Abstract
The serotonin 2A (5-HT2A) and metabotropic glutamate 2 (mGlu2) receptors regulate each other and are associated with schizophrenia. The Roman high- (RHA-I) and the Roman low- (RLA-I) avoidance rat strains present well-differentiated behavioral profiles, with the RHA-I strain emerging as a putative genetic rat model of schizophrenia-related features. The RHA-I strain shows increased 5-HT2A and decreased mGlu2 receptor binding levels in prefrontal cortex (PFC). Here, we looked for differences in gene expression and transcriptional regulation of these receptors. The striatum (STR) was included in the analysis. 5-HT2A, 5-HT1A, and mGlu2 mRNA and [3H]ketanserin binding levels were measured in brain homogenates. As expected, 5-HT2A binding was significantly increased in PFC in the RHA-I rats, while no difference in binding was observed in STR. Surprisingly, 5-HT2A gene expression was unchanged in PFC but significantly decreased in STR. mGlu2 receptor gene expression was significantly decreased in both PFC and STR. No differences were observed for the 5-HT1A receptor. Chromatin immunoprecipitation assay revealed increased trimethylation of histone 3 at lysine 27 (H3K27me3) at the promoter region of the HTR2A gene in the STR. We further looked at the Akt/GSK3 signaling pathway, a downstream point of convergence of the serotonin and glutamate system, and found increased phosphorylation levels of GSK3β at tyrosine 216 and increased β-catenin levels in the PFC of the RHA-I rats. These results reveal region-specific regulation of the 5-HT2A receptor in the RHA-I rats probably due to absence of mGlu2 receptor that may result in differential regulation of downstream pathways.
Keywords: Schizophrenia, 5-HT2A, mGluR2, Roman low-and high-avoidance rats, Epigenetics, Prefrontal cortex, Striatum, GSK3β, β-Catenin
Introduction
The inbred Roman low- and high-avoidance rat strains (RLA-I vs. RHA-I) derive from the Swiss sublines (RHA/Verh and RLA/Verh), which have been bidirectionally selected and bred since 1972 based on their divergent performance on the two-way active avoidance response in the shuttle box [1, 2]. Along the last decades, they have been extensively behaviorally phenotyped with each strain presenting specific characteristics associated with their different coping styles and stress responses: The RLA-I rats are more anxious and fearful, with a more passive coping when facing conflicts, while RHA-I rats display a more active coping response [1–5]. What is moreover interesting is that the RHA-I rats show features which suggest deficits in executive function. They are more prone to substance abuse [6, 7]; show a high degree of impulsivity; and demonstrate attentional deficits [8, 9], worse learning-memory capacity [10], and deficits in sensorimotor gating processes, shown by deficits in prepulse inhibition (PPI) of a startle response [3, 10]. These behavioral deficits are all traits associated with schizophrenia [11–15] and are even present before the manifestation of the disorder [16]. Together, these observations point towards the RHA-I as a good model of schizophrenia-relevant features, which is further supported by additional neurobiological alterations. The RHA-I, when compared to the RLA-I, shows an enhanced dopaminergic response to drugs and anxiogenic stimuli in striatal and prefrontal cortex areas [17, 18], probably due to lower availability of the D2 autoreceptor and thereby increased dopaminergic tonus [19]. Besides, this is accompanied by enhanced serotonin (5-HT) release in cortical areas [20].
The serotonin 2A (5-HT2A) receptor is strongly linked to several of the behavioral manifestations associated with schizophrenia [21]. This receptor is the site of action of hallucinogens, like psilocybin and lysergic acid diethylamide (LSD) [22, 23], as well as of atypical anti-psychotics [23, 24], whereby they exert their effect onto cognitive processes embraced by executive function [21, 25, 26]. The metabotropic glutamate 2 (mGlu2) receptor interacts with the 5-HT2A receptor inducing intracellular Ca2+ release [27] and 5-HT2A receptor-mediated hallucinogenic effects [22, 27, 28], through the formation of heteroreceptor complexes at the postsynaptic site [27]. This functional heterodimer has been implicated in the etiology of schizophrenia [23, 29] and in mediating anti-psychotic responses [30]. The 5-HT2A and mGlu2 receptors modulate each other not only at the membrane level, but also at the transcriptional level by inducing epigenetic modifications at the promoter region [30, 31]. This implies that the 5-HT2A and mGlu2 receptors presumably compensate to innate or developmental-induced receptor alterations through epigenetic control of their expression levels. Abnormal variations in epigenetic regulation have been associated with schizophrenia [32–36]. This is interesting, as epigenetic regulation is reversible and can be modulated with drug treatments, opening up for new treatment strategies in schizophrenia [37].
The RHA-I strain shows increased 5-HT2A receptor binding and decreased mGlu2 receptor protein levels in prefrontal cortical regions compared to RLA-I [9], which corresponds well to that observed in brains of schizophrenia patients [29]. Further, cortical 5-HT2A levels correlate positively with number of premature responses in the five-choice serial reaction time task (a measure of impulsivity) in the RHA-I, which points towards a role of this receptor in some of the behavioral features defining this strain [9]. Here, we wanted to further delve into these observations and investigate to what extent differences in gene expression regulation between the RHA-I and the RLA-I are accountable for the receptor alterations. Besides the prefrontal cortex (PFC), we included striatum (STR) in the analysis, as this is an area highly related to schizophrenia [38]. Our main interest was to look at expression levels and epigenetic modifications of the 5-HT2A and mGlu2 receptor genes in these two regions. The 5-HT1A receptor was also included in order to determine whether changes were 5-HT2A specific or the result of more upstream changes in the serotonergic input. To further investigate whether mutual variations in 5-HT2A and mGluR2 receptors could translate into differences in intracellular transduction mechanisms, we looked at components of the Akt/GSK3 pathway, since this signaling pathway has been proposed as a point of convergence of glutamatergic and serotonergic input and is involved in schizophrenia pathophysiology [39–41].
Materials and Methods
Animals
A total of 20 male Roman high-avoidance (RHA-I) and 20 male Roman low-avoidance (RLA-I) rats were used in this study. All rats came from the breeding colony of inbred Roman strains established in 1996 at the Department of Psychiatry and Forensic Medicine, Autonomous University of Barcelona, Spain. Rats were housed in pairs of the same sex and strain in Makrolon cages (50 × 25 × 14 cm) and maintained with food and water ad libitum under standard conditions of temperature (22 ± 2 °C), humidity (50–70%), and a 12-h light-dark cycle (lights on at 08:30 h). All animals were sacrificed in accordance with the Spanish Royal Decree (RD 53/2013) for the protection of experimental animals and with the European Communities Council Directive (2010/63/EU) at approximately 4 months of age. One group of rats consisting of 12 RHA and 12 RLA rats was used for the gene expression, receptor binding, and western blot analysis. Here, their brains were removed and PFC and STR dissected out into two pieces and immediately frozen in liquid nitrogen and kept at −80 °C until further use. A second group of animals consisting of eight RHA and eight RLA rats was used for the chromatin immunoprecipitation analysis. Here, their brains were removed; PFC and STR were dissected out and immediately processed for protein cross-linking in order to be used for the chromatin immunoprecipitation assay (see further in the following). Further, the hippocampus was dissected for performing the DNA sequencing analysis.
RNA Extraction and Reverse Transcription Synthesis
RNAwas extracted from PFC and STR tissues using the commercially available NucleoSpin®RNA/Protein kit (Macherey-Nagel, cat. no. 740933). Tissue samples (approximately 10 μg) were treated according to the manufacturer’s protocol in an RNase-free environment. Ten millimolar DTT was additionally added to the provided lysis buffer. Additionally, RNA samples were subjected to DNAse treatment using the Turbo DNA-free™ kit (Ambion, cat. no. AM1907). RNA integrity number (RIN) and concentration were determined using the 2100 Agilent Bioanalyzer (Agilent Technologies, Palo Alta, CA, USA). Only samples with a RIN value >5 were included in the analyses following recommendations by [42]. The purified RNA was reverse transcribed into complementary DNA (cDNA) using qScript cDNA SuperMix kit (Quanta Biosciences, cat. no. 95048). In short, 500 ng of purified RNA from each sample, adjusted to a total volume of 16 μL RNase/DNase-free water, was mixed with 4 μL of the qScript cDNA SuperMix (5×). The reverse transcription cDNA synthesis was run according to the manufacturer’s protocol at 25 °C for 5 min, 42 °C for 30 min, and 85 °C for 5 min. To check for gDNA contaminations, a negative control of RNA from which the reverse transcription step had been omitted was included for each sample analysis. One hundred nanograms of Universal Rat Reference RNA (Agilent Technologies, cat. no. 740200) was reverse transcribed along with the samples and used as plate variance control as well as positive control. cDNA samples were kept at −80 °C until further use.
Reverse Transcriptase Semiquantitative RT-qPCR
The real-time polymerase chain reaction (RT-qPCR) experiments were performed according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines [43]. The following primer sets were used: 5-HT2A: F = 5′-CCA CAG CCG CTT CAA CTC-3′ and R = 5′-GCA GCT CCC CTC CTT AAA GA-3′; mGluR2: F = 5′-GTG GTG ACA TTG CGC TGT AA-3′ and R = 5′-GCG ATG AGG AGC ACA TTG TA-3′; 5-HT1A: F = 5′-ATC AGC AAG GAC CAC GGC TAC A-3′ and R = 5′-TGT CCG TTC AGG CTC TTC TTG G-3′; GAPDH: F = 5′-CAT CAA GAA GGT GAA GCA-3′ and R = 5′-CTG TTG AAG TCA CAG GAG ACA-3′; RpL13: F: 5′-AGC AGC TCT TGA GGC TAA GG-3′ and R = 5′-GGG TTC ACA CCA AGA GTC CA-3′. One hundred-nanogram cDNA per sample was mixed with the corresponding concentrations of primer sets (300 nM for 5-HT2A, mGluR2, GAPDH, RpL13, and 500 nM for 5-HT1A), RNase-free water, and Fast SYBR® Green Master Mix (Applied Biosystems, cat. no. 438512) according to the protocol. All samples were run in duplicates. To compare the multiple samples between the assays, a calibrator, Universal Rat Reference cDNA, and a negative control were included in each run. All qPCR reactions were run on a Stratagene Mx3005P qPCR system (Applied Biosystems) using a standardized 40-cycle Fast SYBR Green program with annealing/acquisition segments adjusted for each primer sets: GAPDH and RpL13, 60 + 72 °C; 5-HT1A, 62 + 82 °C; 5-HT2A, 58 + 76 °C; and mGluR2, 60 °C. Expression levels of housekeeping genes (GAPDH and RpL13) did not differ across groups.
