Summary
For many arthropod vectors, the diverse bacteria and fungi that inhabit the gut can negatively impact pathogen colonization. Our attempts to exploit antibiotic treatment of colonized Phlebotomus duboscqi sand flies in order to improve their vector competency for Leishmania major resulted instead in flies that were refractory to the development of transmissible infections that was due to the inability of the parasite to differentiate into the infective, metacyclic stage. The parasite development defect could be overcome by feeding the flies on different symbiont bacteria but not by feeding them on bacterial supernatants or replete medium. The inhibitory effect of the antibiotic treatment was moderated by maintaining the flies on a lower concentration of sucrose in their sugar meals. The data suggest that competing sucrose utilization by the microbiota produces the appropriate nutrient stress and osmotic conditions required for stage differentiation and survival of infectious metacyclic promastigotes in vivo.
Keywords: Leishmania, sand flies, vector competence, microbiota, metacyclogenesis, osmotic stress
Parasites of the genus Leishmania are kinetoplastid protozoans that produce a spectrum of diseases in their mammalian hosts, including humans, ranging from localized cutaneous lesions to fatal, visceral disease. Leishmania have a dimorphic life cycle consisting of extracellular promastigotes that multiply and develop within the digestive tract of their female sand fly vectors, and intracellular amastigotes that multiply within the phagolysosomes of their host cell macrophages. In addition, promastigotes undergo a sequence of development in the vector midgut culminating in their differentiation to infective, metacyclic promastigotes that are uniquely pre-adapted for their transition to life in the vertebrate host. The identification of molecules that enable the parasite to survive within these harsh, diverse environments continues to be the focus of much of the research devoted to these organisms. With respect to the insect promastigotes stages, a family of surface and secreted phosphoglycan-containing molecules have been shown to be essential for the parasite to survive certain natural barriers to midgut colonization, including the digestive enzymes that are induced upon blood feeding, and the excretion of the blood meal remnants that necessitates promastigote attachment to the gut wall to avoid being expelled (Sacks and Kamhawi, 2001). Another potential barrier to parasite development is the natural gut microbiota, which is acquired by adult sand flies from several sources, including during feeding on their respective animal and plant sources of blood and sugar, or from re-colonization of the gut by the microbes ingested by the terrestrial dwelling larval stages (Dillon et al., 1996; Hillesland et al., 2008; Mukhopadhyay et al., 2012; Peterkova-Koci et al., 2012; Sant’Anna et al., 2012). For many arthropod vectors, the diverse bacteria and fungi that inhabit the gut can negatively impact pathogen colonization, either by a direct microbial interaction, by activation of immune mediators, or by competing for essential nutrients (Weiss and Aksoy, 2011). For example, gram-negative bacteria inhibit sporogonic development of Plasmodium in the mosquito midgut (Cirimotich et al., 2011; Pumpuni et al., 1993), tsetse flies lacking certain bacterial endosymbionts are highly susceptible to infection with African trypanosomes (Weiss et al., 2013), and the natural microbiota protects bees against a widespread and highly virulent natural parasite, Crithidia bombi (Koch and Schmid-Hempel, 2011). For sand flies, the few studies that have addressed this relationship also support a role for the natural microbiota in inhibiting parasite development. Mycoses were found in the guts of Phlebotomus papatasi from the Jordan Valley, and fungi-infected laboratory reared P. papatasi were more refractory to infections with L. major (Schlein et al., 1985). Similarly, pre-feeding Lutzomyia longipalpis on commensal yeast or bacterial species reduced the frequency and intensity of L. mexicana infection in these flies (Sant’Anna et al., 2014). The induction of antimicrobial peptides, e.g. defensins, by the introduction of bacteria into sand flies has been shown (Boulanger et al., 2004; Telleria et al., 2012), although what role, if any, these responses play in the inhibition of Leishmania colonization is not known.
One conclusion from these studies is that any manipulation that reduces the size and/or diversity of the natural microbiota should enhance the ability of Leishmania to establish infections in the fly, similar to the increased susceptibility to P. faliciparum or to dengue virus infection observed in antibiotic treated Anopheles gambiae or Aedes aegypti mosquitoes, respectively (Dong et al., 2009; Xi et al., 2008). Our attempts to exploit antibiotic treatment of our colonized P. duboscqi sand flies in order to improve their vector competency for L. major resulted instead in flies that were highly refractory to the development of transmissible infections. We report that the bacterial communities naturally present in the sand fly midgut are essential to the colonization of the midgut by infective stage, metacyclic promastigotes. We attribute this phenomenon to the capacity of the gut bacterial symbionts to generate the appropriate nutrient stress and osmotic conditions required for promastigote differentiation and survival.
Results
Antibiotic treatment prevents late stage development and survival of L. major in P. duboscqi
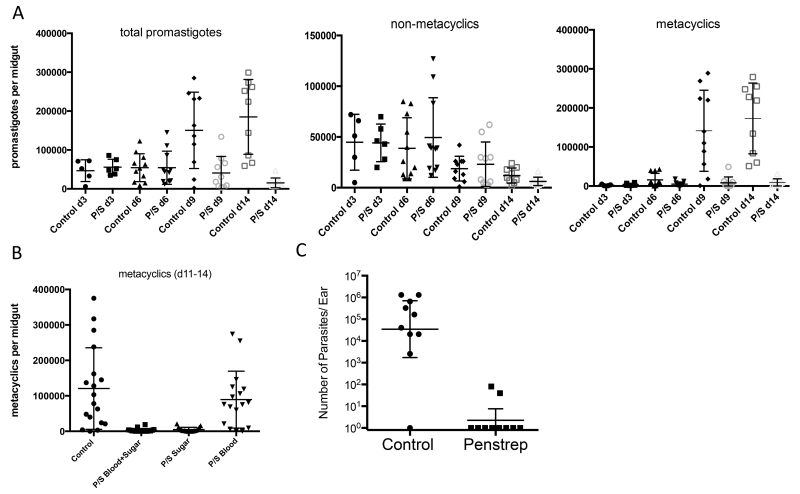
P. duboscqi is the natural vector of L. major transmission in west Africa, and a permissive vector in the laboratory for most L. major strains. Infection with LmRy produces an initial growth of promastigotes in the midgut over the first three days that survive excretion of the digested blood meal (days 4-6), followed by another stage of growth and differentiation into non-dividing, infective stage metacyclic promastigotes (days 9-14) (Fig 1A). Addition of penicillin and streptomycin (P/S) to both the blood meal and sugar meal resulted in normal growth and persistence of promastigotes over the first 6 days, followed by a drastic reduction in the intensity of infection compared to control flies at days 9-14, that was accounted for almost entirely by the absence of metacyclic forms. The dramatic reduction in the number of metacyclic promastigotes was observed when the antibiotic was included in both the blood and subsequent sugar meals, or in the sugar meal only (days 5-14), but not when the antibiotic was confined to the blood meal (Fig. 1B). Our in vitro propagation of Leishmania promastigotes is routinely carried out in the presence of P/S, so we did not believe that there was a direct effect of the antibiotics on the growth and differentiation of the parasite. This was confirmed by the in vitro results showing identical growth and differentiation to metacyclics in the presence or absence of P/S (Sup Fig 1). The reduced number of metacyclics in the antibiotic treated flies resulted in a drastic reduction in their capacity to transmit L. major by bite, both with regard to the frequency of successful transmissions (2 of 10 ears vs 9 of 10 ears) and the inoculated dose when transmissions occurred (Fig. 1C).
