Abstract
Context:
Angiopoietin-like 3 (ANGPTL3) deficiency in plasma due to loss-of-function gene mutations results in familial combined hypobetalipoproteinemia type 2 (FHBL2) in homozygotes. However, the lipid phenotype in heterozygotes is much milder and does not appear to relate directly to ANGPTL3 levels. Furthermore, the low-density lipoprotein (LDL) phenotype in carriers of ANGPTL3 mutations is unexplained.
Objective:
To determine whether reduction below a critical threshold in plasma ANGPTL3 levels is a determinant of lipoprotein metabolism in FHBL2, and to determine whether proprotein convertase subtilisin kexin type 9 (PCSK9) is involved in determining low LDL levels in this condition.
Design:
We studied subjects from 19 families with ANGPTL3 mutations and subjects with familial combined hypobetalipoproteinemia type 1 (FHBL1) due to truncated apolipoprotein B (apoB) species.
Results:
First, total cholesterol, high-density lipoprotein (HDL) cholesterol, triglycerides, and HDL and LDL particle concentration correlated with plasma ANGPTL3 levels but only when the latter was <25% of normal (<60 ng/dL). Second, the very low-density lipoprotein particle concentration correlated strongly with plasma ANGPTL3 when the latter was <58% of normal. Third, both FHBL1 and FHBL2 subjects showed low levels of mature and LDL-bound PCSK9 and higher levels of its furin-cleaved form. Finally, LDL-bound PCSK9 is protected from cleavage by furin and binds to the LDL receptor more strongly than apoB-free PCSK9.
Conclusions:
Our results suggest that the hypolipidemic effects of ANGPTL3 mutations in FHBL2 are dependent on a threshold of plasma ANGPTL3 levels, with differential effects on various lipoprotein particles. The increased inactivation of PCSK9 by furin in FHBL1 and FHBL2 is likely to cause increased LDL clearance and suggests novel therapeutic avenues.
The FHBL2 phenotype seen in carriers of mutations in ANGPTL3 is caused by a critical reduction of its plasma levels of >75%. Low LDL levels inactivate PCSK9, causing increased LDL clearance.
Familial combined hypobetalipoproteinemia type 2 (FHBL2) is characterized by a unique lipid profile, which includes low levels of triglycerides (TGs), low-density lipoprotein (LDL) cholesterol (LDL-c), and high-density lipoprotein (HDL) cholesterol (HDL-c) but unaffected lipoprotein(a) levels (1, 2). However, the concentration and size of each lipoprotein particle in FHBL2 is not well characterized. The full phenotype of FHBL2 is caused by homozygous mutations in angiopoietin-like protein 3 (ANGPTL3), a secreted 460-amino acid protein produced mainly by the liver (3, 4), leading to reduced plasma levels. FHBL2 subjects are healthy, do not have fatty liver disease, and have an apparent lower risk of developing diabetes mellitus (5). It was recently shown that ANGPTL3 deficiency is also associated with protection from coronary artery disease (6). No clear gene–dosage effect of ANGPTL3 mutations on all lipoprotein levels has been found, because heterozygotes show only a mild hypolipidemic phenotype, with LDL mostly in the low-normal range (7). Most of the mutations in ANGPTL3 influence its plasma levels; however, the effect of these changes on plasma lipids and lipoproteins have not been characterized quantitatively. Inhibition of ANGPTL3 either by antisense oligonucleotides (8) or by blocking antibodies (9) is an emerging pharmacological approach to reduce the levels of all atherogenic lipoproteins, with no apparent side effect in phase I trials (8, 9).
The low levels of TG and HDL-c in FHBL2 subjects can be explained by the role of ANGPTL3 as an inhibitor of both lipoprotein lipase (LPL) (10) and endothelial lipase (EL) (11). LPL plays a critical role in hydrolyzing TG carried by very LDL (VLDL) and chylomicrons in the circulation (10, 12). EL binds to heparin-sulfate proteoglycans on the endothelial cell surface to increase HDL retention and delipidation via its phospholipase activity (10–12). Thus, mutations in ANGPTL3 result in increased activity of lipases, an effect that reduces plasma TG and HDL-c levels (12, 13). However, LPL and EL do not affect cholesterol homeostasis; thus, the increased activity of these lipases cannot directly explain the reduced LDL-c levels. Mice without ANGPTL3 demonstrate increased remnant clearance via noncanonical pathways, which might account for the reduction in LDL-c (14–16). It was also shown that homozygous carriers of ANGPTL3 mutations have an increased fractional catabolic rate of LDL, although the mechanism by which ANGPTL3 loss of function mutations promote increased LDL clearance is not clear (2, 4). An increased LDL fractional catabolic rate was also demonstrated in subjects with familial combined hypobetalipoproteinemia type 1 (FHBL1) owing to different truncated apolipoprotein B (apoB) species (17); thus, suggesting a possible common mechanism by which low LDL-c levels, regardless of the primary genetic cause, undergo accelerated LDL receptor clearance.
Proprotein convertase subtilisin kexin type 9 (PCSK9) is a plasma protein that binds to the LDL receptor (LDLR) and targets it for lysosomal degradation (18, 19), leading to reduced cellular uptake of LDL (19–21). Additionally, PCSK9 affect intestinal and hepatic production of TG-rich lipoproteins (22–24). Plasma PCSK9 is in two main monomeric forms, a mature (intact) heterodimer (62+13 kDa) and a shorter heterodimer (55+13 kDa), resulting from the proteolytic cleavage by the enzyme furin (25, 26). Furin-cleaved PCSK9 loses the N-terminal region of the catalytic domain (up to amino acid 218) (26) and has reduced LDLR degradation activity compared with the mature form (26–29). We, and others, have shown that mature PCSK9 circulates predominantly in association with LDL-size particles (24, 25). PCSK9 inhibition with monoclonal antibodies is an emerging therapy to lower LDL-c in patients with atherosclerotic cardiovascular disease or familial hypercholesterolemia (FH) who require additional LDL-c lowering (30). The effect of genetically low LDL levels on plasma PCSK9 and its molecular forms and function has not been described.
