Abstract
The need for drugs with fewer side effects cannot be overemphasized. Today, most drugs modify the actions of enzymes, receptors, transporters and other molecules by directly binding to their active (orthosteric) sites. However, orthosteric site configuration is similar in several proteins performing related functions and this leads to a lower specificity of a drug for the desired protein. Consequently, such drugs may have adverse side effects. A new basis of drug discovery is emerging based on the binding of the drug molecules to sites away (allosteric) from the orthosteric sites. It is possible to find allosteric sites which are unique and hence more specific as targets for drug discovery. Of many available examples, two are highlighted here. The first is caloxins - a new class of highly specific inhibitors of plasma membrane Ca2+ pumps. The second concerns the modulation of receptors for the neurotransmitter acetylcholine, which binds to 12 types of receptors. Exploitation of allosteric sites has led to the discovery of drugs which can selectively modulate the activation of only 1 (M1 muscarinic) out of the 12 different types of acetylcholine receptors. These drugs are being tested for schizophrenia treatment. It is anticipated that the drug discovery exploiting allosteric sites will lead to more effective therapeutic agents with fewer side effects.
Key Words: Adverse effects, Allosterism, Drug specificity, Drug targets, Orthosteric sites, Pharmacology, Side effects
Introduction
History of Drug Discovery
The discovery of drugs for therapies of human diseases likely initiated in various ancient civilizations of India, China and the Middle East [1]. However, one can only conjecture as to how these drugs were discovered. More recently, systematic rational drug design has evolved based on advances in organic chemistry and on the understanding of various aspects of physiology and biochemistry [2]. Advances in the biological sciences led to the concept that all the drugs, including those discovered in ancient times, act on defined biological sites. For example, the foxglove plant used for treating the failing heart contains the active ingredient digoxin, which increases the cardiac tone by inhibiting the enzyme Na+-K+-adenosine triphosphate (ATP)ase [3]. The antibiotic penicillin acts on enzymes to prevent the cell wall formation in bacteria [4]. In essence, rational drug design is based on the idea that if a specific pharmacological target can be identified for a given disease, a chemical that can bind to this site would form the basis of the development of a new therapeutic drug [5,6,7]. This concept has been advanced due to innovations in biochemical and biophysical techniques, i.e. X-ray crystallography and imaging techniques such as magnetic resonance imaging for identifying specific targets, separation sciences to fractionate chemical libraries and technical advances in high-throughput screening. Advances in bioinformatics have also aided in revolutionizing the drug discovery process [8,9]. Drug action at the desired site is important, but an ideal drug should also have a minimum of undesired side effects. One reason for side effects is the interaction with receptors other than the one the drug is targeted for in the treatment of a disease. It is, therefore, paramount that the drug binds predominantly only to its desired site of action.
Basis of Drug Specificity and Selectivity
In the body, the specificity of substances for their sites of action is attained either by extremely high affinities of the substances at these sites, such as for hormones which bind humorally to specific receptors or spatiotemporally as for neurotransmitters [10]. A neurotransmitter is released in a narrow space where the site of action is located within a few nanometers of the release site, limiting the dilution effect due to diffusion over a wider space. The substance is then removed from the site within milliseconds of the release, thereby preventing its action on unintended targets. However, taking full advantage of this sophisticated spatiotemporal compartmentalization is not feasible for drug therapy today, although a certain degree of selectivity can be obtained by advances in drug delivery methods. Therefore, a considerable focus in drug discovery today is on the specificity of a drug for its site of action along with the control of drug distribution in the body. This review focuses on drug specificity.
Allosteric versus Orthosteric Sites
Advances in the studies of protein structure have led to a detailed understanding of the nature of the active sites where substrates bind enzymes or agonists bind receptors [5,6,7,11,12]. A large number of drugs are designed based on the inhibitors that prevent the binding of a ligand to its active site. One main advantage is that a very high affinity and, often, a very high specificity for that site can be attained. However, therapy using such drugs still results in adverse side effects because many enzymes with related functions may have very similar active sites. For example, the transitional state analog of ATP can bind tightly to the orthosteric site of any enzyme that hydrolyzes ATP [13]. Vanadate is then specific in the sense that it inhibits ATPases and not other enzymes, but nonspecific as it cannot distinguish between the different types of ATPases which exist in the body. Similarly, any other drug designed based on its ability to interfere with the binding of a ligand to the ATP binding site will have the same disadvantage of being nonselective. Hence, this method of using orthosteric sites can lead to drugs with side effects that may often be harmful.
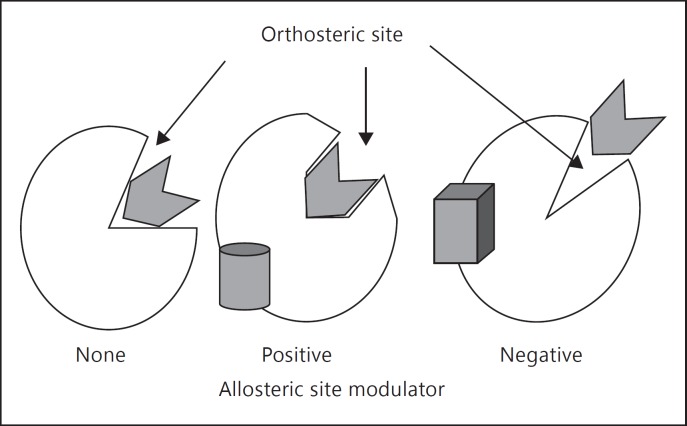
In 1965, Changeux [14] introduced the concept of allosterism. According to this concept, the activity of a target protein can be modulated by ligands that bind to allosteric sites - sites which are located away from the orthosteric sites (fig. 1). The binding of a drug to an allosteric site can alter the macromolecular conformation, thereby either enhancing (positive modulation) or slowing (negative modulation) the reactions carried out at the orthosteric site. This concept was originally introduced to explain the regulatory properties of the bacterial protein aspartate transcarbamylase by a regulatory protein subunit. Today, numerous examples of molecules that can cause modulation by binding to the allosteric sites in their target proteins are available [7]. However, not all modulatory allosteric sites are equally useful as targets for a specific drug design. For example, the regulatory protein calmodulin acts by binding to allosteric sites of proteins, but this binding cannot be considered to be highly specific for a drug design because the sites for calmodulin binding are present in several different proteins [15]. Therefore, one has to search for allosteric sites which are unique to the protein of interest and the properties of the protein are altered when a ligand binds to this site. Thus, there are two challenges to be met. The first is the determination of allosteric sites in the protein of interest. Fortunately, protein sequence data available today can aid in this challenge. The second is finding the molecules that can bind to these sites and change the protein function. Identification of such molecules can be attained by various screening methods, each of which has its own advantages and disadvantages. Here, we describe screening phage display peptide libraries and high-throughput screening using chemical compound libraries. Typically, high-throughput screening can be carried out for a target protein which is abundant, can be purified or is overexpressed in cells. Screening of phage display peptide libraries can be modified to find ligands for proteins of lower abundance which are also difficult to overexpress in cells. Although several alternatives are available, examples of only these two will be discussed in the subsequent sections.