Figure 1.
Effect of penicillin/streptomycin (P/S) on L. major development in P. duboscqi. Flies were artificially fed through a membrane on mouse blood seeded with 4×106/ ml LmRy promastigotes and dissected at the indicated times post-infection and scored for the number and developmental stage of promastigotes under a hemocytometer. Flies were treated or not with (A) P/S in the blood and sugar meals, (B) P/S in the blood and/or sugar meals, (C) Quantification of ear dermal parasite loads in BALB/c mice 4 weeks after exposure to the bites of sand flies infected 2 weeks previously by artificial feeding on mouse blood seeded with 4×106/ ml LmRy promastigotes and treated or not with P/S in the blood and sugar meals. Values shown are parasite numbers per individual ears with geometric means ± 95% confidence interval (CI). Data shown in (A) are representative of more than 5 independent experiments, (B) 3 independent experiments, (C) one experiment involving the same population of infected flies.
Population size and diversity of bacterial communities in P. duboscqi
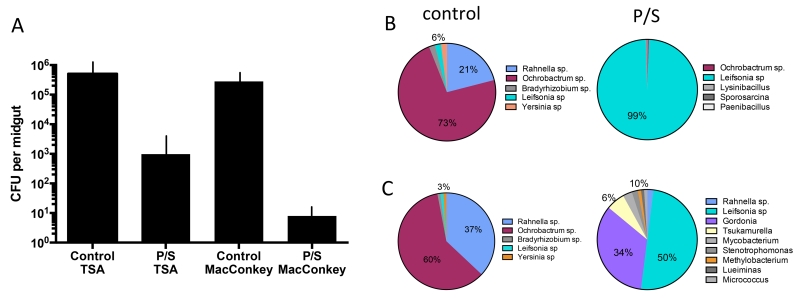
As expected, the P/S treatment greatly reduced the population size of bacteria in the gut of P. duboscqi cultured on TSA and MacConkey agar (Fig. 2A). A meta-analysis of untreated and P/S treated, L. major infected sand flies revealed a highly significant, positive correlation between the population size of the cultivable midgut bacteria and the number of metacyclic promastigotes in individual flies (Sup Fig 2). Approximately 20% of the variance in metacyclic numbers can be explained by the variance in the size of the cultivable bacterial communities, or vice versa.
Figure 2.
Changes in the size and diversity of bacterial communities in P. duboscqi following infection and treatment with P/S. Flies were artificially fed through a membrane on mouse blood seeded with 4×106/ ml LmRy promastigotes and treated or not with P/S in the blood and sugar meals. (A) Dissected midguts at 14 days p.i. were cultured on TSA and MacConkey agar. Bar graphs show mean CFUs + SD, 10 midguts/group. (B) Culture dependent, and (C) culture independent analysis of the gut bacteria at 14 days p.i. based on PCR amplification and cloning of the 16S DNA prepared from pools of 10 flies in each group.
Culture-dependent analysis of the gut bacteria of P. duboscqi revealed a diverse community in the control group dominated by gram negative Ochrobactrum sp. (73%) and Rahnella sp. (21%) members of the Brucellaceae and Enterobacteriaceae families, respectively. In the P/S treated group, these species were replaced by gram positive Leifsonia sp., family Microbacteriaceae (99%) (Fig. 2B). Similar results were obtained using the culture independent method based on cloning and partial sequencing of the 16S rRNA gene, with the exception that a greater diversity of gram positive species such as Gordonia was detected in the P/S treated group (Fig. 2C).
Effect of bacterial reconstitution on parasite development in antibiotic treated flies
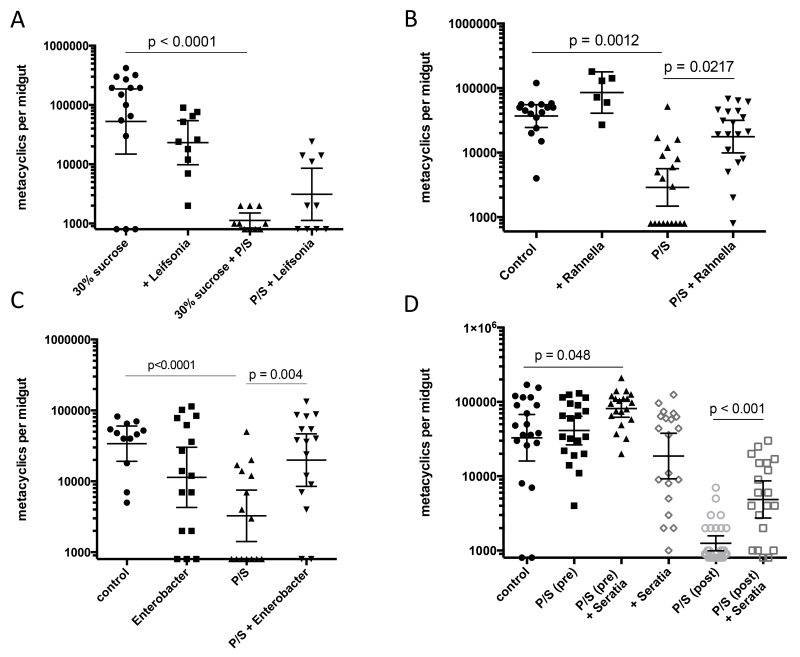
The dominance of Leifsonia sp. in the midguts of the antibiotic treated flies suggested that the growth and differentiation of the parasite is directly inhibited by the outgrowth of this bacterial species. This possibility was addressed by allowing the infected sand flies, with and without P/S treatment, to feed on blood meals containing Leifsonia (103/ml). The bacterial feeds did not significantly inhibit the number and proportion of metacyclics emerging in the infected flies, and actually increased the number of metacyclics in some of the P/S treated flies, though this difference was not significant (Fig 3A). In order to determine if the vector refractory state of the antibiotic treated flies could be reversed by reconstitution of their microbial flora, a P/S resistant strain of Rahnella aquatilis was generated by selection in vitro and added to the infective blood meal. Reconstitution with this single microbial species significantly rescued the ability of the antibiotic treated flies to support the growth and differentiation of L. major (Fig. 3B). While we were unable to generate P/S resistant Ochrobactrum to replicate these findings with the other dominant symbiont species in the gut, we were able to select for P/S resistant clones of two other cultivable Enterobacteriaceae from the gut, Enterobacter cloacae and Serratia rubidaea. In each case, addition of the selected bacteria to the infective blood meal promoted a significant recovery of the number of metacyclics that developed in the flies treated with P/S post-blood meal excretion (Fig. 3 C&D). The feeds on Enterobacter were especially beneficial to the parasite, with the majority of flies harboring infections that were equivalent to controls. Fig. 3D also shows the effect on L. major infection in flies that were antibiotic treated and fed on P/S resistant Serratia rubidaea at 6 days and 3 days prior to receiving their infective blood meal, respectively. While there was no difference in the number of metacyclics between the control and P/S pre-treated groups, the pre-treated flies that were allowed to feed on Serratia had significantly improved infections compared to controls, and were striking with respect to the consistency of the strong, mature infections that were achieved.
