Abstract
TP53 is the most commonly mutated tumor suppressor gene and its mutation drives tumorigenesis. Using ChIP-seq for p53 in the absence of acute cell stress, we found that wild-type but not mutant p53 binds and activates numerous tumor suppressor genes including PTEN, STK11(LKB1), miR-34a, KDM6A(UTX), FOXO1, PHLDA3 and TNFRSF10B through consensus binding sites in enhancers and promoters. Depletion of p53 reduced expression of these target genes, and analysis across 18 tumor types showed that mutation of TP53 associated with reduced expression of many of these genes. Regarding PTEN, p53 activated expression of a luciferase reporter gene containing the p53-consensus site in the PTEN enhancer, and homozygous deletion of this region in cells decreased PTEN expression and increased growth and transformation. These findings show that p53 maintains expression of a team of tumor suppressor genes that may together with the stress-induced targets mediate the ability of p53 to suppress cancer development. p53 mutations selected during tumor initiation and progression thus inactivate multiple tumor suppressor genes in parallel, which could account for the high frequency of p53 mutations in cancer.
Keywords: p53, other tumor suppressor genes, promoter/enhancer analysis, tumor suppression, PTEN, transcriptional regulation
Introduction
TP53 germline mutation is associated with Li-Fraumeni cancer susceptibility syndrome, and its somatic mutation occurs in about half of all cancers (1). p53-null or mutant mice develop spontaneous tumors in multiple organs (2). p53 is a transcription factor that binds DNA in a sequence-specific manner (3), and the majority of missense mutations that inactivate p53 in cancer are clustered within the DNA binding domain (1). p53 is well established to mediate acute stress response in the cell by activating known targets including CDKN1A(p21), GADD45a, BBC3(PUMA), and NOXA (4). Interestingly, p53 stress response targets that regulate apoptosis, cell cycle arrest, and senescence are not required for p53-mediated tumor suppression in genetically engineered model systems (5–7). p53 is expressed at measureable levels in various tissues of the body in the absence of acute stress (8). Basally expressed p53, which has been attributed to various physiologic stresses (9,10), activates the constitutive expression of CDKN1A(p21), MDM2, and miR-34a through consensus binding sites in cis elements (11–15).
Inactivation of both alleles of a tumor suppressor can be necessary for tumor development; but for haploinsufficient tumor suppressors, inactivation of just one allele is sufficient to cause tumor development alone or in cooperation with other gene mutations (16). Here, we show that basally expressed p53 acts as a transcriptional activator of multiple tumor suppressors, several of which are dose responsive in mice (17–20), a finding that suggests that they may play a role in p53-mediated tumor suppression.
Materials and Methods
Cell Culture
MCF10A, MCF7, and HCC38 cell lines were purchased from ATCC. The U2OS cell line was a gift from Dr. James Manfredi (purchased from ATCC). Cell lines were obtained between 2010–2015. ATCC authenticates cell lines using several methods including DNA fingerprinting. Cell lines were clear of mycoplasma as determined by the Lonza kit (LT07-418) within 6 months of their use. Cell lines were further authenticated in 2015 by LabCorp using a short tandem repeat method. All cells were cultured at 37°C and 5% CO2. See the Supplementary Materials and Methods for cell line-specific culturing conditions.
Transient Knockdown of p53
Cells were transfected using the Lipofectamine 2000 reagent (Thermo 11668019) in Opti-MEM media (Invitrogen 11058021). SMARTpool: ON-TARGET plus Human TP53 siRNA and MISSION siRNA Universal Negative Control (Sigma SIC001) were used. The cells were harvested 48–96 hours post-transfection, as indicated. Additional detail is provided in the Supplementary Materials and Methods, including the specific siRNA oligos used.
Luciferase Reporter Assay
H1299 cells were seeded at 2×105 cells/well of Falcon 6-well dishes. The transfections were carried out the following day using Lipofectamine (18324-020) and Plus (11514-015) reagents according to the manufacturer’s instructions. The cells were harvested 24h later using reagents supplied by the Luciferase Assay System (Promega E1910). The total protein concentration was determined using the Bio-Rad Protein Assay reagent (Bio-Rad). The luciferase assays were performed as specified by the manufacturer’s instructions and were quantitated using a TD-20e Luminometer (Turner). Luciferase expression is normalized to protein and expressed as Luciferase units/µg protein. Plasmids for the luciferase reporter assay are shown in the Supplementary Materials and Methods.
CRISPR of Hu PTEN-eP53RE
Cloning of the custom CRISPR guides targeting Hu PTEN-eP53RE was performed according to the LentiCRISPRv2 vector cloning protocol in the Lentiviral CRISPR toolbox from the Zhang lab that uses single guide RNAs (21,22). The custom targeting oligos used are shown in the Supplementary Materials and Methods. The LentiCRISPRv2 (Addgene #52961) plasmid backbone with the custom guide targeting Hu PTEN-eP53RE was named p53-PTEN-LentiCRISPRv2. Lentivirus was produced in HEK-293T cells as previously described (23). Limiting dilution was used to isolate single colonies, DNA from which was amplified and sequenced by Genewiz using the primers listed in the Supplementary Materials and Methods.
Chromatin Immunoprecipitation (ChIP-qPCR and ChIP-sequencing)
ChIP assays were performed as previously described (24). Cells were cross-linked in 1% formaldehyde (J.T. Baker 2106-01) for 5 minutes on ice. After quenching with glycine, the cells were harvested in 1× PBS containing 1× protease inhibitor cocktail (Sigma P8340) and pelleted. For ChIP-qPCR, cells were sonicated for 20 minutes (30s on, 30s off) on the Diagenode Bioruptor Twin (UCD-400) sonicator at 4°C. For ChIP-seq, cells were sonicated similarly for 40 minutes. Lysates were precleared for 1 hour with the appropriate beads (For Chip-qPCR: Protein A Agarose/Salmon Sperm DNA beads Emdmillipore 16–157, and for ChIP-seq: Magna ChIP™ Protein A+G Magnetic Beads Emdmillipore 16–663). Precleared lysates were then incubated with 7µg of antibody overnight at 4°C (3 million cells per antibody). Samples were then incubated with appropriate beads (same as preclear) for at least 2 hours at 4°C and beads were repeatedly washed. The Protein-DNA complexes were eluted, crosslinks were reversed, and DNA was purified using phenol/chloroform extraction followed by sodium acetate/ethanol precipitation. For ChIP-seq, DNA was purified again using the Qiaquick PCR purification kit (Qiagen 28104). Purified DNA was then subject to qPCR (ChIP-qPCR) or high throughput sequencing (ChIP-seq). For ChIP-qPCR, % input was calculated and normalized as a fold change from IgG. Antibodies: p53 (DO1-X, sc-126X), H3K27Ac (Millipore 07-360), H3 (ab 1791), IgG (sc-2025).
ChIP-qPCR primers and details regarding the ChIP-seq data analysis pipeline are located in the Supplementary Materials and Methods.