Radioligand Binding Assay
For obtaining membrane homogenates, tissue samples from PFC and STR were homogenized in 5 mM Tris-HCl buffer containing 0.25 M sucrose. The homogenates were centrifuged at 1000×g for 10 min at 4 °C. The pellet was discarded and the supernatant centrifuged again at 40,000×g for 10 min. The remaining pellet was washed twice in homogenization buffer while spun in between and after at 40,000×g for 20 min and finally resuspended in appropriate amount of homogenization buffer and spun again at 14,000 rpm for 15 min at 4 °C. The final pellet was kept at −80 °C until use. For the radioligand binding assays, a single-concentration assay with [3H]ketanserin (cat. no. NET-791, 63.3 Ci/mmol, 1 mCi/mL) was used for determining maximum 5-HT2A receptor binding. The membrane pellets were thawed on ice, resuspended with a syringe (27G1/2) in 1 mL Tris-HCl, and diluted to a total volume of 13 mL Tris-HCl. Samples were hereafter incubated for 60 min at 37 °C and filtered with polyethylenimide 0.5% (5 mL/500 mL) and washed twice with Tris-HCl. Binding assays were performed on 400 μL of the membrane suspension and 5 nM [3H]ketanserin in triplicates, with or without 50 μL methysergide (10-μM final concentration) to control for non-specific binding. Radioactivity was measured using the Liquid Betaplate Scintillation counter (Wallace SC/9200/21, PerkinElmer 1205-440).
Protein Cross-Linking and Chromatin Immunoprecipitation
Prior to the chromatin immunoprecipitation assay, proteins were cross-linked to the DNA following a specific protocol. In short, once dissected, PFC and STR were placed immediately in a 15-mL centrifuge tube containing a solution of PBS (pH 7.4) with 1% formaldehyde and kept at room temperature (RT) for 20 min. The reaction was stopped by the addition of 10× glycine (G8898-500G) to a final concentration of 0.125 M and 5-min incubation at RT. The samples were centrifuged at 1500×g for 5 min at 4 °C, and the supernatant was discarded. Pellets were resuspended in 1 mL cold PBS, containing a Protease Inhibitor Cocktail (Sigma #P8340, 1000× dilution), and transferred to a 1.5-mL Eppendorf tube. Samples were centrifuged at 1500×g for 5 min at 4 °C, and the supernatant was discarded. Pellets were kept at −80 °C until further use.
The EZ-Magna ChIP™ A kit (cat. no. 17-408, Millipore) was used for the chromatin immunoprecipitation. Samples were thawed and prepared according to the manufacturer’s instructions. The samples were incubated overnight at 4 °C with the A magnetic protein beads (Millipore) and one of the following primary antibodies: rabbit anti-acetyl histone H3 (Millipore, cat. no. 06-599B), rabbit anti-trimethyl histone H3 (Lys27) (Millipore cat. no. 07-449), or normal rabbit IgG (Millipore, cat. no. PP64B). Twenty microliters of each sample was kept as input control and not incubated with antibody. The next day, the samples were placed in a magnetic separator (Millipore cat. no. 20-400), whereby the protein-bound magnetic beads were isolated and washed in Salt Immune Complex Wash Buffer. Both input control and immunoprecipitated samples were then eluted with the ChIP elution buffer containing proteinase K and incubated at 62 °C for 2 h, in order to purify the DNA from the bound proteins. The samples were further incubated at 95 °C for 10 min and kept at RT for 10 min. The magnetic beads were removed from the solution with the magnetic separator and samples transferred to Spin Filters to be further spun for 30 s at 12,000 rpm twice after addition of the elution reagent provided by the kit, in order to elute the DNA. DNA was kept at −20 °C if not analyzed immediately.
qPCR Analysis of Immunoprecipitated DNA
Three primer sets for each gene of interest, covering respectively the 1.4-kb upstream, promoter, and exon regions, were designed (Fig. 1). For the gene of reference, GAPDH, only the promoter area was amplified. Primer sets were as follows: 5-HT2A-1.4 kb upstream: F = 5′-ACT GGT GTG GGC TAG AAG TGC-3′ and R = 5′-GAG GGG CGA AGT GTG AGA AAA-3′; 5-HT2A promoter: F = 5′-CAC GTT TGT TCC CCG AAT TAC-3′ and R = 5′-GAT TAT GCC TCC CTT CAC GGT-3′; 5-HT2A exon 1: F = 5′-TTC GGA AGC ATC GAA CTG GA-3′ and R = 5′-AGA ATG GAG AGG GCA TGT CGG-3′; mGluR2–1.4 kb upstream: F = 5′-GTG TCT GTC TCC CCA GCT TTC-3′ and R = 5′-AAT GGG AGA CAA GGT GGC AG-3′; mGluR2 promoter: F = 5′-ATT CAG CAC CAC AAG GTG GAC A-3′ and R = 5′-CAA TTT GGC CTG CAC CTC TCG C-3′; mGluR2 exon 2: F = 5′-ATG AGC ACC GAG GCATAC AG-3′ and R = 5′-GAT GCG GTC CAG TGC AAA AA-3′; 5-HT1A 1.4 kb upstream: F = 5′-CGG GTG CTG AAC CAA ATT TCA-3′ and R = 5′-TTG GTG GCA TCC CTT GTC TT-3′; 5-HT1A promoter: F = 5′-CTT CGC CCG AGC AAG TAA GA-3′ and R = 5′-TTC AGA GGG AGG GGA TCC AG-3′; 5-HT1A exon 1: F = 5′-TCC ACT TTC GGC GCT TTC TA-3′ and R = 5′-TGA CAG TCT TGC GGA TTC GG-3′; GAPDH promoter: F = 5′-AAC CCT CAT CCG GTC ACT TCC-3′ and R = 5′-CGA GTA GCT GGG CCT CTC TCA 3′. Primers were optimized for use with PowerUp™ SYBR® Green Master Mix (Thermo Fisher Scientific Inc., cat. no. A25742). All qPCR reactions were run with the Applied Biosystems® QuantStudio™ 6 Flex Real-Time PCR System (cat. no. 4485699) on the recommended 40-cycle program for use with the PowerUp™ SYBR® Green Master Mix. Each reaction was performed in quadruplicates along with no template control. All reactions were confirmed to generate a single product by melting curve analysis. Calculations were based on previous publications on ChIP experiments [31, 44].
Fig. 1.
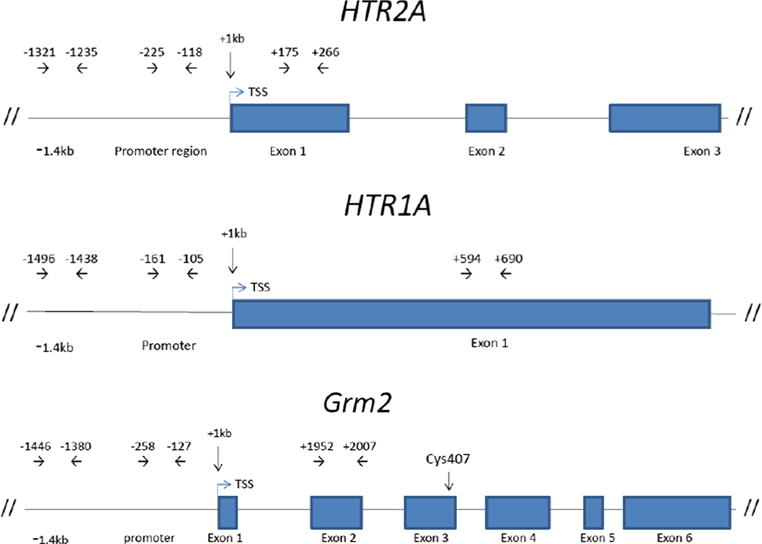
Map of the 5-HT2A (HTR2A), 5-HT1A (HTR1A), and mGluR2 (Grm2) genes showing positions of promoter region and exons (rectangles) and primers used for qPCR analysis of the chromatin immunoprecipitated (ChIP) products. In the Grm2 gene, the position of the cys407 mutation is indicated
Western Blotting
Protein lysates were extracted from PFC and STR tissues using the commercially available NucleoSpin®RNA/Protein kit (Macherey-Nagel, cat. no. 740933). A total volume of 15 μL (containing 10 and 20 μg of protein) from each sample was loaded onto 4–20% 17-well Precast RunBlue SDS gels (Expedeon, cat. no. NXG42027K). An equal amount of protein from a pooled sample was used as a calibrator on each gel. A protein ladder (Novex® Sharp Pre-stained Protein Standard, Life Technologies, cat. no. LC5800) was used as a molecular weight marker. The proteins were transferred to a polyvinylidene fluoride (PVDF) Transfer Membrane (Thermo Scientific, cat. no. 88518), using a Trans-Blot® semidry cell (Bio-Rad, USA) for 60 min, using a 200-mA/membrane constant current. The membranes were blocked for 1 h in PBS pH 7.4 supplemented with 0.1% Tween 20 (PBST) and 5% skimmed milk when detecting non-phosphorylated proteins and in Tris buffer saline with 0.1% Tween 20 (TBST) and 5% BSA when detecting phosphorylated proteins. The membranes were incubated separately overnight at 4 °C on a 3D shaker with the following primary antibodies: GSK3α/β (sc-7291, mouse monoclonal, Santa Cruz Biotechnology Inc., 1:200), p-Tyr216-GSK3β (sc-135653, rabbit polyclonal, Santa Cruz Biotechnology Inc., 1:300), p-GSK3β (ser9) (sc-11757, goat polyclonal, Santa Cruz Biotechnology Inc., 1:500), Akt (#9272, rabbit polyclonal, Cell Signaling Technology Inc., 1:1000), p-Ser 473-Akt (#9271, rabbit polyclonal, Cell Signaling Technology Inc., 1:1000), β-catenin (sc-7963, mouse monoclonal, Santa Cruz Biotechnology Inc., 1:200), and GAPDH (sc-25778, rabbit polyclonal, Santa Cruz Biotechnology Inc., 1:500). The membranes were then washed 3 × 15 min in either PBST or TBST. The membranes were incubated with either goat anti-rabbit (ab97051, Abcam plc., 1:8000) or goat anti-mouse (ab97023, Abcam plc., 1:8000) for 1 h at RT. After incubation, the membranes were washed 3 × 15 min in either PBST or TBST and rinsed in PBS or TBS followed by a rinse in distilled water. For the chemiluminescent development, the HRP reactive SuperSignal® West Dura Extended Duration Substrate (Thermo Scientific, cat. no. 34075) was used. Blots were scanned in the G:BOX iChemi XR Image Analyzer (Syngene, Synoptics Ltd., UK) for the visualization of protein bands. Densitometry quantification was performed using the Syngene GeneTools software (Syngene, Synoptics Ltd., UK). Proteins of interest were normalized to GAPDH and the amount of protein for each sample calculated in relation to total pooled protein used as a calibrator on each gel. GAPDH band intensity did not differ across groups.