Figure 3.
Effect of antibiotic treatment and/or bacterial feeds on the development of metacyclic promastigotes in P. duboscqi. Flies were artificially fed through a membrane on mouse blood seeded with 4×106 / ml LmRy promastigotes. Flies were treated or not with P/S in the blood and sugar meals, and fed or not on blood meals containing 1000 CFU / ml of (A) Leifsonia, (B) Rahnella aquatilis, (C) Enterobacter cloacae, and (D) Seratia rubidaea. Midguts were dissected at 13-15 days p.i. except for flies infected with Rahnella that were dissected 19-20 days p.i., and scored for promastigote numbers and developmental stages under a hemocytometer. D also shows groups of flies that were treated with P/S and fed on Seratia rubidaea (1000 CFU / ml) in the sugar meal at 6 days and 3 days, respectively, prior to blood feeding and infection. Values shown are numbers of metacyclic promastigote per individual flies with geometric means ± 95% CI. Data shown in (A) is one of two representative independent experiments, (B) pooled data from two independent experiments, (C) one of three representative experiments, (D) one of two representative experiments.
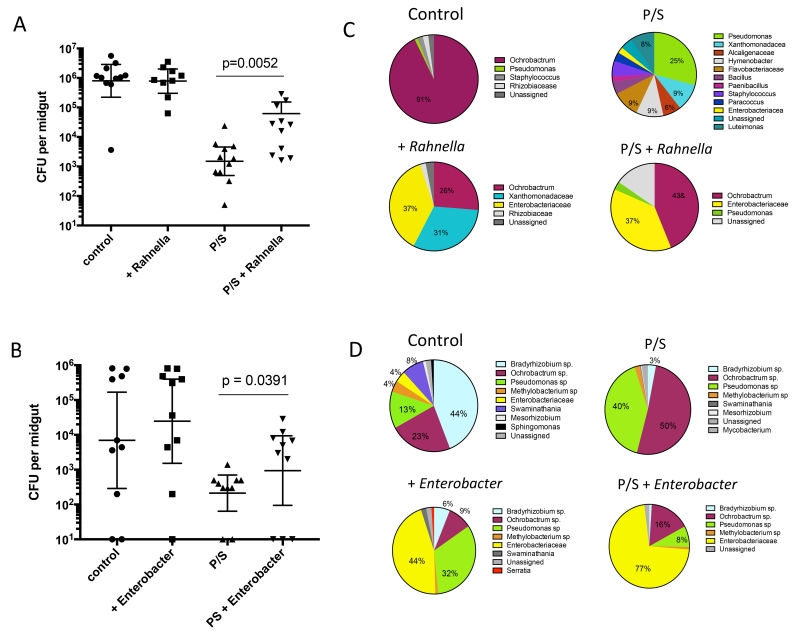
Culture dependent analysis of the size of the bacterial communities in the control, antibiotic treated and/or bacteria fed flies again revealed a greatly reduced number of CFUs in the P/S treated flies that was partially reconstituted in the flies fed on blood meals containing the antibiotic resistant strains of Rahnella or Enterobacter (Fig 4A & B). Monitoring changes in the composition of the microbiota was in this case carried out by next generation sequencing of the 16S amplicon generated from midgut cDNA to confine the analysis to viable bacteria in the gut, particularly in the antibiotic treated flies. Comparison of the 16S sequences from the midguts of the control, antibiotic treated and/or Enterobacter or Rahnella fed flies revealed a microbiome predominated by Enterobacteriaceae genera, to which both Rahnella and Enterobacter belong, in the bacteria fed flies that was observed in the both the control and antibiotic treated groups (Fig 4C & D). The shorter amplicons generated for Illumina sequencing in some cases were only able to provide taxonomy designations at the family level. We also noted a substantial change in the composition of the bacteria comparing midguts from control flies used in the feeds on Enterobacter (Fig 4D) with those from the prior generations of flies obtained 4-12 months previously that were used in the Rahnella feeds (Fig 4C) or the experiments shown in Fig 2. In particular, gram negative, aerobic bacteria in the genera Bradyrhizobium, Pseudomonas, and Methylobacterium were the predominant midgut bacterial communities at this time.
Figure 4.
Population size and diversity of bacterial communities in infected P. duboscqi that were fed live bacteria. Midgut homogenates from infected control or antibiotic treated flies fed on (A) Rahnella aquatilis and dissected at 19 days p.i., or fed on (B) Enterobacter cloacae and dissected at 14 days p.i, were cultured on LB agar. Values shown are CFUs per individual midgut with geometric means ± 95% CI. Culture independent analysis of the gut bacteria in flies fed on (C) Rahnella aquatili, and recovered 19 days p.i, or (D) Enterobacter cloacae, and recovered 14 days p.i, based on 16S amplicon sequencing of cDNA prepared from pools of 10 flies in each group.
Effect of nutrient repletion on parasite development in L. major infected P. duboscqi
A number of experiments were carried out to address the possibility that the bacteria might be providing essential nutrients for the normal growth and differentiation of L. major in the midgut. Supernatants harvested from the bacterial cultures of Ochrobactrum and/or Rahnella were added to the sugar meals starting on day 4 post-infection in antibiotic treated or untreated flies. The antibiotic treatment again had a profound inhibitory effect on metacyclic development that was not reversed by feeding on the supernatants from cultures of either bacterial species (Fig 5A). The partial recovery of metacyclic numbers in some of the flies fed on a combination of the supernatants was not observed in a subsequent experiment (Fig 5B). In so far as the growth and differentiation of L. major proceeds in vitro in the absence of bacteria, we tested whether the promastigote growth medium used for in vitro culture could rescue the differentiation defect observed in the antibiotic treated flies. Using either M199 or Grace’s insect medium supplemented with FBS, glutamine, folate, adenosine, hemin and Basal Medium Eagle (BME) containing eight B vitamins and ten essential amino acids, no significant improvement in the number of metacyclic promastigotes recovered from the antibiotic treated flies was observed (Fig 5 C&D). To the contrary, feeding on the replete medium significantly reduced metacyclic numbers in the non-antibiotic treated flies (Fig 5 C&D).
Figure 5.
Effect of bacterial culture supernatants and replete medium on the development of metacyclic promastigotes in P. duboscqi. Flies were artificially fed through a membrane on mouse blood seeded with 4×106 / ml LmRy promastigotes. Beginning on day 5 p.i., flies were treated or not with P/S and/or culture supernatant from Ochrobactrum anthropi and/or Rahnella aquatilis, in the sugar meal (A & B), or fed on M199 or Grace’s Insect Medium supplemented with BME, folate, hemin, and adenosine with or without P/S (C&D). Values shown are numbers of metacyclic promastigote per individual flies with geometric means ± 95% CI on days 13-14 p.i. Data shown in A,B, &D are each representative of two independent experiments; C is representative of three independent experiments.