Three dimensional cell culture assay
Three dimensional culture assays were performed as previously described (PMID: 12798140) in the 8-well chamber slides (BD Falcon 08-774). Assay medium (DMEM/F12 supplemented with 2% horse serum, 10 µg/mL insulin, 1 ng/mL cholera toxin, 100 µg/mL hydrocortisone, 50 units/mL penicillin, and 50 µg/mL streptomycin) containing 5 ng/mL epidermal growth factor and 2% growth factor-reduced Matrigel (BD Biosciences 356231). Media was replaced every 4 days. Cells were collected using Cell Recovery Solution (BD BioSciences 354253) to remove Matrigel.
Immunofluorescence on 3D cultures
Cultures were fixed with 4% paraformaldehyde/PBS, permeabilized with 0.5% Triton X-100/PBS, and washed 3 times with PBS buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4, Adjust to a final pH of 7.4). Primary antibodies were suspended in IF buffer (3% bovine serum albumin, 0.1% NP-40 in PBS) and incubated overnight. The cultures were washed with 0.1% NP-40/PBS 6 times and incubated with Alexa fluorophore conjugated rabbit or mouse secondary antibodies (Life Technologies) in IF buffer. The slides were washed with 0.1% NP-40/PBS 6 times, counterstained with 4', 6-diamidino-2-phenylindole (DAPI), and mounted with antifade solution (Life Technologies). Confocal microscopy was done using the Zeiss LSM 780 laser scanning confocal microscope (Carl Zeiss AG). p-AKT(S473) (4060) and cleaved Caspase-3 (9664) antibodies were from Cell Signaling Technology. Rabbit polyclonal antibody against Ki67 (ab833) was from Abcam, and Mouse anti-laminin-5 (MAB1947) was from EMD Millipore. Alexa Fluor 488 anti-rabbit (A11008) and Alexa Fluor 594 anti-mouse (A21201) secondary antibodies, and ProLong® Gold antifade reagent with DAPI (P36931) were obtained from Life Technologies.
Results
p53 targets a group of tumor suppressor genes in the absence of acute stress
Breast cancers are known to harbor abnormalities in the tumor suppressors p53 and PTEN (Phosphatase and Tensin homolog, deleted on chromosome 10); hence, we examined their relationship in primary breast tumors. We observed that mutation of TP53 was associated with decreased PTEN transcript (P<.05) and protein levels (P<.01), and enrichment of the previously published PTEN-loss signature (25) (Supplementary Fig. S1A). This result was confirmed by qRT-PCR in tumors from the same cohort (P<.05, Supplementary Fig. S1B). The idea that p53 expressed in breast tissues could regulate baseline expression of PTEN is interesting because PTEN is a dose-dependent tumor suppressor whereby only a 20% deficit in its expression can lead to tumor development in mice(17).
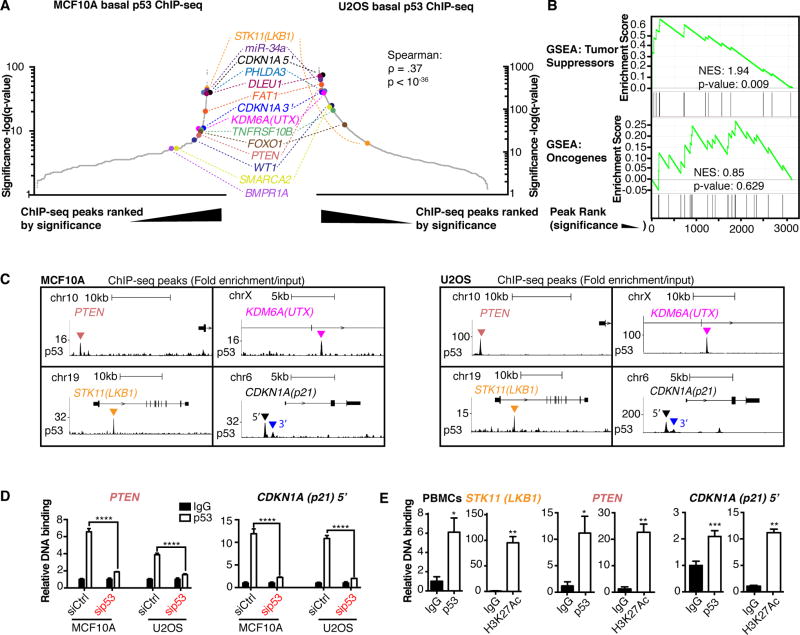
We sought to explore the possibility that basal wild-type p53 could have targets such as PTEN that are important for tumor suppression. In the absence of acute stress, we performed ChIP-seq for basal p53 in non-transformed human mammary epithelial MCF10A cells (wild-type for p53) and ranked the peaks by significance. Examination of the ranked list of peaks revealed that eleven of the first 200 peaks (q<10−8) were within or adjacent to ten known tumor suppressor genes, including PTEN. The number of tumor suppressor genes associated with peaks increased slightly to fourteen when the top 800 peaks (q<10−5) were queried (Fig. 1A left, Supplementary Spreadsheet S1). These tumor suppressor genes were also among the top basal p53 peaks when ranked by sequence read pileup before processing for significance (Supplementary Fig. S1C). The basal p53 binding sites at known tumor suppressors were STK11(LKB1), miR-34a, CDKN1A(p21) both 5’ and 3’ sites, PHLDA3, DLEU1, FAT1, KDM6A(UTX), TNFRSF10B, FOXO1, PTEN, WT1, SMARCA2 (BRM), and BMPR1A (Fig. 1A; Supplementary Fig. S2). These tumor suppressors are highly validated using evidence from mouse models, inherited cancer predisposition syndromes, and somatic mutations in human cancer (Supplementary Table S1). We also observed that basal p53 binds near other genes, including known basal p53 target MDM2 (14) (Supplementary Fig. S2). Others have performed basal wild type p53 ChIP-seq on U2OS cells as a control for comparison to the pattern of peaks detected after DNA damage or p53 stabilization (26). We used their control p53 ChIP-seq data to determine if targets of basal p53 in MCF10A cells were bound in another low-stress context. When comparing the two ranked lists, all of the tumor suppressor targets of basal p53 were present in both cell lines, except BMPR1A (p-value < 10−36, Spearman correlation test, Fig. 1A right, Supplementary Fig. S3A–B, Supplementary Spreadsheet S2).
Fig. 1. Basally expressed p53 binds to multiple other important tumor suppressors.
