. Marwa Heikel and Mr. Abdul-Rahman Rahmatulla for their excellent technical assistance. The work was funded by Kuwait University through the Research Administration (research project NM 02/05).
References
- 1.Nigro G, Adler SP. Cytomegalovirus infections during pregnancy. Curr Opin Obstet Gynecol. 2011;23:123–128. doi: 10.1097/GCO.0b013e328342f1f6. [DOI] [PubMed] [Google Scholar]
- 2.Adler SP. Screening for cytomegalovirus during pregnancy. Infect Dis Obstet Gynecol. 2011;2011:1–9. doi: 10.1155/2011/942937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeckh M, Geballe AP. Cytomegalovirus: pathogen, paradigm, and puzzle. J Clin Invest. 2011;121:1673–1680. doi: 10.1172/JCI45449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Revello MG, Fabbri E, Furione M, Zavattoni M, Lilleri D, Tassis B, Quarenghi A, Cena C, Arossa A, Montanari L, Rognoni V, Spinillo A, Gerna G. Role of prenatal diagnosis and counseling in the management of 735 pregnancies complicated by primary human cytomegalovirus infection: a 20-year experience. J Clin Virol. 2011;50:303–307. doi: 10.1016/j.jcv.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Fabbri E, Revello MG, Furione M, Zavattoni M, Lilleri D, Tassis B, Quarenghi A, Rustico M, Nicolini U, Ferrazzi E, Gerna G. Prognostic markers of symptomatic congenital human cytomegalovirus infection in fetal blood. BJOG. 2011;118:448–456. doi: 10.1111/j.1471-0528.2010.02822.x. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko M, Sameshima H, Ikenoue T, Minematsu T. A two-step strategy for detecting intrauterine cytomegalovirus infection with clinical manifestations in the mother, fetus, and newborn. Jpn J Infect Dis. 2006;59:363–366. [PubMed] [Google Scholar]
- 7.Munro SC, Hall B, Whybin LR, Leader L, Robertson P, Maine GT, Rawlinson WD. Diagnosis of and screening for cytomegalovirus infection in pregnant women. J Clin Microbiol. 2005;43:4713–4718. doi: 10.1128/JCM.43.9.4713-4718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Revello MG, Zavattoni M, Sarasini A, Percivalle E, Simoncini L, Gerna G. Human cytomegalovirus in blood of immunocompetent persons during primary infection: prognostic implications for pregnancy. J Infect Dis. 1998;177:1170–1175. doi: 10.1086/515277. [DOI] [PubMed] [Google Scholar]
- 9.Khare M, Sharland M, Manyonda I, Rice P, Bland JM, Griffiths P. Use of serial maternal urine cytomegalovirus PCR to detect primary CMV infection in seronegative pregnant women. J Virol Methods. 2004;119:31–35. doi: 10.1016/j.jviromet.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Yoshinaga-Itano C. Early intervention after universal neonatal hearing screening: impact on outcomes. Ment Retard Dev Disabil Res Rev. 2003;9:252–266. doi: 10.1002/mrdd.10088. [DOI] [PubMed] [Google Scholar]
- 11.Lum A, Le Marchand L. A simple mouthwash method for obtaining genomic DNA in molecular epidemiological studies. Cancer Epidemiol Biomarkers Prev. 1998;7:719–724. [PubMed] [Google Scholar]
- 12.Dzieciatkowski T, Przybylski M, Tomaszewska A, Rokicka M, Luczak M. Comparison of two methods used for monitoring low-copy cytomegalovirus infection in a patient with chronic myeloid leukemia after unrelated umbilical cord blood transplantation. Arch Immunol Ther Exp (Warsz) 2007;55:199–203. doi: 10.1007/s00005-007-0019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caliendo AM, Ingersoll J, Fox-Canale AM, Pargman S, Bythwood T, Hayden MK, Bremer JW, Lurain NS. Evaluation of real-time PCR laboratory-developed tests using analyte-specific reagents for cytomegalovirus quantification. J Clin Microbiol. 2007;45:1723–1727. doi: 10.1128/JCM.02558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scholz M, Doerr HW, Cinatl J. Human cytomegalovirus retinitis: pathogenicity, immune evasion and persistence. Trends Microbiol. 2003;11:171–178. doi: 10.1016/s0966-842x(03)00066-0. [DOI] [PubMed] [Google Scholar]
- 15.Roback JD, Caliendo AM, Newman JL, Sgan SL, Saakadze N, Gillespie TW, Lane TA, Kurtzberg J, Hillyer CD. Comparison of cytomegalovirus polymerase chain reaction and serology for screening umbilical cord blood components. Transfusion. 2005;45:1722–1728. doi: 10.1111/j.1537-2995.2005.00596.x. [DOI] [PubMed] [Google Scholar]
- 16.el-Mekki A, Deverajan LV, Soufi S, Strannegard O, al-Nakib W. Specific and non-specific serological markers in the screening for congenital CMV infection. Epidemiol Infect. 1988;101:495–501. doi: 10.1017/s0950268800029381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ameen R, Sanad N, Al-Shemmari S, Siddique I, Chowdhury RI, Al-Hamdan S, Al-Bashir A. Prevalence of viral markers among first-time Arab blood donors in Kuwait. Transfusion. 2005;45:1973–1980. doi: 10.1111/j.1537-2995.2005.00635.x. [DOI] [PubMed] [Google Scholar]
- 18.Benoist G, Salomon LJ, Jacquemard F, Daffos F, Ville Y. The prognostic value of ultrasound abnormalities and biological parameters in blood of fetuses infected with cytomegalovirus. BJOG. 2008;115:823–829. doi: 10.1111/j.1471-0528.2008.01714.x. [DOI] [PubMed] [Google Scholar]
- 19.Revello MG, Gerna G. Diagnosis and management of human cytomegalovirus infection in the mother, fetus, and newborn infant. Clin Microbiol Rev. 2002;15:680–715. doi: 10.1128/CMR.15.4.680-715.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto AY, Mussi-Pinhata MM, Cristina P, Pinto G, Moraes Figueiredo LT, Jorge SM. Congenital cytomegalovirus infection in preterm and full-term newborn infants from a population with a high seroprevalence rate. Pediatr Infect Dis J. 2001;20:188–192. doi: 10.1097/00006454-200102000-00014. [DOI] [PubMed] [Google Scholar]
- 21.Gindes L, Teperberg-Oikawa M, Sherman D, Pardo J, Rahav G. Congenital cytomegalovirus infection following primary maternal infection in the third trimester. BJOG. 2008;115:830–835. doi: 10.1111/j.1471-0528.2007.01651.x. [DOI] [PubMed] [Google Scholar]
- 22.Ornoy A, Diav-Citrin O. Fetal effects of primary and secondary cytomegalovirus infection in pregnancy. Reprod Toxicol. 2006;21:399–409. doi: 10.1016/j.reprotox.2005.02.002. [DOI] [PubMed] [Google Scholar]