We used different regression models to study the effect of ANGPTL3 levels on plasma lipids and lipoproteins in 19 families with ANGPTL3 mutations. We demonstrate that ANGPTL3 levels correlate strongly with total cholesterol, HDL-c, TG, HDL particle numbers (HDL-p), and LDL particle numbers (LDL-p) when ANGPTL3 levels are <25% of normal and with VLDL particle numbers (VLDL-p) when ANGPTL3 levels are <58% of normal. These regression models likely represent a biologic threshold for the effect of ANGPTL3 on lipoprotein metabolism. We also show the existence of a reciprocal regulation mechanism, by which low LDL levels result in reduced PCSK9 activity, which can lead to increased LDL particle clearance.
Methods
Subjects and samples
The study cohort was composed of 127 subjects, including 7 homozygous and 62 heterozygous carriers of the ANGPTL3 mutation p.S17* and 58 noncarrier relative controls from 19 different families. The family structure and relationships are shown in Supplemental Fig. 1 (1MB, pdf) . All subjects were identified in the population of Campodimele (Italy), as previously described (5), with institutional review board approval from Sapienza University of Rome. For studies of PCSK9 metabolism, we also used a cohort of FHBL1 subjects carrying different truncated apoB species with institutional review board approval from the University of Palermo.
Plasma lipid and protein determinations
Plasma lipids were measured using standard techniques (5), and LDL-c values were calculated using the Friedewald formula. An enzyme-linked immunosorbent assay (ELISA) method using an assay developed in-house assessed the ANGPTL3 plasma concentration, as previously reported (31), or using a commercially available ELISA kit (R&D Systems, Minneapolis, MN). The lipoprotein particle concentration and size were determined using nuclear magnetic resonance by LipoScience (Raleigh, NC), as previously described (32). The PCSK9 levels were measured using commercially available ELISA kits, according to the manufacturer’s instructions (MBL International, Woburn, MA). LDL-bound PCSK9 levels were measured as recently described (33). In brief, rabbit polyclonal antibodies binding to the C-terminal region of PCSK9 (Abgent, San Diego, CA) were bound to microtiter well plates, and EDTA plasma was added at a 1:50 dilution (40 µL/well) for 75 minutes. For LDL-bound PCSK9 detection, we used biotinylated goat anti-human apoB-100 antibody (Academy Biomedical Co., Houston, TX). The results are reported as relative light units in 100 ms after subtraction of the background relative light units. For size-exclusion chromatography (fast protein liquid chromatography), we used a Superose 6, 10/300GL column (GE Healthcare Life Sciences, Pittsburgh, PA), as previously described (34).
Western blotting
Plasma samples were run through a multiple affinity removal column, Agilent Hu-14 column, to deplete the top 14 major proteins (Hu-14; Agilent, Santa Clara, CA). The samples were loaded onto 4% to 12% Tris-acetate sodium dodecyl sulfate gels (Invitrogen, Carlsbad, CA) for electrophoretic separation. Proteins were transferred onto a nitrocellulose membrane. Rabbit α-PCSK9 primary antibody (MBL International) and goat α-rabbit horseradish peroxidase–conjugated (Sigma-Aldrich, St. Louis, MO) secondary antibodies were used to detect target proteins. The signal was detected using an enhanced chemiluminescent solution made in-house using p-coumaric acid, luminol, and hydrogen peroxide.
PCSK9 cleavage by furin
Recombinant PCSK9 was isolated as previously described (35) and then incubated at 2 μM together with furin (40 nM) for 6 hours at room temperature. Cleavage reaction was performed in 50 mM Tris/150 mM NaCl with 4 mM CaCl2. Reaction products were analyzed using 4% to 12% Tris-acetate sodium dodecyl sulfate gels (Invitrogen) using rabbit α-PCSK9 primary antibody, as described in the previous section.
PCSK9 binding to LDLR
Recombinant, histidine-tagged PCSK9 was competed against increasing concentrations of purified plasma PCSK9 (LDL-bound or apoB-free) for binding to the epidermal growth factor-like AB domain of the LDLR, using a commercially available kit and following the manufacturer’s instructions (MBL International).
Statistical analysis
Continuous variables are summarized as the mean ± standard deviation for each ANGPTL3 mutation group. Multiple regression models and generalized estimating equations (GEEs) were used to estimate the association of plasma measurements to ANGPTL3 levels and the mutation status and tested using Wald statistics. Pearson’s correlation analysis was used for the association of ANGPTL3 with the plasma measurements (36–38). The association of ANGPTL3 and lipoproteins of interest was modeled using the multivariate adaptive regression splines (MARS) methods that assume the expected mean of the lipoprotein is a piecewise linear function of ANGPTL3 and age (39, 40). This model allows the strength of the correlation and regression slope of ANGPLT3 for the lipoprotein to change between different ranges of ANGPTL3 values. MARS performs variable selection and will remove ANGPTL3 and/or age from the model if they are not predictive. Three modeling methods are represented in the scatterplots to visually compare MARS to the GEE linear regression slopes and local polynomial regression loess curves. In most cases, MARS selected a model representing a linear approximation to the loess smoothing curve but provided more interpretability than the nonparametric locally weighted scatterplot smoothing (loess) method. The MARS analysis is based on the best linear approximation of a curve nonparametric model; therefore, the change point for the slopes is considered the best estimate. Statistical analyses were performed using R, version 3.2.2 (R Foundation, Vienna, Austria). Additional details regarding the statistical methods are provided in the Supplemental Data (1MB, pdf) .
Results
Group comparison
The demographic data and clinical characteristics of the carriers and noncarriers of ANGPTL3 mutations are listed in Table 1. Plasma ANGPTL3 levels were undetectable in the homozygotes and averaged 124 ± 100 ng/dL among the heterozygotes and 259 ± 135 ng/dL among the noncarriers. Homozygotes had lower levels of all plasma lipid and lipoprotein parameters measured, except for lipoprotein(a) level and HDL size, and heterozygotes had only a mild hypolipidemic phenotype (Table 1; Supplemental Fig. 2 (1MB, pdf) ). A summary of P values from Wald test statistics for the group comparisons is given in Supplemental Table 1 (1MB, pdf) .
Table 1.