(A) Basal p53 ChIP-seq peaks ordered by significance in MCF10A (left) and U2OS cells (right). ρ is Spearman coefficient, and corresponding p-value is reported. (B) GSEA of basal MCF10A p53 ChIP-seq list (ordered by significance) for TSGs and Oncogenes as classified in the Cancer Gene Census from COSMIC. Enrichment scores and p-values are indicated. (C) ChIP-seq traces show fold enrichment over input for genes of interest in MCF10A (left) and U2OS (right) cells. Location of peak is indicated by triangle. Scales are indicated and start at zero. (D) ChIP-qPCR of p53 binding to PTEN (left) and 5’ CDKN1A(p21) (right) loci in MCF10A and U2OS cells transfected with control or p53-targeting siRNA (pool of 4 siRNAs). Relative DNA Binding is % input (normalized to IgG). Error bars: mean ± s.d. of representative experiment (performed twice), triplicate measurements. Significance: two-way ANOVA, Sidak’s correction. (E) ChIP-qPCR for basal p53 (left) and H3K27Ac (right) on the STK11(LKB1), PTEN, and 5’ CDKN1A (p21) loci in PBMCs from a healthy donor. Error bars: mean ± s.d., triplicate measurements. Significance (over IgG): paired t-tests. (****p≤.0001, ***p≤.001, **p≤.01, *p≤.05)
To better understand the potential function of the basal p53 peaks, we performed Gene Ontology (GO) for biological processes on the top 200 genes from the MCF10A list and discovered that only negative regulation of the G1/S phase transition of the cell cycle (GO: 2000134 and GO: 1902807) and signal transduction by p53 (GO: 0072331) were significantly enriched (Supplementary Fig. S3C). To evaluate the basal p53 binding targets for the enrichment of tumor suppressor genes (TSGs), we used the gene classifications (TSG or oncogene) in the Cancer Gene Census from COSMIC (27) (Supplementary Spreadsheet S3) and found that the ranked MCF10A basal p53 ChIP-seq list was enriched for TSGs but not oncogenes (p-value for TSGs = 0.009, GSEA, Fig. 1B). We also found significant enrichment of homozygously deleted genes across multiple cancers (frequency of 0.5% or greater, Supplementary Spreadsheet S4) (p-value = 0.001, GSEA, Supplementary Fig. S3D), independently suggesting that the p53-binding peak list is enriched for tumor suppressors. This enrichment was not present for frequently amplified genes (Supplementary Fig. S3E, Supplementary Spreadsheet S5). ChIP-seq peaks for select TSGs in both cell lines are shown (Fig. 1C). Using ChIP-qPCR in U2OS and MCF10A cells, we confirmed that p53 binds to the PTEN and 5’ CDKN1A sites identified by ChIP-seq, and found that this signal was diminished by p53 knockdown (Fig. 1D). In uncultured primary peripheral blood mononuclear cells (PBMCs) isolated from whole blood, basal p53 was bound to the STK11, PTEN, and CDKN1A loci, all of which had the histone 3 lysine 27 acetyl (H3K27Ac) mark of active enhancers and promoters (Fig. 1E).
p53 maintains expression of a group of tumor suppressor genes through consensus p53 binding sites in enhancers and promoters
p53 ChIP-seq studies have been published to investigate properties of wild type p53 after its induction or baseline mutant p53, but the baseline wild-type peaks were not analyzed (26,28–30). We examined these basal p53 ChIP-seq datasets from three nontumorigenic normal cell lines containing wild-type p53 (MCF10A, IMR90, and HFKs), three cancer cell lines containing wild-type p53 (U2OS, MDA-MB-175-VII, and MCF7) and three cancer cell lines containing mutant p53 (BT-549, HCC-70, and MDA-MB-468) and found that both normal and cancer cell lines containing wild-type p53 had significantly more basal p53 binding peaks near TSGs than the p53-mutant group, and the p53-mutant group did not have any basal p53 peaks near TSGs (Table 1, *P<.05) (26,28–30). We also determined that the R273L mutation abrogated p53 binding to PTEN or 5’CDKN1A in HCC38 cells (Supplementary Fig. S4A).
Table 1. Mutation of p53 ablates binding to tumor suppressor targets of basal p53.
Basal p53 ChIP-seq peaks near tumor suppressor genes in published datasets. Table contains (left to right) Gene Name, Peak Location (Hg19 location from MCF10A basal p53 ChIP-seq dataset), and whether there is a basal p53 ChIP-seq peak in each location (yes or no, Y/N) for the p53 ChIP-seq datasets for the cell lines: MCF10A immortalized non-transformed human mammary epithelial cells, IMR90 lung fibroblasts (28), human foreskin keratinocytes (HFKs) (29), the U2OS osteosarcoma cell line (26), and the breast cancer cell lines MDA-MB-175-VII, MCF7, BT-549, HCC-70, and MDA-MB-468 (30). TP53 status (WT or specific p53 mutation) as well as normal or cancer status (N or C) are listed.
| Gene Name | Peak Location (MCF10A) | MCF10A (WT-N) |
IMR90 (WT-N) |
HFK (WT-N) |
U2OS (WT-C) |
MDA-MB- 175-VII (WT-C) |
MCF7 (WT-C) |
BT-549 (R249S-C) |
HCC-70 (R248Q-C) |
MDA-MB- 468 (R273H-C) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| CDKN1A(p21) 5’ | Chr6: 36644096–36644510 | Y | Y | Y | Y | Y | Y | N | N | N | |
| DLEU1 | Chr13: 51103523–51103774 | Y | Y | Y | Y | Y | Y | N | N | N | |
| SMARCA2 | Chr9: 1707411–1707559 | Y | N | Y | Y | Y | N | N | N | N | |
| PTEN | Chr10: 89602953–89603111 | Y | Y | Y | Y | Y | Y | N | N | N | |
| KDM6A(UTX) | ChrX: 44835018–44835187 | Y | Y | Y | Y | N | N | N | N | N | |
| miR-34 | Chr1: 9242010–9242446 | Y | Y | Y | Y | Y | N | N | N | N | |
| TNFRSF10B | Chr8: 22926070–22926238 | Y | Y | Y | Y | Y | N | N | N | N | |
| STK11(LKB1) | Chr19: 1210487–1210722 | Y | N | Y | Y | N | N | N | N | N | |
| PHLDA3 | Chr1: 201438170–201438422 | Y | Y | Y | Y | Y | N | N | N | N | |
| WT1 | Chr11: 32325276–32325457 | Y | N | N | Y | N | N | N | N | N | |
| FOXO1 | Chr13: 40907488–40907636 | Y | N | Y | Y | N | N | N | N | N | |
| CDKN1A(p21) 3’ | Chr6: 36644988–36645255 | Y | Y | Y | Y | Y | Y | N | N | N | |
| FAT1 | Chr4: 187841713–187841958 | Y | Y | Y | Y | Y | N | N | N | N | |
| BMPR1A | Chr10: 88566579–88566727 | Y | N | N | N | N | N | N | N | N | |
| Total TS Peaks | 14/14 | 9/14 | 11/14 | 13/14 | 9/14 | 4/14 | 0/14 | 0/14 | 0/14 | ||
| Group significance | * | * | |||||||||
P-value: One-way ANOVA, Sidak’s correction. (*p≤.05, MUT-C group different than both WT-N and WT-C groups)
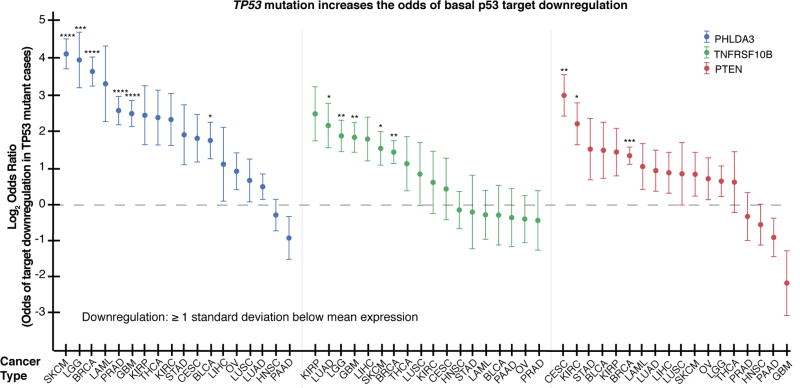
We next explored the possibility that p53 mutation is associated with lower target gene expression in cancer. Using data from The Cancer Genome Atlas (TCGA) from 4899 cases across 18 types of cancer, we found that TP53 mutation significantly increased the odds of transcriptional downregulation (by ≥ 1 standard deviation) of tumor suppressor targets of basal p53 (PHLDA3, TNFRSF10B, and PTEN, Fig. 2) in cases with normal copy number, and most of the tumor suppressor targets showed this trend to some degree. For example, STK11 had a weaker association (Supplementary Fig. S4B). This association was also detected in many cancer types when this analysis was performed without correcting for copy number change (Supplementary Fig. S4C).