Fig. 1.
A schematic representation of allosterism. The orthosteric sites are the sites for binding of the substrates or competitive inhibitors of enzymes and agonists or competitive antagonists of receptors. Allosteric sites are away from these sites but their binding to the protein can change its conformation. Binding of positive allosteric modulators leads to either an increase in the affinity of the substrates/agonists or it speeds up the subsequent actions. Negative allosteric modulators act as inhibitors by either altering the affinities at the orthosteric sites or by preventing changes in the protein conformation needed for the subsequent steps.
Allosteric Inhibitors of Plasma Membrane Ca2+ Pumps
Plasma Membrane Ca2+ Pumps
Maintenance of low cytosolic Ca2+ concentration during the resting state is pivotal to the survival of mammalian cells. Although other pathways play a role during signal transduction cycles, there are two types of Ca2+ pumps, which use the energy of ATP hydrolysis to transport Ca2+ ions against an electrochemical gradient [16,17,18]. One type of Ca2+ pump is located in the internal cellular organelle sarco/endoplasmic reticulum (SERCA) and transports cytosolic Ca2+ into its lumen. The other type is located in the plasma membrane (PMCA) and expels Ca2+ from the cells into the exoplasm. SERCA pumps are abundant in the skeletal and cardiac muscles and their structure has been examined by X-ray crystallography [19]. They play a major role in lowering cytosolic Ca2+ immediately at the end of the cell excitation state. In contrast, PMCA have higher affinity for Ca2+ and can maintain low cytosolic Ca2+ levels even in the resting state. PMCA are low-abundance proteins, and unlike SERCA, their overexpression at high levels has been problematic. As a result, the crystal structure of the PMCA proteins has not been established. Only a hypothetical structure of PMCA computed from the homology with the structure of SERCA is available. Based on this structure, the protein has 10 transmembrane domains, the N- and C-terminals of the protein are cytoplasmic and there are 5 extracellular domains.
PMCA function is important in maintaining cellular Ca2+ homeostasis. Defects in PMCA are associated with heart failure, hypertension and other disorders, and hence PMCA may be potential therapeutic targets in the management of these diseases [16]. PMCA are encoded by 4 genes (PMCA1-4), which are differently expressed in various tissues with PMCA1 and PMCA4 being most ubiquitous [20]. The unique expression pattern of the 4 PMCA genes may reflect their roles in tissue-specific physiology. In pig coronary arteries, an increase in cytosolic Ca2+ concentration in smooth muscle cells leads to vasoconstriction, whereas a similar increase in endothelial cells leads to vasodilation. Thus, an inhibition of PMCA4 in smooth muscle cells is anticipated to cause coronary vasoconstriction, while a similar inhibition in endothelial cells is likely to lead to vasodilation. The two tissues also differ in the PMCA gene expression: smooth muscle cells express more PMCA4 than PMCA1 while endothelial cells have more PMCA1 than PMCA4 [21,22]. The above example illustrates the uniqueness in the functions of the PMCA isoforms in the physiology of different tissues. In order to understand the role of these isoforms in the coronary artery physiology, we have invented allosteric inhibitors which are selective for the isoforms PMCA1 and PMCA4.
Extracellular Domains as Potential Allosteric Sites
At the time we started the work to develop selective inhibitors of PMCA, vanadate and eosin were the two commonly used inhibitors to study PMCA physiology [10,21,23,24,25,26,27,28,29]. Both compounds are orthosteric inhibitors of the ATP binding site found in PMCA proteins. These sites are similar for all ATPases and hence both vanadate and eosin inhibit all ATPases that had been tested. Thus, these inhibitors were not selective for PMCA.
PMCA and SERCA, like other ion pumps, shuttle between two different conformational states during their reaction cycle - E1 and E2 (fig. 2a). Several allosteric inhibitors of SERCA which interfere with the E1-E2 transition have been discovered. For example, thapsigargin, which has a very high affinity for SERCA, is an allosteric inhibitor. It binds tightly to the E2 form of the pump in the cavity surrounded by the transmembrane domains 3, 5 and 7 and prevents it from reverting to the E1 form. Thus, the reaction cycle of SERCA cannot be completed. In order to invent selective allosteric inhibitors of PMCA, we decided to use the extracellular domains of the protein as targets. Based on the protein sequence, PMCA have 5 short extracellular domains, while the bulk of the protein is on the cytosolic side of the membrane [16,20,30]. The cytosolic side contains the sites for ATP binding, high-affinity Ca2+ binding and various other regulatory sites. Thus, the extracellular domains are allosteric in that they are far away from the orthosteric sites, which lie in the cytosolic side of the protein. We decided to explore the use of the extracellular domains as the targets for obtaining selective inhibitors of PMCA. This concept is now supported by the differences observed in the conformations of the extracellular domains in the hypothetical structures of the E1 and E2 states of PMCA (fig. 2b). Although any of the extracellular domains can be exploited since their sequences are unique to PMCA, we decided to use extracellular domain 1 as the target for the allosteric inhibitors. The amino acid sequences of the extracellular domain 1 in the PMCA isoforms 1-4 and the most abundant ion transporting ATPase, Na+-K+-ATPase, are compared in figure 2c. Ouabain, an allosteric inhibitor of Na+-K+-ATPase, also interferes with its E1-E2 transition. First, it is noted that there are no similarities between the sequence of the Na+-K+-ATPase and those of any of the PMCA isoforms. Furthermore, the sequences of the extracellular domain 1 also differ between individual PMCA isoforms. Of particular interest are the sequences of PMCA1 and PMCA4 which differ significantly from each other and hence can be exploited as potential targets to obtain isoform-selective inhibitors for PMCA1 and PMCA4. An additional advantage of using the extracellular domains as targets would be that the created inhibitors would not need to cross the plasma membranes for use in the subsequent physiological experiments.
