Viral activity exerts a particularly important role in the dark ocean across the global tropical and subtropical oceans.
Abstract
Viruses are a key component of marine ecosystems, but the assessment of their global role in regulating microbial communities and the flux of carbon is precluded by a paucity of data, particularly in the deep ocean. We assessed patterns in viral abundance and production and the role of viral lysis as a driver of prokaryote mortality, from surface to bathypelagic layers, across the tropical and subtropical oceans. Viral abundance showed significant differences between oceans in the epipelagic and mesopelagic, but not in the bathypelagic, and decreased with depth, with an average power-law scaling exponent of −1.03 km−1 from an average of 7.76 × 106 viruses ml−1 in the epipelagic to 0.62 × 106 viruses ml−1 in the bathypelagic layer with an average integrated (0 to 4000 m) viral stock of about 0.004 to 0.044 g C m−2, half of which is found below 775 m. Lysogenic viral production was higher than lytic viral production in surface waters, whereas the opposite was found in the bathypelagic, where prokaryotic mortality due to viruses was estimated to be 60 times higher than grazing. Free viruses had turnover times of 0.1 days in the bathypelagic, revealing that viruses in the bathypelagic are highly dynamic. On the basis of the rates of lysed prokaryotic cells, we estimated that viruses release 145 Gt C year−1 in the global tropical and subtropical oceans. The active viral processes reported here demonstrate the importance of viruses in the production of dissolved organic carbon in the dark ocean, a major pathway in carbon cycling.
INTRODUCTION
Viruses are abundant, diverse, and dynamic components of marine ecosystems (1–3), where they play a pivotal role in controlling microbial mortality (4). In particular, virus-mediated cell lysis promotes fluxes from particulate to dissolved organic matter (5), decreasing the availability of carbon to higher trophic levels (6). However, most studies to date have been carried out in coastal environments or in the epipelagic and mesopelagic zones, with only a few studies reporting on viral abundance and dynamics in the deep ocean (7–16). Therefore,evaluation of the role of viruses in the global ocean is precluded by a paucity of information on the deep sea. Particularly, estimates of viral abundance and carbon flux supported by viruses in the dark ocean are still sparse (2). Addressing this gap will be an important contribution to microbial oceanography, for which a robust account of viral abundance and dynamics should be a pivotal underpinning. The bathypelagic zone (1000 to 4000 m), with an estimated volume of 1.3 × 1018 m3, is the largest and least explored habitat in the biosphere (17). Available evidence indicates that viral abundance in the bathypelagic realm is 0.5 to 2 orders of magnitude lower than that in surface waters (18, 19). However, attenuation of viral abundance with depth appears to be smoother than that of prokaryotes, suggesting increasing virus-to-prokaryote ratios (VPRs) in the deep sea (8, 14, 17, 20, 21). Thus, these findings would seem to support the generality of the recently proposed Piggyback-the-Winner model: “more microbes, fewer viruses” (22).
The proportionally elevated viral abundance and VPRs could be explained by a long viral turnover time in deep waters and/or by the input of allochthonous viruses attached to sinking particles (8, 15). In addition, whether viruses exert a similar mortality pressure on prokaryotes in the dark ocean as in the upper ocean remains unknown because of the lack of viral abundance and dynamics estimates in the dark realm (2).
Here, we report viral abundance and patterns of changes in abundance with depth across the global tropical and subtropical oceans. We explore patterns in viral abundance in relation to a number of possible drivers, including water mass properties (as characterized by temperature, salinity, and apparent oxygen utilization); the abundance of phototrophs in the photic layer, as represented by chlorophyll a concentration and the abundance of Synechococcus and Prochlorococcus, as important contributors to the phototrophic prokaryote community; and the abundance of heterotrophic components, as represented by heterotrophic prokaryotes and protist abundance. We quantified viral production (VP) rates and assessed the importance of viral processes from the upper to the dark ocean, and compared the magnitude of prokaryote losses through viral lysis with that by protist grazing. We do so on the basis of the Malaspina 2010–2011 Circumnavigation Expedition (Fig. 1A), which explored pelagic microbial processes across the Atlantic, Indian, and Pacific tropical and subtropical oceans (23).
Fig. 1. World map showing the location of the Malaspina sampling stations and depth-integrated viral abundance.
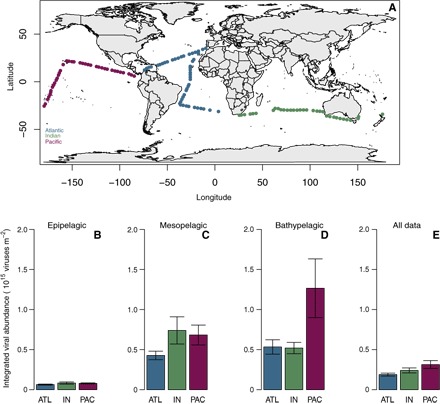
(A) Map of the Malaspina 2010 cruise showing the 120 sampling stations. Colors indicate the three different oceanic regions (Atlantic, Indian, and Pacific). Average values of depth-integrated viral abundance and their SEs are shown for the three main oceanic regions in the (B) epipelagic (0 to 200 m), (C) mesopelagic (200 to 1000 m), and (D) bathypelagic (1000 to 4000 m) layers. In (E), all depth average data are summarized. ATL, Atlantic; IN, Indian; PAC, Pacific.
RESULTS
The average viral abundance in the tropical and subtropical oceans decreased 10-fold with depth, from 7.76 × 106 viruses ml−1 in the epipelagic layer (0 to 200 m) to 0.62 × 106 viruses ml−1 in the bathypelagic layer (1000 to 4000 m) (Table 1 and fig. S1), corresponding to an average power-law scaling exponent of −1.03 km−1 (confidence interval, −1.09 to −0.97; Fig. 2A and Table 2). Significant differences were found in viral abundance between oceans for the epipelagic and mesopelagic layers, whereas no significant inter-ocean differences existed for the bathypelagic layer (Table 3). The highest average viral abundance was observed in the Pacific Ocean, whereas the lowest average viral abundance was found in the Atlantic Ocean (Table 1). The viral stock integrated between 0 and 4000 m averaged 2.41 ± 0.19 × 1014 viruses m−2 across the tropical and subtropical oceans, corresponding to an estimated carbon stock of about 0.004 to 0.044 g C m−2. Integrated viral stocks were one order of magnitude higher in the mesopelagic (200 to 1000 m) and bathypelagic layers than in the epipelagic layer (Fig. 1, B to D), with half of the viral stock integrated between 0 and 4000 m below 775 m (range, 410 to 1300 m among basins; Fig. 2B).
Table 1. Mean, SE, median, minimum, and maximum values of viral abundance in the different layers and oceans.
| n |
Mean (106 viruses ml−1) |
SE (105 viruses ml−1) |
Median (106 viruses ml−1) |
Minimum (105 viruses ml−1) |
Maximum (107 viruses ml−1) |
|
| Epipelagic | ||||||
| Atlantic | 241 | 6.60 | 3.19 | 4.83 | 4.34 | 2.59 |
| Indian | 168 | 8.31 | 4.00 | 7.70 | 2.93 | 3.31 |
| Pacific | 193 | 8.72 | 4.71 | 6.89 | 0.85 | 4.39 |
| All epipelagic data | 602 | 7.76 | 2.30 | 6.52 | 0.85 | 4.39 |
| Mesopelagic | ||||||
| Atlantic | 88 | 0.10 | 0.64 | 0.88 | 0.85 | 0.32 |
| Indian | 60 | 1.89 | 4.22 | 1.35 | 1.22 | 2.51 |
| Pacific | 80 | 1.67 | 2.15 | 1.09 | 0.13 | 1.33 |
| All mesopelagic data | 228 | 1.47 | 1.38 | 1.55 | 0.13 | 2.51 |
| Bathypelagic | ||||||
| Atlantic | 90 | 0.51 | 0.57 | 0.47 | 0.36 | 0.47 |
| Indian | 63 | 0.52 | 0.56 | 0.47 | 0.38 | 0.25 |
| Pacific | 58 | 0.89 | 2.29 | 0.35 | 0.05 | 1.11 |
| All bathypelagic data | 211 | 0.62 | 0.70 | 0.45 | 0.05 | 1.11 |
Fig. 2. Viral abundance and depth.
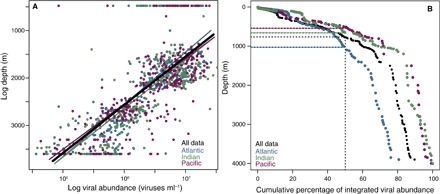
(A) Power-law fit between log-transformed viral abundance and depth in the Atlantic, Indian, and Pacific oceans and in all the data. Lines denote the best power-law model regression. (B) Cumulative percentage curves of integrated viral abundance according to depth for the three oceanic regions and for all the data. Dashed lines in (B) show the depth where half of the integrated viral stock between 0 and 4000 m is contained in each oceanic region and for all the data.
