Abstract
Targeting the tumor microenvironment (TME) through which cancer stem cells (CSCs) crosstalk for cancer initiation and progression, may open up new treatments different from those centered on the original hallmarks of cancer genetics thereby implying a new approach for suppression of TME-driven activation of CSCs. Cancer is dynamic, heterogeneous, evolving with the TME and can be influenced by tissue-specific elasticity. One of the mediators and modulators of the crosstalk between CSCs and mechanical forces is miRNA, which can be developmentally regulated, in a tissue- and cell-specific manner. Here, based on our previous data, we provide a framework through which such gene expression changes in response to external mechanical forces can be understood during cancer progression. Recognizing the ways mechanical forces regulate and affect intracellular signals with applications in cancer stem cell biology. Such TME-targeted pathways shed new light on strategies for attacking cancer stem cells with fewer side effects than traditional gene-based treatments for cancer, requiring a “watch-and-wait” approach. We attempt to address both normal brain microenvironment and tumor microenvironment as both works together, intertwining in pathology and physiology – a balance that needs to be maintained for the “watch-and-wait” approach to cancer. Thus, this review connected the subjects of tissue elasticity, tumor microenvironment, epigenetic of miRNAs, and stem-cell biology that are very relevant in cancer research and therapy. It attempts to unify apparently separate entities in a complex biological web, network, and system in a realistic and practical manner, i.e., to bridge basic research with clinical application.
Keywords: Tissue elasticity, miRNAs, gene expression, cancer stem cells
INTRODUCTION
Cancer research has traditionally centered around tumor development exploring genetics behind unhinged malignant cells [1]. This simplistic view has generated a remarkable database of information, leading to the creation of the cancer genome project [2]. Conventional cancer therapies have been rooted in the concept of genetic alternations or around faulty DNA repair [1]. Failure to eradicate all cancer cells during treatments prompted the emergence of non-traditional cancer gene based approaches. A striking advancement has occurred over the course of recent years as the cancer-gene-only approach has failed in clinical trials to affect survival for cancer patients. The discovery of the tumor microenvironment (TME), by which cancer cells crosstalk for cancer initiation and progression, implies the possibility of new treatments that move beyond the paradigm of cancer genetics, which focuses on cancer cells only.
TME consists of cancer stem cells (CSCs), tumor cells, immune cells, endothelium, fibroblasts, extracellular matrix (ECM), chemical factors, and the physical factors including tissue elasticity. Physical factors such as mechanical forces, both external and internal, can affect cell behavior. Forms of mechanical forces such as tissue elasticity support cells within the local cellular environment. Tissue elasticity conveys mechanical resistance to the cell’s cytoskeleton, the inner framework of protein tubes that give cellular shape, which in turn formats signals to the nucleus affecting gene expression. These findings indicate a force based mechanism by which cells sense their environment; however, the molecules that mediate this process remain undefined. The question remains as to whether the elasticity itself regulates gene expression through mediator molecules, or if instead the forces trigger a chemical signaling pathway within the cell. Cancer stem cells secrete biomolecules (proteins, DNA fragments, cytokines/chemokines, and microRNAs)), which may serve as biomarkers for sensitive non-invasive diagnostics of cancer, with a potential for clinical use in screening and monitoring of cancer progression and response to therapeutics. MicroRNAs (miRNAs) are developmentally regulated and are tissue specific. Altered miRNA expression is frequently observed in human cancers, though the underlying regulatory mechanism driving this change in expression is largely unknown. Our previous findings show that tissue-specific elasticity-dependent changes during cancer progression regulate genome-scale gene expression via miRNAs.