Fig. 2.
Extracellular domains of PMCA as potential allosteric sites. a Reaction cycle of PMCA. PMCA can exist in 2 conformational states: E1 and E2. Transition between these states is mandatory for its ability to pump Ca2+ out of the cells. b E1 and E2 differ in their structures. The orthosteric sites for binding to ATP, high-affinity Ca2+, release of ADP and inorganic phosphate and for modulation by calmodulin and protein kinases are all on the cytosolic side [figure taken with permission from ref. [31]]. E1-E2 transition results in a change in the conformations of the extracellular domains which are circled. c Extracellular domain 1 sequences of PMCA1 to PMCA4 differ from those of any other ATPases - the sequence of the extracellular domain 1 of Na+-K+-ATPase is shown for comparison and it has no similarities with the corresponding sequences of PMCA1-PMCA4. Even the sequences of PMCA1-PMCA4 differ from each other.
Phage Display for Screening Peptides That Bind Extracellular Domains
In phage display screening, exogenous (poly)peptides are expressed and presented by phage particles to bind to the specified target molecules [31]. Completely random or biased peptide libraries of different lengths (typically 7 or 12 residues) are used for the display. Phage that bind the target are selected. By analyzing the gene sequences in specific regions of the phage genome, one can determine the sequences of the peptides which bound to the target. Phage display has evolved as a powerful technology for drug discovery and for identifying and engineering polypeptides with novel functions. The technology and its applications have been described recently in detail [31,32,33].
Phage display peptide libraries were screened using the extracellular domains of PMCA as target. This method allowed for the use of the target in two forms: first as synthetic peptides based on the amino acid sequences of the extracellular domains and in the second step as part of the native PMCA protein in the membrane. This was important particularly because PMCAs are low abundant proteins which are difficult to overexpress in a cell line. Details of this protocol and its merits have recently been discussed [34].
Examples of PMCA-Specific Inhibitors
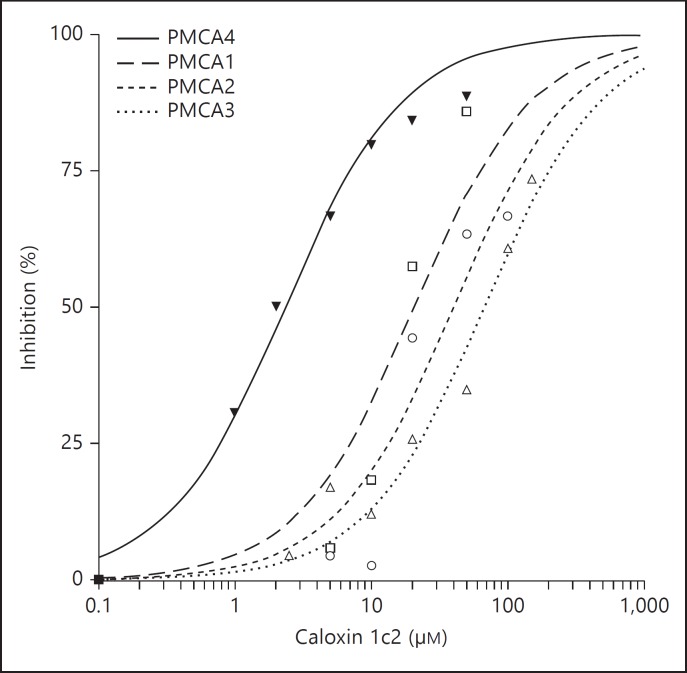
Phage display methods were modified as needed to obtaintheselective inhibitors of PMCA based on extracellular domains 1, 2 and 3 [10,21,23,24,25,26,27,28,29]. This class of inhibitors was termed caloxins. Of particular interest are the caloxins 1c2 and 1b3, which used the N-terminal half of extracellular domain 1 of PMCA4 and PMCA1 as targets, respectively. Caloxin 1c2 is a PMCA4-selective inhibitor with its affinity for PMCA4 being approximately 10× higher than for PMCA1, 20× higher than for PMCA2 and 30× higher than for PMCA3 (fig. 3) [24]. The affinity of caloxin 1b3 is only marginally greater (3×) for PMCA1 than that of PMCA4. However, when added to cultured cells, it increased cytosolic Ca2+ concentration in the endothelial cells [26]. It is significant that neither caloxin 1c2 nor 1b3 inhibits any other ATPases which were tested.
Fig. 3.
Inhibition of PMCA1-PMCA4 by the PMCA4-specific caloxin 1c2 [based on data from ref. [24]].
Thus, phage display led to the identification of extracellular domain 1 as an allosteric target to obtain the PMCA isoform-selective peptide inhibitors. Further screening of this target for nonpeptide inhibitors remains to be conducted. It is anticipated that further screening combined with appropriate drug targeting technology will aid in obtaining a new class of drugs for hypertension and other diseases.