Fig. 2. TP53 mutation increases relative risk for basal p53 target downregulation.
RNA-seq data from TCGA shows that TP53 mutation increases the relative risk for p53 target downregulation (defined as ≥ 1 s.d. below mean RNA-seq z-score for expression). All cases included have normal copy number. Acronyms for TCGA cancer types expanded in Supplementary Materials and Methods. Error bars: log2OR ± s.d. of dataset. Significance: Fisher’s exact test. (****p≤.0001, ***p≤.001, **p≤.01, *p≤.05)
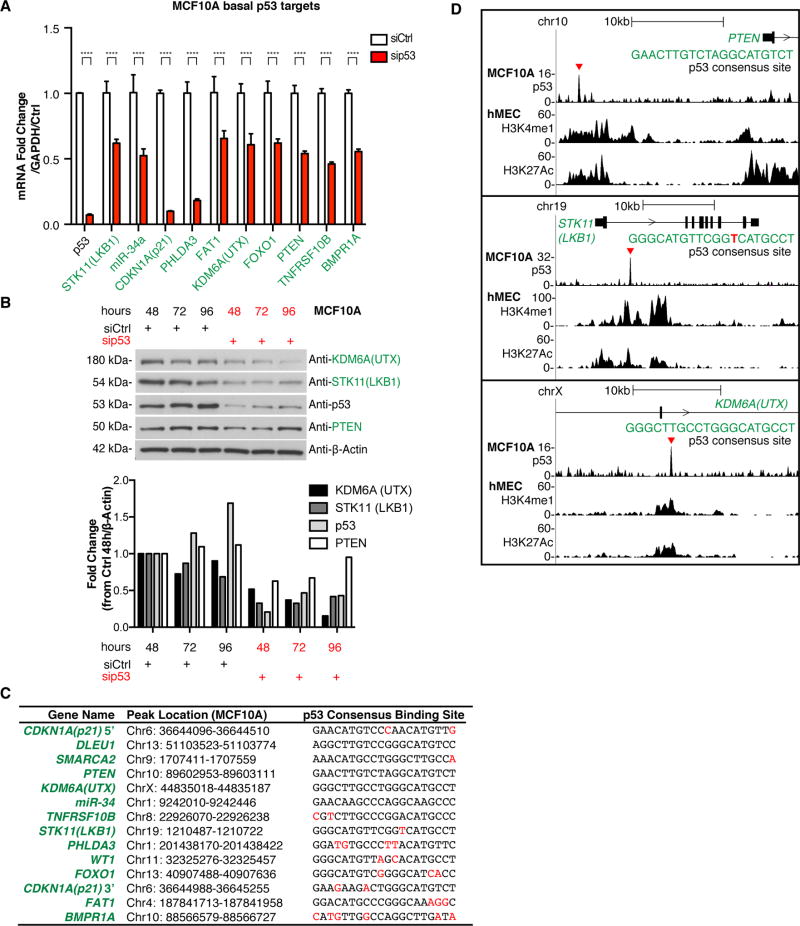
To determine if basal p53 regulates the mRNA expression of the identified binding targets, we performed a siRNA knockdown of p53 in MCF10A cells and measured transcript levels of select targets. With the exception of DLEU1, WT1, and SMARCA2, knockdown of p53 in MCF10A cells decreased mRNA of all of the identified binding targets and lowered protein levels for the three candidates that were tested (Fig. 3A–B, validated with individual siRNAs in Supplementary Fig. S4D). Knockdown of p53 in primary human mammary epithelial cells (hMECs) (31) also decreased mRNA for six out of seven of the tested TSGs (Supplementary Fig. S4E). Analysis of eleven of the top 400 (q<10−6) non-tumor suppressor peaks with similar consensus binding sites showed that p53 activated all of these genes in MCF10A cells with the exception of PLK2, which it repressed (Supplementary Fig. S4F).
Fig. 3. p53 maintains baseline expression of its tumor suppressor target genes.
(A) MCF10A cells were transfected with control or p53-targeting siRNAs (pool of 4 siRNAs), transcript levels were measured by qRT-PCR. Error bars: mean ± s.d. of representative experiment (performed twice), triplicate measurements. Significance: two-way ANOVA, Sidak’s correction (****p≤.0001). (B) Western blots measuring KDM6A(UTX), STK11(LKB1), p53, and PTEN at 48h, 72h, and 96h post-transfection. β-actin is loading control. Blot is quantified below (signal normalized to β -Actin, shown as fold change from siCtrl 48h). (C) Alignment of p53 consensus DNA binding sites. Includes gene, location of p53 peak, and p53 consensus binding site sequence. Deviations from the known p53 consensus are highlighted in red. (D) p53-bound DNA near PTEN, STK11, and KDM6A has H3K27Ac and H3K4me1 enhancer marks. For each gene, top: basal p53 ChIP-seq in MCF10A cells (fold enrichment over input), and bottom (2 tracks): published data of enrichment of H3K4me1 and H3K27Ac ChIP-seq in hMEC cells(33,34). p53 peaks are indicated (red triangle). Sequence of p53 consensus site is shown.
Using JASPAR software, which predicts transcription factor DNA binding sites, we found that the basal p53 ChIP-seq peak for all of the tumor suppressors contains a predicted p53 response element (Fig. 3C, Supplementary Table S2). These sites have no spacer DNA and obey the classic p53 consensus site (3) (RRRCWWGYYY-RRRCWWGYYY, where R=purine, Y=pyrimidine, W=adenine or thymine), suggesting that binding is mediated by the DNA sequence motif. Furthermore, we examined previously published Hi-C (32) and chromatin state segmentation using the ChromHMM method (33) and determined that physical interaction exists between basal p53 binding sites distal to TSGs (20 kb or more from the transcriptional start site, TSS) and the gene promoter/TSS of the TSGs (Supplementary Fig. S5). Collectively, our results suggest that these tumor suppressors are direct targets of p53 in a basal, low-stress context.
To examine the epigenetic landscape near the basal p53 binding sites, we used previously published ChIP-seq data from hMEC cells (33,34). Chromatin marks histone 3 lysine 4 monomethylation (H3K4me1), histone 3 lysine 4 trimethylation (H3K4me3), and histone 3 lysine 27 acetylation (H3K27Ac) in combination with the distance from the TSS were used to classify each basal p53 binding region as a promoter or an enhancer. We found that 9/14 of the tumor suppressor binding sites were located in enhancers (7 intergenic, 2 intragenic), 4/14 were in promoters; CDKN1A(p21) had both promoter and enhancer peaks, and BMPR1A did not fit either category (Supplementary Table S3). The intergenic enhancer elements were located between 2–450 kb from the TSS of TSGs. These results show that basal p53 binds to DNA elements that activate transcription of tumor suppressors. For example, the basal p53 binding sites near PTEN, STK11, and KDM6A are all located in active enhancers (Fig. 3D).