Table 2. Log-log slopes and fits between viral and prokaryotic abundance in the epipelagic, mesopelagic, and bathypelagic layers in the three different oceans and in all the subtropical ocean and power-law log-log slopes and fits of the relationship between depth and viral abundance in the three different oceans and in all the tropical and subtropical oceans.
| Viral abundance–prokaryotic abundance | |||||||
| n | Estimated intercept | Intercept SE | Slope | Slope SE | r2 | P | |
| Epipelagic | |||||||
| Atlantic | 240 | 1.7 | 0.54 | 0.87 | 0.09 | 0.26 | <0.0001 |
| Indian | 168 | 4.72 | 0.45 | 0.36 | 0.08 | 0.12 | <0.0001 |
| Pacific | 193 | 6.36 | 0.42 | 0.08 | 0.07 | 0.007 | 0.25 |
| All epipelagic data | 593 | 4.93 | 0.27 | 0.32 | 0.05 | 0.07 | <0.0001 |
| Mesopelagic | |||||||
| Atlantic | 87 | 2.19 | 0.56 | 0.75 | 0.11 | 0.35 | <0.0001 |
| Indian | 60 | 3.76 | 1.27 | 0.45 | 0.24 | 0.05 | 0.07 |
| Pacific | 80 | 3.89 | 0.67 | 0.41 | 0.13 | 0.12 | <0.0001 |
| All mesopelagic data | 227 | 3.31 | 0.38 | 0.52 | 0.07 | 0.18 | <0.0001 |
| Bathypelagic | |||||||
| Atlantic | 89 | 2.45 | 0.85 | 0.7 | 0.19 | 0.13 | <0.0001 |
| Indian | 63 | 3.31 | 1.03 | 0.48 | 0.22 | 0.07 | 0.03 |
| Pacific | 58 | 2.98 | 0.92 | 0.53 | 0.19 | 0.12 | 0.007 |
| All bathypelagic data | 210 | 3.68 | 0.44 | 0.41 | 0.09 | 0.08 | <0.0001 |
| All data | 1030 | 1.44 | 0.12 | 0.91 | 0.02 | 0.62 | <0.0001 |
| Viral abundance–depth (power-law function) | |||||||
| Atlantic | 419 | 9.26 | 0.32 | −1.12 | 0.05 | 0.54 | <0.0001 |
| Indian | 291 | 8.77 | 0.37 | −1.02 | 0.06 | 0.52 | <0.0001 |
| Pacific | 331 | 8.3 | 0.33 | −0.96 | 0.05 | 0.51 | <0.0001 |
| All data | 1040 | 8.75 | 0.2 | −1.03 | 0.03 | 0.52 | <0.0001 |
Table 3. Analysis of variance (ANOVA) and post hoc Tukey results testing for significant differences in viral abundance and VPR among layers [epipelagic (EPI), mesopelagic (MESO), and bathypelagic (BATHY)] and oceans [Atlantic (ATL), Indian (IN), and Pacific (PAC)].
Lower panel: ANOVA and post hoc Tukey results testing for significant differences in viral abundances among water masses in the mesopelagic and bathypelagic layers. Only significant values are shown (P < 0.05).
| ANOVA | Tukey’s test | ||||
| Viral abundance | |||||
| Layers | |||||
| n | F | P | Layers | P | |
| 1040 | 1040.16 | <0.0001 | EPI-BATHY | <0.0001 | |
| EPI-MESO | <0.0001 | ||||
| MESO-BATHY | <0.0001 | ||||
| Oceans | |||||
| Layers | n | F | P | Oceans | P |
| Epipelagic | 601 | 10.59 | <0.0001 | PAC-ATL | 0.0002 |
| IN-ATL | 0.0004 | ||||
| Mesopelagic | 227 | 5.12 | 0.007 | ATL-IN | 0.01 |
| ATL-PAC | 0.05 | ||||
| Bathypelagic | 210 | 0.005 | 0.1 | ||
| VPR | |||||
| Layers | |||||
| n | F | P | Layers | P | |
| 1030 | 2.93 | 0.05 | EPI-MESO | 0.05 | |
| Oceans | |||||
| Layers | n | F | P | Oceans | P |
| Epipelagic | 601 | 17.71 | <0.0001 | PAC-ATL | <0.0001 |
| PAC-IN | 0.004 | ||||
| Mesopelagic | 227 | 1.14 | 0.321 | ||
| Bathypelagic | 210 | 3.1 | 0.05 | ATL-IN | 0.04 |
| Water masses | |||||
| n | F | P | Water masses | P | |
| Mesopelagic | 228 | 3.08 | 0.001 | AAIW-13EqPac | 0.04 |
| AAIW-ICW_13 | 0.02 | ||||
| ENACW-ICW_13 | 0.03 | ||||
| Bathypelagic | 211 | 1.53 | 0.16 | ||
Viral abundance in the mesopelagic ocean differed across water masses (Table 3 and table S1), with the highest abundance detected in the Indian Central Water of 13°C (ICW_13), with a mean value of 3.93 × 106 viruses ml−1; the lowest abundance was found in the Antarctic Intermediate Water (AAIW) waters, with a mean value of 0.81 × 106 viruses ml−1 (table S1). However, no significant differences in viral abundance were detected among bathypelagic water masses (Table 3 and table S2). The highest bathypelagic abundance was observed in the Circumpolar Deep Water (CDW), with an average abundance of 0.60 × 106 viruses ml−1; the lowest abundance was observed in the Antarctic Bottom Water (AABW), with an average abundance of 0.35 × 106 viruses ml−1 (table S2).
The average VPR was similar in epipelagic (14.2 ± 0.6) and bathypelagic (14.4 ± 1.6) layers of tropical and subtropical oceans, with somewhat lower values (12.5 ± 1.8) in the mesopelagic layer (Fig. 3, A to D, and Table 3). In the epipelagic layer of the Atlantic Ocean, however, VPR was significantly lower than in the Indian and Pacific oceans (Fig. 3A and Table 3), whereas in the bathypelagic layer, VPR values in the Atlantic were higher than those in other oceanic regions (Fig. 3C). Viral abundance scaled as the 0.91 ± 0.02 power of prokaryotic abundance across the tropical and subtropical oceans (r2 = 0.62, P < 0.001, n = 1030; Table 2 and Fig. 3H), with the strength of this relationship varying across basins and depth layers (Fig. 3, E to G, and Table 2).
Fig. 3. VPR and relationships between viral and prokaryotic abundances.
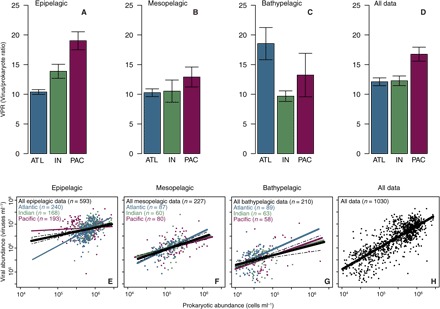
Averages values of the VPR and their SEs in the three oceanic regions (Atlantic, Indian, and Pacific) sampled during the cruise in the (A) epipelagic (0 to 200 m), (B) mesopelagic (200 to 1000 m), and (C) bathypelagic (1000 to 4000 m) layers and in (D) all the data. Relationships between viral and prokaryotic abundances in the (E) epipelagic, (F) mesopelagic, and (G) bathypelagic layers from the Atlantic, Indian, and Pacific oceans and in (H) all the data. Dashed lines show the 95% prediction intervals from linear regressions of all data in the epipelagic, mesopelagic, and bathypelagic layers.
Because the relationship between viral and prokaryotic abundance was significant but weak in the different layers (Fig. 3, E to G, and Table 2), multivariate multiple regression analysis was used to identify potential variables explaining viral abundance distribution and variability between the different layers and between oceanic regions. Viral abundance in the epipelagic layer was negatively related to salinity and AOU (apparent oxygen utilization) but increased with increasing chlorophyll a concentration and Synechococcus abundance across the tropical and subtropical oceans. In contrast to Synechococcus, Prochlorococcus abundance was not significantly related to viral abundance in our data set. Prokaryotic abundance did not explain viral abundance variability for the entire epipelagic layer, although present in the Atlantic and Indian multiple regression models (table S3). In the mesopelagic layer, the main variables contributing to explain viral abundance variability were prokaryotic production and abundance and heterotrophic protist (HP) abundance (table S4), whereas in the bathypelagic layer, prokaryotic abundance and temperature contributed similarly, yet the best model explained only the 11% of the viral abundance variability (table S5).