Tumours represent mind-boggling ecologies containing various cell types, and that tumor development and metastasis require a favorable “tumor microenvironment” (TME). Cancer (malignant) treatments often fail because a hostile (non-malignant) TME, comprised of microvasculature (endothelial cells, pericytes, angioblasts and endothelial forebear cells), fibroblasts, mesenchymal stem cells and other stromal components, and pro-inflammatory leukocytes, as well as physical scaffold composed of extracellular and intracellular matrix structure), has developed [3]. Malignant cancer cells do not live in isolation, not on Petri dishes (conventional study matrix for drug screening), not in mouse models, but reside in a complex TME, constantly recruiting non-malignant cells and non-cellular components. Identification of genetic variants associated with adaptations to regional living conditions and dietary practices [4] indicates that changing the tissue microenvironment causes genomescale changes. Further understanding of the role of tumor-TME interplay in acquired resistance to conventional cancer therapies (surgery, radiotherapy and chemotherapy) and “targeted” anticancer agents (i.e., antiangiogenic therapy) may open up a new avenue to target cancer stem cell niche within TME. In general, a normal stem cell niche is not yet well defined [5]. Less is known about cancer stem cells (CSCs) and their role (niche) in TME. Some glimpse that stemness of colon cancer cells is maintained by the cancer microenvironment [6, 7]. Discovery of endothelial cells interacted closely with self-renewing brain tumor stem cells led to define a perivascular niche for brain tumor stem cells [8]. In reality, stem cells dynamically shuttle between stem cells niche and progeny in the body as stem cells are preserved for tissue injury repair, and are mobilized to injury as needed [9].
The local environment cues changes in genomic profiling of transcriptional [10] and epigenetic [11] states and feeds back to regulatory intracellular circuitry, epigenetic memory, cell type fluidity, and reuse of regulatory modules, allowing the body to achieve and maintain appropriate responses to a changing environment [12]. The changing environment includes the surrounding extracellular matrix (ECM) effecting changes in elasticity. Figure 1, as per previous description [13]. We found that the extracellular matrix (ECM) forms a mechanical scaffold with a certain degree of elasticity (brain like), inducing global changes in gene expression via changes in this mechanical property [14]. Specifically, of note, majority of laboratory grow the cells on tissues-treated PS, rather than on standard plastic - Please note the distinction between “tissues-treated polystyrene and standard plastic are versus our “tissue-elasticity based culture system,” which is with 0.1 kPa (brain microenvironment) of elasticity, while both tissues-treated PS and standard plastic have 100 kPa of elasticity [14]. Studies of the TME have led to immunotherapy [15] (e.g., adoptive immunotherapy CAR T cells [16]), cancer vaccines [17, 18], and immune-based cancer prevention [19]. TME-associated cancer cells are predicted to originate from normal cells, and then convert to malignancy upon cancer initiation, by a mechanism that is yet to be elucidated. How does the mechanical scaffold (elasticity) affect a TME-associated tumor or normal cell, and vice versa? Emerging evidence shows that tissue elasticity and cells intertwine and evolve through time. In a multiethnic population-based sample, Blacks and Hispanics had higher proximal aortic stiffness compared with Whites, independent of blood pressure and relevant risk factors [20]. The underlying mechanisms of how forces influence nuclear events leading to gene regulation are beginning to surface in recent publications.
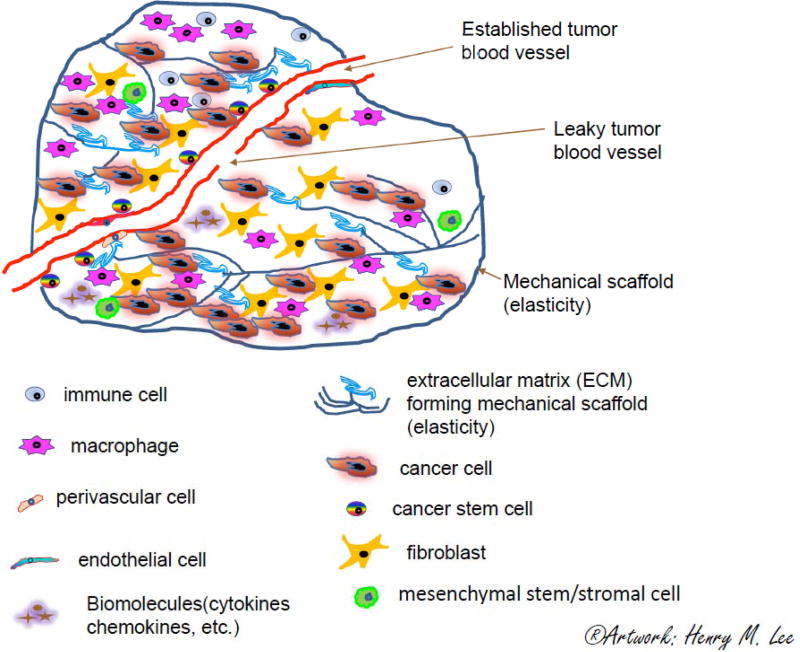
Fig 1. Schematic diagram of a tumor microenvironment.