A p53-dependent enhancer maintains expression of PTEN
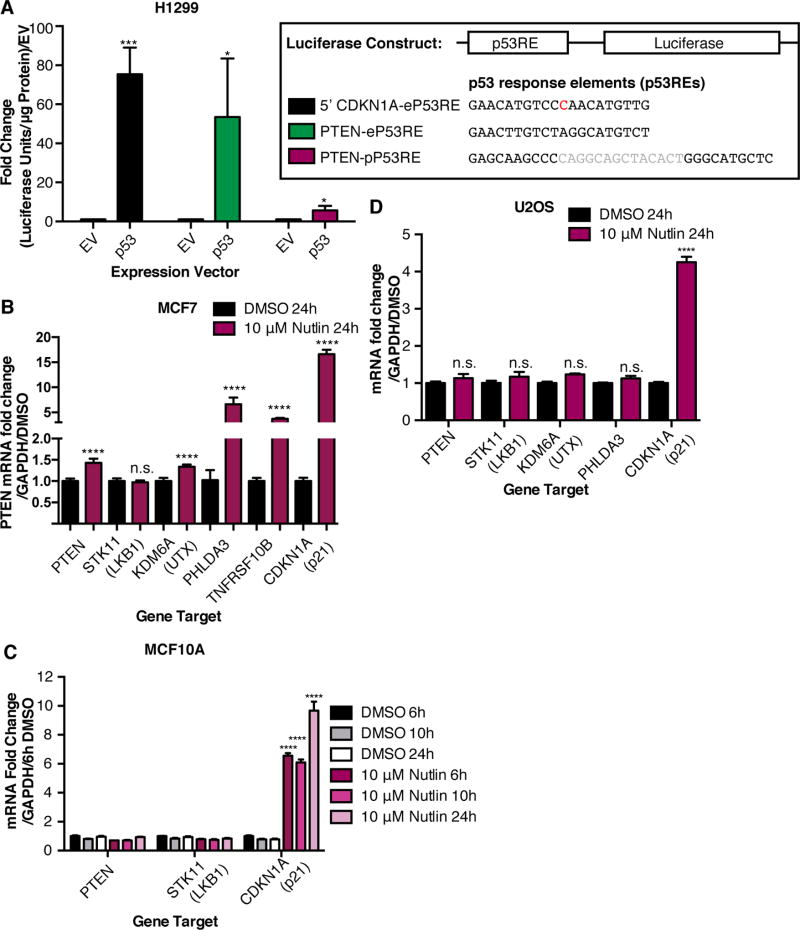
The basal p53 binding site upstream of PTEN is representative because the predicted binding site has no spacer DNA, it was classified as an enhancer, and it is distal (~20 kb upstream) to the TSS of its target gene. In fact, H3K27Ac and H3K4me1 peaks occur with this site across many cell types (Supplementary Fig. S6). Since the p53 binding site on PTEN is in an enhancer based on histone marks, we named this site PTEN-eP53RE and a previously characterized p53 response element in the PTEN promoter PTEN-pP53RE (35,36) (Supplementary Fig. S7A). p53 is known to bind to PTEN-pP53RE in response to stress, resulting in a subtle increase in PTEN levels in certain contexts (35,36). The PTEN gene locus is also in proximity with the ATAD1, CFL1P1, and KLLN loci (Supplementary Fig. S7A), and as shown in Supplementary Fig. S5, physical interaction occurs between the basal p53 binding site and these genes in lymphoblastoid cells. We found that p53 knockdown in MCF10A and U2OS cells led to decreased transcript levels of ATAD1 and KLLN, while CFL1P1, a pseudogene, was not expressed (Supplementary Fig. S7B). Therefore, the p53-bound enhancer for PTEN also regulates other genes in the locus, as is typical for enhancers. In freshly isolated PBMCs, p53 is strongly bound to PTEN-eP53RE but not PTEN-pP53RE (Supplementary Fig. S7C). Furthermore, PTEN-eP53RE is highly conserved in many mammals (Supplementary Fig. S8A–B). Interestingly, p53 knockdown led to a decreased H3K27Ac signal at PTEN-eP53RE in U2OS and MCF10A cells suggesting loss of p53-dependent enhancer activity (Supplementary Fig. S8C–D). To investigate whether the sequence of PTEN-eP53RE can activate transcription, we overexpressed empty vector or p53 along with luciferase reporter plasmids containing 5’CDKN1A-eP53RE (a known p53 response element located in an intergenic enhancer), PTEN-eP53RE, or PTEN-pP53RE. 5’CDKN1A-eP53RE and PTEN-eP53RE were able to strongly activate transcription, whereas PTEN-pP53RE activates transcription to a lesser extent (Fig. 4A).
Fig. 4. Transcriptional activation of tumor suppressor targets of p53 at baseline and after p53 induction.
(A) Luciferase reporter assay showing luciferase/renilla activity (fold change/empty vector) in H1299 (p53-null) cells overexpressing p53 with a luciferase reporter vector containing 5’CDKN1A-eP53RE, PTEN-eP53RE, or PTEN-pP53RE (constructs shown). Error bars: mean ± s.e.m.; n = 3 experiments. Significance: two-tailed Student’s t-test. (B) MCF7 cells treated with Nutlin-3 for 24 hours. Error bars: mean ± s.d., repeated twice. Transcripts measured by qRT-PCR. Significance: two-tailed students t-test. (C) MCF10A cells treated with 10 µM Nutlin-3 for 6, 10, and 24 hours. Error bars: mean ± s.d., triplicate measurements. (D) U2OS cells treated with 10 µM Nutlin-3 for 24 hours. Error bars: mean ± s.e.m, n = 4 experiments, triplicate measurements. Significance: one-way ANOVA, Dunnett’s correction. (****p≤.0001,***p≤.001,*p≤.05, n.s. p>.05)
We then investigated the dynamics of regulation of the basal p53 tumor suppressor targets using the p53-stabilizing drug Nutlin-3 in wild-type p53 MCF10A, U2OS and MCF7 cells. MCF7 cells, which had low basal p53 binding to tumor suppressor targets (Table 1), displayed significantly increased expression of PTEN, KDM6A, PHLDA3, TNFRSF10B, and CDKN1A after Nutlin-3 treatment (Fig. 4B). However, induction was limited to CDKN1A in MCF10A and U2OS cell lines with higher basal binding to tumor suppressor targets, possibly due to saturation of the targets at baseline (Table 1, Fig. 4C–D). MCF7 cells harbor a SNP in MDM2 that leads to increased transcriptional activation of MDM2 (37). This SNP could cause reduced basal p53 levels and binding to tumor suppressors (37), allowing for restoration of tumor suppressor expression upon Nutlin-3 treatment.