Characteristics of ANGPTL3 Cohort (n = 127)
| Characteristic | Noncarriers (n = 58) | Heterozygotes (n = 62) | Homozygotes (n = 7) |
|---|---|---|---|
| Age, y | 61 ± 21 | 51 ± 20 | 48 ± 20 |
| Female sex, % | 39.7 | 40.3 | 42.8 |
| ANGPTL3, ng/dL | 259.4 ± 134.9 | 123.9 ± 100.1 | 0 ± 0 |
| Total cholesterol, mg/dL | 187.5 ± 25.5 | 167.2 ± 31.6 | 82.4 ± 12.1 |
| HDL-c, mg/dL | 69.6 ± 15.1 | 55.4 ± 13.7 | 27.8 ± 8.1 |
| TG, mg/dL | 80.8 ± 39.1 | 74.3 ± 40.6 | 32.4 ± 4.1 |
| VLDL-p concentration, nmol/L | 45.5 ± 17.5 | 34.9 ± 13.0 | 11.9 ± 4.2) |
| HDL-p concentration, nmol/L | 21.3 ± 5.9 | 19.1 ± 5.0 | 12.4 ± 2.7) |
| LDL-p concentration, nmol/L | 886 ± 210 | 766 ± 250 | 563 ± 75) |
| LDL-c, mg/dL | 102.2 ± 24.7 | 97.3 ± 28.1 | 48.3 ± 15.3 |
| Lipoprotein(a), mg/dL | 21.3 ± 30.7 | 23.0 ± 28.4 | 20.7 ± 25.1 |
| VLDL size, nm | 51.7 ± 5.0 | 52.7 ± 4.9 | NA |
| LDL size, nm | 20.4 ± 0.7 | 20.5 ± 0.6 | 19.8 ± 0.2 |
| HDL size, nm | 9.2 ± 0.5 | 8.9 ± 0.5 | 9.4 ± 0.2 |
| Creatinine, U/L | 1.10 ± 0.19 | 1.12 ± 0.18 | 1.03 ± 0.13 |
| Glucose, mg/dL | 101.5 ± 18.8 | 99.2 ± 28.7 | 86.8 ± 5.9 |
| ALT, U/L | 30.9 ± 8.7 | 28.7 ± 6.0 | 29.7 ± 5.2 |
| AST, U/L | 29.1 ± 11.9 | 29.0 ± 11.9 | 36.5 ± 4.9 |
| ApoB, mg/dL | 87.6 ± 22.9 | 84.4 ± 20.7 | 47.1 ± 13.3 |
| Apolipoprotein AI, mg/dL | 179.9 ± 23.9 | 163.7 ± 27.8 | 77.2 ± 18.9 |
| Weight, kg | 68.1 ± 12.2 | 73.2 ± 15.5 | 70.1 ± 12.8 |
| Height, m | 1.57 ± 0.1 | 1.62 ± 0.09 | 1.58 ± 0.08 |
| BMI, kg/m2 | 27.6 ± 4.9 | 27.9 ± 5.0 | 28.1 ± 7.3 |
| PCSK9, ng/mL | 207.1 ± 59.1 | 173.1 ± 63.3 | 144.3 ± 30.9 |
| LDL-bound PCSK9, RLU | 12,608 ± 5204 | 12,358 ± 5168 | 6131 ± 2008 |
Data presented as mean ± standard deviation.
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; NA, data not available; RLU, relative light units.
Multiple GEE linear regression analyses
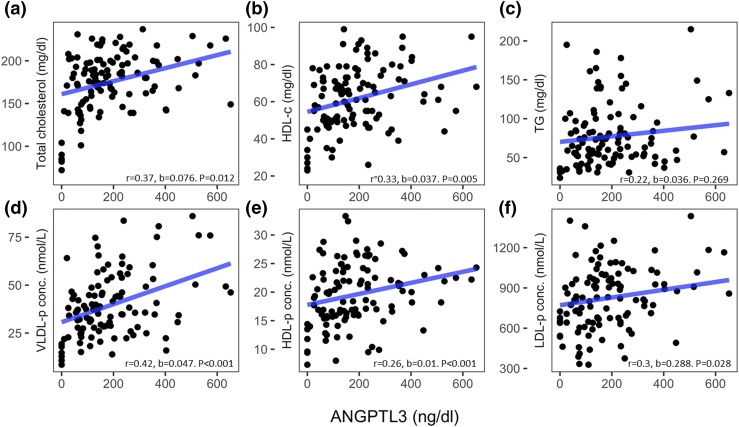
We used Pearson’s correlation coefficients and GEE regression coefficients with plasma measurements as dependent variables and ANGPTL3 as the independent variable to study whether ANGPTL3 levels correlate with the changes in plasma lipids levels and lipoprotein particle concentration. Figure 1(a) and 1(b) demonstrates that the total cholesterol and HDL-c levels correlate with the ANGPTL3 levels (r = 0.37, P = 0.012, and r = 0.33, P = 0.005, respectively), but the TG levels do not correlate with the ANGPTL3 levels [Fig. 1(c)]. Figure 1(d–f) shows that VLDL-p, HDL-p, and LDL-p also correlate with the ANGPTL3 levels (r = 0.42, P < 0.001, r = 0.26, P = < 0.001, and r = 0.3, P = 0.028, respectively). A summary of Pearson’s correlation coefficients, regression coefficient, and P values is presented in Supplemental Table 2 (1MB, pdf) . Similar results were obtained when the homozygotes were excluded from the regression model (Supplemental Fig. 3 (1MB, pdf) ).
Figure 1.
Scatterplot with univariate GEE linear regression slopes (blue) of ANGPTL3 levels against (a) total cholesterol, (b) HDL-c, (c) TG, (d) VLDL-p, (e) HDL-p, and (f) LDL-p. P values from Wald test for the slope. b, univariate GEE regression coefficients; conc., concentration; r, Pearson’s correlation coefficients.