In regard of the mGlu2 receptor western blot experiments, these were performed as previously reported with minor modifications [30]: briefly, frontal cortex membrane homogenates (see previously mentioned) were loaded onto a polyacrylamide gel (12%) and submitted to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After transfer to nitrocellulose membranes, blocking with 2.5% non-fat dry milk and 0.5% BSA in TBST buffer (Tris-buffered saline and 0.05% Tween 20) was followed by incubation with primary antibody (mGlu2: Abcam ab15672; 1:1000) for 90 min at RT. Incubation with the secondary antibody coupled to peroxidase (Amesham Biosciences) was performed at RT for 90 min. Immunoreactive proteins were visualized with enhanced chemiluminiscence (Thermo Scientific) according to the manufacturer’s instructions. In each case, the blots were stripped and reprobed for a control protein (β-actin: Abcam ab8227; 1:3000) to control for loading amounts.
DNA Sequencing
Genomic DNA was extracted from hippocampal tissue using REDExtract-N-Amp™ Tissue PCR Kit, Sigma-Aldrich. The DNA quality and concentration were measured using a UV-Vis spectrophotometer (NanoDrop 2000c, Thermo Scientific). PCR primers (mGlur2_3.2 F: 5′-CGTGAGTTCTGGGAGGAGAG-3′ and mGlur2_3.2R: 5′-TTGTGGTCCATCCTGTTTCC-3′) were designed with the online Primer3 (v. 0.4.0) software (http://bioinfo.ut.ee/primer3-0.4.0/primer3//) to flank the cys407* Grm2 mutation (exon 3, nucleotide 1419, NM 001105711.1). PCR reactions were performed in the Stratagene Mx3005P qPCR system, Agilent Technologies, using standard thermal cycling. Activation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 30 s, a final extension at 72 °C for 10 min, and termination at 25 °C, was performed. The PCR fragments were run on a 1% agarose gel and visualized under UV light in the presence of GelGreen™ Nucleic Acid Gel Stain 10,000× (in water) from Biotium, Hayward, CA, USA. The mGluR2_3.2 forward primer was also used to sequence the amplified PCR products (Lightrun Sanger Sequencing at GATC Biotech AG, Germany (www.gatc-biotech.com)). The cys407* Grm2 sequencing results were visualized using the Chromas software (www.technelysium.com.au), version 2.6, and compared to the reference sequence of the Grm2 gene (www.ensembl.org, Transcript ID: ENSRNOT00000017607. 3) using NCBI nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical Analyses
A two-way ANOVA with Sidak’s multiple comparison tests was applied for analyzing the receptor messenger RNA (mRNA) levels and DNA product levels after ChIP. An RM two-way ANOVA was applied when comparing the levels of specific DNA product along the different areas in the same gene. Unpaired Student’s t test, with Welch correction in case of unequal variance, was applied for analyzing the receptor binding levels and DNA product after ChIP for the promoter region. Western results were analyzed by Sidak’s multiple comparison test when comparing strains within each region. Before analysis, the Grubbs outlier test was run and all significant outliers were removed. All statistical analysis and graphs were done in GraphPad Prism 6.07 (GraphPad Software, Inc., USA). Data is presented as mean ± standard error of the mean (SEM) with p < 0.05 considered statistically significant.
Results
Decreased Prefrontal and Striatal mGlu2 and Striatal 5-HT2A Receptor Gene Expression in RHA-I Rats
When looking at mRNA levels for the 5-HT2A, 5-HT1A, and mGlu2 receptors in PFC and STR for both strains, there was a significant effect of receptor by region (F(5,117) = 5.061, p < 0.001), of strain (F(1,117) = 21.29, p < 0.0001), and of the interaction between these two factors (F(5,117) = 4.705, p < 0.001). Post hoc analysis revealed a significant decrease of the mGlu2 receptor in PFC (p < 0.01) and STR (p < 0.001) and of the 5-HT2A receptor in STR (p < 0.05) in the RHA-I compared to the RLA-I (Fig. 2a). When comparing the relative expression of 5-HT2A versus mGlu2, we found in the RHA-I a significant difference between these two receptors in the PFC (p < 0.05) that was not present in the STR (Fig. 2b). No significant differences were observed for 5-HT1A receptor mRNA expression levels in either PFC or STR, though a trend was seen for decreased expression in RHA in STR.
Fig. 2.
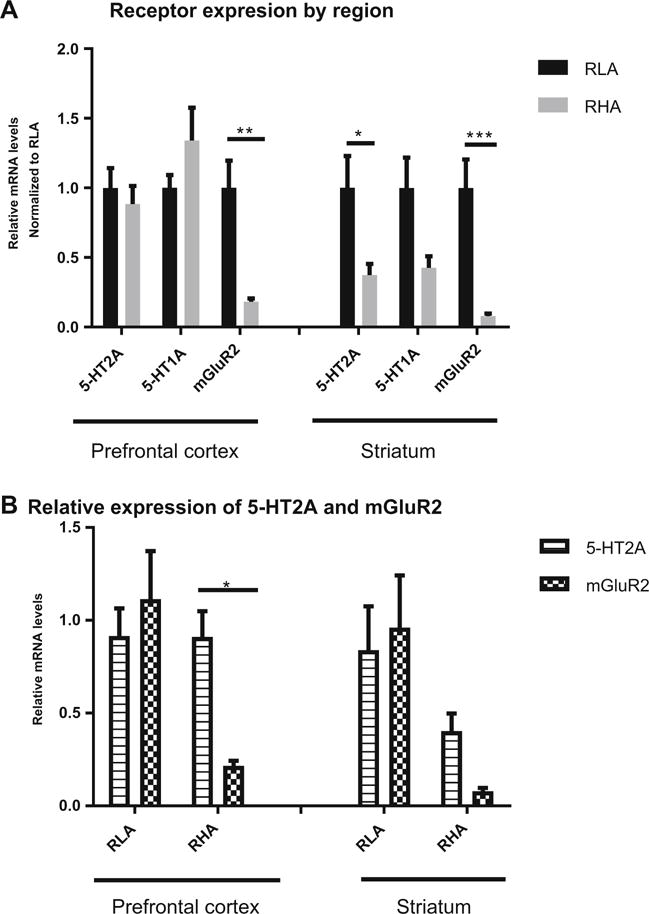
a Receptor expression in prefrontal cortex and striatum normalized to RLA-I. Both in prefrontal and striatum, mGluR2 levels are significantly lower in RHA-I when compared to RLA-I. For the 5-HT2A receptor only in the striatum, the RHA-I strain presented lower expression levels of this receptor. No difference was obtained for the 5-HT1A receptor. b When comparing the relative expression of 5-HT2A versus mGluR2, there was a significant difference in prefrontal cortex of the RHA-I strain that was diminished in the striatum as a result of the lower expression of 5-HT2A receptor of this strain in this region. Two-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001
Higher 5-HT2A Receptor Binding in PFC of RHA-I Rats
When incubating membrane homogenates isolated from PFC and STR, in a single-concentration assay, with a saturating concentration of 5 nM of the 5-HT2A receptor ligand [3H]ketanserin, receptor binding in PFC was significantly higher in the RHA-I when compared to the RLA-I strain (p < 0.05). For STR, no difference in 5-HT2A receptor binding was observed between the two strains (Fig. 3).
Fig. 3.