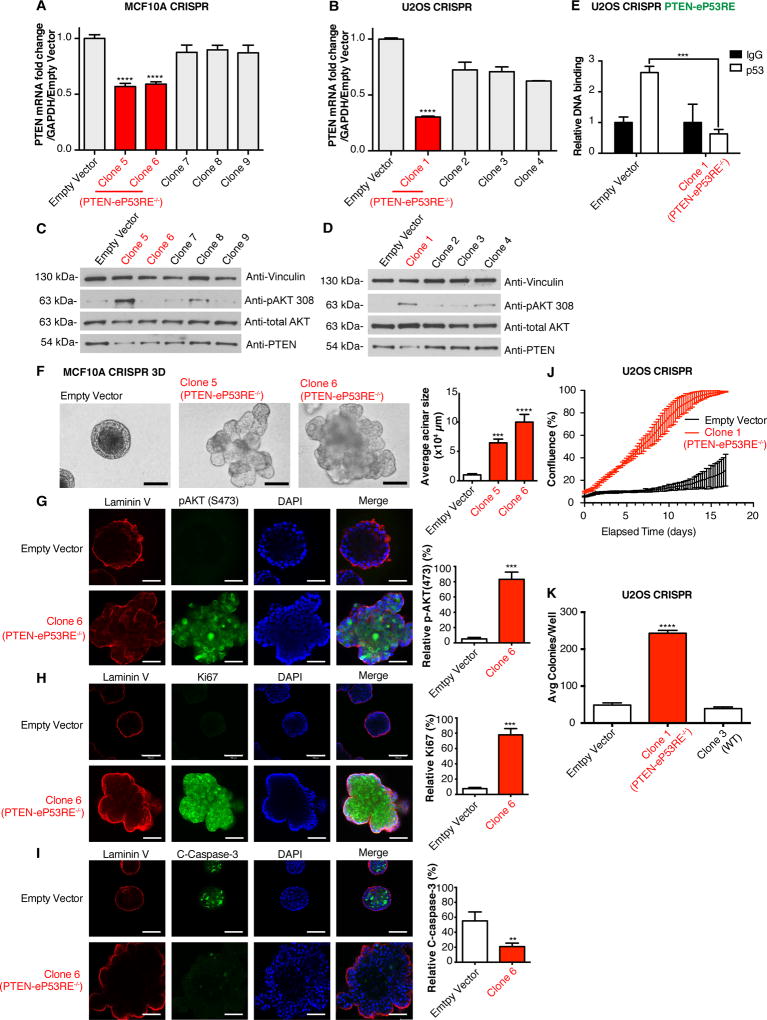
We discovered two cancer cases from TCGA that harbor somatic focal deletions of the enhancer containing PTEN-eP53RE (called as deep deletions, likely homozygous), both of which had downregulated PTEN transcript levels by RNA-seq (Supplementary Fig. S9A). To test the enhancer function of the region containing PTEN-eP53RE, we transduced MCF10A and U2OS cells with a CRISPR/Cas9 lentiviral vector containing a sgRNA targeting PTEN-eP53RE (Supplementary Fig. S9B). After extensive screening, we found three clones containing homozygous deletions of all or part of PTEN-eP53RE (MCF10A: Clones 5 and 6, U2OS: Clone 1, labeled as PTEN-eP53RE−/−, Supplementary Fig. S9C). We measured PTEN expression (Fig. 5A–D), where the PTEN-eP53RE−/− clones had significantly lower expression of PTEN transcript and protein when compared to all other clones in both cell lines. In MCF10A cells, an increase in pAKT(Thr308) was only present in Clone 5 (PTEN- eP53RE−/−) (Fig. 5C). In U2OS cells, we also observed increased pAKT (Thr308) in Clone 1 (PTEN-eP53RE−/−) (Fig. 5D). In U2OS Clone 1 (PTEN-eP53RE−/−), because the deletion includes only half of PTEN-eP53RE, we used ChIP-qPCR to test whether this prevents p53 binding. As expected, p53 no longer binds at PTEN-eP53RE in Clone 1 (Fig. 5E). To confirm that the decrease in PTEN expression in the PTEN-eP53RE−/− clones is dependent on PTEN-eP53RE, we performed a p53 knockdown in U2OS Empty Vector cells and Clone 1 (PTEN-eP53RE−/−) and discovered that a decrease in PTEN expression only occurred when PTEN-eP53RE was present (Supplementary Fig. S10A–B).
Fig. 5. Loss of PTEN-eP53RE results in downregulation of PTEN and increased tumor cell phenotypes.
(A–B) qRT-PCR of CRISPR/Cas9 clones in (A) MCF10A and (B) U2OS cells (PTEN-eP53RE−/− clones in red). Error bars: mean ± s.d., triplicate measurements. Significance: one-way ANOVA, Sidak’s correction. (C–D) Western blots of (C) MCF10A and (D) U2OS clones. Vinculin and total-AKT were loading controls. (E) ChIP-qPCR for p53 in U2OS clones on PTEN-eP53RE. Relative DNA Binding is % input (normalized to IgG). Error bars: mean ± s.d. of representative experiment (performed twice), triplicate measurements. Significance: two-way ANOVA, Sidak’s correction. (F) MCF10A clones grown in 3D culture for 20 days, representative photos of acini shown, acinar size is quantified. Error bars: mean ± s.d. of representative experiment, triplicate measurements. Significance: one-way ANOVA, Dunnett’s correction. (G–I) Representative immunofluorescence staining for Laminin V (red, all rows), (G) pAKT(Ser473), (H) Ki67, and (I) cleaved Caspase-3 (green), DAPI (blue, all rows), and merge (right, all rows). Scale bars: 100µm. Quantifications on right. Error bars: mean ± s.d. of representative experiment, triplicate measurements. Significance: two-tailed t-test. (J) Proliferation assay of U2OS clones in low serum showing % confluence over time (days). Triplicate readings taken every 6 hours. Error bars: mean ± s.d. (K) Soft agar colony formation assay of U2OS clones showing colonies/well after 3 weeks (triplicate measurements). Error bars: mean ± s.d. of representative experiment (performed twice), significance: one-way ANOVA, Sidak’s correction. (****p≤.0001, ***p≤.001, **p≤.01)
Deletions in the p53-dependent enhancer for PTEN induce cell transformation phenotypes
Given that the region of chromatin containing PTEN-eP53RE is an active enhancer for PTEN, we wanted to characterize its effect on growth properties of the cells. MCF10A clones showed no differences in proliferation under low serum conditions and did not grow in soft agar (Supplementary Fig. S10C); hence, we grew MCF10A acini for 20 days in 3D matrigel culture. Compared to Empty vector, the acini from Clones 5 and 6 (both PTEN-eP53RE−/−) were overgrown and lacked normal morphology (Fig. 5F). Furthermore, immunofluorescence staining for pAKT(S473), Ki67, and cleaved Caspase-3 revealed that pAKT (S473) and Ki67 levels were increased in acini from Clone 6 (Fig. 5G–H), whereas cleaved Caspase-3 was decreased (Fig. 5I) compared to Empty Vector. Similar results were obtained for Clone 5 (Supplementary Fig. S10D–F), and these results are consistent with published data on acini from PTEN−/− MCF10A cells (38). We performed a proliferation assay in U2OS cells in low serum and found that Clone 1 (PTEN-eP53RE−/−) grew faster than Empty Vector (Fig. 5J). Clone 1 also formed more colonies in soft agar when compared to both Empty Vector and Clone 3 (wild-type, expressing sgRNA) (Fig. 5K).