Allosteric Modulation of M1 Muscarinic Receptors
Cholinergic Receptors
Cholinergic receptors bind to the neurotransmitter acetylcholine. They are broadly classified as nicotinic and muscarinic. There are 7 subtypes of nicotinic and 5 subtypes of muscarinic receptors [35]. The agonist for all the cholinergic receptors is the neurotransmitter acetylcholine. The selectivity of action of acetylcholine in the body is attained as follows [10]: acetylcholine is released from the presynaptic nerves into a cleft and it acts on postsynaptic receptors which lie within a few nanometers of the release site. It is then quickly degraded by acetylcholine esterase so that it does not diffuse to other sites. The sequence of events is so rapid that it allows a hummingbird to flap its wings up to a thousand times per second. However, the technology available today does not permit the use of any drugs with this high degree of spatiotemporal selectivity. Due to this type of limitation, any drugs designed to bind the orthosteric site for the acetylcholine binding cannot be very specific since this site is highly conserved among all the nicotinic and the muscarinic receptors. Allosteric sites, however, may be more diverse in the individual cholinergic receptor types.
Schizophrenia and M1 Receptors
Schizophrenia is a mental disorder whose symptoms are the breakdown of rational thought processes or the presence of very poor emotional responses. The symptoms are termed positive or negative (deficit) [36,37,38,39,40,41]. Positive symptoms are those which are observed in schizophrenia patients but not in normal persons. These include delusions, disordered thought and speech, and tactile, auditory, visual, olfactory and gustatory hallucinations, and are typically regarded as manifestations of psychosis. In contrast, the negative symptoms are those which are absent in the patients but present in normal persons. An example would be the normal emotional responses or other thought processes. The exact cause of schizophrenia is not known. However, there is a large body of evidence to suggest that dopamine plays a significant role and the disease may well be associated with dopamine fluxes. Some of the drugs used in the treatment are olanzapine, clozapine and risperidone, which are highly nonselective antagonists for γ-aminobutyric acid, 5HT2, dopamine and adrenergic receptors [39,40,41]. These drugs do not work completely and also have severe adverse effects. Behavior therapies are also not as useful.
In the treatment of schizophrenia, it may be beneficial to use choline esterase inhibitors together with antipsychotics such as olanzapine, risperidone or clozapine [36,37,38]. This use of choline esterase inhibitors led to the idea that cholinergic receptors may play a role in this disease. The activation of M1 muscarinic receptors potentiates N-methyl-D-aspartate activation-induced currents. Since schizophrenia may reflect a decrease in the N-methyl-D-aspartate function, the M1 receptors may rectify some of the symptoms associated with schizophrenia via a correction of this deficit [42,43]. Pirenzipine binding is decreased in the cortical tissue of at least some schizophrenia patients [36]. Pirenzipine binds M1 muscarinic receptors although not very selectively. Various neurological studies conducted on schizophrenia patients also suggest that M1 and possibly M4 muscarinic receptors may be involved in this disease. Xanomeline, an agonist somewhat selective for M1/M4 receptors, also has good antipsychotic activity. Moreover, transgenic mice lacking the M1 receptors are more prone to schizophrenia [44]. This knowledge formed the impetus for invention of selective modulators of M1 muscarinic receptors.
Allosteric Sites as Basis for M1 Muscarinic Selective Modulators
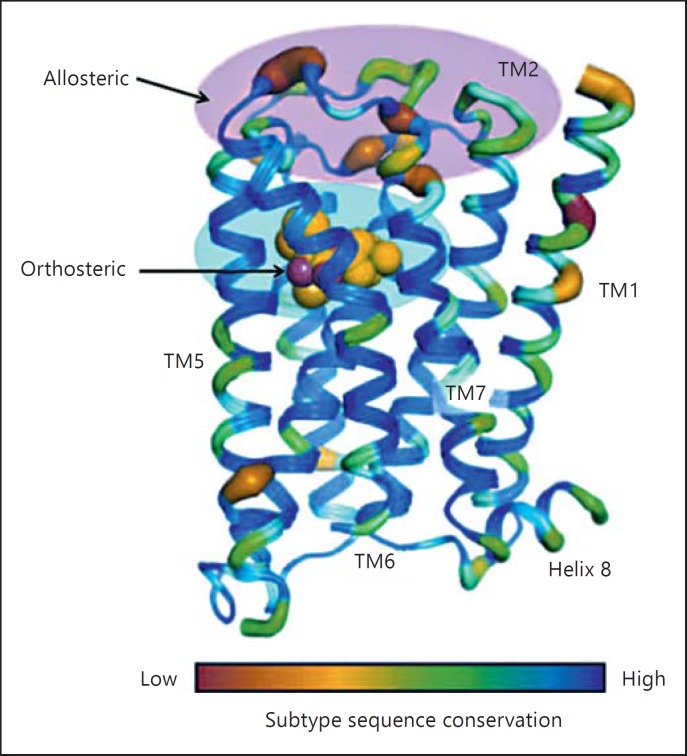
The mechanism of action of the muscarinic receptors involves acetylcholine binding to the receptor molecules followed by coupling via binding to Gi or Gq proteins which in turn leads to subsequent metabolic or ion conductance changes [35]. A schematic representation of the receptor structure with a color scheme based on the sequence similarities between the 5 subtypes of muscarinic receptors is shown in figure 4. The maximum sequence similarity is in the orthosteric site for acetylcholine binding and the minimum similarities are in the allosteric extracellular domains of the receptors [45]. Ligands that bind to these extracellular domains and can modulate the receptor function would be the ideal choice for selective modulators.
Fig. 4.
Subtype sequence conservation in the muscarinic receptor subtypes. The sequences for the orthosteric site for acetylcholine binding are highly conserved. There are very few sequences which are clearly different between M1 and M5 and these can be used as potential allosteric targets. TM = Transmembrane domain [taken with permission from ref. [45]].