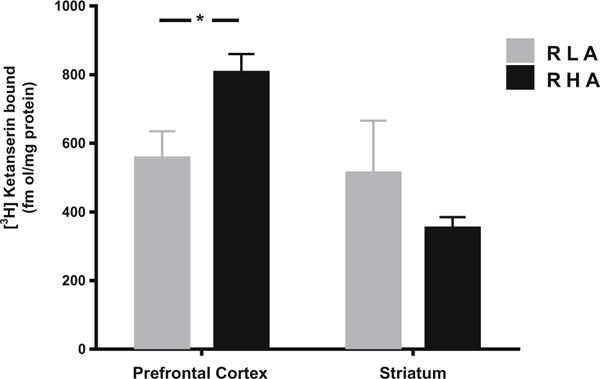
Maximum binding levels of 5-HT2A receptor by [3H]ketanserin are higher in prefrontal cortex of the RHA-I when compared to RLA-I. In striatum, there is no difference in binding levels between the two strains. Student’s t test, *p < 0.05
No Strain Differences in Binding of H3K27me3 to 5-HT2A, 5-HT1A, and Grm2 Genes in the PFC Area
In PFC, chromatin immunoprecipitation results of histone H3 showed a significant effect of epigenetic modifications for both the HTR2A (F(8,115) = 13.55, p < 0.0001), HTR1A (F(8,116) = 20.81, p < 0.0001), and Grm2 (F(8,117) = 25.67, p < 0.0001) genes. Post hoc analysis when grouping both strains showed H3K27me3 binding significantly higher than background for the HTR2A gene at the promoter (p < 0.0001) and exon 1 (p < 0.001) regions, for the HTR1A gene in the −1.4 kb upstream (p < 0.0001), promoter (p < 0.0001), and exon 1 (p < 0.001) regions, and for the Grm2 gene in the −1.4 kb upstream (p < 0.001) and promoter (p < 0.0001) regions. Sidak’s multiple comparison tests taking both strains into account revealed no significant difference in H3K27me3 binding between RHA-I and RLA-I (Fig. 4a). Binding of acetylated H3 (H3ac) was not significantly different from background levels for the three genes but was at the GAPDH gene promoter region (F(2,42) = 15.13, p < 0.0001), validating the immunoprecipitation assay (Fig. 4c).
Fig. 4.
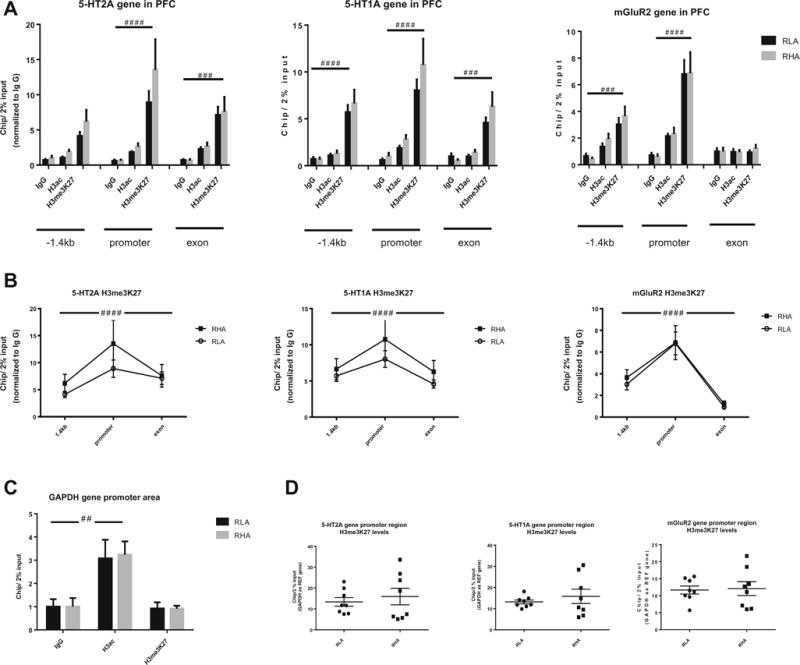
a Trimethylation of histone H3 at lysine 27 (H3me3K27) of the 5-HT2A, 5-HT1A, and mGluR2 genes at the −1.4 kb upstream, promoter, and exon 1 regions in the prefrontal cortex (PFC). There was no acetylation of the histone H3 in any of the regions of the three genes. The gene of reference GAPDH showed acetylation of H3 at the promoter region, validating the chromatin immunoprecipitation assay (c). b When comparing the different regions within the same gene for H3me3K27 levels, the promoter region peaked significantly for all three genes. d When comparing individually the promoter region for differences in H3me3K27 levels between the two strains, no differences were observed for any of the three genes. Two-way ANOVA #p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001
When looking specifically at differences in methylated H3 binding throughout the different gene regions, H3K27me3 binding differed significantly for the HTR2A (F(2,26) = 13.72, p < 0.0001) with a within-subject matching effect across regions (F(13,26) = 10.28, p < 0.0001), the HTR1A (F(2,26) = 20.13, p < 0.0001) with a within-subject matching effect across regions (F(13,26) = 16.02, p < 0.0001), and the mGluR2 (F(2,26) = 45.54, p < 0.0001) with a within-subject matching effect across regions (F(13,26) = 4.395, p < 0.001) and with the peak for H3K27me3 binding at the promoter region (Fig. 4b). When looking specifically at the promoter region, no difference in H3K27me3 binding to this region was found between the RHA-I and RLA-I for any of the genes (Fig. 4d).
Increased Binding of H3K27me3 at the Promoter Region of the 5-HT2A Gene in the STR of RHA-I Rats
In STR, chromatin immunoprecipitation results of histone H3 showed a significant effect of epigenetic modifications for HTR1A (F(8,114) = 76.89, p < 0.0001) and the Grm2 gene (F(8,118) = 40.44, p < 0.0001). Post hoc analysis when grouping both strains showed H3K27me3 binding significantly higher than background for the HTR1A gene in the −1.4 kb upstream (p < 0.0001), promoter (p < 0.0001), and exon 1 (p < 0.0001) areas. H3ac binding was not significantly different from background levels. For the Grm2 gene, post hoc analysis when grouping both strains showed significant increased H3K27me3 binding in the −1.4 kb upstream (p < 0.001) and at promoter (p < 0.0001) areas and increased H3ac binding at the promoter region (p < 0.001). Sidak’s multiple comparison tests taking both strains into account revealed no significant difference in H3K27me3 binding levels between RHA-I and RLA-I for either the HTR1A or the Grm2 gene and in H3ac binding levels for the Grm2 gene. In regard of the HTR2A gene, there was a significant effect of H3K27me3 binding levels (F(8,115) = 49.42, p < 0.0001), of strain (F(1,115) = 8.030, p < 0.01), and of their interaction (F(8,115) = 2.886, p < 0.01). Post hoc analysis when grouping both strains showed H3K27me3 binding levels significantly higher than background in the −1.4 kb upstream (p < 0.0001), promoter (p < 0.0001), and exon 1 (p < 0.0001) regions. H3ac binding levels were not significantly different from background levels. When performing Sidak’s multiple comparison tests between both strains, there was a significant difference in H3K27me3 binding levels at the promoter region, with the RHA-I showing higher binding (p < 0.0001) (Fig. 5a). The GAPDH gene promoter region showed a significant effect of the epigenetic modifications (F(2,40) = 31.01, p < 0.0001), with higher binding than background for both H3K27me3 (p < 0.05) and H3ac (p < 0.0001) (Fig. 5c).
Fig. 5.
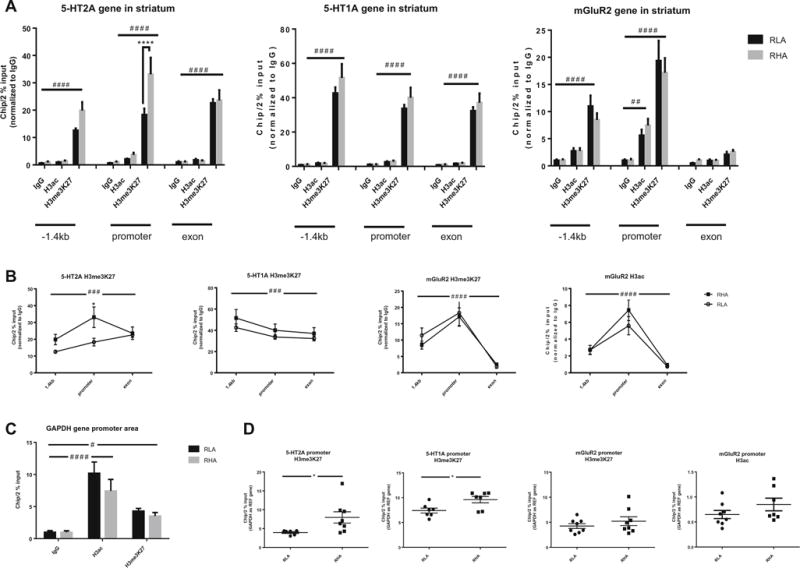
a Trimethylation of histone H3 at lysine 27 (H3me3K27) of the 5-HT2A, 5-HT1A, and mGluR2 genes at the −1.4 kb upstream, promoter, and exon 1 regions in the striatum. The mGluR2 gene showed acetylation of histone H3 in promoter region. There was no acetylation of the histone H3 in any of the regions of 5-HT2A and 5-HT1A. Post hoc analysis showed significant higher levels of H3me3K27 at the promoter region of the 5-HT2A gene in the RHA-I rats. The gene of reference GAPDH showed H3me3K27 and acetylation of H3 at the promoter region, validating the chromatin immunoprecipitation assay (c). b When comparing the different regions within the same gene for H3me3K27 levels, the promoter region peaked significantly for all three genes. In the case of the mGluR2 gene, acetylation of H3 was also significantly higher in the promoter region. For the 5-HT2A gene, there was furthermore a significant difference between the RHA-I and RLA-I strains in the promoter region. d When comparing individually the promoter region for differences in H3me3K27 levels between the two strains, significant difference was observed not only for the 5-HT2A gene but also for the 5-HT1A. No difference in H3me3K27 or H3ac levels was observed for the mGluR2 between the two strains. Two-way ANOVA, #p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001; Sidack’s post hoc multiple comparison test, ****p < 0.0001; Student’s t test *p < 0.05
When looking specifically at H3K27me3 binding levels throughout the different gene regions, levels differed significantly for the HTR2A (F(2,26) = 10.03, p < 0.001), with a within-subject matching effect across regions (F(13,26) = 6.024, p < 0.0001), the 5-HT1A (F(2,26) = 15.38, p < 0.0001) with a within-subject matching effect across regions (F(13,26) = 12.73, p < 0.0001), and the Grm2 (F(2,24) = 51.95, p < 0.0001), with a within-subject matching effect across regions (F(12,24) = 4.141, p < 0.01) with the peak of H3K27me3 binding being at the promoter region (Fig. 5b). When looking at binding of H3ac at the Grm2 gene, levels differed significantly through the gene regions (F(2,24) = 53.33, p < 0.0001), with a within-subject matching effect across regions (F(12,24) = 3.287, p < 0.01). For the HTR2A gene, a significant effect of interaction between subjects and gene area was obtained (F(2,26) = 5.008, p < 0.05). After multiple comparison test, the difference is shown for the promoter region between RHA-I and RLA-I (p < 0.05). When looking specifically at the promoter region, differences in H3K27me3 binding levels were found between the RHA-I and RLA-I for the HTR2A gene and also for the HTR1A gene. No differences were found in either H3K27me3 and H3ac levels for the Grm2 gene (Fig. 5d).