Discussion
Here we show that p53 binds cis regulatory regions of a group of thirteen highly validated tumor suppressor genes at baseline: STK11(LKB1), miR-34a, CDKN1A(p21) both 5’ and 3’ sites, PHLDA3, DLEU1, FAT1, KDM6A(UTX), TNFRSF10B, FOXO1, PTEN, WT1, SMARCA2(BRM), and BMPR1A. The tumor suppressors PTEN, STK11, KDM6A, and FAT1 are among the most genetically altered genes in cancer based on a systematic analysis of 21 types of cancer (39). Germline mutations of the tumor suppressors PTEN, STK11, WT1, and BMPR1A cause inherited cancer predisposition syndromes (Supplementary Table S1). Furthermore, all of the members of this group, with the exception of FAT1, suppress development of tumors or other neoplasia in genetically engineered mouse models (Supplementary Table S1).
Lowering the expression of some tumor suppressors via haploinsufficiency and hypomorphic mutations is sufficient to stimulate cancer development. Importantly, among the group of tumor suppressors targeted by p53--PTEN, STK11, PHLDA3, and TNFRSF10B--are haploinsufficient and/or hypomorphic tumor suppressors in mice (17–20). Three of these tumor suppressors target the PI3K/AKT/mTOR signaling axis. PTEN directly antagonizes PI3K by dephosphorylating PIP3, STK11 phosphorylates AMPK leading to inhibition of mTOR, and PHLDA3 binds PIP3 to prevent the activation of AKT (40–42). TNFRSF10B is a cell death receptor that binds TRAIL and mediates apoptosis (43). In our pan-cancer genomic analysis of human cancer, we found that TP53 mutation is significantly associated with PHLDA3, TNFRSF10B, and PTEN transcript downregulation, demonstrating the transcriptional changes that occur to these haploinsufficient tumor suppressors in the TP53-mutant setting. Mice heterozygous for PTEN and hypomorphic for STK11 develop tumors in multiple organs at a much shorter latency than alteration of either locus alone (44), suggesting that partial loss of multiple members of this group of tumor suppressors in parallel, which we predict would happen in the p53-mutant setting, could hasten tumor development.
Our findings show that wild-type p53 binds enhancer and promoter elements to activate baseline expression of a team of tumor suppressor genes. Mutation of p53 may cause spontaneous tumorigenesis due to the loss of basal p53 target expression combined with the inability of p53 to appropriately activate stress-responsive targets (summarized in Fig. 6). Certain p53 mutations also have gain-of-function properties that can contribute to tumorigenesis (30,45). p53-dependent regulation of basal targets could vary depending on the enhancer and promoter activity controlled by tissue and cell lineage, a feature that could contribute to the proclivity of p53 to suppress cancer development in some tissues more than others, and could explain the spectrum of tumors observed in Li-Fraumeni syndrome. In breast cancer, p53 mutations likely occur early in tumor development (46) where they would coordinately downregulate p53 target tumor suppressors and could select for chromosome losses that would further the loss of expression of these genes.
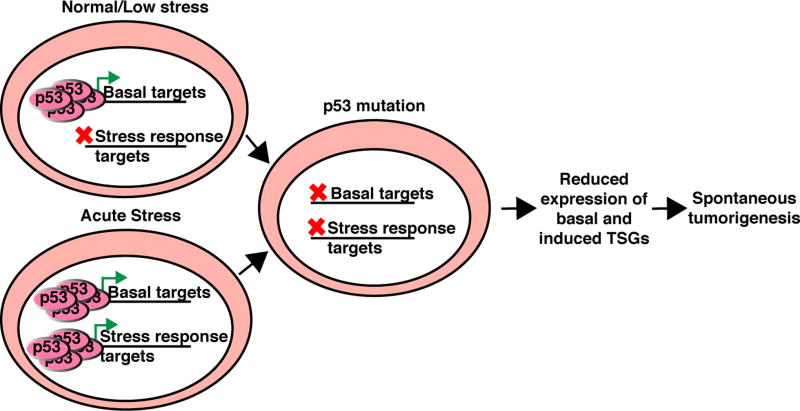
Fig. 6. The basal targets of p53 may contribute to p53-mediated tumor suppression.
The low levels of p53 present under normal physiologic stress activate a set of basal targets. Mutation of p53 could cause spontaneous tumorigenesis through the loss of expression of tumor suppressor targets of basal p53, combined with the inability of p53 to appropriately activate stress responsive targets.
Studies aimed to identify targets of p53 using ChIP-seq have focused on response to extracellular stresses or targets of mutant p53; thus, the targets of basal wild type p53 have not been comprehensively studied. The group of tumor suppressor genes regulated by basal p53 comprises a distinct set of targets that only has some overlap with those genes activated by p53 in response to stress (overlap includes CDKN1A (47,48), TNFRSF10B (49), PTEN (35,36), PHLDA3 (42), and mIR-34a (50)). With the exception of known p53 stress responsive genes CDKN1A, TNFRSF10B, PHLDA3, and mIR-34a, the basal ChIP-seq p53 binding sites for the remaining tumor suppressors, including PTEN, have not been previously described. Notably, all of these stress responsive genes have promoter p53 response elements, whereas tumor suppressors not induced by stress tend to have basal p53 binding sites in enhancers but not promoters. We hypothesize that the difference between basal and induced activities of p53 not only depends on p53 expression level and binding affinity to the site, but also depends on other factors that can mediate preferential binding to one site over another, such as the presence of repressor proteins or repressive chromatin states. Furthermore, the level of basal p53 activity may dictate the ability of Nutlin-3 to restore target tumor suppressor expression in cells. In this study, we determined that deletion of the p53-dependent enhancer for PTEN (PTEN-eP53RE) lowers its expression and induces transformation phenotypes. Mutation of the p53-binding site should be performed for the other basal p53 target tumor suppressors, alone or in combination, to determine their respective roles in tumor development. Overall, our findings provide a model of p53-mediated tumor suppression through activation of a team of tumor suppressor genes when the level of p53 is limited.
Supplementary Material
Statement of Significance.
In this study, we investigate the activities of p53 under normal low-stress conditions and discover that p53 is capable of maintaining the expression of a group of important tumor suppressor genes at baseline, many of which are haploinsufficient, which could contribute to p53-mediated tumor suppression.
Acknowledgments
We thank the members of Kyrie Pappas’ Thesis Advisory Committee: Timothy Wang (CUMC), Raul Rabadan (CUMC), and Hanina Hibshoosh (CUMC). We thank Emily Bernstein (MSSM) and Dan Hasson (E. Bernstein Lab, MSSM) for advice on ChIP-seq. We thank Ravi Sachidanandam (MSSM) and laboratory members for sequencing. We thank the Icahn School of Medicine Human Immune Monitoring Core for providing PBMCs and the Tumor Bank at the Columbia University Herbert Irving Comprehensive Cancer Center Molecular Pathology Shared Resource for providing primary breast tumor tissue. We would also like to acknowledge the ENCODE Consortium, the Broad Institute, and the B. Bernstein laboratory at Massachusetts General Hospital/Harvard Medical School for generating the histone modification ChIP-seq datasets, and the MIT Computational Biology Group (Kellis lab) by Jason Ernst for producing the ChromHMM tracks. Raw data for MCF10A ChIP-seq was tabulated in the Supplementary Materials has been deposited into the NCBI Sequence Read Archive under the study accession number SRP073955.