Effect of sucrose concentration on parasite development in L. major infected P. duboscqi
In vitro, metacyclogenesis is confined to the stationary phase of promastigote growth. The inability of the bacterial supernatants or replete medium to rescue the parasite differentiation defect in antibiotic treated flies suggests that it is not essential nutrients that the microbiota provides, but its competition for and depletion of specific nutrients that signal for metacyclogenesis in vivo. The colonized sand flies are maintained on a continuously available source of 30% sucrose, meant to mimic the concentration and availability of plant sugars on which they naturally feed. Reducing the sucrose concentration of the sugar meals to 15%, and 10%, partially rescued the parasite differentiation defect in the antibiotic treated flies (Fig. 6A & B). The mortality of the flies maintained on <10% sucrose exceeded 85% and were not used for infection. The size of the cultivable gut bacterial communities in these flies did not show any significant differences, though there was a trend toward fewer CFUs in the surviving flies maintained on 7.5% sucrose (Sup Fig. 3).
Figure 6.
Effect of reduced sucrose concentration on the development of metacyclic promastigotes in P. duboscqi. Flies were artificially fed through a membrane on mouse blood seeded with 4×106 / ml LmRy promastigotes. Beginning on day 5 p.i., flies were treated or not with P/S in sugar meals composed of (A) 30% or 15% sucrose, or (B) 30% or 10% sucrose. Values shown are numbers of metacyclic promastigote per individual flies with geometric means ±95% CI on days 13-14 p.i. Data in (A) and (B) are representative of three and two independent experiments, respectively.
Effect of sucrose as a sole carbon source on promastigote growth and differentiation in vitro
While Leishmania promastigotes grown in replete medium differentiate into metacyclic forms during stationary growth, this critical process is relatively inefficient under these in vitro growth conditions (<20%, see Fig. S1) as compared to the sand fly midgut (>70 %, see Fig. 1A). In an effort to better reproduce the conditions influencing promastigote growth and development in vivo, L. major promastigotes harvested from mid-logarithmic phase growth in complete medium, corresponding to the early procyclic forms replicating in the blood meal following an infective feed, were washed and resuspended in water containing different concentrations of sucrose as the sole carbon source. This was meant to simulate the change in nutrient availability following excretion of the digested blood meal. Optimal promastigote survival over 68 hr. was observed at or near iso-osmolar concentrations of sucrose (7.5-12.5%) (Fig. 7A). Promastigotes survived poorly at hypo- (≤5%) or hyper-osmolar concentrations of sucrose (≥15%). Most organisms died within 1 hr. of exposure to ≥25% sucrose, and few remained viable after 40 hr. exposure ≥15% sucrose. When the stage differentiation of the viable cells was scored, 63-87% of the promastigotes cultured in 7.5-12.5% sucrose were metacyclics by 68 hr (Fig. 7B). The metacyclics produced under these starvation conditions behaved as fully infectious forms for macrophages in vitro (Sup Fig. 4). While the striking effects of these conditions on in vitro metacyclogenesis appear to replicate the highly efficient stage differentiation that occurs in the vector, the sensitivity of the promastigotes to killing by sucrose concentrations of ≥15% was unexpected given their presumed exposure to the high concentration (30%) of sucrose that is provided in the sugar feeds. This exposure would seem especially relevant to the promastigotes that migrate to and accumulate in the thoracic midgut where the sugars meals that are stored in the crop will be delivered at high concentration prior to diffusion to the more posterior aspects of the midgut (see Sup Fig. 5). The findings suggest that a crucial role of the microbiota for Leishmania development in the fly may be in sucrose utilization that reduces the osmolality of the sugar meal to a sustainable level for the parasite.
Figure 7.
Survival and differentiation of L. major promastigotes cultured in sucrose solutions in vitro. LmRy promastigotes were cultured at 26°C in complete medium 199 to mid-logarithmic phase, washed × 3 in PBS and resuspended to 2 × 108 / ml in an isosmotic sucrose solution (10.4%). Ten μl of the parasite suspension was added to 500 μl of different concentrations of sucrose in microwell plates, and the number and developmental stage of viable promastigotes were scored under a hemocytometer after different times in culture at 26°C. Data shown are the total number of viable promastigotes (A) or percent metacyclics (B). The data are representative of three independent experiments.
Discussion
Symbiont microorganisms of disease vectors influence many aspects of insect biology, including nutrition, reproduction, and immune system homeostasis. The microbiota can affect vector competency itself, in most instances having a negative impact on pathogen transmission via induction of innate defenses or the direct inhibitory actions of secreted enzymes or toxins (reviewed in (Weiss and Aksoy, 2011). In the case of the phlebotomine vectors of Leishmania that may acquire exogenous microorganisms from soil or plants, the fact that the entirety of the parasite life cycle in the invertebrate is confined to the digestive tract provides ample opportunities for interactions between the gut microbiota and Leishmania developmental stages. We report that reducing the size of and altering the diversity of the bacterial communities in the gut by antibiotic treatment profoundly impairs sand fly vector competence. The gut bacteria appear essential to the development of transmissible infections by promoting the appropriate nutrient stress and osmotic conditions required for the differentiation to and survival of infective stage, metacyclic promastigotes.
In the only prior studies to explore the role of the gut microbiota in sand fly vector competence, pre-feeding flies on certain fungi and bacteria reduced the ability of Leishmania to colonize the midgut (Sant’Anna et al., 2014; Schlein et al., 1985). These studies employed extremely high concentrations of bacteria (>107 CFU/ml), and assessed infections at only one early time point. Effects at later time points and on stage differentiation were not explored. In our studies, feeds employing a low dose (103 CFU/ml) of gram negative, aerobic bacteria, including Enterobacter cloacae, Serratia rubidaea, or Rahnella aquatilis, isolated from a laboratory reared colony of P. duboscqi and selected for resistance to P/S, had minimal impact on L. major infection in the untreated flies, but in each case rescued at least partially the parasite developmental defect in the antibiotic treated flies. Our efforts to understand the nature of the role played by the normal bacterial communities in promoting L. major infection addressed two main possibilities: 1) they provide nutrients essential to parasite growth and development, and 2) their metabolic demands help to promote the conditions of nutrient deprivation and reduced osmotic stress required for stage differentiation and survival of metacyclic promastigotes. Using multiple approaches, we failed to provide evidence for the former. Supernatant material from cultures of two dominant bacterial species in the midgut, Ochrobactrum and Rahnella, as well as replete medium that supports metacyclogenesis during growth in vitro, in each case failed to reconstitute the ability of L. major to differentiate and survive in the antibiotic treated flies. Experimental support for the second possibility is indirect, but we believe strong. Phlebotomine sand flies obtain energy from a staple diet of sucrose rich sugar meals obtained from floral nectars, plant leaves and stems, and from honeydews excreted by aphids and coccids (Killick-Kendrick and Killick-Kendrick, 1987; Schlein and Jacobson, 1999). These sugar meals are also the source of energy for the natural microbiota and Leishmania sp. that can colonize the midgut. Reducing the sucrose concentration in the sugar meal moderated the effect of antibiotic treatment on parasite development, suggesting that the competing sucrose utilization by the microbiota contributes to the conditions necessary to promote metacyclogenesis, and/or the survival of metacyclic forms in the midgut. The fact that feeds on three different genera of live bacteria could partially rescue parasite development supports a more generalizable effect of bacterial colonization related to nutrient utilization.