Lytic viral production (VPL) and lysogenic viral production (VPLG) were determined in 11 selected stations at the surface, the deep chlorophyll maximum (DCM), and at 4000-m depth. Although significant statistical differences were not detected, VPL production decreased, on average, almost threefold from 9.12 ± 3.0 × 106 viruses ml−1 day−1 in surface waters to 3.44 ± 1.29 × 106 viruses ml−1 day−1 at 4000-m depth, whereas VPLG decreased almost 20-fold from the surface (22.4 ± 10.0 × 106 viruses ml−1 day−1) to 4000-m depth (1.36 ± 0.49 × 106 viruses ml−1 day−1), with bathypelagic VPL being twice as high as VPLG (Fig. 4A, fig. S2, and table S6). Mortality by viruses [virus-mediated mortality (VMM)] and grazing by protists [protist-mediated mortality (PMM)] accounted for a similar fraction of total prokaryotic mortality in surface waters, whereas protistan grazing prevailed at the DCM layer (Fig. 4B, fig. S2, and table S6). In contrast, viral lysis was, by far, the dominant source of prokaryote mortality at 4000-m depth (Fig. 4B, fig. S2, and table S6), but it did not show significant differences from surface values. Similarly, in surface waters fluxes of C, N, and P from prokaryotes to the dissolved organic pool by viral lysis were similar to C, N, and P levels incorporated by protist grazers (table S7). In contrast, virus-mediated C, N, and P fluxes were lower than those associated with protistan grazing at the DCM (table S7). At 4000-m depth, the C, N, and P released by viruses, while being much lower than those at the surface and DCM layers, exceeded by 60 times the C, N, and P incorporated by grazing at the same deep layer.
Fig. 4. VPL and VPLG and prokaryotic mortality due to viruses.
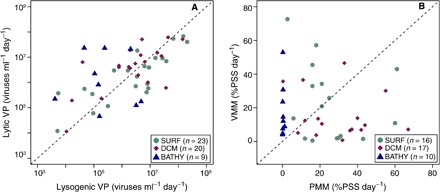
(A) VPL against VPLG and (B) VMM against PMM detected at the surface (SURF; 3-m depth), DCM, and bathypelagic (BATHY; 4000-m depth) layers from 11 selected stations: 4 in the Atlantic Ocean, 3 in the Indian Ocean, and 4 in the Pacific Ocean. Three replicates were performed in each experiment. Dashed line is the 1:1 line. Lytic/lysogenic ratios at the surface, DCM, and bathypelagic layers were 4.63 ± 2.76, 10.00 ± 7.23, and 53.89 ± 39.17, respectively. Viral/grazing-mediated mortality ratios were 2.35 ± 1.47 at the surface layer, 7.15 ± 6.52 at the DCM layer, and 173.11 ± 128.91 at the bathypelagic layer.
DISCUSSION
The results presented here provide a global assessment of the viral abundance, the role of viruses in prokaryote mortality, and the associated flows of carbon and nutrients in the global tropical and subtropical oceans, the largest biome on Earth. The global attenuation of viral abundance with depth reported here, with an average power-law scaling exponent of −1.03 km−1 (Table 2), extends patterns previously reported for stations sampled in the Atlantic Ocean (8, 13, 15, 21), Pacific Ocean (7, 16, 20), and Indian Ocean (21). We found that, despite the observed decrease with depth, the mesopelagic and bathypelagic layers host, on average, 94.7% of the total viruses present down to 4000 m in the tropical and subtropical oceans (Fig. 2B), depicting the dark ocean as the largest reservoir of marine viruses. We estimated a total number of 5.34 × 1028 viruses in the global tropical and subtropical oceans. This is only 4% of the value (1.29 × 1030) estimated by Cobián Güemes et al. (24) for the global ocean, despite the tropical and subtropical oceans representing 75% of the global ocean surface and our estimate of integrated water column microbes down to the mean depth of the ocean. In the absence of previous data on viral abundance on the deep sea, Cobián Güemes et al. (24) used an indirect approach to estimate global viral abundance in the ocean, as the product of the abundance of prokaryote cells and the median VPR. Their assumed median VPR, 12.76, is close to that derived here in our data set. Hence, the 20-fold difference between our direct estimate and the indirect estimate provided by Cobián Güemes et al. (24) is likely explained by an overestimate of the global abundance of prokaryote cells used by Cobián Güemes et al. (24).
Here, viral abundance was higher in the Pacific region and particularly in bathypelagic waters (Fig. 1D and fig. S1). This result agrees with previous findings, from the same expedition, that indicated a higher abundances of planktonic HPs in this region (25). We also report comparable VPR values between epipelagic and bathypelagic waters (Fig. 3, A to D), indicating that viruses are likely to play an important role also in the deep sea (8, 15, 20).
Viral abundance differed significantly among water masses, particularly in the mesopelagic region. However, we found that viral abundance was relatively homogeneous in the bathypelagic layer, regardless of water mass type (Table 3). In addition to factors defining water masses, such as thermohaline and AOU properties, the abundance of potential hosts, including prokaryotes and protists, partially explained differences in viral abundance in the tropical and subtropical oceans. The importance of autotroph abundance, as represented by chlorophyll a concentration and Synechococcus abundance, as predictors of viral abundance in the epipelagic ocean agrees with previous reports of relationships between viral and photoautotrophic picoplankton abundance in the euphotic zone (16, 20, 21, 26) and the dominance of cyanophages in metagenomic analyses on phage sequences in euphotic layers (27–29).
The scaling of total viral abundance as the 0.91 power of total prokaryote abundance (Table 2) in the tropical and subtropical oceans is consistent with previous reports (17, 21). However, the scaling of viral to prokaryote abundance varied greatly across layers and basins, and most of the regression analysis for different layers and ocean basins yielded power slopes between log viral abundance and log bacterial abundance <1. Our results are consistent with the recent findings by Wigington et al. (30) and Knowles et al. (22), who also suggested a global pattern in which viral abundance decreases with increasing prokaryotic abundance.
The rates of VP (both lytic and lysogenic) decreased from the surface to 4000 m, as shown in previous reports (8, 14, 15, 31, 32). Our data also revealed that lysogeny is more important than lysis in the photic layer (Fig. 4 and fig. S2), consistent with the oligotrophic conditions prevailing in the surface of the open ocean, which may favor the lysogenic over the lytic viral cycle (33, 34). Conversely, we demonstrate that lytic production rates are not statistically dissimilar, on average, to lysogenic rates in the dark ocean, despite lytic rates in the bathypelagic slightly predominating over the lysogenic ones. This finding, together with the finding of similar values of VPR in the epipelagic and bathypelagic layers, suggests that viruses are more active components of the deep-sea microbial food web than previously thought. Thus, our results show higher rates of lysogeny in the oligotrophic surface ocean, which displayed the higher host densities compared with the deeper layers. Therefore, in our data set, there is potential support for both the promotion of lysogeny by low growth rates and low nutrient concentrations as wells as for the Piggyback-the-Winner dynamics (24), wherein lysogeny is promoted by higher host density. Our findings suggest that, in marine ecosystems, the lytic-lysogenic switch is probably more complex than so far believed and does not depend simply on nutrient concentration, growth rates, and host densities. This novel conclusion calls for further investigations to elucidate the factors that trigger the lytic-lysogenic switch.
One possible explanation for the similar rates of lysis and lysogeny in the dark ocean is that sinking particles may act as hot spots of VP. The deep-sea pelagic ecosystem contains a variety of particles that represent important sources of organic matter fueling the dark ocean food web (35). Diversity and composition of free-living (FL) and particle-attached (PA) bacterial communities have been studied in some of the same stations during the Malaspina cruise using high-throughput sequencing of their 16S ribosomal DNA (36). The results indicated that the FL and PA bacterial communities had diversity differences at the global ocean scale (36). Therefore, this nonrandom distribution of bacterial communities is expected to affect viral abundance. A predominately prokaryotic PA life strategy might sustain a relatively high abundance of viruses in the deep ocean (37). Proctor and Fuhrman (38) estimated that 2 to 37% of the particulate-associated bacteria may be killed by viral lysis, and Riemann and Grossart (39) demonstrated elevated lytic phage production associated with model particles. Hence, future studies should quantify the dynamics and patterns of PA viruses as opposed to their FL counterpart. These results are important because they come to clarify an ongoing debate resulting from the hitherto scarcity of data on viruses in the deep sea. Weinbauer et al. (11) reported an increase of lysogeny in deep waters in the Mediterranean Sea, a pattern that our results do not support for the global ocean. However, our results are consistent with the findings by Winter et al. (40) and Boras et al. (41) who did not find an evident pattern with depth in the subtropical Atlantic Ocean.