Screening Strategy for Positive Allosteric Modulators of M1 Muscarinic Receptors
All the muscarinic receptor subtypes can be overexpressed in cultured cell lines and their activation results in an increase in cytosolic Ca2+ concentration [39,46,47,48,49]. For high-throughput screening, the increase in the cytosolic Ca2+ concentration can be monitored with fluorescent probes in cells cultured in multiwell plates. However, if an added substance alone increases this signal, it is likely that it acts as an agonist due to its binding at the orthosteric site. Therefore, the strategy used in this case was to select a substance that produced no effects on its own but increased the affinity for acetylcholine. A series of synthetic chemical libraries were first screened in the presence of a submaximal concentration of acetylcholine. Next, the compounds that produced a response were tested in the complete absence of acetylcholine and those that produced a response were rejected. The chosen compounds were then selected based on their selectivity for M1 receptors over the other receptor subtypes. This protocol was then repeated using more refined chemical libraries based on the selected compounds in order to obtain the optimum positive allosteric modulator molecules.
Example of a Positive Allosteric Modulator of M1 Muscarinic Receptors
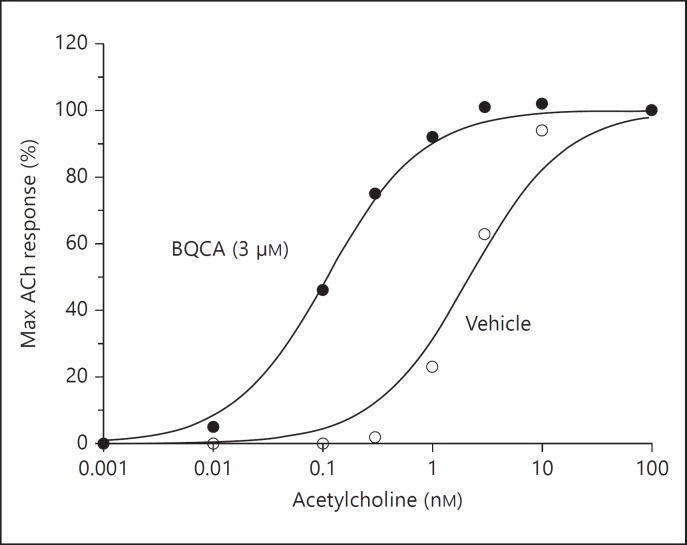
Using the strategy described above, a positive allosteric modulator (benzylquinolone carboxylic acid, BQCA) has been identified and characterized [48]. The effects of different concentrations of BQCA on the affinity of acetylcholine for M1 receptors are shown in figure 5. Acetylcholine alone stimulated Ca2+ mobilization via the M1 receptors with an EC50 value of 2.42 ± 0.34 nM. In the presence of 3 μM BQCA, the EC50 value for acetylcholine decreased to 0.12 ± 0.03 nM, representing a 21× increase in the affinity of acetylcholine for the M1 receptors. The same concentration of BQCA (3 μM) did not alter the EC50 values of acetylcholine for M2, M3, M4 or M5 receptors and this established the specificity of this agent for the M1 subtype. BQCA did not produce any effects on the M1 receptors in the absence of acetylcholine. Whereas the muscarinic antagonist atropine inhibited the binding of the M1 receptor orthosite antagonist L-N-methyl-scopolamine methyl chloride, BQCA had no effect. These properties established BQCA as a positive allosteric modulator of the M1 receptor with no direct effect on the orthosteric site. The effects of BQCA have now been examined in different animal models of schizophrenia [39]. In mice, BQCA reversed the amphetamine-induced hyperlocomotion. It also affected memory, hallucination-like responses and cerebral blood flow in anesthetized rats. A different allosteric site has also been exploited in M1 muscarinic receptors to produce positive allosteric modulators [46,47,50].
Fig. 5.
The allosteric modulator increases the affinity of M1 receptors for acetylcholine (ACh) by 21×. Although only one concentration of BQCA is shown here, the determined EC50 value of BQCA for the M1 receptors was 267 ± 31 nM [the figure is based on data from ref. [48]].
Other Examples of Drug Discovery Based on Allosterism
Although only two examples are discussed above, allosteric modulators have been invented for a number of receptors, enzymes and regulatory proteins. For example, a number of negative allosteric modulators of the metabotropic GLUTR receptors, such as fenobam, raseglurant, dipraglurant, mavoglurant, RG7090, STX107, AZD9272, AZD2066 and AZD2516 [51]. These agents are under phase I and II clinical trials for pain, anxiety, gastroesophageal reflux disease, Parkinson's disease, levodopa-induced dyskinesia or fragile X syndrome. Allosteric modulators of the following 7 transmembrane-spanning receptors have also been developed: adenosine (A1, A2, A3), adrenergic (α1, α2a, α2b, β2), cannabinoid (CB1), chemokines (several types), dopamine (D1, D2), endothelin (A), muscarinic (all subtypes), neurokinin, opioid (μ and δ), serotonin (several subtypes), γ-amino butyric acid (B), glutamate (several subtypes) and corticotropin-releasing factor [11,52,53,54]. Allosteric targets of regulatory proteins such as calmodulin have also been designed based on changes in their conformation. These may be useful in the control of virulence of bacteria such as Bacillus anthracis[37].
Allosteric sites have also been exploited to develop modulators of enzymes to control various diseases. Some of the examples are the HCV NS5B polymerase inhibitors to control chronic infection with hepatitis C virus [3,55] and the HIV-1 reverse transcriptase inhibitors for the treatment of AIDS [12]. Recently, proteasomes have been implicated in several diseases and hence used as targets for rational drug design. An example is Velcade, which has now been approved to treat multiple myeloma and other types of cancers [7]. Activation of human liver pyruvate kinase, in an effort to create a glycolytic/gluconeogenic futile cycle, is one potential mechanism to counteract hyperglycemia. Allosteric activators of this enzyme are being developed in various laboratories [56]. Protein kinases play regulatory roles for various pathways and hence allosteric regulators of these enzymes may be useful in understanding the signal transduction pathways and the disorders associated with their defects. The list of protein kinases for which allosteric modulators have been discovered includes PDK1, mitogen-activated protein kinase, Rap R kinases and p21-activated kinase [57,58,59,60,61,62]. The above list is by no means complete and is also expanding rapidly.