The behavior of promastigotes in culture conditions that better reflect the post-blood meal nutrient availability in the fly also support a role for sucrose consumption by the microbiota in producing a differentiation and survival niche for the parasite. A starvation medium containing near iso-osmolar concentrations of sucrose as the sole energy source permitted survival and differentiation to metacyclics of the majority of promastigotes over 3 days in culture, reproducing the highly efficient transformation that occurs in vivo. Acidic pH has also been shown to be an effective cue for metacyclogenesis in vitro (Bates and Tetley, 1993). While there was no drop in pH associated with the transformation that occurred in the sucrose starvation medium in vitro, we cannot rule out the possibility that products of both promastigote and bacterial metabolism contribute to a drop in pH that provides an additional signal for metacyclogenesis in vivo. Our in vitro studies furthermore revealed that the high sucrose concentration used to feed the flies (30%) was lethal to the promastigotes, as were sucrose concentrations down to 15%, suggesting a critical role for sucrose utilization by the bacteria to reduce the osmolyte concentration to a tolerable level for the parasite. The use of 30% sucrose for colony maintenance is within the reported range of natural nectar sucrose concentrations of 20-40% (600 – 1200 milliosmolar; iso-osmolar condition = 305 mOsM) (Chalcoff et al., 2006; Gottsberger et al., 1984; Marcum and Murdoch, 1992). Most of the sugar meal is stored in the crop, a large ventral diverticulum of the foregut that is not colonized by the parasite but is used to store food before it enters the midgut. Its contents are passively delivered through a duct into the thoracic midgut for digestion and absorption (Schlein and Warburg, 1986). Thus, the anterior migration of promastigotes that is essential to their differentiation and positioning for successful transmission by bite results in exposure to an especially high sugar concentration in the thoracic midgut, and may explain why the early growth of promastigotes in the abdominal midgut was normal in the antibiotic treated flies. While an osmotic stress response has been characterized in Leishmania promastigotes that may be relevant to their adaption to the wide ranging concentrations of inorganic and organic solutes to which they are exposed in the fly (Inbar et al., 2013; LeFurgey et al., 2001; Suescun-Bolivar and Thome, 2015), this response may be inadequate to withstand the hyperosmotic conditions present in the anterior gut. The ability of sugar utilization by the symbiont bacteria to moderate these effects in natural vectors will vary according to the concentration and constituents of the particular sugar meal, and the size and localization of the bacterial communities within the microanatomy of the gut. It is interesting that plant diet has been shown to be a potential cause of L. major mortality in the sand fly midgut (Schlein and Jacobson, 1994), with certain plant species found to produce a high proportion of deformed promastigotes or membrane ghosts in flies with mature infections, consistent with an osmolytic effect of the sugar meals on promastigotes stationed in the anterior midgut.
While our findings represent a departure from the negative associations between symbiont microbes and pathogen transmission that have been typically described, a recent study also revealed a beneficial role of the gut microbiota of the Lyme disease vector, Ixodes scapularis, in promoting colonization of the gut epithelium by Borrelia burgdorferi, in this case by reinforcing the integrity of the peritrophic matrix in larval ticks (Narasimhan et al., 2014). And while experimental co-infections of bacteria with Plasmodium sp. have consistently been shown to reduce the number of oocysts in the mosquito midgut, a positive correlation between the abundance of Enterobacteriaceae sp. and P. falciparum infection in Anopheles gambiae collected from natural breeding sites has been reported (Boissiere et al., 2012). Thus, the invertebrate midgut microbiota in some cases provides a favorable niche for pathogen transmission. For Leishmania transmission studies involving colonized sand flies, dysbiosis followed by reconstitution with a single bacterial species (Serratia rubidaea) prior to infection allowed us to substantially improve the proportion of flies with the high number of metacyclic promastigotes that is required for transmission by bite (Stamper et al., 2011). While the species, dose and timing of bacterial feeds will undoubtedly need to be customized to the sand fly and Leishmania species involved, deliberate manipulation of the microbiota offers a general experimental approach to establish reproducible, mature infections that has so far been extraordinarily difficult to achieve. In field settings, manipulations that alter the size and/or the diversity of the microbiota might be exploited to reduce disease transmission.
Methods
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Animal Care and Use Committee of the NIAID, NIH (protocol number LPD 68E). Invertebrates are not covered under NIH guidelines.
Parasites
L. major Ry was derived from a strain originally isolated from a lesion biopsy of a laboratory worker accidentally exposed to sand flies that were experimentally infected with a strain of L. major (WR2885) originating in Iraq (Stamper et al., 2011). Parasites were cultured as promastigotes in vitro at 26°C in complete medium 199 (CM199) supplemented with 20% heat-inactivated FCS (Gemini Bio-products), 100 U/ml penicillin, 100μg/ml streptomycin, 2mM L-glutamine, 40mM Hepes, 0.1 mM adenine (in 50mM Hepes), 5mg/ml hemin (in 50% triethanolamine), and 1mg/ml 6-biotin. To explore the effect of sucrose as the sole carbon source for promastigote growth and differentiation in vitro, mid-logarithmic phase cultures of LmRy growing in CM199 were washed × 3 in PBS and resuspended to 2 × 108/ ml in an isosmotic sucrose solution (305 mOsM). Ten μl of the parasite suspension were added in 5 ml polypropylene tubes to 500 μl of a range of sucrose concentrations (2.5-30%) prepared in de-ionized H2O. Promastigote viability and the number and frequency of metacyclic promastigotes were scored under a hemocytometer after different times in culture at 26°C.
Infection of mouse peritoneal macrophages in vitro
Normal peritoneal resident cells were obtained from C57Bl/6 mice by lavage, washed twice in RPMI 1640 medium (Advanced Biotechnologies, Silver Spring, MD), and were resuspended at 5.0 × 106cells/ml in RPMI 1640 medium containing 10%(v/v) fetal calf serum, 12 mM HEPES, 20 mM L-glutamine, 100 U/ml penicillin, and 100μg/ml streptomycin. The cells were plated in 0.4 ml on eight-chamberLab-Tek tissue culture slides (Miles Laboratories, Elkhart, IL) and were allowed to adhere for 4 hr at 35°C in 5% CO2. Nonadherent cells were removed by washing, and the remaining adherent macrophages were infected with 2 × 106 promastigotes at an approximate ratio of two parasites per macrophage. After 2 hr, free promastigotes were removed by repeated washings with RPMI 1640 medium, and the cultures were additionally incubated for 3 days at 35°C in 5% CO2. The slides were fixed and stained with Hema 3 solutions (Fisher Scientific, Kalmazoo, MI) to visualize intracellular parasites by microscopy.