MARS analyses
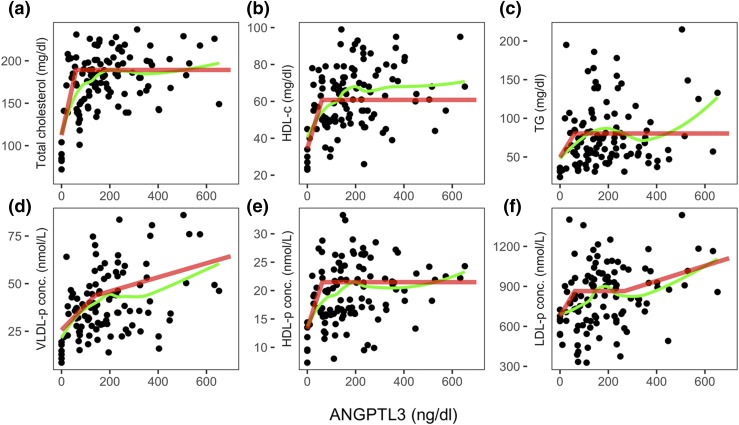
Despite the relevant correlations between ANGPTL3 levels and plasma lipid or lipoprotein concentrations (Fig. 1), heterozygotes exhibit normal levels of many of these measurements (Supplemental Fig. 2 (1MB, pdf) ), representing a lack of a clear gene–dosage effect of ANGPTL3, as previously reported (5). A curve fit model (Fig. 2, green lines) suggests that, at low levels, ANGPTL3 strongly correlates with the plasma lipid levels and lipoprotein concentrations but that at higher values, these correlations are lost or become weaker. To approximate the curve fit shown in green into linear slopes, we used a model that allows for different correlation coefficients at low and high ANGPTL3 levels, known as MARS. Thus, instead of just one regression line with one slope, this model allows for several regression lines at a range of ANGPTL3 levels, as explained in the Supplemental Data (1MB, pdf) . MARS analysis (Fig. 2, red line) demonstrated that the levels of plasma lipids and lipoprotein (total cholesterol, HDL-c, TG, HDL-p, and LDL-p) correlate strongly with the ANGPTL3 level when the latter is <60 ng/dL (~25% of normal ANGPTL3 levels), and VLDL-p correlates strongly with the ANGPTL3 level when the latter is <138 ng/dL (~58% of normal ANGPTL3 levels). All plasma measurements, aside from TG levels, showed a threshold effect, even when the homozygotes were excluded from the regression model, with the main change being a slightly higher threshold levels of ANGPTL3 (Supplemental Fig. 4 (1MB, pdf) ). The MARS model fit R-squared (r2) and equations of regression lines denoting the red slopes are summarized in Supplemental Table 3 (1MB, pdf) .
Figure 2.
Scatterplot with nonparametric smoothed local polynomial regression fit (green) and MARS model fit (red) of ANGPTL3 levels against (a) total cholesterol, (b) HDL-c, (c) TG, (d) VLDL-p, (e) HDL-p, and (f) LDL-p. b, univariate GEE regression coefficients; conc., concentration; r, Pearson’s correlation coefficients.
Effect of low LDL on plasma PCSK9
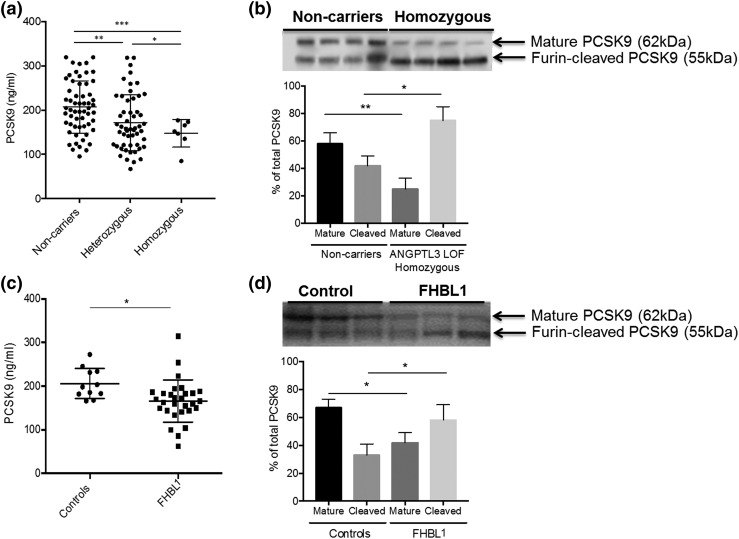
Figure 3(a) shows a moderate reduction in plasma PCSK9 levels, a known modulator of LDL metabolism, in heterozygotes compared with noncarriers (−16.4%; P = 0.025), in homozygotes compared with noncarriers (−30.5%; P = 0.001), and in heterozygotes compared with homozygotes (−16.6%; P = 0.0069). The PCSK9 levels correlated with both LDL-c (r = 0.245, P = 0.015) and apoB (r = 0.41; P < 0.001) levels (Supplemental Fig. 5 (1MB, pdf) ). Immunoblotting of the molecular forms of PCSK9 in plasma [Fig. 3(b)] showed that homozygotes have reduced levels of the mature, intact form of PCSK9 and increased levels of its furin-cleaved form. This finding was not explained by alterations in plasma furin function, because no differences were found in furin activity between the carriers and noncarriers of the ANGPTL3 mutations (data not shown). To study whether this phenomenon depends on low ANGTPL3 levels or low LDL levels (regardless of its primary genetic cause), we analyzed the total PCSK9 levels and its molecular forms in hypolipidemic subjects with FHBL1 due to apoB truncations (range, 1% to 80% of normal apoB), low LDL-c levels, and normal ANGPTL3 levels (the clinical characteristics of this cohort are listed in Supplemental Table 4 (1MB, pdf) ). Figure 3(c) shows a moderate reduction in plasma PCSK9 levels in subjects with truncated apoB compared with controls (−16.0%; P = 0.017). Analysis of the molecular forms of PCSK9 in plasma [Fig. 3(d)] demonstrated that subjects with truncated apoB have reduced levels of the mature form of PCSK9 and increased levels of the furin-cleaved form.
Figure 3.
Effect of low LDL levels on plasma PCSK9: (a) group comparison for total PCSK9 levels in the ANGPTL3 cohort; (b) immunoblot (upper) and quantification (lower) for plasma PCSK9 molecular forms in the ANGPTL3 cohort; (c) group comparison for total PCSK9 levels in the FHBL1 cohort; and (d) Immunoblot (upper) and quantification (lower) for plasma PCSK9 molecular form in the FHBL1 cohort. *P < 0.05, **P < 0.01,***P < 0.001.