The tumor ecosystem consists of different cell types (tumor cells, non-tumor cells), chemical and physical factors. Tumor growth and metastasis requires an appropriate support structure, known as the tumor microenvironment (TME). A typical TME includes microvasculature (endothelial cells, pericytes, angioblasts and endothelial progenitor cells), fibroblasts, mesenchymal stem/stromal cell and other stromal elements, pro-inflammatory leukocytes (immune cells, lymphocytes, tumor associated macrophages), and a surrounding extracellular matrix (ECM) that forms a mechanical scaffold and defines the tissue elasticity. Cancer progression involves tissue elasticity changes through time. Tumor cells metastasize through the leaky tumor vasculature and migrate through blood vessels to other organs. Disruption therapy may break down the physical barrier (ECM mechanical scaffold), leading to imbalance of the cancer ecosystem. Certain stages of cancer should be managed with a “watch-and-wait” approach to balance risks and benefits by stabilizing and regulating the TME through lifestyle and risk-factor management (Artwork: Henry M. Lee).
Collaboration between Ning Wang’s and Andrew Belmont’s laboratories yielded answers to cellular mechanosensing from the outside of the cell into the nucleus. They applied mechanical forces generated from Arg-Gly-Aspcoated magnetic beads to cells, which are sensed by cellular membrane-integrins attached through the tensed actin cytoskeleton, transmitting to the LINC complex and then through lamina-chromatin interactions to directly stretch chromatin and upregulate transcription [21]. Thus, the tension exerted by the beads transmits to chromatin [22]. This chromatin stretch increased transcription of the genes in the stretched regions of the chromosome, perhaps answering a phenomenon known as chromosome decondensation, which correlates with increased gene expression. This force-induced chromatin deformation and transcription process can be visualized through a green fluorescent protein (GFP)-tagged bacterial-chromosome dihydrofolate reductase (DHFR) transgene in a living cell. Utilizing three-dimensional magnetic twisting cytometry to apply local stresses on the cell surface to disrupt filamentous actin or by inhibiting actomyosin contraction alters force-induced DHFR transcription, whereas activating endogenous cellular contraction upregulates force-induced DHFR transcription. This force tension mechanism resonates with a previous report that tissue-level elasticity determines the fate of naïve mesenchymal stem cells (MSCs): “Soft matrices that mimic brain are neurogenic, stiffer matrices that mimic muscle are myogenic, and comparatively rigid matrices that mimic collagenous bone prove osteogenic” [10]. Surprisingly, matrix elasticity alone can fix the lineage of MSCs, underlining the physical effects of the in vivo microenvironment on MSCs, and may allow for therapeutic uses of mechanical force. All of these findings indicate a mechanically based mechanism by which cells sense the mechanical forces of their environment; however, the molecules that mediate this cross-talk between cells and mechanical forces remain elusive.
Recent discoveries show that cancer initiation progresses through intercellular communication between normal cells (non-malignant cells) and malignant cells via functional molecules, including proteins, mRNA and microRNAs (miRNAs) [23]. MicroRNAs are small non-coding RNAs that play a major role in posttranscriptional gene regulation in diverse biological processes. They function as both tumor suppressors and promoters of many aspects of the autonomous behavior of cancer cells [24]. Theoretically, dysfunction in the gene regulatory networks of cancer cells is one of the major driving forces for alterations of ostensibly normal surrounding cells. In this context, the core targets of miRNAs, termed miRNA regulons, are currently being expanded to include various modulators of the TME [25]. Recent advances have highlighted two important roles played by miRNAs in the evolution of TMEs: miRNAs in tumor cells transform the microenvironment via non-cell-autonomous mechanisms, and miRNAs in neighboring cells stabilize cancer hallmark traits [25, 26]. MicroRNAs have been shown to serve as a bridge between breast cancer cells and their neighboring cells [27] through the membrane-derived microRNAs-containing vesicles for exosome-mediated intercellular communication within the tumor microenvironment in breast cancers [28], or acting as delivery vehicles for pancreatic cancer [29], or serving as immunotherapeutic targets in colorectal carcinoma [30]. The expression patterns of miRNAs, which normally govern by negatively regulating the expression of protein-coding genes through either translational repression or RNA degradation, are frequently observed in human cancers, though the underlying regulatory mechanism is largely unknown. Furthermore, miRNAs are developmentally regulated and/or tissue specific, tissue plasticity changes bridge cancer progression to the TME, which manifests in miRNA-mediated gene expression. Here, we discuss the challenges or bottlenecks in the field, including: 1) determining which species of miRNA in cancer stem cells (CSCs) respond to tissue mechanics; 2) what are the mechanisms of miRNA-mediated gene expression that affect tissue elasticity in regulation of CSC growth; and 3) implications for clinical applications. We postulate that we can design better therapeutics if we can determine the role of nonmalignant cells and the role of malignant cells as well as how these two types of cells cross-talk in TME. This can help understand why breakdown of TME-scaffold helps tumor cells metastasize. Facilitating TME-scaffold repair may yield a new therapy for cancer such as a watch-and-wait approach that lets patients avoid a therapy’s side effect until they need treatment.