Financial Support: This research was supported by the NCI at the NIH (R01 CA82783 PI: R. Parsons, P01 97403 PI: R. Parsons) and the Komen Foundation (SAC110028, PI: R. Parsons). K. Pappas was supported by the Ruth L. Kirschstien National Research Service Award for Individual Predoctoral Fellows (F31 CA183268). J. Xu was supported by the T32 posdoctoral fellowship (T32 CA078207). The work of R. Rabadan, F. Abate and S. Zairis was partially supported by the NIH (U54 CA193313) and TL1 personalized medicine fellowship (5TL1TR000082). L.H. Saal was supported by the Swedish Cancer Society and the Swedish Research Council. L. Resnick-Silverman and J. Manfredi were supported by the NCI at the NIH (R01 CA200256). The Tisch Cancer Institute was supported by the NCI Cancer Center Support Grant (P30 CA196521)
References
- 1.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Jr, CAM. Butel JS, et al. Mice deficient for p53 are developmentally normal but suceptible to spontaneous tumors. Nature. 1992;356:215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 3.El-Deiry SEK Wafik S, Pietenpol Jennifer A, Kinzler Kenneth W, Vogelstein Bert. Definition of consensus binding site for p53. Nature Genetics. 1992;1:45–9. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 4.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–70. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–83. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–83. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valente LJ, Gray DH, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL, et al. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep. 2013;3:1339–45. doi: 10.1016/j.celrep.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Seim I, Ma S, Gladyshev VN. Gene expression signatures of human cell and tissue longevity. npj Aging and Mechanisms of Disease. 2016;2:16014. doi: 10.1038/npjamd.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–13. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang H-y, Zhao K, Pizzolato JF, Fonarev M, Langer JC, Manfredi JJ. Constitutive Expression of the Cyclin-dependent Kinase Inhibitor p21 Is Transcriptionally Regulated by the Tumor Suppressor Protein p53. J Biol Chem. 1998;273:29156–63. doi: 10.1074/jbc.273.44.29156. [DOI] [PubMed] [Google Scholar]
- 12.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamard PJ, Barthelery N, Hogstad B, Mungamuri SK, Tonnessen CA, Carvajal LA, et al. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev. 2013;27:1868–85. doi: 10.1101/gad.224386.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barak Y, Juven T, Haffner R, Oren M. Mdm2 Expression Is Induced by Wild Type-P53 Activity. Embo Journal. 1993;12:461–8. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Resnick-Silverman L, Clair SS, Maurer M, Kathy Zhao, Manfredi JJ. Identification of a novel class of genomic DNA binding sites suggests a mechanism for selectivity in target gene activation by the tumor suppressor protein p53. Genes & Development. 1998;12:2102–7. doi: 10.1101/gad.12.14.2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santarosa M, Ashworth A. Haploinsufficiency for tumour suppressor genes: when you don't need to go all the way. Biochim Biophys Acta. 2004;1654:105–22. doi: 10.1016/j.bbcan.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42:454–8. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morton JP, Jamieson NB, Karim SA, Athineos D, Ridgway RA, Nixon C, et al. LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21-dependent growth arrest. Gastroenterology. 2010;139:586–97. 97 e1–6. doi: 10.1053/j.gastro.2010.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohki R, Saito K, Chen Y, Kawase T, Hiraoka N, Saigawa R, et al. PHLDA3 is a novel tumor suppressor of pancreatic neuroendocrine tumors. Proc Natl Acad Sci U S A. 2014;111:E2404–13. doi: 10.1073/pnas.1319962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finnberg N, Klein-Szanto AJ, El-Deiry WS. TRAIL-R deficiency in mice promotes susceptibility to chronic inflammation and tumorigenesis. J Clin Invest. 2008;118:111–23. doi: 10.1172/JCI29900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott David A, Mikkelsen TS, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2014;343:84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lois C, Hong EJ, Shirley Pease, Brown EJ, Baltimore D. Germline Transmission and Tissue-Specific Expression of Transgenes Delivered by Lentiviral Vectors. Science. 2002;295:868–72. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 24.Niu H, Cattoretti G, Dalla-Favera R. BCL6 controls the expression of the B7-1/CD80 costimulatory receptor in germinal center B cells. The Journal of experimental medicine. 2003;198:211–21. doi: 10.1084/jem.20021395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proceedings of the National Academy of Sciences. 2007;104:7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menendez D, Nguyen TA, Freudenberg JM, Mathew VJ, Anderson CW, Jothi R, et al. Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucleic Acids Res. 2013;41:7286–301. doi: 10.1093/nar/gkt504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sammons MA, Zhu J, Drake AM, Berger SL. TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res. 2015;25:179–88. doi: 10.1101/gr.181883.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDade SS, Patel D, Moran M, Campbell J, Fenwick K, Kozarewa I, et al. Genome-wide characterization reveals complex interplay between TP53 and TP63 in response to genotoxic stress. Nucleic Acids Res. 2014;42:6270–85. doi: 10.1093/nar/gku299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525:206–11. doi: 10.1038/nature15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ENCODE. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–39. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, et al. Regulation of PTEN transcription by p53. Molecular Cell. 2001;8:317–25. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 36.Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–53. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 37.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh S, Varela L, Sood A, Park BH, Lotan TL. mTOR signaling feedback modulates mammary epithelial differentiation and restrains invasion downstream of PTEN loss. Cancer Res. 2013;73:5218–31. doi: 10.1158/0008-5472.CAN-13-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maehama T, Dixon J. The Tumor Suppresor, PTEN/MMAC1, Dephosphorylates the Lipid Second Messenger, Phosphatadylnositol 3,4,5-triphosphate. J Biol Chem. 1998;273:13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 41.Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LGD, Neumann D, et al. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Current Biology. 2003;13:2004–8. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 42.Kawase T, Ohki R, Shibata T, Tsutsumi S, Kamimura N, Inazawa J, et al. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell. 2009;136:535–50. doi: 10.1016/j.cell.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Sheridan JP, Marsters SA, Pitti RM, Gurney A, Skubatch M, Baldwin D, et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277:818–21. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 44.Huang X, Wullschleger S, Shpiro N, McGuire VA, Sakamoto K, Woods YL, et al. Important role of the LKB1-AMPK pathway in suppressing tumorigenesis in PTEN-deficient mice. Biochem J. 2008;412:211–21. doi: 10.1042/BJ20080557. [DOI] [PubMed] [Google Scholar]
- 45.Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–58. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a Potential Mediator of p53 Tumor Suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 48.El-Deiry WS, Tokino T, WaldInan T, Oliner JD, Velculescu VE, Burrell M, et al. Topological Control of p21wAF1/CIP1 expression in Normal and Neoplastic Tissues. Cancer Research. 1995;55:2910–9. [PubMed] [Google Scholar]
- 49.Wu GS, Burns TF, McDonald ER, 3rd, Jiang W, Meng R, Krantz ID, et al. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet. 1997;17:141–3. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 50.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.