The VP rates reported here imply, on average (±SE), turnover rates for the free viruses of 1.71 ± 0.88 day−1 in the photic layer (0.98 ± 0.40 day−1 at the surface layer and 2.44 ± 1.36 day−1 at the DCM layer), compared to 11.67 ± 5.51 day−1 at 4000-m depth. These correspond to turnover times of ca. 0.6 days in the photic layer and only 0.1 days in the bathypelagic layer, revealing that viruses in the bathypelagic layer are highly dynamic, exhibiting a much faster turnover than viral communities of surface waters or deep-sea sediments (42). The fast viral turnover in the bathypelagic ocean not only results from high rates of VPL but also points at high loss rates of the viral particles. Whereas these loss factors remain unknown, we speculate that these may involve adsorption of viruses to particles, a possibility that remain to be tested. Moreover, the relatively high VPL at depth suggests that a high infection rate occurs. Although infection rates were not determined directly, this pattern is consistent with the somewhat higher median VPR in the deep sea, suggesting a higher encounter probability between viruses and hosts.
Our results may also contribute to resolve the role of virus-induced mortality for prokaryotes. The percentage of prokaryotic standing stock (%VMMPSS) shunted by viral lysis was, on average, 22.50 ± 5.54% day−1 (fig. S2), which agrees with the values reported by Fuhrman (43) and Suttle (4) for the ocean. This requires, to maintain prokaryote biomass in steady state, a minimum of one duplication around every 5 days, on average. This compares to an estimated prokaryote doubling time, in the experimental samples where mortality was measured, of 1.33 ± 0.32 days, sufficient to support viral prokaryotic lysis rates along with other losses, such as grazing losses. In particular, our results indicate that, on average, 26.6% of prokaryote cells are removed by viruses in the bathypelagic ocean, nonsignificantly more than those removed in the epipelagic (22.76%) waters. Protistan grazing rates comprised a similar percentage of prokaryotic mortality in the epipelagic layer (surface and DCM layers, 23.61 and 33.51%, respectively). However, the estimated pressure on prokaryote populations from protist grazing declined markedly in the dark ocean, where protist grazing was found to be negligible, removing only, on average, 0.33% of the prokaryote biomass per day. These results are against the hypothesized major role of flagellates as a source of bacterial mortality in bathypelagic waters postulated by Arístegui et al. (17) on the basis of the observed decline of the prokaryote/flagellate ratio with depth, whereas the VPR did not decline with depth. Arístegui et al. (17) provided a set of hypotheses to address the lack of data precluding progress in microbial oceanography research of the deep, dark ocean, some of which we intended to test in our research. For instance, Pernice et al. (25) suggest the increase, not the decrease, of the prokaryote/flagellate ratio with depth in the tropical and subtropical oceans. Our results are also consistent with recent findings in the subtropical Atlantic Ocean, in which the fraction of the prokaryote standing stocks consumed by grazing decline with depth (44) and viral lysis exceeds grazing rates in bathypelagic communities (30). Hence, our results, which provide the first global survey of the deep tropical and subtropical oceans, confirm that viral lysis in the bathypelagic ocean is overwhelmingly far more important than grazing as a cause of mortality of prokaryote populations. Elucidating which environmental drivers are able to switch top-down control on prokaryotes, from grazing to infection, cannot be resolved here. Although our data set offers a unique description of deep-sea viral abundance and dynamics, it does have limitations to resolve the mechanisms underlying the contrasting patterns found with depth and across ocean basins. The data set has opened up new relevant questions, such as the control of virus infectivity and the mechanisms that lead to the lytic-lysogenic switch in pelagic communities. The broad-scale patterns provided here for the tropical and subtropical oceans have opened a number of key questions to guide future efforts that could not have been formulated before because of the previous scarcity of empirical information on deep-sea viruses.
The finding that VP and lysis dominate material cycling in the tropical and subtropical oceans points at these processes as a significant source of dissolved organic carbon (DOC) and nutrients for the microbial food web. In particular, the fluxes of carbon, nitrogen, and phosphorus released by viruses in the bathypelagic layer, which far exceeded those incorporated by protists via prokaryote grazing, could fuel prokaryotic activity in the deep ocean. Recent results, also derived from the Malaspina Circumnavigation Expedition, demonstrated that the low concentration of different compounds within the DOC pool, rather than their possible recalcitrant nature, limits microbial growth in the deep sea (45). Because dissolved organic matter released from viral lysis is easily assimilated by prokaryotes (43, 46–48), the viral shunt may play a key role in fueling prokaryotic growth and production through a recycling loop in the deep sea. Because DOC is in near steady state in the bathypelagic ocean, the DOC released by the viral shunt is likely to be rapidly reused by prokaryotes. A first-order estimate, derived by extrapolating our results to the global tropical and subtropical oceans, suggests that the viral shunt could release about 145 Gt of C, 27.6 Gt of N, and 4.6 Gt of P annually. This compares to an estimated oceanic net primary production ranging from 35 to 65 Gt of C per year (49). The carbon release by the viral shunt represents a transfer from particulate to dissolved organic form, likely fueling significant recycling by prokaryotes, which may take up the released products, thereby supporting recycled prokaryotic production. However, each iteration of this cycle would result in respiratory losses contributing to increased community respiration (4), a large fraction of which occurs in the dark ocean (49). Although the empirical basis for the estimates of carbon released by viruses from our data needs to be expanded, they are consistent with previous results. Suttle (50) estimated that the viral shunt releases 150 Gt of C per year for the photic zone of the global ocean, 70% of which is comprised (by surface) by the tropical and subtropical oceans sampled here. Hence, considering Suttle’s estimate of 150 Gt of C per year released by the viral shunt across the photic zone of the global ocean and our estimate of the carbon released through this process in the dark ocean along the tropical and subtropical oceans, we conclude that the dark ocean is likely to contribute approximately one-fourth of all carbon recycled through the viral shunt.
In conclusion, our results confirm that viruses play a key role in the food web and biogeochemical fluxes of the global tropical and subtropical oceans. Although viral abundance and production decrease with depth, viruses play a particularly important role in the dark ocean, which contains the highest integrated viral abundance and where viral lysis is the dominant cause of prokaryote mortality. The viral activity represents a globally significant vector of regenerated organic carbon and nutrients across the global tropical and subtropical oceans as an ecosystem with virioplankton prevalence over other planktonic compartments. The causes for the higher dominance of viruses and viral lytic processes in the dark ocean relative to the overlying water column, possibly linked to particle association and the absence of other sources of prokaryote mortality as protistan grazing, should be specifically addressed in future studies.
MATERIALS AND METHODS
Sample collection
The Malaspina 2010 Circumnavigation Expedition sailed the tropical and subtropical oceans on board R/V Hespérides (23), sampling a total of 120 stations along the Atlantic, Indian, and Pacific oceans (Fig. 1A). At each station, water samples from 1 to 21 depths were collected from the surface down to 4000 m using Niskin bottles attached to a rosette sampling system equipped with a Seabird 911plus CTD probe.
Chlorophyll a concentration
Chlorophyll a concentration was determined in water samples within the top 200 m by filtering 200 to 500 ml of seawater on Whatman GF/F filters, extracting the pigment in acetone (90%, v/v) in the dark at 4°C for 24 hours, and measuring fluorescence with a Turner Designs fluorometer, as described by Estrada et al. (51).
Viruses, prokaryotes, protistan, and picophytoplankton abundances
A total of 1040 estimates from each sampled station and depth of viral abundance were conducted by flow cytometry. Immediately after collection, 2 ml of seawater was fixed with glutaraldehyde (0.5% final concentration), refrigerated, quick-frozen in liquid nitrogen, and stored at −80°C (52). Counts were made using a FACSCalibur flow cytometer (BD Biosciences) with a blue laser emitting at 488 nm. We chose flow cytometry as a method to perform viral counts because previous studies have shown strong correlation with epifluorescence microscopy (EFM) to detect large double-stranded DNA viruses (52) and lower fluorescent bacteriophages (53, 54). Several months after collection (ca. 12 months), the samples were unfrozen, stained with SYBR Green I (Molecular Probes, Invitrogen), and run at a medium flow speed (55). The samples were diluted with tris-EDTA buffer such that the event rate was between 100 and 800 viruses per second. The data obtained for green fluorescence (FL1) and side scatter (SSC) were collected for analysis using CellQuest software (Becton Dickinson). Viral abundance, expressed as the number of viruses per milliliter, was integrated over depth to calculate the total number of viruses per square meter. The depth-integrated viral abundance was calculated by multiplying the distance between the different sampling points integrated, and the viral abundance was expressed in cubic meters. This resulted in depth-integrated virus abundances from 0 to 200 m (epipelagic layer), 200 to 1000 (mesopelagic layer), and 1000 to 4000 m (bathypelagic layer) and for the whole total water column (0 to 4000 m) expressed as virus per square meter. Viral C biomass was estimated using a range of carbon factor of 0.02 to 0.2 fg of C per virus (56).