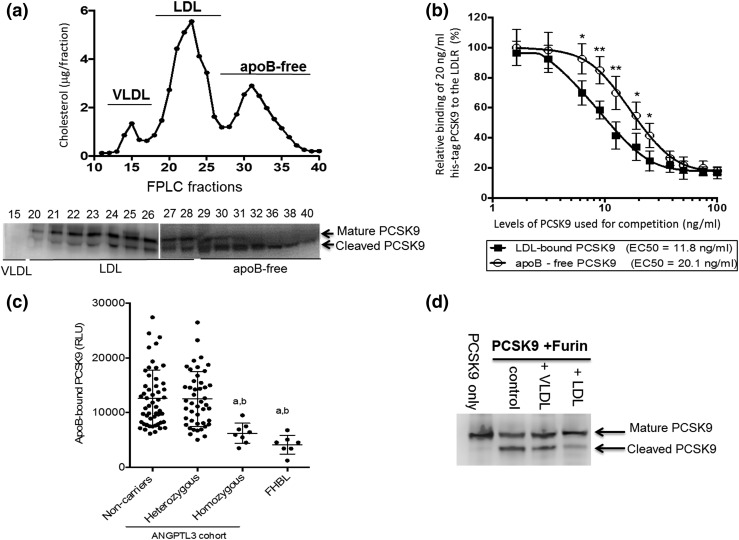
To further characterize the effect of LDL on PCSK9 function, we analyzed the distribution of PCSK9 molecular forms in plasma. Figure 4(a) shows the mature form of PCSK9 is found predominantly in LDL fractions, and the furin-cleaved PCSK9 is found mostly as apoB-free. Our results further showed that LDL-bound PCSK9 has a 70% stronger affinity toward the LDLR than does apoB-free PCSK9 [Fig. 4(b)]. In addition, subjects with either truncated apoB or homozygous ANGPTL3 mutations showed lower levels of LDL-bound PCSK9 compared with the heterozygotes and noncarriers [Fig. 4(c)], and the levels of LDL-bound PCSK9 correlated with the plasma apoB levels (Supplemental Fig. 5 (1MB, pdf) ). Finally, we tested the direct effect of LDL on the ability of furin to cleave PCSK9 as a possible explanation for why low LDL levels are coupled with increased furin-cleaved PCSK9 in both ANGPTL3 and apoB mutation carriers. Figure 4(d) demonstrates that furin efficiently cleaves PCSK9 in vitro (control lane) and that LDL (but not VLDL) inhibits this cleavage reaction.
Figure 4.
Effect of LDL binding on PCSK9: (a) size exclusion chromatography analysis of cholesterol (upper) and PCSK9 molecular forms (immunoblot; lower) of a normolipidemic subject. (b) Competitive binding of PCSK9 to epidermal growth factor-like AB domain of the LDLR; 20 ng/mL recombinant histidine-tag PCSK9 (y-axis) was competed against increasing concentrations of LDL-bound or apoB-free PCSK9 (x-axis). (c) Group comparison for LDL-bound PCSK9 levels. (d) In vitro reaction of PCSK9 cleavage by furin in the presence or absence of lipoproteins. *P < 0.05, **P < 0.01, aP < 0.01 compared with noncarriers, bP < 0.01 compared with heterozygotes. EC50, half maximal effective concentration; FPLC, fast protein liquid chromatography; RLU, relative light units.
Discussion
We analyzed a unique cohort composed of individuals carrying ANGPTL3 mutations and noncarrier relatives as controls to gain insight into the effect of ANGPTL3 levels on the phenotypic expression of FHBL2. Our cohort showed a nonlinear pattern, as the homozygotes had plasma lipid and lipoprotein particle concentrations 43% to 70% lower than the concentrations in noncarriers, and heterozygotes exhibited levels that were only 10% to 20% lower than those in noncarriers, as previously described (5, 41). Our cohort also showed a weak correlation between plasma ANGPTL3 levels and plasma lipids and lipoprotein levels. We hypothesized that the weak correlation between the ANGPTL3 levels and plasma lipid/lipoprotein parameters is linked to a critical concentration of plasma ANGPTL3 that triggers the combined hypolipidemia trait. Our results have demonstrated that ANGPTL3 levels correlate strongly and linearly with plasma lipids, HDL-p, and LDL-p for ANGPTL3 levels less than ∼25% of normal and with VLDL-p at levels less than ∼58% of normal. The varying ANGPTL3 concentrations associated with plasma lipid and lipoprotein modulation can explain the lack of a linear gene–dosage effect of ANGPTL3 mutations. The FHBL2 phenotype seems to be dependent, not only on the presence of mutations in ANGPTL3, but also, even more to a critical reduction in plasma concentrations of ANGPTL3. In support of a threshold effect of ANGPTL3 levels, it was recently shown that heterozygotes with ANGPTL3 levels in the lowest tertile had reduced myocardial infarction rates compared with subjects with the highest tertile, who had similar event rates as noncarriers (6).
In addition to the role of ANGPTL3 as an LPL and EL inhibitor and its consequent effect on TGs and HDL-c levels, animal and human studies have consistently reported that ANGPTL3 deficiency and blockade are associated with lower LDL-c levels (4, 6–9, 42). Although LDLR is not required for the ANGPTL3 effect on LDL-c levels, the absence of LDLR in homozygous FH subjects attenuates the LDL lowering effect of ANGPTL3 inhibition compared with the results in FH subjects with residual LDLR function (43) or in normal volunteers (9). Therefore, LDLR contributes to the plasma LDL lowering caused by reduced ANGPTL3 levels.