Sand fly infections
Briefly, 2 to 4-day-old P. duboscqi females were obtained from a colony initiated from field specimens collected in Mali in 2010 and have been maintained in continuous cycle in the Laboratory of Parasitic Diseases, NIAID since that time. The larvae are fed a mixture of 50% rabbit droppings and 50% rabbit chow. The flies were infected by artificial feeding through a chick skin membrane on heparinized mouse blood reconstituted with heat inactivated serum and seeded with 4 × 106 / ml logarithmic phase promastigotes. Blood engorged flies were separated and maintained at 26°C and 75% humidity and were provided 30% sucrose ad libitum. Additional groups of infected flies were treated with 50 U/ml penicillin plus 50μg/ml streptomycin in the blood meal and sugar meal, in the blood meal only, or in the sugar meal only starting on day 5 post-infection (pi). At various days after the infective feed, 8–20 flies per experimental group were anesthetized with CO2, killed in 5% soap solution, and whole midguts were dissected and transferred individually into a microfuge tube containing 100 μl phosphate buffered saline pH 7.4 (PBS). The guts were macerated briefly using a plastic pestle, a 10-μl sample of the supernatant was counted under a hemocytometer and the numbers of metacyclic promastigotes, non-metacyclic forms, and total parasite number, as determined by morphology and movement, were counted.
Exposure of mice to infected sand flies
L. major infections were allowed to mature for 14-16 days within the sand fly midgut, and BALB/c mice were exposed to the bites of 4-5 infected flies as previously described (Stamper et al., 2011). One day before transmission the sucrose diet was removed. On the day of transmission, 4–5 flies were transferred to small plastic vials covered at one end with a 0.25-mm nylon mesh. Mice were anesthetized by intraperitoneal injection of 30 ul of ketamine/xylazine (100 mg/ml). Specially designed clamps were used to bring the mesh end of each vial flat against the ear, allowing flies to feed on exposed skin for a period of 2–3 hours in the dark at 23°C and 50% humidity. Following exposure to the ear, the number of flies per vial with a blood meal was determined using a dissecting microscope. Six weeks following transmission, parasite loads in the ears were determined as previously described (Stamper et al., 2011).
Isolation and identification of bacteria
The population size and diversity of the gut bacteria in P. duboscqi were determined by both culture-dependent and culture independent methods as previously described (Hubert et al., 2012). For culture, bacteria from surface sterilized and homogenized whole flies, or from dissected and homogenized midguts, were grown on tryptic soy agar (TSA), MacConkey agar (MAC), or LB agar. Plates were then incubated aerobically at 26 °C (TSA & LB), or 37 °C (MAC) for 48–72 h. Colony forming units (CFU) per fly were determined and bacterial colonies with distinct morphologies were sub-cultured on TSA or LB and stored at 4 °C until further analysis. DNA from single isolates was used in a polymerase chain reaction (PCR) to amplify the variable region of the 16S ribosomal RNA gene using universal eubacterial primers: 8F (5′-AGAGTTTGATCC TGGCT CAG-3′) and 806R (5′- CTACCAGGGTATCTAAT-3′) or 27F (5´-AGAGTTTGATCA/CTGGCTCAG-3’) and 1492R (5´-TACGGT/CTACCTTGTTACGACTT-3’) following standard protocols (Hugenholtz et al., 1998). The resulting products were purified, sequenced and identified by BLAST search of the NCBI GenBank database as previously described (Hubert et al., 2012). For culture-independent method, DNA was extracted from homogenates prepared from pools of surface sterilized whole flies (10 per group) and partial sequences of the 16S rRNA gene were amplified and cloned using pGEM®-T Easy Vector (Promega). Selected clones (100 per sample) were sequenced in the UC-Riverside Core Instrumentation facility on the ABI 3730xl sequencer. Nearly full-length sequences were assembled with CodonCode Aligner, version 1.5.2 (CodonCode Corporation, Dedham, MA, USA) and assigned to bacterial taxonomy using Ribosomal Database Project naïve Bayesian rRNA classifier.
For determination of the taxonomy and relative abundance of the gut bacteria in experiments involving bacterial feeding, next generation sequencing of cDNA prepared from dissected midguts was carried out. RNA was extracted from a pool of 10 dissected midguts by lysing and homogenizing in Trizole (Molecular research center INC #TR-118). Following the addition of 0.2ml of chloroform and centrifugation at 15,000g, 4°C, the upper aqueous phase was collected, mixed with 70% Ethanol, 30% DEPC-treated H2O, and added to an RNeasy (qiagen #74106) column for RNA purification as per manufacturer’s instructions. cDNA was produced with random primers using High-Capacity cDNA Reverse Transcription kit (Thermo fisher # 4368813). Complete removal of DNA was confirmed by preforming the cDNA reaction without reverse transcriptase, which yielded no PCR-amplification of the 16S rDNA. Samples were submitted to 16S amplicon seqquencing at the DNA Services facility at the University of Illinois at Chicago (UIC). cDNA was PCR amplified with primers 27F and 534R targeting the V1-V3 16S variable regions using a two-stage “targeted amplicon sequencing (TAS)” protocol (Bybee et al., 2011). PCR cycling conditions were preformed according to the Human Microbiome Project consortium 16S rRNA gene amplicon sequencing protocol (http://www.hmpdacc.org/doc/16S_Sequencing_SOP_4.2.2.pdf). Sequence data were processed using the software package QIIME (Caporaso et al., 2010).
Sand fly feeds on bacteria, bacterial supernatants, and replete medium
Enterobacter cloacae, Serratia rubidaea, and Rahnella aquatilis were selected for antibiotic resistance by sequential selection of colonies grown on LB agar containing increasing concentrations of P/S (25U/mL penicillin and 25 μg/ml streptomycin to 100 U/ml penicillin and 100 μg/ml streptomycin). The antibiotic resistant clones were cultured in LB medium to an OD600 value defined for each isolate to yield a known number of CFUs per ml, pelleted, washed and resuspended in PBS. A suspension of naturally resistant Leifsonia was similarly prepared. The bacterial suspensions were added to the blood meal in a volume of 20 μl to give a final concentration of 1000 CFU/ml, and fed to sand flies by membrane feeding as described above. Bacterial feeds in blood fed flies were evaluated by culture on LB agar and by sequencing of the 16S amplicon generated from cDNA prepared from infected midguts. Supernatant material from stationary cultures of Rahnella aquatilis and Ochrobactrum anthropic growing in minimal medium (42mM Na2HPO4, 22mM KH2PO4, 19mM NH4Cl, 8.5 mM NaCl, 1mM MgSO4, 100μM CaCl2) with 10% sucrose was obtained by centrifugation and filter sterilization. The concentrations of residual sucrose in the Rahnella and Ochrobactrum supernatants were determined individually or after combining at a 1:1 ratio using a commercial sucrose assay kit (Sigma, St. Louis, MO, product# SCA-20). The supernatants were added in equal volumes to a sucrose solution adjusted so that the final sucrose concentration of the mixture was 30%, and used as a source of sugar meals that were replaced daily. For feeds on replete medium, the sugar meal was replaced with either M199 or Grace’s Insect Medium (Sigma, St. Louis, MO) with or without P/S and supplemented with Basal Medium Eagle (BME, 200× stock,1× final) containing eight B vitamins, the ten essential amino acids, plus cysteine, tyrosine, and glutamine (Life Technologies, Grand Island, NY), 10 mM folate, 5mg/ml hemin and 0.1 mM adenosine.