The abundance of heterotrophic prokaryotes from the same samples than viral abundance was measured by flow cytometry. Immediately after collection, 1.5 to 2 ml of seawater samples were fixed with 1% paraformaldehyde and 0.05% glutaraldehyde and, after 15 min at room temperature in the dark, were deep-frozen in liquid nitrogen. A few days after sampling, the samples were unfrozen, and the prokaryotes were stained with SYBR Green I (Molecular Probes, Invitrogen) at a 1:10,000 dilution and run in a FACSCalibur flow cytometer (BD Biosciences) equipped with a 488-nm, 15-mW argon-ion blue laser, as explained elsewhere (57). At least 100,000 events were recorded, and photosynthetic prokaryotes were distinguished in a DNA-derived green fluorescence against a chlorophyll-derived red fluorescence scatterplot. The flow rate ranged between 35 μl min−1 for samples above 1000-m depth and 150 μl min−1 for deeper samples, whereas the acquisition time ranged from 30 to 260 s depending on cell concentration in each sample. Data were collected in an FL1 (green fluorescence) versus SSC plot and analyzed as detailed by Gasol and del Giorgio (57). The calibration of the instrument for absolute counts was done daily by measuring the exact volume being analyzed with a calibrated automatic pipette. Fluorescent beads (1 μm; Fluoresbrite carboxylate microspheres, Polysciences Inc.) were added at a known density as internal standards.
Picophytoplankton abundance and identification of Prochlorococcus, Synechococcus, and picoeukaryotes were also determined on board in fresh samples from each surface sampled station by flow cytometry (BD FACSCalibur). An aliquot of a calibrated solution of 1-μm-diameter fluorescent spheres (Polysciences) was added to 1-ml replicated samples as an internal standard for the quantification of cell concentration (58). Red (FL3; band-pass filter, >670 nm), green (FL1; band-pass filter, 530 nm), and orange (FL2; band-pass filter, 585 nm) fluorescence, as well as the forward and side scattering signals of the cells and beads, were used to detect picoplanktonic populations of Synechococcus sp., Prochlorococcus sp., and eukaryotes (59).
HPs were measured only for mesopelagic and bathypelagic layers from, at least, five depths between 200 and 4000 m. Seawater (4.8 ml) was fixed with 25% glutaraldehyde (electron microscopy grade; 1% final concentration), deep-frozen in liquid nitrogen, and stored at −80°C until it was analyzed in the laboratory within 7 months after the end of the cruise. Samples were processed with a FACSCalibur flow cytometer (BD Biosciences) with a blue laser emitting at 488 nm using the settings described by Christaki et al. (60), adapted from the protocol of Zubkov et al. (61). Each sample was stained for at least 10 min in the dark with dimethyl sulfoxide–diluted SYBR Green I (Molecular Probes, Invitrogen) at a final concentration of 1:10,000. The flow rate was established at about 250 ml min−1, with data acquisition for 5 to 8 min depending on cell abundance. Samples showing more than one 200 events per second were diluted. Filtered samples (that is, blanks) never had any event in the flow cytometrically defined area of interest. The flow cytometer output was analyzed using CellQuest software (Becton Dickinson), initially visualized as a cloud of points in a window showing SSC versus green fluorescence (FL1), which contained all cells stained by SYBR Green I. From this plot, target cells were identified after excluding the remaining noise, autofluorescent particles, and heterotrophic prokaryotes, using different displays of the optical properties of the detected particles, as explained by Christaki et al. (60). Measurements were repeated several times in three random stations, and the calculated SEs corresponded to 1.5% of the average.
Viral and prokaryotic production
Prokaryotic heterotrophic production (PHP) rates were estimated from each station and depth sampled from [3H]leucine incorporation (62) using the centrifugation method (63) with four 1.2-ml replicate Eppendorfs and two blank controls for waters <1000 m. Seawater was incubated in water baths or thermostatic chambers maintained at the in situ temperature ±1°C for a varying amount of time from 2 (surface) to 12 (4000 m) hours. Leucine was used at 20 nM and was diluted 10× with cold leucine. In bathypelagic waters, leucine incorporated rates were measured by incubating 40 ml in Falcon tubes and filtration on 0.2-μm Polycarbonate (Millipore) filters using 5 nM and nondiluted hot leucine. The filters were rinsed three times in trichloroacetic acid (final concentration, 50%) and dissolved in scintillation vials with FilterCount. Because no empirical leucine-to-carbon conversion factors were available for the entire water column (64), we converted leucine uptake rates to PHP using the theoretical value of 1.5 kg of C per mol of Leu.
Samples for VP were collected from the surface, DCM, and 4000-m depth at 11 selected stations: 4 in the Atlantic Ocean, 3 in the Indian Ocean, and 4 in the Pacific Ocean. We used the virus reduction approach to determine VP and prokaryotic losses due to phages (65). Briefly, 1 liter of seawater was prefiltered through a 0.8–μm–pore size cellulose filter (Whatman) and then concentrated by a spiral wound cartridge (0.22 μm pore size; VivaFlow 200) to obtain 100 ml of prokaryotic cell concentrate. Virus-free water was prepared by filtering 0.5 liter of seawater using a 30-kDa VivaFlow 200 cartridge. A mixture of virus-free water (210 ml) and prokaryotic concentrate (70 ml) was prepared and distributed into six sterile 50-ml Falcon plastic tubes. Three of the tubes were kept without any manipulations as controls, whereas in the other three, mitomycin C (Sigma-Aldrich) was added (final concentration of 1 μg ml−1) as inducing agent of the lytic cycle. All tubes were incubated at in situ temperature in the dark for 12 to 24 hours. Samples for viral and prokaryotic abundances were collected at time 0 and every 4 hours, fixed with glutaraldehyde (0.5% final concentration), stored, and counted by flow cytometry, as described above. The number of viruses released by prokaryotic cells (burst size) was estimated from VP measurements (66, 67) as the increase in viral abundance during the 4-hour time intervals divided by the decrease in prokaryotic abundance in the same time period. We assumed that the increase in prokaryotic production and viral decay in this time interval were negligible. VMM was estimated as previously described (68, 69). Briefly, an increase in viral abundance in the control Falcon tubes represents VPL, and the difference between the viral increase in the mitomycin C treatments and VPL gives the VPLG. Because a fraction of the prokaryotes are lost during the concentration process, VPL and VPLG were multiplied by a correction factor to compare the VP values from different incubations. This factor was calculated by dividing the in situ prokaryotic abundance from the corresponding station by the prokaryotic abundances at T0 in the VP measurement tubes (70). We then calculated the rate of lysed cells (RLC; cells per milliliter per day) by dividing VPL by BS, as described by Guixa-Boixereu (71). RLC was used to calculate VMM as VMMPSS as
where PA0 is the initial prokaryotic abundance in the corresponding natural sample.
Grazing rates
Samples for prokaryotic mortality due to protists were collected from the surface and DCM in the same stations where VP rates were measured. For samples at 4000 m, grazing rates were estimated on the basis of the study by Vaqué et al. (72). Grazing rates (G) were measured following the fluorescently labeled bacteria (FLB) disappearance method (73, 74). For each measurement, triplicated 1.5-liter sterile bottles were filled with 0.5-liter aliquots of seawater from each sample, and a fourth bottle was filled with 0.5 liter of grazer-free seawater as a control. Each triplicate and control was inoculated with FLB at 20% of the natural prokaryotic abundance. The FLB were prepared from a culture of Brevundimonas diminuta (www.cect.org), as described by Vázquez-Domínguez et al. (74). Bottles were incubated at in situ temperature in the dark for 48 hours. For assessing the prokaryotic and FLB abundances, samples were taken at the beginning and at the end of the grazing assay. Abundances of total prokaryotes and FLB were obtained by EFM. Natural prokaryotes were identified by their blue fluorescence when excited with ultraviolet radiation, whereas FLB were identified by their yellow-green fluorescence when excited with blue light. Control bottles showed no decrease in FLB at the end of the incubation time. Grazing rates of prokaryotes were obtained using the equations of Salat and Marrasé (75) [for more details, see the study of Lara et al. (76)]. Grazing rates from bathypelagic depths were estimated following the model by Vaqué et al. (72), which calculates grazing rates (G) as a function of temperature (T) and heterotrophic nanoflagellate (HF) abundance (log G = −0.64 + 1.2 log HF + 0.07 T).