Allosterism and Future of Drug Design
A large number of drugs with fewer side effects are being developed using allosteric targets. Only two types of screening strategies were discussed above - phage display and high-throughput screening. It is foreseen that complex computations can be conducted about pockets within the macromolecules for which such modulators can be designed [58,60,63]. This would depend on the availability of information on X-ray structures of molecules and on the molecular motions involved upon a ligand binding to them. Such studies would further aid the rational design of allosteric modulators. Thus, advances in drug design using allosteric modulators have taken place in the last 2 decades and are likely to become more impressive in the near future. Novel drug delivery methods may aid in the spatial selectivity of the drugs even further, thereby increasing the chances of obtaining more effective drugs with fewer side effects [64,65]. However, no matter how specific such drugs are, they cannot match the spatiotemporal basis of specificity that occurs naturally in our body. Yet every advance is helpful and adds to the hope for more effective drugs with fewer adverse effects.
References
- 1.Patwardhan B. Ethnopharmacology and drug discovery. J Ethnopharmacol. 2005;100:50–52. doi: 10.1016/j.jep.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Pina AS, Hussain A, Roque AC. An historical overview of drug discovery. Methods Mol Biol. 2009;572:3–12. doi: 10.1007/978-1-60761-244-5_1. [DOI] [PubMed] [Google Scholar]
- 3.Warren JV. William Withering revisited: 200 years of the foxglove. Am J Cardiol. 1986;58:189–190. doi: 10.1016/0002-9149(86)90276-6. [DOI] [PubMed] [Google Scholar]
- 4.Lederberg J. Mechanism of action of penicillin. J Bacteriol. 1957;73:144. doi: 10.1128/jb.73.1.144-144.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mavromoustakos T, Durdagi S, Koukoulitsa C, et al. Strategies in the rational drug design. Curr Med Chem. 2011;18:2517–2530. doi: 10.2174/092986711795933731. [DOI] [PubMed] [Google Scholar]
- 6.Mandal S, Moudgil M, Mandal SK. Rational drug design. Eur J Pharmacol. 2009;625:90–100. doi: 10.1016/j.ejphar.2009.06.065. [DOI] [PubMed] [Google Scholar]
- 7.Tan X, Osmulski PA, Gaczynska M. Allosteric regulators of the proteasome: potential drugs and a novel approach for drug design. Curr Med Chem. 2006;13:155–165. doi: 10.2174/092986706775197926. [DOI] [PubMed] [Google Scholar]
- 8.Chen YP, Chen F. Using bioinformatics techniques for gene identification in drug discovery and development. Curr Drug Metab. 2008;9:567–573. doi: 10.2174/138920008784892056. [DOI] [PubMed] [Google Scholar]
- 9.Blundell TL, Sibanda BL, Montalvao RW, et al. Structural biology and bioinformatics in drug design: opportunities and challenges for target identification and lead discovery. Philos Trans R Soc Lond B Biol Sci. 2006;361:413–423. doi: 10.1098/rstb.2005.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funder JW. Receptors, hummingbirds and refrigerators. News Physiol Sci. 1987;2:231–232. [Google Scholar]
- 11.Harms JE, Benveniste M, Maclean JK, et al. Functional analysis of a novel positive allosteric modulator of AMPA receptors derived from a structure-based drug design strategy. Neuropharmacology. 2013;64:45–52. doi: 10.1016/j.neuropharm.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mager PP. A check on rational drug design: molecular simulation of the allosteric inhibition of HIV-1 reverse transcriptase. Med Res Rev. 1997;17:235–276. doi: 10.1002/(sici)1098-1128(199705)17:3<235::aid-med2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 13.Hansen O. Vanadate and phosphotransferases with special emphasis on ouabain/NA, K-ATPase interaction. Acta Pharmacol Toxicol (Copenh) 1983;52(suppl 1):1–19. doi: 10.1111/j.1600-0773.1983.tb02475.x. [DOI] [PubMed] [Google Scholar]
- 14.Changeux JP. Allostery and the Monod-Wyman-Changeux model after 50 years. Annu Rev Biophys. 2012;41:103–133. doi: 10.1146/annurev-biophys-050511-102222. [DOI] [PubMed] [Google Scholar]
- 15.Brostrom CO, Wolff DJ. Properties and functions of calmodulin. Biochem Pharmacol. 1981;30:1395–1405. doi: 10.1016/0006-2952(81)90358-0. [DOI] [PubMed] [Google Scholar]
- 16.Pande J, Grover AK. Plasma membrane calcium pumps in smooth muscle: from fictional molecules to novel inhibitors. Can J Physiol Pharmacol. 2005;83:743–754. doi: 10.1139/y05-075. [DOI] [PubMed] [Google Scholar]
- 17.Misquitta CM, Mack DP, Grover AK. Sarco/endoplasmic reticulum Ca2+ (SERCA)-pumps: link to heart beats and calcium waves. Cell Calcium. 1999;25:277–290. doi: 10.1054/ceca.1999.0032. [DOI] [PubMed] [Google Scholar]
- 18.Grover AK, Khan I. Calcium pump isoforms: diversity, selectivity and plasticity. Review article. Cell Calcium. 1992;13:9–17. doi: 10.1016/0143-4160(92)90025-n. [DOI] [PubMed] [Google Scholar]
- 19.Toyoshima C, Nakasako M, Nomura H, et al. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 20.Strehler EE, Filoteo AG, Penniston JT, et al. Plasma-membrane Ca(2+) pumps: structural diversity as the basis for functional versatility. Biochem Soc Trans. 2007;35:919–922. doi: 10.1042/BST0350919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pande J, Mallhi KK, Sawh A, et al. Aortic smooth muscle and endothelial plasma membrane Ca2+ pump isoforms are inhibited differently by the extracellular inhibitor caloxin 1b1. Am J Physiol Cell Physiol. 2006;290:C1341–C1349. doi: 10.1152/ajpcell.00573.2005. [DOI] [PubMed] [Google Scholar]