Increased Levels of Phospho-Tyr216-GSK3β and β-Catenin in PFC of RHA-I Rats
Both phospho-Tyr216-GSK3β (p < 0.05) and β-catenin (p < 0.01) levels were increased in PFC of RHA-I rats (Fig. 6). Levels of GSK3α/β did not differ between regions in either the RLA and RHA-I rats. No differences in phosphorylated Akt and phospho-ser9-GSK3 β were found among regions and strains (Fig. 6).
Fig. 6.
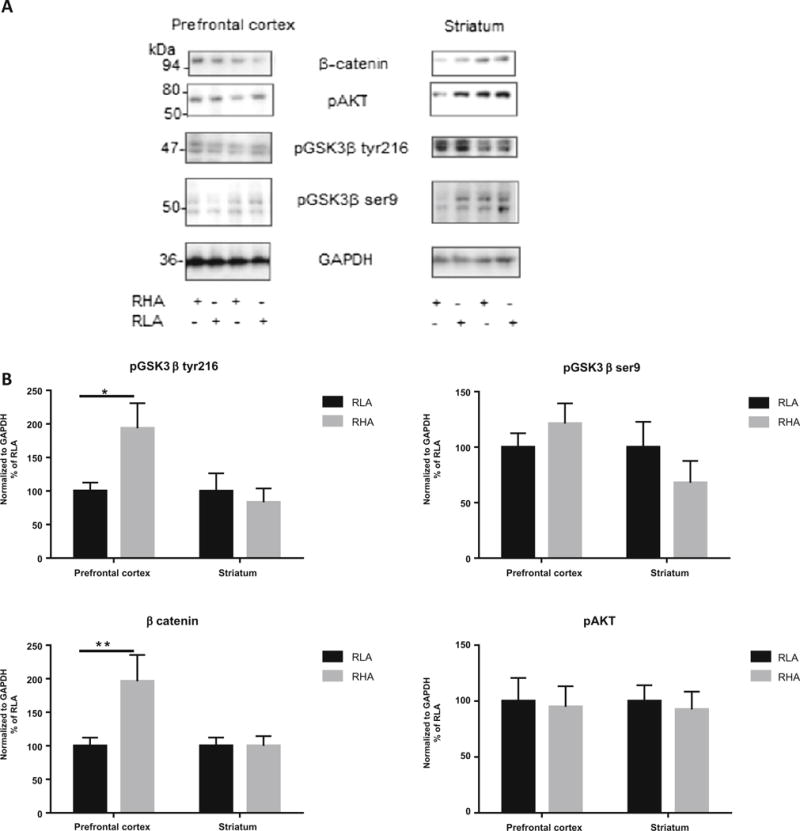
a Representative western blots for β-catenin, phosphorylated GSK3β at tyrosine 216 site (pGSK3β tyr216), phosphorylated GSK3β at serine 9 (pGSK3β ser9), phosphorylated Akt (pAkt), and GAPDH in prefrontal cortex and striatum for both RLA-I and RHA-I rats. b Levels of pGSK3β tyr216 and β-catenin are increased in prefrontal cortex of RHA-I compared to RLA-I rats. No differences were observed in striatum or in levels of pGSK3β ser9 or pAkt. Sidak’s multiple comparison test, *p < 0.05, **p < 0.01
Grm2 Gene Sequencing Showed RHA-I Rats to Carry the cys407* Grm2 Mutation
A previous study by [45] has in some Wistar strains, including the Roman rat strains, reported a common stop codon mutation at cysteine 407 in the Grm2 gene. To investigate the prevalence of this mutation in our RHA-I and RLA-I rats, we sequenced in a subsample of our rats the Grm2 gene according to protein position 407. Homozygous cys407* Grm2 (c.1429C>A, p.Cys/TGC407Stop/TGA) in exon 3 of the Grm2 gene was found in all the investigated RHA-I rats (n = 8). RLA-I rats (n = 7) were Grm2 wild type (TGC) except for one RLA-I rat that was found to be heterozygous for the cys407* Grm2 mutation (TGC/TGA) (Fig. 7a). Hence, our results support previous findings and emphasize the distribution of the cys407* Grm2 mutation in RHA-I rats. Further, we confirmed the absence of receptor protein in this strain by western analysis on PFC membrane extracts of a subsample of the RLA-I (n = 3) and RHA-I (n = 3) rats. No mGluR2 band was visible in the RHA-I rats (Fig. 7b).
Fig. 7.
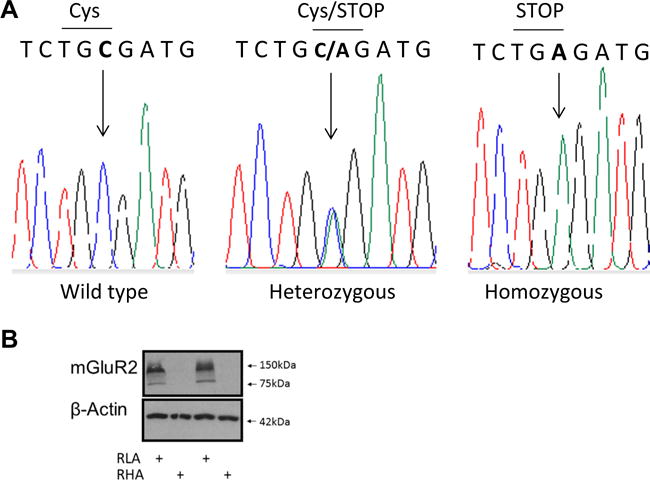
a Sanger sequencing of the Grm2 gene illustrating the area in exon 3 for the Cys407* mutation. The nucleotide cytosine (C) is substituted by the nucleotide adenine (a) (arrow) leading to a stop codon (TGA). RLA rats (n = 7) were found to be Cys407* Grm2 wild type except one RLA rat that was Cys407* Grm2 heterozygous. All RHA (n = 8) rats were found to be Cys407* Grm2 homozygous. b Representative western blots for mGlu2 receptor in prefrontal cortex for both RLA-I and RHA-I rats. No protein bands were detectable in the RHA-I rat samples
Discussion
The present results confirm the Roman rat strain differences in 5-HT2A and mGlu2 receptor levels previously described by us [9]. Further, we report how the inexistent receptor protein levels found for the mGlu2 receptor in the RHA-I [9] are accompanied by low expression of the Grm2 gene, most likely as a result of the presence of a stop codon mutation at cysteine 407 previously identified in this strain [45] and confirmed by us here. For the 5-HT2A receptor, the increase in receptor binding in PFC in the RHA-I was not accompanied by a corresponding increase in mRNA levels. As we only measure membrane-bound receptors by the binding assay, total 5-HT2A protein levels may not differ between strains. The higher 5-HT2A receptor binding could therefore reflect increased receptor functionality and/or ligand-binding affinity probably as a consequence of the absence of the mGlu2 receptor. The 5-HT2A and the mGlu2 are G protein-coupled receptors (GPCRs), both normally highly expressed in the PFC [46–48]. They interact with each other in a functional cross talk through heteromeric complexes [27, 29, 49]. The mGlu2 is also capable of forming heterodimers with the mGlu3 receptor [50]. Pharmacological activation of the mGlu2/3 receptor complex suppresses the intracellular and behavioral effects of the hallucinogenic 5-HT2A receptor agonist 1-(2,5-dimethoxy-4-iodophenyl)-2-aminopropane (DOI) [29]. The 5-HT2A-dependent polyphosphoinositide (PI) hydrolysis intracellular cascade is further attenuated when activating the mGlu2/3 receptor [51]. Recent studies have established that this functional cross talk between the 5-HT2A and mGlu2 receptors takes place at the submembrane level, where the Gi/0-coupled mGlu2 and the Gq/11-coupled 5-HT2A receptor modulate each other’s G protein-dependent signaling cascade [27]. Changes in mGlu2 receptor levels affect 5-HT2A receptor function. Indeed, 21 days of treatment with the mGlu2/3 receptor antagonist LY341495 causes downregulation of 5-HT2A receptor [3H]ketanserin binding in somatosensory cortex, which was shown to be specifically mediated by the mGlu2 receptor [28]. However, in mGlu2 receptor knockout mice, 5-HT2A receptor [3H]ketanserin binding is preserved [52], indicating that the 5-HT2A receptor reacts differently to a sudden decrease in mGlu2 receptor levels than to the total absence of the mGlu2 receptor. In the RHA-I, the mGlu2 receptor is probably already missing during development and the adaptive response of the 5-HT2A receptor would be comparable to that seen in the mGlu2 receptor knockout mice. The increased 5-HT2A PFC binding observed here can also be an adaptive response to more general alterations in glutamatergic tonus in the RHA-I, as mGlu2/3 receptors function as autoreceptors and are involved in modulating excitatory neurotransmission [53]. The absence of the mGlu2 receptor probably has an impact on basal glutamate levels in the RHA-I, even though this has not been investigated yet. We know that 5-HT2A receptor functionality and downstream signaling responses are influenced by changes in glutamatergic tonus [54, 55].