Estimations of carbon, nitrogen, and phosphorus from viral lysis and grazing rates
To estimate the carbon, nitrogen, and phosphorus released by prokaryotic viral lysis or incorporated by protists grazing on prokaryotes, the average rate of lysed prokaryotes (cells per milliliter per day) by viruses and the average grazed prokaryotes by protists (cells per milliliter per day) were multiplied by the average prokaryotic cell content of C, N, and P determined as a function of average cell volume (77). Prokaryote cellular volume was estimated from bead-standardized SSC flow cytometric measurements following Calvo-Díaz and Morán (78). All rates were expressed as micrograms of C per liter per day. With this, we provided a first-order estimate of the total carbon, nitrogen, and phosphorus released by virus or incorporated by protist grazing across the global tropical and subtropical oceans as the product of the geometric mean of the integrated rates of C, N, and P per total area of the tropical and subtropical oceans (2.40 × 108 km2).
Identification of water masses
The water masses in the dark ocean (mesopelagic and bathypelagic layers) were defined by the mixing of distinct water types (WTs), which are characterized on the basis of salinity and potential temperature (79). A robust calculation of the different WT proportions found in the sampling stations was performed by means of a classical water mass analysis, leading to the identification of 22 WTs during the Malaspina cruise (79). Here, the water masses ENACW_12 (Eastern North Atlantic Central Water of 12°C) and ENACW_15, defined by two WTs of 12° and 15°C, were grouped in only one water mass (named ENACW) through the addition of their respective percentages. Likewise, we grouped water masses SACW_12 (South Atlantic Central Water of 12°C) and SACW_18, AAIW_3.1 (AAIW of 3.1°C) and AAIW_5.0, and NADW_2.0 (North Atlantic Deep Water of 2.0°C) and NADW_4.6, which were converted into SACW, AAIW, and NADW, respectively. The AOU was calculated for each sample as the difference between the saturation and measured dissolved oxygen using data from the oxygen sensor mounted on the CTD, calibrated with estimates derived from high-precision Winkler titration (79). Oxygen saturation was obtained from potential temperature and salinity using the equation reported by Benson and Krause (80). The data presented here are available in data file S1.
Statistical analyses
Shapiro-Wilk W tests for data normality were applied before analysis, and the data were logarithmically transformed if necessary. ANOVA tests were used to statistically detect significant differences (at the significance level of P < 0.05) of viral abundance and VPR among the different layers, oceanic regions, and water masses. If significant differences were observed, the post hoc Tukey’s test was also performed. A power-law regression model was used to characterize the relationship between viral abundance and depth as in the study by Wigington et al. (30). These calculations were performed on nontransformed data. For easier graphical representation of the data, cell counts were plotted on a logarithmic scale. Regression analysis was used to describe the relationship between log-transformed viral and prokaryotic abundance. To test whether viral abundance was related to environmental and biological variables such as temperature, salinity, AOU, chlorophyll a concentration, total prokaryotic abundance and PHP, heterotrophic picoflagellate/nanoflagellate abundance, Synechococcus abundance, Prochlorococcus abundance, and picoeukaryote abundance, we carried out a distance-based multivariate analysis for a linear model using forward selection (DISTLM forward). All statistical analyses were performed with JMP (version 8.0.2, SAS Institute Inc.) and the R studio statistical software (version 0.99.491) (81).
Supplementary Material
Acknowledgments
We thank all the scientists, chief scientists in the cruise, technicians, and the crew of the different cruise legs for their helpful collaboration. Funding: This work was supported by the Spanish Ministry of Science and Innovation through project Consolider-Ingenio Malaspina 2010 (CSD2008-00077) and funding received through grant FCS/1/2449-01-01 of the Office of Sponsored Research of the King Abdullah University of Science and Technology. Author contributions: E.L., D.V., C.M.D., and J.M.G. designed the study. E.L.S., J.A.B., A.G., E.B., C.D.-V., E.T., M.C.P., F.C.G., I.F., Y.M.C., and A.P. contributed extensively to sampling collection. G.S., X.A.G.M., R.M., T.S.C., G.M.L., S.A., and M.E. helped in the analysis of the samples. All the authors provided constructive comments and revised and edited the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/9/e1602565/DC1
table S1. Mean, SE, median, minimum, and maximum values of viral abundance in each of the water masses identified in the mesopelagic layer (200 to 1000 m).
table S2. Mean, SE, median, minimum, and maximum values of viral abundance according to the water masses in the bathypelagic layer (1000 to 4000 m).
table S3. Results of the multivariate multiple regression analyses to explain variability of viral abundance in the epipelagic layer (0 to 200 m).
table S4. Results of the multivariate multiple regression analyses to explain variability of viral abundance in the mesopelagic layer (200 to 1000 m).
table S5. Results of the multivariate multiple regression analyses to explain variability of viral abundance in the bathypelagic layer (1000 to 4000 m).
table S6. Values of VPL and VPLG.
table S7. Mean and SE of the estimated C, N, and P released by viruses and that incorporated by grazers in the three layers (surface, DCM, and bathypelagic).
fig. S1. Viral abundance with depth along the entire cruise visualized with Ocean Data View.
fig. S2. VPL and VPLG and prokaryotic mortality due to viruses.
data file S1. List of all the environmental and biological variables used in this study.
REFERENCES AND NOTES
- 1.Weinbauer M. G., Rassoulzadegan F., Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6, 1–11 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Breitbart M., Marine viruses: Truth or dare. Annu. Rev. Mar. Sci. 4, 425–448 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Brum J. R., Sullivan M. B., Rising to the challenge: Accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 13, 147–159 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Suttle C. A., Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Proctor L. M., Fuhrman J. A., Viral mortality of marine bacteria and cyanobacteria. Nature 343, 60–62 (1990). [Google Scholar]
- 6.Weinbauer M. G., Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28, 127–181 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Hara S., Koike I., Terauchi K., Kamiya H., Tanoue E., Abundance of viruses in deep oceanic waters. Mar. Ecol. Prog. Ser. 145, 269–277 (1996). [Google Scholar]
- 8.Parada V., Sintes E., van Aken H. M., Weinbauer M. G., Herndl G. J., Viral abundance, decay, and diversity in the meso- and bathypelagic waters of the North Atlantic. Appl. Environ. Microbiol. 73, 4429–4438 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Umani S. F., Malisana E., Focaracci F., Magagnini M., Corinaldesi C., Danovaro R., Disentangling the effect of viruses and nanoflagellates on prokaryotes in bathypelagic waters of the Mediterranean Sea. Mar. Ecol. Prog. Ser. 418, 73–85 (2010). [Google Scholar]
- 10.Magiopoulos I., Pitta P., Viruses in a deep oligotrophic sea: Seasonal distribution of marine viruses in the epi-, meso- and bathypelagic waters of the Eastern Mediterranean Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 66, 1–10 (2012). [Google Scholar]
- 11.Weinbauer M. G., Brettar I., Höfle M. G., Lysogeny and virus-induced mortality of bacterioplankton in surface, deep, and anoxic marine waters. Limnol. Oceanogr. 48, 1457–1465 (2003). [Google Scholar]
- 12.Magagnini M., Corinaldesi C., Monticelli L. S., De Domenico E., Danovaro R., Viral abundance and distribution in mesopelagic and bathypelagic waters of the Mediterranean Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 54, 1209–1220 (2007). [Google Scholar]
- 13.Winter C., Kerros M.-E., Weinbauer M. G., Seasonal changes of bacterial and archaeal communities in the dark ocean: Evidence from the Mediterranean Sea. Limnol. Oceanogr. 54, 160–170 (2009). [Google Scholar]
- 14.De Corte D., Sintes E., Winter C., Yokokawa T., Reinthaler T., Herndl G. J., Links between viral and prokaryotic communities throughout the water column in the (sub)tropical Atlantic Ocean. ISME J. 4, 1431–1442 (2010). [DOI] [PubMed] [Google Scholar]