It was recently suggested that increased clearance of remnants results in decreased production of LDL in ANGPTL3-deficient animals, leading to a reduced LDL-c level (14). If preferential clearance of large- and medium-size LDL occurs, LDL-c, LDL-p, and LDL size should all be reduced, a phenomenon that was recently described in subjects treated with anti-PCSK9 antibodies (44) and was also seen in the homozygotes of our cohort (Table 1). Our results have demonstrated mildly reduced plasma PCSK9 levels in ANGPTL3 mutation homozygotes and heterozygotes compared with noncarriers. We further established that LDL-bound PCSK9, which accounts for most of the mature PCSK9 in plasma (25, 35), is dramatically reduced in ANGPTL3 mutation homozygotes and also in FHBL1 subjects carrying truncated apoB. We have also shown that LDL-bound PCSK9 has a stronger binding affinity toward the LDLR compared with apoB-free PCSK9. Kinetic studies in humans have shown that both FHBL2 and FHBL1 subjects have increased LDL clearance rates (2, 4, 17) and support the notion of a common mechanism whereby low LDL levels, regardless of their initial genetic cause, reduce PCSK9 activity to further promote LDL clearance. Finally, we propose the following mechanism to explain the link between LDL levels and PCSK9 molecular forms and function. We envision a scenario in which PCSK9 binding to LDL protects it from cleavage by furin and preserve its LDLR-binding capacity but that low LDL levels result in less binding of PCSK9 to the particle, increased cleavage of PCSK9 by furin, and reduced LDLR binding.
In conclusion, we have demonstrated that a critical threshold of ANGPTL3 reduction is associated with marked reductions in plasma lipid and lipoprotein concentration and that changes in compartmentalization of plasma PCSK9 in plasma might contribute to the low LDL phenotype by causing increased LDL clearance. Our results suggest that ongoing therapies (8, 9) directed at inhibiting ANGPTL3 should target reductions of ≥75% in plasma ANGPTL3 to replicate the full FHBL2-like phenotype.
Acknowledgments
The authors thank Dr. Ujwal Shinde for helpful discussion and assistance in the analysis of furin activity.
Acknowledgments
The present study was supported by National Institutes of Health (National Heart, Lung, and Blood Institute) Grants R01-HL132985 (to S.F.) and R01-HL119828 (to S.T.) and a Medical Research Foundation of Oregon New Investigator Grant 1011656 (to H.T.).
Disclosure Summary: S.T. is a co-inventor of, and receives royalties from, patents or patent applications owned by the University of California, San Diego, on antibodies used in biotheranostic applications and has a dual appointment at Ionis Pharmaceuticals, Inc. (Carlsbad, CA) and University of California, San Diego. The remaining authors have nothing to disclose.
Footnotes
- ANGPTL3
- angiopoietin-like 3
- apoB
- apolipoprotein B
- EL
- endothelial lipase
- ELISA
- enzyme-linked immunosorbent assay
- FH
- familial hypercholesterolemia
- FHBL1
- familial combined hypobetalipoproteinemia type 1
- FHBL2
- familial combined hypobetalipoproteinemia type 2
- GEE
- generalized estimating equation
- HDL
- high-density lipoprotein
- HDL-c
- high-density lipoprotein cholesterol
- HDL-p
- high-density lipoprotein particle numbers
- LDL
- low-density lipoprotein
- LDL-c
- low-density lipoprotein cholesterol
- LDL-p
- low-density lipoprotein particle numbers
- LDLR
- low-density lipoprotein receptor
- LPL
- lipoprotein lipase
- MARS
- multivariate adaptive regression splines
- PCSK9
- proprotein convertase subtilisin kexin type 9
- TG
- triglyceride
- VLDL
- very low-density lipoprotein.
References
- 1.Fazio S, Sidoli A, Vivenzio A, Maietta A, Giampaoli S, Menotti A, Antonini R, Urbinati G, Baralle FE, Ricci G. A form of familial hypobetalipoproteinaemia not due to a mutation in the apolipoprotein B gene. J Intern Med. 1991;229:41–47. [DOI] [PubMed] [Google Scholar]
- 2.Elias N, Patterson BW, Schonfeld G. In vivo metabolism of ApoB, ApoA-I, and VLDL triglycerides in a form of hypobetalipoproteinemia not linked to the ApoB gene. Arterioscler Thromb Vasc Biol. 2000;20:1309–1315. [DOI] [PubMed] [Google Scholar]
- 3.Pisciotta L, Favari E, Magnolo L, Simonelli S, Adorni MP, Sallo R, Fancello T, Zavaroni I, Ardigo D, Bernini F, Calabresi L, Franceschini G, Tarugi P, Calandra S, Bertolini S. Characterization of three kindreds with familial combined hypolipidemia caused by loss-of-function mutations of ANGPTL3. Circ Cardiovasc Genet. 2012;5:42–50. [DOI] [PubMed] [Google Scholar]
- 4.Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, Fennell T, Banks E, Ambrogio L, Cibulskis K, Kernytsky A, Gonzalez E, Rudzicz N, Engert JC, DePristo MA, Daly MJ, Cohen JC, Hobbs HH, Altshuler D, Schonfeld G, Gabriel SB, Yue P, Kathiresan S. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minicocci I, Santini S, Cantisani V, Stitziel N, Kathiresan S, Arroyo JA, Marti G, Pisciotta L, Noto D, Cefalu AB, Maranghi M, Labbadia G, Pigna G, Pannozzo F, Ceci F, Ciociola E, Bertolini S, Calandra S, Tarugi P, Averna M, Arca M. Clinical characteristics and plasma lipids in subjects with familial combined hypolipidemia: a pooled analysis. J Lipid Res. 2013;54:3481–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stitziel NO, Khera AV, Wang X, Bierhals AJ, Vourakis AC, Sperry AE, Natarajan P, Klarin D, Emdin CA, Zekavat SM, Nomura A, Erdmann J, Schunkert H, Samani NJ, Kraus WE, Shah SH, Yu B, Boerwinkle E, Rader DJ, Gupta N, Frossard PM, Rasheed A, Danesh J, Lander ES, Gabriel S, Saleheen D, Musunuru K, Kathiresan S; PROMIS and Myocardial Infarction Genetics Consortium Investigators . ANGPTL3 deficiency and protection against coronary artery disease. J Am Coll Cardiol. 