In the STR, contrary to PFC, 5-HT2A mRNA levels were significantly reduced in the RHA-I when compared to the RLA-I. mGlu2 receptor expression was likewise drastically reduced in STR of the RHA-I. Receptor binding by [3H]ketanserin, however, was not different from RLA-I despite the decrease in gene expression. Thus, here again, this may indicate separate regulation of the total receptor protein pool and 5-HT2A ligand affinity and functionality at the membrane level. Whether this is a response to specific neurotransmitter and other receptor alterations in STR we can only infer. In STR, mGlu2/3 receptors are primarily present on terminals of the corticostriatal afferents [56]. A deficit in the glutamatergic corticostriatal input to the STR has been proposed as a central factor in the etiology of schizophrenia [57, 58]. According to this hypothesis, a deficit in this pathway would lead to an increased dopaminergic tonus in the STR as a result of increased disinhibition of the nigrostriatal dopaminergic projecting neurons [57]. This corresponds well with the higher dopaminergic tonus and decreased D2 receptor level and functional sensitivity reported for the RHA-I in this area [19]. Thus, the decrease in striatal 5-HT2A receptor expression could be a compensatory response to dopaminergic alteration. Anti-psychotic-induced dopaminergic supersensitivity by the D2 antagonist haloperidol affects 5-HT2A receptor density [59], and the 5-HT2A also seems to interact with the D2 receptor into functional heterodimers in this area [39, 60, 61].
Interestingly, we found that striatal 5-HT2A expression in the RHA-I is regulated by epigenetic control of the promoter region of the HTR2A gene. We looked at histone modifications as these are dynamic and reversible regulatory mechanisms of gene expression that reveal adaptive responses of gene × environmental interactions [33]. We focused on his-tone H3, as previous studies have described H3 modifications in relation to 5-HT2A and mGlu2 receptor interactions [30, 31, 62]. No differences in acetylated H3 binding levels for any of the gene regions included in this study were observed for the two strains. Histone acetylation enables gene transcription by unfolding the chromatin structure through neutralization of the basic charges of the lysine [63]. This is in concordance with our observations of no increased expression of the HTR2A, HTR1A, or Grm2 genes. On the contrary, when looking at H3K27 methylation, we found increased binding levels at the promoter region of the HTR2A gene in the STR of the RHA-I rats. Histone lysine methylation is more specific and site directed than histone acetylation and implicated in gene silencing [63, 64]. Even though still not fully elucidated what the exact roles of the different histone methylation sites are in the regulation of transcriptional repression/activation, it is believed that H3K27me is implicated in facilitating the recruitment of methylating enzymes to the promoter of the repressed genes [63]. More specifically, H3K27me at the promoter region may act as “dials” or “switches” of gene expression by fine-tuning expression levels from active to poised to inactive [64]. Indeed, H3K27me has been reported to be involved in regulating DNA methylation during brain development [65, 66]. On the other side, alterations in the epigenetic regulation of the HTR2A gene during gestation have been associated with neurobehavioral deficits related to schizophrenia [67, 68]. Inferring from this, we can say that the lower expression of striatal 5-HT2A in the RHA-I is very likely reflecting a developmental adaptive response at the gene transcriptional level. Still, whether this particular gene regulatory effect is crucial for the expression of the behavioral phenotype of the RHA-I we cannot conclude. Other gene sites may show similar epigenetic adaptations, and the 5-HT2A receptor changes may be an epiphenomena of other more central changes not investigated yet. Nevertheless, the importance of these observations relies in the fact that strains selected based on their different behavioral and cognitive strategies show differences in epigenetic modifications. These modifications are reversible and by that open up for the possibility of adjusting behavior through pharmacological modulation of epigenetic regulation.
In addition to the previous observations, we wanted to further investigate whether the strain and region differences in 5-HT2A and mGlu2 receptor regulation are accompanied by differences in downstream intracellular pathways. We focused on the Akt/GSK3 pathway as both the mGlu2/3 receptor complex and the 5-HT2A receptor have a direct regulatory effect on this pathway [40, 69]. In the study by Sutton and Rushlow [69], they reported how repeated agonist stimulation of the mGlu2/3 receptor resulted in increased phosphorylated Akt, phosphoser9-GSK3β, and β-catenin levels in both PFC and STR, with an opposite effect when administering an antagonist. Our observations do not fully match these, as we did not see differences in Akt or Ser9-GSK3β phosphorylation, though we found increased levels of β-catenin and phosphorylation at Tyr216-GSK3β in PFC of RHA-I rats, despite the absence of the mGlu2 receptor. This divergence is not surprising. As discussed earlier, effects obtained from pharmacological manipulation of the mGlu2/3 receptor complex and from blocking mGlu2 receptor expression during development show conflicting results, which probably are due to compensatory mechanisms in the second situation. No study has yet looked at the Akt/GSK3 pathway in mGlu2 knockout mouse, and therefore, we do not know which effect the absence of the mGlu2 receptor, like is the case in the RHA-I rats, may have on this pathway. Our observations of a region-specific regulation of this pathway in the RHA-I regardless of the low presence of the mGlu2 receptor in both PFC and STR lead us to believe that the differential regulation in the mGlu2–5-HT2A receptor balance in STR versus PFC may somehow be involved in this. Overactivation of the 5-HT2A receptor in the PFC may explain this, though further studies should clarify this. Phosphorylation at ser9 inactivates GSK3β enzymatic function, while phosphorylation at Tyr216 stabilizes the protein facilitating its activation [70]. According to this, GSK3 constitutive activity would be enhanced in PFC of the RHA-I rats. Which specific upstream pathway regulates Tyr216 phosphorylation is not known yet. In our study, we looked at Akt, as this kinase is downstream of 5-HT2A and 5-HT1A and upstream of GSK3 [71, 72], but we found no differences in phosphorylation levels or protein levels (results not shown), indicating that other upstream effectors are acting upon this activation. In relation to schizophrenia, there is still some controversy on whether GSK3 activation and β-catenin levels are decreased or not in cortical areas of subjects with schizophrenia [73, 74]. However, there is strong evidence of GSK3β and β-catenin being involved in spine stability and synaptic plasticity both during development and in adulthood [75–77]. Based on this and our observations, we believe that looking into differences in spine density in the PFC of the RHA-I is warranted and of great importance for understanding the neurobiological substrates behind their different behavioral phenotype.
Summarizing, we have further characterized the receptor differences in 5-HT2A and mGlu2 previously reported by us in the Roman rat strains. We found in the RHA-I strain no presence of mGlu2 receptor protein, distinctive low gene expression in both PFC and STR, most likely as a direct consequence of the mutation at the Gmr2 gene. Interestingly, we found an epigenetically controlled silencing of the HTR2A gene in the STR of the RHA-I strain, which is probably a developmental-induced adaptive response to the absence of the mGlu2 and may be due to the consequent region-specific alterations in neurotransmitter balance in this strain. This differential regulation is reflected on the intracellular signaling cascades responsible for synaptic maturation and regulation during development, further supporting the biological validity of the RHA-I strain as a putative rat model of schizophrenia-related features.
Acknowledgments
This work has received partial support from grants PSI2013-41872-P (MINECO), 2014SGR-1587 (DGR), and “ICREA-Academia 2013” (to AF-T) and NIH R01MH084894. I.O. is recipient of a PhD FI fellowship (DGR 2014). Travel grants for L.F. were provided by A.P. Moeller Foundation and the Danish Foundation of Medical Science (Fonden til Lægevidenskabens Fremme).