- 15.De Corte D., Sintes E., Yokokawa T., Reinthaler T., Herndl G. J., Links between viruses and prokaryotes throughout the water column along a North Atlantic latitudinal transect. ISME J. 6, 1566–1577 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y., Yokokawa T., Motegi C., Nagata T., Large-scale distribution of viruses in deep waters of the Pacific and Southern Oceans. Aquat. Microb. Ecol. 71, 193–202 (2014). [Google Scholar]
- 17.Arístegui J., Gasol J. M., Duarte C. M., Herndl G. J., Microbial oceanography of the dark ocean’s pelagic realm. Limnol. Oceanogr. 54, 1501–1529 (2009). [Google Scholar]
- 18.Nagata T., Tamburini C., Arístegui J., Baltar F., Bochdansky A. B., Fonda-Umani S., Fukuda H., Gogou A., Hansell D. A., Hansman R. L., Herndl G. J., Panagiotopoulos C., Reinthaler T., Sohrin R., Verdugo P., Yamada N., Yamashita Y., Yokokawa T., Bartlett D. H., Emerging concepts on microbial processes in the bathypelagic ocean—Ecology, biogeochemistry, and genomics. Deep Sea Res. Part II Top. Stud. Oceanogr. 57, 1519–1536 (2010). [Google Scholar]
- 19.Mizuno C. M., Ghai R., Saghaï A., López-García P., Rodriguez-Valera F., Genomes of abundant and widespread viruses from the deep ocean. mBio 7, e00805-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Luo T., Sun J., Cai L., Liang Y., Jiao N., Zhang R., Lytic viral infection of bacterioplankton in deep waters of the western Pacific Ocean. Biogeosciences 11, 2531–2542 (2014). [Google Scholar]
- 21.Liang Y., Li L., Luo T., Zhang Y., Zhang R., Jiao N., Horizontal and vertical distribution of marine virioplankton: A basin scale investigation based on a global cruise. PLOS ONE 9, e111634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knowles B., Silveira C. B., Bailey B. A., Barott K., Cantu V. A., Cobián-Güemes A. G., Coutinho F. H., Dinsdale E. A., Felts B., Furby K. A., George E. E., Green K. T., Gregoracci G. B., Haas A. F., Haggerty J. M., Hester E. R., Hisakawa N., Kelly L. W., Lim Y. W., Little M., Luque A., McDole-Somera T., McNair K., de Oliveira L. S., Quistad S. D., Robinett N. L., Sala E., Salamon P., Sanchez S. E., Sandin S., Silva G. G. Z., Smith J., Sullivan C., Thompson C., Vermeij M. J. A., Youle M., Young C., Zgliczynski B., Brainard R., Edwards R. A., Nulton J., Thompson F., Rohwer F., Lytic to temperate switching of viral communities. Nature 531, 466–470 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Duarte C. M., Seafaring in the 21st century: The Malaspina 2010 circumnavigation expedition. Limnol. Oceanogr. Bull. 24, 11–14 (2015). [Google Scholar]
- 24.Cobián Güemes A. G., Cantú V. A., Felts B., Nulton J., Rohwer F., Viruses as winners in the game of life. Annu. Rev. Virol. 3, 197–214 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Pernice M. C., Forn I., Gomes A., Lara E., Alonso-Sáez L., Arrieta J. M., del Carmen Garcia F., Hernando-Morales V., MacKenzie R., Mestre M., Sintes E., Teira E., Valencia J., Varela M. M., Vaqué D., Duarte C. M., Gasol J. M., Massana R., Global abundance of planktonic heterotrophic protists in the deep ocean. ISME J. 9, 782–792 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsons R. J., Breitbart M., Lomas M. W., Carlson C. A., Ocean time-series reveals recurring seasonal patterns of virioplankton dynamics in the northwestern Sargasso Sea. ISME J. 6, 273–284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLong E. F., Preston C. M., Mincer T., Rich V., Hallam S. J., Frigaard N.-U., Martinez A., Sullivan M. B., Edwards R., Brito B. R., Chisholm S. W., Karl D. M., Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311, 496–503 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Brum J. R., Ignacio-Espinoza J. C., Roux S., Doulcier G., Acinas S. G., Alberti A., Chaffron S., Cruaud C., de Vargas C., Gasol J. M., Gorsky G., Gregory A. C., Guidi L., Hingamp P., Iudicone D., Not F., Ogata H., Pesant S., Poulos B. T., Schwenck S. M., Speich S., Dimier C., Kandels-Lewis S., Picheral M., Searson S. Tara Oceans Coordinators, Bork P., Bowler C., Sunagawa S., Wincker P., Karsenti E., Sullivan M. B., Patterns and ecological drivers of ocean viral communities. Science 348, 1261498 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Roux S., Brum J. R., Dutilh B. E., Sunagawa S., Duhaime M. B., Loy A., Poulos B. T., Solonenko N., Lara E., Poulain J., Pesant S., Kandels-Lewis S., Dimier C., Picheral M., Searson S., Cruaud C., Alberti A., Duarte C. M., Gasol J. M., Vaqué D.; Tara Oceans Coordinators, Bork P., Acinas S. G., Wincker P., Sullivan M. B., Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537, 689–693 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Wigington C. H., Sonderegger D., Brussaard C. P. D., Buchan A., Finke J. F., Fuhrman J. A., Lennon J. T., Middelboe M., Suttle C. A., Stock C., Wilson W. H., Wommack K. E., Wilhelm S. W., Weitz J. S., Re-examination of the relationship between marine virus and microbial cell abundances. Nat. Microbiol. 1, 15024 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Bongiorni L., Magagnini M., Armeni M., Noble R., Danovaro R., Viral production, decay rates, and life strategies along a trophic gradient in the North Adriatic Sea. Appl. Environ. Microbiol. 71, 6644–6650 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rowe J. M., DeBruyn J. M., Poorvin L., LeCleir G. R., Johnson Z. I., Zinser E. R., Wilhelm S. W., Viral and bacterial abundance and production in the Western Pacific Ocean and the relation to other oceanic realms. FEMS Microbiol. Ecol. 79, 359–370 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Weinbauer M. G., Suttle C. A., Lysogeny and prophage induction in coastal and offshore bacterial communities. Aquat. Microb. Ecol. 18, 217–225 (1999). [Google Scholar]
- 34.Brum J. R., Hurwitz B. L., Schofield O., Ducklow H. W., Sullivan M. B., Seasonal time bombs: Dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 10, 437–449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herndl G. J., Reinthaler T., Microbial control of the dark end of the biological pump. Nat. Geosci. 6, 718–724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salazar G., Cornejo-Castillo F. M., Borrull E., Díez-Vives C., Lara E., Vaqué D., Arrieta J. M., Duarte C. M., Gasol J. M., Acinas S. G., Particle-association lifestyle is a phylogenetically conserved trait in bathypelagic prokaryotes. Mol. Ecol. 24, 5692–5706 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Weinbauer M. G., Bettarel Y., Cattaneo R., Luef B., Maier C., Motegi C., Peduzzi P., Mari X., Viral ecology of organic and inorganic particles in aquatic systems: Avenues for further research. Aquat. Microb. Ecol. 57, 321–341 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Proctor L. M., Fuhrman J. A., Roles of viral infection in organic particle flux. Mar. Ecol. Prog. Ser. 69, 133–142 (1991). [Google Scholar]
- 39.Riemann L., Grossart H.-P., Elevated lytic phage production as a consequence of particle colonization by a marine Flavobacterium (Cellulophaga sp.). Microb. Ecol. 56, 505–512 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Winter C., Kerros M.-E., Weinbauer M. G., Seasonal and depth-related dynamics of prokaryotes and viruses in surface and deep waters of the northwestern Mediterranean Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 56, 1972–1982 (2009). [Google Scholar]
- 41.Boras J. A., Sala M. M., Baltar F., Arístegui J., Duarte C. M., Vaqué D., Effect of viruses and protists on bacteria in eddies of the Canary Current region (subtropical northeast Atlantic). Limnol. Oceanogr. 55, 885–898 (2010). [Google Scholar]
- 42.Danovaro R., Dell’Anno A., Corinaldesi C., Magagnini M., Noble R., Tamburini C., Weinbauer M., Major viral impact on the functioning of benthic deep-sea ecosystems. Nature 454, 1084–1087 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Fuhrman J. A., Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Rocke E., Pachiadaki M. G., Cobban A., Kujawinski E. B., Edgcomb V. P., Protist community grazing on prokaryotic prey in deep ocean water masses. PLOS ONE 10, e0124505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arrieta J. M., Mayol E., Hansman R. L., Herndl G. J., Dittmar T., Duarte C. M., Dilution limits dissolved organic carbon utilization in the deep ocean. Science 348, 331–333 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Thingstad T. F., Heldal M., Bratbak G., Dundas I., Are viruses important partners in pelagic fend webs? Trends Ecol. Evol. 8, 209–213 (1993). [DOI] [PubMed] [Google Scholar]