Statistical analysis
Student t-test was used to assess significant differences in parasite counts between control and treated flies. ANOVA was used in cases where more than two groups wee compared.
Supplementary Material
Sup Figure 1. Effect of antibiotics on L. major growth and differentiation in vitro. LmRy promastigotes were grown in CM199 with or without P/S. The numbers of promastigotes and metacyclic stage promastigotes were determined by counting under a hemocytometer after various days in culture.
Sup Figure 2. Meta-analysis of the relationship between midgut bacterial load and metacyclogenesis. Data are pooled from 3 independent experiments in which infected P. duboscqi treated or not with P/S in the blood and sugar meals were dissected at 13-14 days p.i. and their homogenized midguts were scored for parasites and plated on LB agar. Values shown are for individual midguts.
Sup Figure 3. Population size of the bacterial communities in infected P. duboscqi fed on different concentrations of sucrose. Flies were infected with LmRy and fed on different sucrose concentrations starting at 4 days p.i. At 13 days p.i. bacteria from dissected, homogenized midguts were grown on LB agar. Data shown are CFUs from individual midguts with geometric means ±95% CI.
Sup Figure 4. Infectivity of metacyclic promastigotes recovered from starvation conditions for normal mouse resident peritoneal macrophages in vitro. Log phase promastigotes obtained from cultures of LmRy growing in CM199 were washed × 3 in 10% sucrose in dH2O and resuspended into 10% sucrose in dH2O at 2x106 / ml. Promastigotes recovered from cultures at days 1-3 were added to macrophages at a 1:1 ratio, and the free promastigotes removed after 2 hr. The percent of the original intracellular inoculum remaining at 72 hr was calculated as follows: (percent macrophages infected at 72 hr/percent macrophages infected at 2 hr) × 100. The results shown are means of duplicate cultures.
Sup Figure 5. Light micrographs of dissected midguts showing the enlarged foregut diverticulum of the foregut (crop). Figure on the left is a low power image showing the entire midgut and crop with a blood engorged abdominal midgut immediately following an infective feed. Image on the right is a higher resolution of a 12 day infected gut showing distention of the thoracic midgut caused by large numbers of promastigotes, and an enlarged crop with a duct connecting it to the proventriculus. The plug of promastigotes protruding out from the thoracic midgut is an artifact of the dissection, and would normally be contained behind the cardiac valve. Scale bar = 50 μm.
Acknowledgments
This work was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. C.C.M. was supported by a CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior) sandwich Ph.D. fellowship. We thank Kimberly Beacht for assistance with the mouse experiments.
Footnotes
Author Contributions
C.C.M., E.I., L.Z. and D.S. conceived and performed experiments, and wrote the manuscript, K.G., R.M., P.L., A.P., M.L., and D.E. performed experiments, N.S. provided expertise.
References
- Bates PA, Tetley L. Leishmania mexicana: induction of metacyclogenesis by cultivation of promastigotes at acidic pH. Exp Parasitol. 1993;76:412–423. doi: 10.1006/expr.1993.1050. [DOI] [PubMed] [Google Scholar]
- Boissiere A, Tchioffo MT, Bachar D, Abate L, Marie A, Nsango SE, Shahbazkia HR, Awono-Ambene PH, Levashina EA, Christen R, et al. Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 2012;8:e1002742. doi: 10.1371/journal.ppat.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger N, Lowenberger C, Volf P, Ursic R, Sigutova L, Sabatier L, Svobodova M, Beverley SM, Spath G, Brun R, et al. Characterization of a defensin from the sand fly Phlebotomus duboscqi induced by challenge with bacteria or the protozoan parasite Leishmania major. Infection and immunity. 2004;72:7140–7146. doi: 10.1128/IAI.72.12.7140-7146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bybee SM, Bracken-Grissom H, Haynes BD, Hermansen RA, Byers RL, Clement MJ, Udall JA, Wilcox ER, Crandall KA. Targeted amplicon sequencing (TAS): a scalable next-gen approach to multilocus, multitaxa phylogenetics. Genome Biol Evol. 2011;3:1312–1323. doi: 10.1093/gbe/evr106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalcoff VR, Aizen MA, Galetto L. Nectar concentration and composition of 26 species from the temperate forest of South America. Ann Bot-London. 2006;97:413–421. doi: 10.1093/aob/mcj043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, Mulenga M, Dimopoulos G. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332:855–858. doi: 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RJ, el Kordy E, Shehata M, Lane RP. The prevalence of a microbiota in the digestive tract of Phlebotomus papatasi. Ann Trop Med Parasitol. 1996;90:669–673. doi: 10.1080/00034983.1996.11813102. [DOI] [PubMed] [Google Scholar]
- Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottsberger G, Schrauwen J, Linskens HF. Amino-Acids and Sugars in Nectar, and Their Putative Evolutionary Significance. Plant Syst Evol. 1984;145:55–77. [Google Scholar]
- Hillesland H, Read A, Subhadra B, Hurwitz I, McKelvey R, Ghosh K, Das P, Durvasula R. Identification of aerobic gut bacteria from the kala azar vector, Phlebotomus argentipes: a platform for potential paratransgenic manipulation of sand flies. Am J Trop Med Hyg. 2008;79:881–886. [PubMed] [Google Scholar]
- Hubert J, Kopecky J, Perotti MA, Nesvorna M, Braig HR, Sagova-Mareckova M, Macovei L, Zurek L. Detection and identification of species-specific bacteria associated with synanthropic mites. Microb Ecol. 2012;63:919–928. doi: 10.1007/s00248-011-9969-6. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar E, Schlisselberg D, Suter Grotemeyer M, Rentsch D, Zilberstein D. A versatile proline/alanine transporter in the unicellular pathogen Leishmania donovani regulates amino acid homoeostasis and osmotic stress responses. Biochem J. 2013;449:555–566. doi: 10.1042/BJ20121262. [DOI] [PubMed] [Google Scholar]
- Killick-Kendrick R, Killick-Kendrick M. Honeydew of aphids as a source of sugar for Phlebotomus ariasi. Med Vet Entomol. 1987;1:297–302. doi: 10.1111/j.1365-2915.1987.tb00358.x. [DOI] [PubMed] [Google Scholar]
- Koch H, Schmid-Hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci U S A. 2011;108:19288–19292. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeFurgey A, Ingram P, Blum JJ. Compartmental responses to acute osmotic stress in Leishmania major result in rapid loss of Na+ and Cl. Comp Biochem Physiol A Mol Integr Physiol. 2001;128:385–394. doi: 10.1016/s1095-6433(00)00319-6. [DOI] [PubMed] [Google Scholar]