Footnotes
Compliance with Ethical Standards All animals were sacrificed in accordance with the Spanish Royal Decree (RD 53/2013) for the protection of experimental animals and with the European Communities Council Directive (2010/63/EU) at approximately 4 months of age
References
- 1.Driscoll P, Escorihuela RM, Fernandez-Teruel A, Giorgi O, Schwegler H, Steimer T, Wiersma A, Corda MG, et al. Genetic selection and differential stress responses. The Roman lines/strains of rats. Ann N Y Acad Sci. 1998;851:501–510. doi: 10.1111/j.1749-6632.1998.tb09029.x. [DOI] [PubMed] [Google Scholar]
- 2.Escorihuela RM, Fernandez-Teruel A, Gil L, Aguilar R, Tobena A, Driscoll P. Inbred Roman high- and low-avoidance rats: differences in anxiety, novelty-seeking, and shuttlebox behaviors. Physiol Behav. 1999;67:19–26. doi: 10.1016/s0031-9384(99)00064-5. [DOI] [PubMed] [Google Scholar]
- 3.Esnal A, Sanchez-Gonzalez A, Rio-Alamos C, Oliveras I, Canete T, Blazquez G, Tobena A, Fernandez-Teruel A. Prepulse inhibition and latent inhibition deficits in Roman high-avoidance vs. Roman low-avoidance rats: modeling schizophrenia-related features. Physiol Behav. 2016;163:267–273. doi: 10.1016/j.physbeh.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 4.Steimer T, Driscoll P. Divergent stress responses and coping styles in psychogenetically selected Roman high-(RHA) and low-(RLA) avoidance rats: behavioural, neuroendocrine and developmental aspects. Stress. 2003;6:87–100. doi: 10.1080/1025389031000111320. [DOI] [PubMed] [Google Scholar]
- 5.Diaz-Moran S, Palencia M, Mont-Cardona C, Canete T, Blazquez G, Martinez-Membrives E, Lopez-Aumatell R, Tobena A, et al. Coping style and stress hormone responses in genetically heterogeneous rats: comparison with the Roman rat strains. Behav Brain Res. 2012;228:203–210. doi: 10.1016/j.bbr.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Giorgi O, Piras G, Corda MG. The psychogenetically selected Roman high- and low-avoidance rat lines: a model to study the individual vulnerability to drug addiction. Neurosci Biobehav Rev. 2007;31:148–163. doi: 10.1016/j.neubiorev.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Manzo L, Gomez MJ, Callejas-Aguilera JE, Donaire R, Sabariego M, Fernandez-Teruel A, Canete A, Blazquez G, et al. Relationship between ethanol preference and sensation/novelty seeking. Physiol Behav. 2014;133:53–60. doi: 10.1016/j.physbeh.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Moreno M, Cardona D, Gomez MJ, Sanchez-Santed F, Tobena A, Fernandez-Teruel A, Campa L, Sunol C, et al. Impulsivity characterization in the Roman high- and low-avoidance rat strains: behavioral and neurochemical differences. Neuropsychopharmacology. 2010;35:1198–1208. doi: 10.1038/npp.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein AB, Ultved L, Adamsen D, Santini MA, Tobena A, Fernandez-Teruel A, Flores P, Moreno M, et al. 5-HT(2A) and mGlu2 receptor binding levels are related to differences in impulsive behavior in the Roman low- (RLA) and high- (RHA) avoidance rat strains. Neuroscience. 2014;263:36–45. doi: 10.1016/j.neuroscience.2013.12.063. [DOI] [PubMed] [Google Scholar]
- 10.Oliveras I, Rio-Alamos C, Canete T, Blazquez G, Martinez-Membrives E, Giorgi O, Corda MG, Tobena A, et al. Prepulse inhibition predicts spatial working memory performance in the inbred Roman high- and low-avoidance rats and in genetically heterogeneous NIH-HS rats: relevance for studying pre-attentive and cognitive anomalies in schizophrenia. Front Behav Neurosci. 2015;9:213. doi: 10.3389/fnbeh.2015.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luck SJ, Gold JM. The construct of attention in schizophrenia. Biol Psychiatry. 2008;64:34–39. doi: 10.1016/j.biopsych.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes NF, Carrick LA, McIntosh AM, Lawrie SM. Working memory in schizophrenia: a meta-analysis. Psychol Med. 2009;39:889–905. doi: 10.1017/S0033291708004558. [DOI] [PubMed] [Google Scholar]
- 13.Avsar KB, Weller RE, Cox JE, Reid MA, White DM, Lahti AC. An fMRI investigation of delay discounting in patients with schizophrenia. Brain Behav. 2013;3:384–401. doi: 10.1002/brb3.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weller RE, Avsar KB, Cox JE, Reid MA, White DM, Lahti AC. Delay discounting and task performance consistency in patients with schizophrenia. Psychiatry Res. 2014;215:286–293. doi: 10.1016/j.psychres.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan EJ, Rossell SL. Building a neurocognitive profile of thought disorder in schizophrenia using a standardized test battery. Schizophr Res. 2014;152:242–245. doi: 10.1016/j.schres.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Barch DM, Cohen R, Csernansky J. Altered cognitive development in the siblings of individuals with schizophrenia. Clinical psychological science : a journal of the Association for Psychological Science. 2014;2:138–151. doi: 10.1177/2167702613496244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lecca D, Piras G, Driscoll P, Giorgi O, Corda MG. A differential activation of dopamine output in the shell and core of the nucleus accumbens is associated with the motor responses to addictive drugs: a brain dialysis study in Roman high- and low-avoidance rats. Neuropharmacology. 2004;46:688–699. doi: 10.1016/j.neuropharm.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Giorgi O, Lecca D, Piras G, Driscoll P, Corda MG. Dissociation between mesocortical dopamine release and fear-related behaviours in two psychogenetically selected lines of rats that differ in coping strategies to aversive conditions. Eur J Neurosci. 2003;17:2716–2726. doi: 10.1046/j.1460-9568.2003.02689.x. [DOI] [PubMed] [Google Scholar]
- 19.Tournier BB, Steimer T, Millet P, Moulin-Sallanon M, Vallet P, Ibanez V, Ginovart N. Innately low D2 receptor availability is associated with high novelty-seeking and enhanced behavioural sensitization to amphetamine. Int J Neuropsychopharmacol. 2013;16:1819–1834. doi: 10.1017/S1461145713000205. [DOI] [PubMed] [Google Scholar]
- 20.Giorgi O, Piras G, Lecca D, Hansson S, Driscoll P, Corda MG. Differential neurochemical properties of central serotonergic transmission in Roman high- and low-avoidance rats. J Neurochem. 2003;86:422–431. doi: 10.1046/j.1471-4159.2003.01845.x. [DOI] [PubMed] [Google Scholar]
- 21.Aznar S, Hervig M. The 5-HT2A serotonin receptor in executive function: implications for neuropsychiatric and neurodegenerative diseases. Neurosci Biobehav Rev. 2016;64:63–82. doi: 10.1016/j.neubiorev.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Maeso J, Weisstaub NV, Zhou M, Chan P, Ivic L, Ang R, Lira A, Bradley-Moore M, et al. Hallucinogens recruit specific cortical 5-HT(2A) receptor-mediated signaling pathways to affect behavior. Neuron. 2007;53:439–452. doi: 10.1016/j.neuron.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Maeso J, Sealfon SC. Psychedelics and schizophrenia. Trends Neurosci. 2009;32:225–232. doi: 10.1016/j.tins.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Ebdrup BH, Rasmussen H, Arnt J, Glenthøj B. Serotonin 2A receptor antagonists for treatment of schizophrenia. Expert Opin Investig Drugs. 2011;20:1211–1223. doi: 10.1517/13543784.2011.601738. [DOI] [PubMed] [Google Scholar]
- 25.Desamericq G, Schurhoff F, Meary A, Szoke A, Macquin-Mavier I, Bachoud-Levi AC, Maison P. Long-term neurocognitive effects of antipsychotics in schizophrenia: a network meta-analysis. Eur J Clin Pharmacol. 2014;70:127–134. doi: 10.1007/s00228-013-1600-y. [DOI] [PubMed] [Google Scholar]
- 26.Vollenweider FX, Csomor PA, Knappe B, Geyer MA, Quednow BB. The effects of the preferential 5-HT2A agonist psilocybin on prepulse inhibition of startle in healthy human volunteers depend on interstimulus interval. Neuropsychopharmacology. 2007;32:1876–1887. doi: 10.1038/sj.npp.1301324. [DOI] [PubMed] [Google Scholar]
- 27.Moreno JL, Miranda-Azpiazu P, Garcia-Bea A, Younkin J, Cui M, Kozlenkov A, Ben-Ezra A, Voloudakis G, et al. Allosteric signaling through an mGlu2 and 5-HT2A heteromeric receptor complex and its potential contribution to schizophrenia. Sci Signal. 2016;9:ra5. doi: 10.1126/scisignal.aab0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno JL, Holloway T, Rayannavar V, Sealfon SC, Gonzalez-Maeso J. Chronic treatment with LY341495 decreases 5-HT(2A) receptor binding and hallucinogenic effects of LSD in mice. Neurosci Lett. 2013;536:69–73. doi: 10.1016/j.neulet.2012.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez-Maeso J, Ang RL, Yuen T, Chan P, Weisstaub NV, Lopez-Gimenez JF, Zhou M, Okawa Y, et al. Identification of a serotonin/glutamate receptor complex implicated in psychosis. Nature. 2008;452:93–97. doi: 10.1038/nature06612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurita M, Holloway T, Garcia-Bea A, Kozlenkov A, Friedman AK, Moreno JL, Heshmati M, Golden SA, et al. HDAC2 regulates atypical antipsychotic responses through the modulation of mGlu2 promoter activity. Nat Neurosci. 2012;15:1245–1254. doi: 10.1038/nn.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurita M, Moreno JL, Holloway T, Kozlenkov A, Mocci G, Garcia-Bea A, Hanks JB, Neve R, et al. Repressive epigenetic changes at the mGlu2 promoter in frontal cortex of 5-HT2A knockout mice. Mol Pharmacol. 2013;83:1166–1175. doi: 10.1124/mol.112.084582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma RP, Grayson DR, Gavin DP. Histone deactylase 1 expression is increased in the prefrontal cortex of schizophrenia subjects: analysis of the National Brain Databank microarray collection. Schizophr Res. 2008;98:111–117. doi: 10.1016/j.schres.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ibi D, Gonzalez-Maeso J. Epigenetic signaling in schizophrenia. Cell Signal. 2015;27:2131–2136. doi: 10.1016/j.cellsig.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holloway T, Gonzalez-Maeso J. Epigenetic mechanisms of serotonin signaling. ACS Chem Neurosci. 2015;6:1099–1109. doi: 10.1021/acschemneuro.5b00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akbarian S, Ruehl MG, Bliven E, Luiz LA, Peranelli AC, Baker SP, Roberts RC, Bunney WE, Jr, et al. Chromatin alterations associated with down-regulated metabolic gene expression in the prefrontal cortex of subjects with schizophrenia. Arch Gen Psychiatry. 2005;62:829–840. doi: 10.1001/archpsyc.62.8.829. [DOI] [PubMed] [Google Scholar]