- 47.Wommack K. E., Colwell R. R., Virioplankton: Viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64, 69–114 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilhelm S. W., Suttle C. A., Viruses and nutrient cycles in the sea: Viruses play critical roles in the structure and function of aquatic food webs. BioScience 49, 781–788 (1999). [Google Scholar]
- 49.del Giorgio P. A., Duarte C. M., Respiration in the open ocean. Nature 420, 379–384 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Suttle C. A., Viruses in the sea. Nature 437, 356–361 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Estrada M., Delgado M., Blasco D., Latasa M., Cabello A. M., Benítez-Barrios V., Fraile-Nuez E., Mozetič P., Vidal M., Phytoplankton across tropical and subtropical regions of the Atlantic, Indian and Pacific Oceans. PLOS ONE 11, e0151699 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marie D., Brussaard C. P. D., Thyrhaug R., Bratbak G., Vaulot D., Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl. Environ. Microbiol. 65, 45–52 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.C. P. D. Brussaard, J. P. Payet, C. Winter, M. G. Weinbauer, Quantification of aquatic viruses by flow cytometry, in Manual of Aquatic Viral Ecology, S. W. Wilhelm, M. G. Weinbauer, C. A. Suttle, Eds. (ASLO, 2010), pp. 102–109. [Google Scholar]
- 54.Martínez J. M., Swan B. K., Wilson W. H., Marine viruses, a genetic reservoir revealed by targeted viromics. ISME J. 8, 1079–1088 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brussaard C. P. D., Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 70, 1506–1513 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jover L. F., Effler T. C., Buchan A., Wilhelm S. W., Weitz J. S., The elemental composition of virus particles: Implications for marine biogeochemical cycles. Nat. Rev. Microbiol. 12, 519–528 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Gasol J. M., del Giorgio P. A., Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64, 197–224 (2000). [Google Scholar]
- 58.L. M. Lubián, Determinación de la abundancia de nano y picofitoplancton mediante citometría de flujo, in Expedición de Circunnavegación Malaspina 2010: Cambio Global y Exploración de la Biodiversidad del Océano. Libro Blanco de Métodos y Técnicas de Trabajo Oceanográfico, E. Moreno-Ostos, Ed. (Consejo Superior de Investigaciones Cientificas, 2012), pp. 381–385. [Google Scholar]
- 59.D. Marie, N. Simon, D. Vaulot, Phytoplankton cell counting by flow cytometry, in Algal Culturing Techniques, R. A. Anderson, Ed. (Elsevier Academic Press, 2005), pp. 253–268. [Google Scholar]
- 60.Christaki U., Courties C., Massana R., Catalá P., Lebaron P., Gasol J. M., Zubkov M. V., Optimized routine flow cytometric enumeration of heterotrophic flagellates using SYBR Green I. Limnol. Oceanogr. Meth. 9, 329–339 (2011). [Google Scholar]
- 61.Zubkov M. V., Burkill P. H., Topping J. N., Flow cytometric enumeration of DNA-stained oceanic planktonic protists. J. Plankton Res. 29, 79–86 (2007). [Google Scholar]
- 62.Kirchman D., K’nees E., Hodson R., Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49, 599–607 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith D. C., Azam F., A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6, 107–114 (1992). [Google Scholar]
- 64.Teira E., Hernando-Morales V., Cornejo-Castillo F. M., Alonso-Sáez L., Sarmento H., Valencia-Vila J., Catalá T. S., Hernández-Ruiz M., Varela M. M., Ferrera I., Morán X. A. G., Gasol J. M., Sample dilution and bacterial community composition influence empirical leucine-to-carbon conversion factors in surface waters of the world’s oceans. Appl. Environ. Microbiol. 81, 8224–8232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilhelm S. W., Brigden S. M., Suttle C. A., A dilution technique for the direct measurement of viral production: A comparison in stratified and tidally mixed coastal waters. Microb. Ecol. 43, 168–173 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Middelboe M., Lyck P. G., Regeneration of dissolved organic matter by viral lysis in marine microbial communities. Aquat. Microb. Ecol. 27, 187–194 (2002). [Google Scholar]
- 67.Wells L. E., Deming J. W., Significance of bacterivory and viral lysis in bottom waters of Franklin Bay, Canadian Arctic, during winter. Aquat. Microb. Ecol. 43, 209–221 (2006). [Google Scholar]
- 68.Weinbauer M. G., Winter C., Höfle M. G., Reconsidering transmission electron microscopy based estimates of viral infection of bacterio-plankton using conversion factors derived from natural communities. Aquat. Microb. Ecol. 27, 103–110 (2002). [Google Scholar]
- 69.Winter C., Herndl G. J., Weinbauer M. G., Diel cycles in viral infection of bacterioplankton in the North Sea. Aquat. Microb. Ecol. 35, 207–216 (2004). [Google Scholar]
- 70.Winget D. M., Williamson K. E., Helton R. R., Wommack K. E., Tangential flow diafiltration: An improved technique for estimation of virioplankton production. Aquat. Microb. Ecol. 41, 221–232 (2005). [Google Scholar]
- 71.M. Guixa-Boixereu, thesis, University of Barcelona, Barcelona, Spain (1997). [Google Scholar]
- 72.Vaqué D., Gasol J. M., Marrasé C., Grazing rates on bacteria: The significance of methodology and ecological factors. Mar. Ecol. Prog. Ser. 109, 263–274 (1994). [Google Scholar]
- 73.Sherr B. F., Sherr E. B., Fallon R. D., Use of monodispersed, fluorescently labeled bacteria to estimate in situ protozoan bacterivory. Appl. Environ. Microbiol. 53, 958–965 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vázquez-Domínguez E., Peters F., Gasol J. M., Vaqué D., Measuring the grazing losses of picoplankton: Methodological improvements in the use of fluorescently labeled tracers combined with flow cytometry. Aquat. Microb. Ecol. 20, 119–128 (1999). [Google Scholar]
- 75.Salat J., Marrasé C., Exponential and linear estimations of grazing on bacteria: Effects of changes in the proportion of marked cells. Mar. Ecol. Prog. Ser. 104, 205–209 (1994). [Google Scholar]
- 76.Lara E., Arrieta J. M., Garcia-Zarandona I., Boras J. A., Duarte C. M., Agustí S., Wassmann P. F., Vaqué D., Experimental evaluation of the warming effect on viral, bacterial and protistan communities in two contrasting Arctic systems. Aquat. Microb. Ecol. 70, 17–32 (2013). [Google Scholar]
- 77.Gundersen K., Heldal M., Norland S., Purdie D. A., Knap A. H., Elemental C, N, and P cell content of individual bacteria collected at the Bermuda Atlantic time-series study (BATS) site. Limnol. Oceanogr. 47, 1525–1530 (2002). [Google Scholar]
- 78.Calvo-Díaz A., Morán X. A. G., Seasonal dynamics of picoplankton in shelf waters of the southern Bay of Biscay. Aquat. Microb. Ecol. 42, 159–174 (2006). [Google Scholar]
- 79.Catalá T. S., Reche I., Álvarez M., Khatiwala S., Guallart E. F., Benítez-Barrios V. M., Fuentes-Lema A., Romera-Castillo C., Nieto-Cid M., Pelejero C., Fraile-Nuez E., Ortega-Retuerta E., Marrasé C., Álvarez-Salgado X. A., Water mass age and aging driving chromophoric dissolved organic matter in the dark global ocean. Glob. Biogeochem. Cycles 29, 917–934 (2015). [Google Scholar]
- 80.Benson B. B., Krause D. Jr, The concentration and isotopic fractionation of oxygen dissolved in freshwater and seawater in equilibrium with the atmosphere. Limnol. Oceanogr. 29, 620–632 (1984). [Google Scholar]
- 81.RStudio Team, RStudio: Integrated Development for R (RStudio Inc., 2015); www.rstudio.com/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/9/e1602565/DC1
table S1. Mean, SE, median, minimum, and maximum values of viral abundance in each of the water masses identified in the mesopelagic layer (200 to 1000 m).
table S2. Mean, SE, median, minimum, and maximum values of viral abundance according to the water masses in the bathypelagic layer (1000 to 4000 m).
table S3. Results of the multivariate multiple regression analyses to explain variability of viral abundance in the epipelagic layer (0 to 200 m).
table S4. Results of the multivariate multiple regression analyses to explain variability of viral abundance in the mesopelagic layer (200 to 1000 m).
table S5. Results of the multivariate multiple regression analyses to explain variability of viral abundance in the bathypelagic layer (1000 to 4000 m).
table S6. Values of VPL and VPLG.
table S7. Mean and SE of the estimated C, N, and P released by viruses and that incorporated by grazers in the three layers (surface, DCM, and bathypelagic).
fig. S1. Viral abundance with depth along the entire cruise visualized with Ocean Data View.
fig. S2. VPL and VPLG and prokaryotic mortality due to viruses.
data file S1. List of all the environmental and biological variables used in this study.