2017;69:2054–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minicocci I, Montali A, Robciuc MR, Quagliarini F, Censi V, Labbadia G, Gabiati C, Pigna G, Sepe ML, Pannozzo F, Lutjohann D, Fazio S, Jauhiainen M, Ehnholm C, Arca M. Mutations in the ANGPTL3 gene and familial combined hypolipidemia: a clinical and biochemical characterization. J Clin Endocrinol Metab. 2012;97:E1266–E1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham MJ, Lee RG, Brandt TA, Tai LJ, Fu W, Peralta R, Yu R, Hurh E, Paz E, McEvoy BW, Baker BF, Pham NC, Digenio A, Hughes SG, Geary RS, Witztum JL, Crooke RM, Tsimikas S. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides [published online ahead of print May 24, 2017]. N Engl J Med. doi: 10.1056/NEJMoa1701329. [DOI] [PubMed]
- 9.Dewey FE, Gusarova V, Dunbar RL, O’Dushlaine C, Schurmann C, Gottesman O, McCarthy S, Van Hout CV, Bruse S, Dansky HM, Leader JB, Murray MF, Ritchie MD, Kirchner HL, Habegger L, Lopez A, Penn J, Zhao A, Shao W, Stahl N, Murphy AJ, Hamon S, Bouzelmat A, Zhang R, Shumel B, Pordy R, Gipe D, Herman GA, Sheu WHH, Lee IT, Liang KW, Guo X, Rotter JI, Chen YI, Kraus WE, Shah SH, Damrauer S, Small A, Rader DJ, Wulff AB, Nordestgaard BG, Tybjaerg-Hansen A, van den Hoek AM, Princen HMG, Ledbetter DH, Carey DJ, Overton JD, Reid JG, Sasiela WJ, Banerjee P, Shuldiner AR, Borecki IB, Teslovich TM, Yancopoulos GD, Mellis SJ, Gromada J, Baras A. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease [published online ahead of print May 24, 2017]. N Engl J Med doi:10.1056/NEJMoa1612790. [DOI] [PMC free article] [PubMed]
- 10.Shimizugawa T, Ono M, Shimamura M, Yoshida K, Ando Y, Koishi R, Ueda K, Inaba T, Minekura H, Kohama T, Furukawa H. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. J Biol Chem. 2002;277:33742–33748. [DOI] [PubMed] [Google Scholar]
- 11.Shimamura M, Matsuda M, Yasumo H, Okazaki M, Fujimoto K, Kono K, Shimizugawa T, Ando Y, Koishi R, Kohama T, Sakai N, Kotani K, Komuro R, Ishida T, Hirata K, Yamashita S, Furukawa H, Shimomura I. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler Thromb Vasc Biol. 2007;27:366–372. [DOI] [PubMed] [Google Scholar]
- 12.Nakajima K, Kobayashi J, Mabuchi H, Nakano T, Tokita Y, Nagamine T, Imamura S, Ai M, Otokozawa S, Schaefer EF. Association of angiopoietin-like protein 3 with hepatic triglyceride lipase and lipoprotein lipase activities in human plasma. Ann Clin Biochem. 2010;47:423–431. [DOI] [PubMed] [Google Scholar]
- 13.Minicocci I, Tikka A, Poggiogalle E, Metso J, Montali A, Ceci F, Labbadia G, Fontana M, Di Costanzo A, Maranghi M, Rosano A, Ehnholm C, Donini LM, Jauhiainen M, Arca M. Effects of angiopoietin-like protein 3 deficiency on postprandial lipid and lipoprotein metabolism. J Lipid Res. 2016;57:1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Gusarova V, Banfi S, Gromada J, Cohen JC, Hobbs HH. Inactivation of ANGPTL3 reduces hepatic VLDL-triglyceride secretion. J Lipid Res. 2015;56:1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ando Y, Shimizugawa T, Takeshita S, Ono M, Shimamura M, Koishi R, Furukawa H. A decreased expression of angiopoietin-like 3 is protective against atherosclerosis in apoE-deficient mice. J Lipid Res. 2003;44:1216–1223. [DOI] [PubMed] [Google Scholar]
- 16.Lee EC, Desai U, Gololobov G, Hong S, Feng X, Yu XC, Gay J, Wilganowski N, Gao C, Du LL, Chen J, Hu Y, Zhao S, Kirkpatrick L, Schneider M, Zambrowicz BP, Landes G, Powell DR, Sonnenburg WK. Identification of a new functional domain in angiopoietin-like 3 (ANGPTL3) and angiopoietin-like 4 (ANGPTL4) involved in binding and inhibition of lipoprotein lipase (LPL). J Biol Chem. 2009;284:13735–13745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabelli C, Bilato C, Martini S, Tennyson GE, Zech LA, Corsini A, Albanese M, Brewer HB Jr, Crepaldi G, Baggio G. Homozygous familial hypobetalipoproteinemia: increased LDL catabolism in hypobetalipoproteinemia due to a truncated apolipoprotein B species, apo B-87Padova. Arterioscler Thromb Vasc Biol. 1996;16:1189–1196. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Z, Tuakli-Wosornu Y, Lagace TA, Kinch L, Grishin NV, Horton JD, Cohen JC, Hobbs HH. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am J Hum Genet. 2006;79:514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maxwell KN, Breslow JL. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci USA. 2004;101:7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjannet S, Rhainds D, Essalmani R, Mayne J, Wickham L, Jin W, Asselin MC, Hamelin J, Varret M, Allard D, Trillard M, Abifadel M, Tebon A, Attie AD, Rader DJ, Boileau C, Brissette L, Chretien M, Prat A, Seidah NG. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J Biol Chem. 2004;279:48865–48875. [DOI] [PubMed] [Google Scholar]
- 21.Park SW, Moon YA, Horton JD. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in Mouse Liver. J Biol Chem. 2004;279:50630–50638. [DOI] [PubMed] [Google Scholar]
- 22.Le May C, Kourimate S, Langhi C, Chetiveaux M, Jarry A, Comera C, Collet X, Kuipers F, Krempf M, Cariou B, Costet P. Proprotein convertase subtilisin kexin type 9 null mice are protected from postprandial triglyceridemia. Arterioscler Thromb Vasc Biol. 2009;29:684–690. [DOI] [PubMed] [Google Scholar]