- 36.Abdolmaleky HM, Zhou JR, Thiagalingam S. An update on the epigenetics of psychotic diseases and autism. Epigenomics. 2015;7:427–449. doi: 10.2217/epi.14.85. [DOI] [PubMed] [Google Scholar]
- 37.Kurita M, Holloway T, Gonzalez-Maeso J. HDAC2 as a new target to improve schizophrenia treatment. Expert Rev Neurother. 2013;13:1–3. doi: 10.1586/ern.12.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goghari VM, Sponheim SR, Macdonald AW., III The functional neuroanatomy of symptom dimensions in schizophrenia: a qualitative and quantitative review of a persistent question. Neurosci Biobehav Rev. 2010;34:468–486. doi: 10.1016/j.neubiorev.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Bartolomeis A, Buonaguro EF, Iasevoli F. Serotonin-glutamate and serotonin-dopamine reciprocal interactions as putative molecular targets for novel antipsychotic treatments: from receptor heterodimers to postsynaptic scaffolding and effector proteins. Psychopharmacology. 2013;225:1–19. doi: 10.1007/s00213-012-2921-8. [DOI] [PubMed] [Google Scholar]
- 40.Polter AM, Li X. Glycogen synthase kinase-3 is an intermediate modulator of serotonin neurotransmission. Front Mol Neurosci. 2011;4:31. doi: 10.3389/fnmol.2011.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svenningsson P, Tzavara ET, Carruthers R, Rachleff I, Wattler S, Nehls M, McKinzie DL, Fienberg AA, et al. Diverse psychotomimetics act through a common signaling pathway. Science. 2003;302:1412–1415. doi: 10.1126/science.1089681. [DOI] [PubMed] [Google Scholar]
- 42.Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol Lett. 2006;28:1601–1613. doi: 10.1007/s10529-006-9127-2. [DOI] [PubMed] [Google Scholar]
- 43.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, et al. The MIQE guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 44.Huang HS, Matevossian A, Jiang Y, Akbarian S. Chromatin immunoprecipitation in postmortem brain. J Neurosci Methods. 2006;156:284–292. doi: 10.1016/j.jneumeth.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 45.Wood CM, Nicolas CS, Choi SL, Roman E, Nylander I, Fernandez-Teruel A, Kiianmaa K, Bienkowski P, et al. Prevalence and influence of cys407* Grm2 mutation in Hannover-derived Wistar rats: mGlu2 receptor loss links to alcohol intake, risk taking and emotional behaviour. Neuropharmacology pii. 2016;(16):30092–30092. doi: 10.1016/j.neuropharm. pii S0028–3908. [DOI] [PubMed] [Google Scholar]
- 46.Lopez-Gimenez JF, Mengod G, Palacios JM, Vilaro MT. Selective visualization of rat brain 5-HT2A receptors by autoradiography with [3H]MDL 100,907. Naunyn Schmiedeberg’s Arch Pharmacol. 1997;356:446–454. doi: 10.1007/pl00005075. [DOI] [PubMed] [Google Scholar]
- 47.Erritzoe D, Frokjaer VG, Haugbol S, Marner L, Svarer C, Holst K, Baare WF, Rasmussen PM, et al. Brain serotonin 2A receptor binding: relations to body mass index, tobacco and alcohol use. NeuroImage. 2009;46:23–30. doi: 10.1016/j.neuroimage.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 48.Wright RA, Johnson BG, Zhang C, Salhoff C, Kingston AE, Calligaro DO, Monn JA, Schoepp DD, et al. CNS distribution of metabotropic glutamate 2 and 3 receptors: transgenic mice and [(3)H]LY459477 autoradiography. Neuropharmacology. 2013;66:89–98. doi: 10.1016/j.neuropharm.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 49.Delille HK, Mezler M, Marek GJ. The two faces of the pharmacological interaction of mGlu2 and 5-HT(2)A—relevance of receptor heterocomplexes and interaction through functional brain pathways. Neuropharmacology. 2013;70:296–305. doi: 10.1016/j.neuropharm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Levitz J, Habrian C, Bharill S, Fu Z, Vafabakhsh R, Isacoff EY. Mechanism of assembly and cooperativity of homomeric and heteromeric metabotropic glutamate receptors. Neuron. 2016;92:143–159. doi: 10.1016/j.neuron.2016.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molinaro G, Traficante A, Riozzi B, Di ML, Curto M, Pallottino S, Nicoletti F, Bruno V, et al. Activation of mGlu2/3 metabotropic glutamate receptors negatively regulates the stimulation of inositol phospholipid hydrolysis mediated by 5-hydroxytryptamine2A serotonin receptors in the frontal cortex of living mice. Mol Pharmacol. 2009;76:379–387. doi: 10.1124/mol.109.056580. [DOI] [PubMed] [Google Scholar]
- 52.Moreno JL, Holloway T, Albizu L, Sealfon SC, Gonzalez-Maeso J. Metabotropic glutamate mGlu2 receptor is necessary for the pharmacological and behavioral effects induced by hallucinogenic 5-HT2A receptor agonists. Neurosci Lett. 2011;493:76–79. doi: 10.1016/j.neulet.2011.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grueter BA, Winder DG. Group II and III metabotropic glutamate receptors suppress excitatory synaptic transmission in the dorsolateral bed nucleus of the stria terminalis. Neuropsychopharmacology. 2005;30:1302–1311. doi: 10.1038/sj.npp.1300672. [DOI] [PubMed] [Google Scholar]
- 54.Santini MA, Ratner C, Aznar S, Klein AB, Knudsen GM, Mikkelsen JD. Enhanced prefrontal serotonin 2A receptor signaling in the subchronic phencyclidine mouse model of schizophrenia. J Neurosci Res. 2013;91:634–641. doi: 10.1002/jnr.23198. [DOI] [PubMed] [Google Scholar]
- 55.Santini MA, Balu DT, Puhl MD, Hill-Smith TE, Berg AR, Lucki I, Mikkelsen JD, Coyle JT. D-serine deficiency attenuates the behavioral and cellular effects induced by the hallucinogenic 5-HT(2A) receptor agonist DOI. Behav Brain Res. 2014;259:242–246. doi: 10.1016/j.bbr.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Testa CM, Friberg IK, Weiss SW, Standaert DG. Immunohistochemical localization of metabotropic glutamate receptors mGluR1a and mGluR2/3 in the rat basal ganglia. J Comp Neurol. 1998;390:5–19. [PubMed] [Google Scholar]
- 57.Carlsson M, Carlsson A. Interactions between glutamatergic and monoaminergic systems within the basal ganglia—implications for schizophrenia and Parkinson’s disease. Trends Neurosci. 1990;13:272–276. doi: 10.1016/0166-2236(90)90108-m. [DOI] [PubMed] [Google Scholar]
- 58.Carlsson M, Carlsson A. Schizophrenia: a subcortical neurotransmitter imbalance syndrome? Schizophr Bull. 1990;16:425–432. doi: 10.1093/schbul/16.3.425. [DOI] [PubMed] [Google Scholar]
- 59.Charron A, Hage CE, Servonnet A, Samaha AN. 5-HT2 receptors modulate the expression of antipsychotic-induced dopamine supersensitivity. Eur Neuropsychopharmacol. 2015;25:2381–2393. doi: 10.1016/j.euroneuro.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 60.Borroto-Escuela DO, Romero-Fernandez W, Narvaez M, Oflijan J, Agnati LF, Fuxe K. Hallucinogenic 5-HT2AR agonists LSD and DOI enhance dopamine D2R protomer recognition and signaling of D2-5-HT2A heteroreceptor complexes. Biochem Biophys Res Commun. 2014;443:278–284. doi: 10.1016/j.bbrc.2013.11.104. [DOI] [PubMed] [Google Scholar]
- 61.Borroto-Escuela DO, Pintsuk J, Schafer T, Friedland K, Ferraro L, Tanganelli S, Liu F, Fuxe K. Multiple D2 heteroreceptor complexes: new targets for treatment of schizophrenia. Ther Adv Psychopharmacol. 2016;6:77–94. doi: 10.1177/2045125316637570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moreno JL, Kurita M, Holloway T, Lopez J, Cadagan R, Martinez-Sobrido L, Garcia-Sastre A, Gonzalez-Maeso J. Maternal influenza viral infection causes schizophrenia-like alterations of 5-HT(2)A and mGlu(2) receptors in the adult offspring. J Neurosci. 2011;31:1863–1872. doi: 10.1523/JNEUROSCI.4230-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 64.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 65.Chen Y, Damayanti NP, Irudayaraj J, Dunn K, Zhou FC. Diversity of two forms of DNA methylation in the brain. Front Genet. 2014;5:46. doi: 10.3389/fgene.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu H, Chen Y, Lv J, Liu H, Zhu R, Su J, Liu X, Zhang Y, et al. Quantitative epigenetic co-variation in CpG islands and co-regulation of developmental genes. Sci Rep. 2013;3:2576. doi: 10.1038/srep02576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paquette AG, Marsit CJ. The developmental basis of epigenetic regulation of HTR2A and psychiatric outcomes. J Cell Biochem. 2014;115:2065–2072. doi: 10.1002/jcb.24883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paquette AG, Lesseur C, Armstrong DA, Koestler DC, Appleton AA, Lester BM, Marsit CJ. Placental HTR2A methylation is associated with infant neurobehavioral outcomes. Epigenetics. 2013;8:796–801. doi: 10.4161/epi.25358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sutton LP, Rushlow WJ. Regulation of Akt and Wnt signaling by the group II metabotropic glutamate receptor antagonist LY341495 and agonist LY379268. J Neurochem. 2011;117:973–983. doi: 10.1111/j.1471-4159.2011.07268.x. [DOI] [PubMed] [Google Scholar]
- 70.Jope RS, Johnson GV. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem Sci. 2004;29:95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 71.Sutton LP, Rushlow WJ. The dopamine D2 receptor regulates Akt and GSK-3 via Dvl-3. Int J Neuropsychopharmacol. 2012;15:965–979. doi: 10.1017/S146114571100109X. [DOI] [PubMed] [Google Scholar]
- 72.Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 73.Pandey GN, Rizavi HS, Tripathi M, Ren X. Region-specific dysregulation of glycogen synthase kinase-3beta and beta-catenin in the postmortem brains of subjects with bipolar disorder and schizophrenia. Bipolar Disord. 2015;17:160–171. doi: 10.1111/bdi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kozlovsky N, Belmaker RH, Agam G. GSK-3 and the neurodevelopmental hypothesis of schizophrenia. Eur Neuropsychopharmacol. 2002;12:13–25. doi: 10.1016/s0924-977x(01)00131-6. [DOI] [PubMed] [Google Scholar]
- 75.Ochs SM, Dorostkar MM, Aramuni G, Schon C, Filser S, Poschl J, Kremer A, Van LF, et al. Loss of neuronal GSK3beta reduces dendritic spine stability and attenuates excitatory synaptic transmission via beta-catenin. Mol Psychiatry. 2015;20:482–489. doi: 10.1038/mp.2014.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maguschak KA, Ressler KJ. The dynamic role of beta-catenin in synaptic plasticity. Neuropharmacology. 2012;62:78–88. doi: 10.1016/j.neuropharm.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mills F, Bartlett TE, Dissing-Olesen L, Wisniewska MB, Kuznicki J, Macvicar BA, Wang YT, Bamji SX. Cognitive flexibility and long-term depression (LTD) are impaired following beta-catenin stabilization in vivo. Proc Natl Acad Sci U S A. 2014;111:8631–8636. doi: 10.1073/pnas.1404670111. [DOI] [PMC free article] [PubMed] [Google Scholar]