- Marcum KB, Murdoch CL. Salt tolerance of the coastal salt marsh grass, Sporobulus virginicus (L.) kunth. New Phytology. 1992;120:281–288. [Google Scholar]
- Mukhopadhyay J, Braig HR, Rowton ED, Ghosh K. Naturally occurring culturable aerobic gut flora of adult Phlebotomus papatasi, vector of Leishmania major in the Old World. PLoS One. 2012;7:e35748. doi: 10.1371/journal.pone.0035748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisig J, Pan J, Eppler-Epstein R, Deponte K, Fish D, Fikrig E. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe. 2014;15:58–71. doi: 10.1016/j.chom.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterkova-Koci K, Robles-Murguia M, Ramalho-Ortigao M, Zurek L. Significance of bacteria in oviposition and larval development of the sand fly Lutzomyia longipalpis. Parasit Vectors. 2012;5:145. doi: 10.1186/1756-3305-5-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumpuni CB, Beier MS, Nataro JP, Guers LD, Davis JR. Plasmodium falciparum: inhibition of sporogonic development in Anopheles stephensi by gram-negative bacteria. Exp Parasitol. 1993;77:195–199. doi: 10.1006/expr.1993.1076. [DOI] [PubMed] [Google Scholar]
- Sacks D, Kamhawi S. Molecular aspects of parasite-vector and vector-host interactions in leishmaniasis. Annu Rev Microbiol. 2001;55:453–483. doi: 10.1146/annurev.micro.55.1.453. [DOI] [PubMed] [Google Scholar]
- Sant’Anna MR, Darby AC, Brazil RP, Montoya-Lerma J, Dillon VM, Bates PA, Dillon RJ. Investigation of the bacterial communities associated with females of Lutzomyia sand fly species from South America. PLoS One. 2012;7:e42531. doi: 10.1371/journal.pone.0042531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant’Anna MR, Diaz-Albiter H, Aguiar-Martins K, Al Salem WS, Cavalcante RR, Dillon VM, Bates PA, Genta FA, Dillon RJ. Colonisation resistance in the sand fly gut: Leishmania protects Lutzomyia longipalpis from bacterial infection. Parasit Vectors. 2014;7:329. doi: 10.1186/1756-3305-7-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlein Y, Jacobson RL. Mortality of Leishmania major in Phlebotomus papatasi caused by plant feeding of the sand flies. Am J Trop Med Hyg. 1994;50:20–27. [PubMed] [Google Scholar]
- Schlein Y, Jacobson RL. Sugar meals and longevity of the sandfly Phlebotomus papatasi in an arid focus of Leishmania major in the Jordan Valley. Med Vet Entomol. 1999;13:65–71. doi: 10.1046/j.1365-2915.1999.00138.x. [DOI] [PubMed] [Google Scholar]
- Schlein Y, Polacheck I, Yuval B. Mycoses, bacterial infections and antibacterial activity in sandflies (Psychodidae) and their possible role in the transmission of leishmaniasis. Parasitology. 1985;90(Pt 1):57–66. doi: 10.1017/s0031182000049015. [DOI] [PubMed] [Google Scholar]
- Schlein Y, Warburg A. Phytophagy and the feeding cycle of Phlebotomus papatasi (Diptera: Psychodidae) under experimental conditions. Journal of medical entomology. 1986;23:11–15. doi: 10.1093/jmedent/23.1.11. [DOI] [PubMed] [Google Scholar]
- Stamper LW, Patrick RL, Fay MP, Lawyer PG, Elnaiem DE, Secundino N, Debrabant A, Sacks DL, Peters NC. Infection parameters in the sand fly vector that predict transmission of Leishmania major. PLoS Negl Trop Dis. 2011;5:e1288. doi: 10.1371/journal.pntd.0001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suescun-Bolivar LP, Thome PE. Osmosensing and osmoregulation in unicellular eukaryotes. World J Microbiol Biotechnol. 2015;31:435–443. doi: 10.1007/s11274-015-1811-8. [DOI] [PubMed] [Google Scholar]
- Telleria EL, Sant’Anna MR, Ortigao-Farias JR, Pitaluga AN, Dillon VM, Bates PA, Traub-Cseko YM, Dillon RJ. Caspar-like gene depletion reduces Leishmania infection in sand fly host Lutzomyia longipalpis. J Biol Chem. 2012;287:12985–12993. doi: 10.1074/jbc.M111.331561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B, Aksoy S. Microbiome influences on insect host vector competence. Trends in parasitology. 2011;27:514–522. doi: 10.1016/j.pt.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss BL, Wang J, Maltz MA, Wu Y, Aksoy S. Trypanosome infection establishment in the tsetse fly gut is influenced by microbiome-regulated host immune barriers. PLoS Pathog. 2013;9:e1003318. doi: 10.1371/journal.ppat.1003318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sup Figure 1. Effect of antibiotics on L. major growth and differentiation in vitro. LmRy promastigotes were grown in CM199 with or without P/S. The numbers of promastigotes and metacyclic stage promastigotes were determined by counting under a hemocytometer after various days in culture.
Sup Figure 2. Meta-analysis of the relationship between midgut bacterial load and metacyclogenesis. Data are pooled from 3 independent experiments in which infected P. duboscqi treated or not with P/S in the blood and sugar meals were dissected at 13-14 days p.i. and their homogenized midguts were scored for parasites and plated on LB agar. Values shown are for individual midguts.
Sup Figure 3. Population size of the bacterial communities in infected P. duboscqi fed on different concentrations of sucrose. Flies were infected with LmRy and fed on different sucrose concentrations starting at 4 days p.i. At 13 days p.i. bacteria from dissected, homogenized midguts were grown on LB agar. Data shown are CFUs from individual midguts with geometric means ±95% CI.
Sup Figure 4. Infectivity of metacyclic promastigotes recovered from starvation conditions for normal mouse resident peritoneal macrophages in vitro. Log phase promastigotes obtained from cultures of LmRy growing in CM199 were washed × 3 in 10% sucrose in dH2O and resuspended into 10% sucrose in dH2O at 2x106 / ml. Promastigotes recovered from cultures at days 1-3 were added to macrophages at a 1:1 ratio, and the free promastigotes removed after 2 hr. The percent of the original intracellular inoculum remaining at 72 hr was calculated as follows: (percent macrophages infected at 72 hr/percent macrophages infected at 2 hr) × 100. The results shown are means of duplicate cultures.
Sup Figure 5. Light micrographs of dissected midguts showing the enlarged foregut diverticulum of the foregut (crop). Figure on the left is a low power image showing the entire midgut and crop with a blood engorged abdominal midgut immediately following an infective feed. Image on the right is a higher resolution of a 12 day infected gut showing distention of the thoracic midgut caused by large numbers of promastigotes, and an enlarged crop with a duct connecting it to the proventriculus. The plug of promastigotes protruding out from the thoracic midgut is an artifact of the dissection, and would normally be contained behind the cardiac valve. Scale bar = 50 μm.