- 23.Rashid S, Tavori H, Brown PE, Linton MF, He J, Giunzioni I, Fazio S. Proprotein convertase subtilisin kexin type 9 promotes intestinal overproduction of triglyceride-rich apolipoprotein B lipoproteins through both low-density lipoprotein receptor-dependent and -independent mechanisms. Circulation. 2014;130:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tavori H, Giunzioni I, Predazzi IM, Plubell D, Shivinsky A, Miles J, Devay RM, Liang H, Rashid S, Linton MF, Fazio S. Human PCSK9 promotes hepatic lipogenesis and atherosclerosis development via apoE- and LDLR-mediated mechanisms. Cardiovasc Res. 2016;110:268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavori H, Giunzioni I, Linton MF, Fazio S. Loss of plasma proprotein convertase subtilisin/kexin 9 (PCSK9) after lipoprotein apheresis. Circ Res. 2013;113:1290–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjannet S, Rhainds D, Hamelin J, Nassoury N, Seidah NG. The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: functional consequences of natural mutations and post-translational modifications. J Biol Chem. 2006;281:30561–30572. [DOI] [PubMed] [Google Scholar]
- 27.Han B, Eacho PI, Knierman MD, Troutt JS, Konrad RJ, Yu X, Schroeder KM. Isolation and characterization of the circulating truncated form of PCSK9. J Lipid Res. 2014;55:1505–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Essalmani R, Susan-Resiga D, Chamberland A, Abifadel M, Creemers JW, Boileau C, Seidah NG, Prat A. In vivo evidence that furin from hepatocytes inactivates PCSK9. J Biol Chem. 2011;286:4257–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipari MT, Li W, Moran P, Kong-Beltran M, Sai T, Lai J, Lin SJ, Kolumam G, Zavala-Solorio J, Izrael-Tomasevic A, Arnott D, Wang J, Peterson AS, Kirchhofer D. Furin-cleaved proprotein convertase subtilisin/kexin type 9 (PCSK9) is active and modulates low density lipoprotein receptor and serum cholesterol levels. J Biol Chem. 2012;287:43482–43491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein EA, Raal FJ. New therapies for reducing low-density lipoprotein cholesterol. Endocrinol Metab Clin North Am. 2014;43:1007–1033. [DOI] [PubMed] [Google Scholar]
- 31.Robciuc MR, Tahvanainen E, Jauhiainen M, Ehnholm C. Quantitation of serum angiopoietin-like proteins 3 and 4 in a Finnish population sample. J Lipid Res. 2010;51:824–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mora S, Otvos JD, Rosenson RS, Pradhan A, Buring JE, Ridker PM. Lipoprotein particle size and concentration by nuclear magnetic resonance and incident type 2 diabetes in women. Diabetes. 2010;59:1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tavori H, Christian D, Minnier J, Plubell D, Shapiro MD, Yeang C, Giunzioni I, Croyal M, Duell PB, Lambert G, Tsimikas S, Fazio S. PCSK9 association with lipoprotein(a). Circ Res. 2016;119:29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tavori H, Su YR, Yancey PG, Giunzioni I, Wilhelm AJ, Blakemore JL, Zabalawi M, Linton MF, Sorci-Thomas MG, Fazio S. Macrophage apoAI protects against dyslipidemia-induced dermatitis and atherosclerosis without affecting HDL. J Lipid Res. 2015;56:635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hori M, Ishihara M, Yuasa Y, Makino H, Yanagi K, Tamanaha T, Kishimoto I, Kujiraoka T, Hattori H, Harada-Shiba M. Removal of plasma mature and furin-cleaved proprotein convertase subtilisin/kexin 9 by low-density lipoprotein-apheresis in familial hypercholesterolemia: development and application of a new assay for PCSK9. J Clin Endocrinol Metab. 2015;100:E41–E49. [DOI] [PubMed] [Google Scholar]
- 36.Højsgaard S, Halekoh U, Yan J. The R package geepack for Generalized Estimating Equations. J Stat Software. 2006;15:1–11. [Google Scholar]
- 37.Yan J, Fine JP. Estimating equations for association structures. Stat Med. 2004;23:859–880. [DOI] [PubMed] [Google Scholar]
- 38.Yan J. geepack: yet another package for generalized estimating equations. R-News. 2002. 2:12–14. [Google Scholar]
- 39.Friedman JH, Roosen CB. An introduction to multivariate adaptive regression splines. Stat Methods Med Res. 1995;4:197–217. [DOI] [PubMed] [Google Scholar]
- 40.Milborrow S. Derived from mda:mars by Trevor Hastie and Rob Tibshirani. Uses Alan Miller’s Fortran utilities with Thomas Lumley’s leaps wrapper. earth: Multivariate Adaptive Regression Splines. R package version 4.4.4. Available at: http://CRAN.R-project.org/package=earth. Accessed 21 April 2017.
- 41.Noto D, Cefalu AB, Valenti V, Fayer F, Pinotti E, Ditta M, Spina R, Vigna G, Yue P, Kathiresan S, Tarugi P, Averna MR. Prevalence of ANGPTL3 and APOB gene mutations in subjects with combined hypolipidemia. Arterioscler Thromb Vasc Biol. 2012;32:805–809. [DOI] [PubMed] [Google Scholar]
- 42.Gusarova V, Alexa CA, Wang Y, Rafique A, Kim JH, Buckler D, Mintah IJ, Shihanian LM, Cohen JC, Hobbs HH, Xin Y, Valenzuela DM, Murphy AJ, Yancopoulos GD, Gromada J. ANGPTL3 blockade with a human monoclonal antibody reduces plasma lipids in dyslipidemic mice and monkeys. J Lipid Res. 2015;56:1308–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaudet D, Gipe D, Hovingh K, Ahmad Z, Cuchel M, Shah P, Chyu K-U, Pordy R, Sasiela W, Chan K-C, Khoury E. Safety and efficacy of evinacumab, a monoclonal antibody to ANGPTL3, in homozygous familial hypercholesterolemia. J Clin Lipidol. 2017;11(3): 837–838. [Google Scholar]
- 44.Toth PP, Hamon SC, Jones SR, Martin SS, Joshi PH, Kulkarni KR, Banerjee P, Hanotin C, Roth EM, McKenney JM. Effect of alirocumab on specific lipoprotein non-high-density lipoprotein cholesterol and subfractions as measured by the vertical auto profile method: analysis of 3 randomized trials versus placebo. Lipids Health Dis. 2016;15:28. [DOI] [PMC free article] [PubMed] [Google Scholar]