- 22.Szewczyk MM, Davis KA, Samson SE, et al. Ca2+-pumps and Na2+-Ca2+-exchangers in coronary artery endothelium versus smooth muscle. J Cell Mol Med. 2007;11:129–138. doi: 10.1111/j.1582-4934.2007.00010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhary J, Walia M, Matharu J, et al. Caloxin: a novel plasma membrane Ca2+ pump inhibitor. Am J Physiol Cell Physiol. 2001;280:C1027–C1030. doi: 10.1152/ajpcell.2001.280.4.C1027. [DOI] [PubMed] [Google Scholar]
- 24.Pande J, Szewczyk MM, Kuszczak I, et al. Functional effects of caloxin 1c2, a novel engineered selective inhibitor of plasma membrane Ca(2+)-pump isoform 4, on coronary artery. J Cell Mol Med. 2008;12:1049–1060. doi: 10.1111/j.1582-4934.2008.00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szewczyk MM, Pande J, Grover AK. Caloxins: a novel class of selective plasma membrane Ca2+ pump inhibitors obtained using biotechnology. Pflugers Arch. 2008;456:255–266. doi: 10.1007/s00424-007-0348-6. [DOI] [PubMed] [Google Scholar]
- 26.Szewczyk MM, Pande J, Akolkar G, et al. Caloxin 1b3: a novel plasma membrane Ca(2+)-pump isoform 1 selective inhibitor that increases cytosolic Ca(2+) in endothelial cells. Cell Calcium. 2010;48:352–357. doi: 10.1016/j.ceca.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Pande J, Mallhi KK, Grover AK. A novel plasma membrane Ca(2+)-pump inhibitor: caloxin 1A1. Eur J Pharmacol. 2005;508:1–6. doi: 10.1016/j.ejphar.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 28.Pande J, Mallhi KK, Grover AK. Role of third extracellular domain of plasma membrane Ca2+-Mg2+-ATPase based on the novel inhibitor caloxin 3A1. Cell Calcium. 2005;37:245–250. doi: 10.1016/j.ceca.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Holmes ME, Chaudhary J, Grover AK. Mechanism of action of the novel plasma membrane Ca(2+)-pump inhibitor caloxin. Cell Calcium. 2003;33:241–245. doi: 10.1016/s0143-4160(02)00207-5. [DOI] [PubMed] [Google Scholar]
- 30.Lushington GH, Zaidi A, Michaelis ML. Theoretically predicted structures of plasma membrane Ca(2+)-ATPase and their susceptibilities to oxidation. J Mol Graph Model. 2005;24:175–185. doi: 10.1016/j.jmgm.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Pande J, Szewczyk MM, Grover AK. Phage display: concept, innovations, applications and future. Biotechnol Adv. 2010;28:849–858. doi: 10.1016/j.biotechadv.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Bratkovic T. Progress in phage display: evolution of the technique and its application. Cell Mol Life Sci. 2010;67:749–767. doi: 10.1007/s00018-009-0192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss G, von Haeseler A. A coalescent approach to the polymerase chain reaction. Nucleic Acids Res. 1997;25:3082–3087. doi: 10.1093/nar/25.15.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pande J, Szewczyk MM, Grover AK. Allosteric inhibitors of plasma membrane Ca pumps: invention and applications of caloxins. World J Biol Chem. 2011;2:39–47. doi: 10.4331/wjbc.v2.i3.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caulfield MP, Birdsall NJ. International Union of Pharmacology. XVII. Classification of muscarinic acetylcholine receptors. Pharmacol Rev. 1998;50:279–290. [PubMed] [Google Scholar]
- 36.Scarr E, Dean B. Muscarinic receptors: do they have a role in the pathology and treatment of schizophrenia? J Neurochem. 2008;107:1188–1195. doi: 10.1111/j.1471-4159.2008.05711.x. [DOI] [PubMed] [Google Scholar]
- 37.Raedler TJ, Bymaster FP, Tandon R, et al. Towards a muscarinic hypothesis of schizophrenia. Mol Psychiatry. 2007;12:232–246. doi: 10.1038/sj.mp.4001924. [DOI] [PubMed] [Google Scholar]
- 38.Jones CK, Byun N, Bubser M. Muscarinic and nicotinic acetylcholine receptor agonists and allosteric modulators for the treatment of schizophrenia. Neuropsychopharmacology. 2012;37:16–42. doi: 10.1038/npp.2011.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas SP, Nandhra HS, Singh SP. Pharmacologic treatment of first-episode schizophrenia: a review of the literature. Prim Care Companion CNS Disord. 2012;14 doi: 10.4088/PCC.11r01198. E-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleischhacker WW, Uchida H. Critical review of antipsychotic polypharmacy in the treatment of schizophrenia. Int J Neuropsychopharmacol. 2012:1–11. doi: 10.1017/S1461145712000399. [DOI] [PubMed] [Google Scholar]
- 41.Miyamoto S, Miyake N, Jarskog LF, et al. Pharmacological treatment of schizophrenia: a critical review of the pharmacology and clinical effects of current and future therapeutic agents. Mol Psychiatry. 2012;17:1206–1207. doi: 10.1038/mp.2012.47. [DOI] [PubMed] [Google Scholar]
- 42.Marino MJ, Rouse ST, Levey AI, et al. Activation of the genetically defined m1 muscarinic receptor potentiates N-methyl-D-aspartate (NMDA) receptor currents in hippocampal pyramidal cells. Proc Natl Acad Sci USA. 1998;95:11465–11470. doi: 10.1073/pnas.95.19.11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marino MJ, Conn PJ. Direct and indirect modulation of the N-methyl D-aspartate receptor. Curr Drug Targets CNS Neurol Disord. 2002;1:1–16. doi: 10.2174/1568007023339544. [DOI] [PubMed] [Google Scholar]
- 44.Gerber DJ, Sotnikova TD, Gainetdinov RR, et al. Hyperactivity, elevated dopaminergic transmission, and response to amphetamine in M1 muscarinic acetylcholine receptor-deficient mice. Proc Natl Acad Sci USA. 2001;98:15312–15317. doi: 10.1073/pnas.261583798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kruse AC, Hu J, Pan AC, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bridges TM, Reid PR, Lewis LM, et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda: National Center for Biotechnology Information; 2010. Discovery and Development of a Second Highly Selective M1 Positive Allosteric Modulator (PAM). http://www.ncbi.nlm.nih.gov/books/NBK50704/2010. [PubMed] [Google Scholar]
- 47.Bridges TM, Lewis LM, Dawson ES, et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda: National Center for Biotechnology Information; 2010. Discovery and Development of a Highly Selective M1 Positive Allosteric Modulator (PAM). http://www.ncbi.nlm.nih.gov/books/NBK50695/2010. [PubMed] [Google Scholar]
- 48.Shirey JK, Brady AE, Jones PJ, et al. A selective allosteric potentiator of the M1 muscarinic acetylcholine receptor increases activity of medial prefrontal cortical neurons and restores impairments in reversal learning. J Neurosci. 2009;29:14271–14286. doi: 10.1523/JNEUROSCI.3930-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Digby GJ, Shirey JK, Conn PJ. Allosteric activators of muscarinic receptors as novel approaches for treatment of CNS disorders. Mol Biosyst. 2010;6:1345–1354. doi: 10.1039/c002938f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Espinoza-Fonseca LM, Trujillo-Ferrara JG. The existence of a second allosteric site on the M1 muscarinic acetylcholine receptor and its implications for drug design. Bioorg Med Chem Lett. 2006;16:1217–1220. doi: 10.1016/j.bmcl.2005.11.097. [DOI] [PubMed] [Google Scholar]
- 51.Emmitte KA. mGlu(5) negative allosteric modulators: a patent review (2010-2012) Expert Opin Ther Pat. 2013;23:393–408. doi: 10.1517/13543776.2013.760544. [DOI] [PubMed] [Google Scholar]
- 52.Huggins DJ, Sherman W, Tidor B. Rational approaches to improving selectivity in drug design. J Med Chem. 2012;55:1424–1444. doi: 10.1021/jm2010332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grigoriadis DE, Hoare SR, Lechner SM, et al. Drugability of extracellular targets: discovery of small molecule drugs targeting allosteric, functional, and subunit-selective sites on GPCRs and ion channels. Neuropsychopharmacology. 2009;34:106–125. doi: 10.1038/npp.2008.149. [DOI] [PubMed] [Google Scholar]
- 54.Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov. 2009;8:41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barreca ML, Iraci N, Manfroni G, et al. Allosteric inhibition of the hepatitis C virus NS5B polymerase: in silico strategies for drug discovery and development. Future Med Chem. 2011;3:1027–1055. doi: 10.4155/fmc.11.53. [DOI] [PubMed] [Google Scholar]
- 56.Fenton AW. Identification of allosteric-activating drug leads for human liver pyruvate kinase. Methods Mol Biol. 2012;796:369–382. doi: 10.1007/978-1-61779-334-9_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viaud J, Peterson JR. Identification of allosteric inhibitors of p21-activated kinase. Methods Mol Biol. 2012;928:67–79. doi: 10.1007/978-1-62703-008-3_6. [DOI] [PubMed] [Google Scholar]
- 58.Wilhelm A, Lopez-Garcia LA, Busschots K, et al. 2-(3-Oxo-1,3-diphenylpropyl)malonic acids as potent allosteric ligands of the PIF pocket of phosphoinositide-dependent kinase-1 development and prodrug concept. J Med Chem. 2012;55:9817–9830. doi: 10.1021/jm3010477. [DOI] [PubMed] [Google Scholar]
- 59.Wang L, Perera BG, Hari SB, et al. Divergent allosteric control of the IRE1alpha endoribonuclease using kinase inhibitors. Nat Chem Biol. 2012;8:982–989. doi: 10.1038/nchembio.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hindie V, Lopez-Garcia LA, Biondi RM. Use of a fluorescent ATP analog to probe the allosteric conformational change in the active site of the protein kinase PDK1. Methods Mol Biol. 2012;928:133–141. doi: 10.1007/978-1-62703-008-3_10. [DOI] [PubMed] [Google Scholar]
- 61.Heald RA, Jackson P, Savy P, et al. Discovery of novel allosteric mitogen-activated protein kinase kinase (MEK) 1,2 inhibitors possessing bidentate Ser212 interactions. J Med Chem. 2012;55:4594–4604. doi: 10.1021/jm2017094. [DOI] [PubMed] [Google Scholar]
- 62.Karginov AV, Hahn KM. Allosteric activation of kinases: design and application of RapR kinases. Curr Protoc Cell Biol. 2011 doi: 10.1002/0471143030.cb1413s53. chapt 14: unit 14.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laine E, Martinez L, Ladant D, et al. Molecular motions as a drug target: mechanistic simulations of anthrax toxin edema factor function led to the discovery of novel allosteric inhibitors. Toxins (Basel) 2012;4:580–604. doi: 10.3390/toxins4080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalia YN, Perozzo R, Scapozza L. The pharmaceutical biochemistry group: where pharmaceutical chemistry meets biology and drug delivery. Chimia (Aarau) 2012;66:313–319. doi: 10.2533/chimia.2012.313. [DOI] [PubMed] [Google Scholar]
- 65.Allemann E, Delie F, Lange N. Pharmaceutical technology at the service of targeted drug delivery. Chimia (Aarau) 2012;66:308–312. doi: 10.2533/chimia.2012.308. [DOI] [PubMed] [Google Scholar]