WHICH SPECIES OF miRNA IN CSCs RESPOND TO TISSUE MECHANICS?
CSCs are self-renewing cells that are thought to be the cause of tumorigenesis and cancer metastasis. The role of miRNAs includes directing and regulating the gene expression of the CSCs. Aberrant expression of miRNA is commonly observed in many types of tumors [31, 32]. One such miRNA, let-7, was found to help control the self-renewal and tumorigenicity in breast, lung, and many other types of cancers [33, 34]. Let-7 was previously found to be important during the regulation of embryonic development; it acts like a tumor suppressor, with lowered expression of let-7 correlating with poor survival and less differentiated tumor [32, 35]. Cancer stem cells may express miRNA profiles similar to those seen during the embryonic stages to gain the traits associated with stem cells. Some miRNAs are abundant in a few types of tumors but not in others. One explanation is that some miRNAs, like let-7, are needed to maintain the “stemness” of cancer stem cells, while other miRNAs are responsible for adapting to and maintaining the extracellular environment or taking advantage of the unique traits of the surrounding tissue type in order to survive. MicroRNAs in the miR-181 family, which are overexpressed in breast, pancreas, and prostate cancers, may promote tumorigenesis through their role in regulating MYCN gene (N-Myc) [36]. However, low miR-181 expression can also be a marker for poor prognosis in chronic lymphocytic leukemia, possibly because it may act via regulatory paths that are open because of the 11q chromosome deletion commonly found in this type of cancer [32]. Lin’s laboratory shows that elimination of microRNA miR-34a preserves embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) to restore the potential to retain both embryonic and extra-embryonic lineages [37]. It’s worth looking at CSCs to see if it is relevant to cancer.
Colorectal cancer (CRC) patients’ blood plasma prior to surgery contains elevated levels of 13 epithelial cell adhesion molecule extracellular vesicle EpCAM(+)-EV miRNAs compared with healthy individuals. Surgical tumor removal downregulates the plasma levels of 8 of these miRNAs (miR-16-5p, miR-23a-3p, miR-23b-3p, miR-27a-3p, miR-27b-3p, miR-30b-5p, miR-30c-5p and miR-222-3p) [38]. Both miR-205 and miR-373 contribute to the aggressive phenotype of MAC in CRC - Mucinous adenocarcinoma (MAC) is a distinct subtype of colorectal cancer (CRC) [39]. Consistent with this finding, miRNAs plays an essential role in crosstalk between the TME and cancer cells [40]. Blockage of such crosstalk has emerged as a new therapeutic target. Multiple myeloma (MM) drug resistance (DR) is due to the bone marrow microenvironment in both soluble factor-mediated drug resistance (SFM-DR) and cell adhesion-mediated drug resistance (CAM-DR); both are mediated by miRNAs, exosomes, and cancer-associated fibroblasts [41]. The bone marrow microenvironment niche regulates miR-221/222 in Acute Lymphoblastic Leukemia (ALL) [42], suggesting that therapeutic modulation of these miRNAs may affect cancer progression.
Specifically, which species of miRNA in CSCs respond to tissue mechanics? The species of miRNAs appear to be tissue-elasticity specific. Recent report shows that increased matrix stiffness of tumour-promoting tissue mechanic induces miR-18a to reduce levels of the tumor suppressor phosphatase and tensin homolog (PTEN), both directly and indirectly by decreasing levels of homeobox A9 (HOXA9) [43]. More importantly, extracellular matrix stiffness correlates with miR-18a expression in human breast tumor biopsies: miR-18a expression is highest in basal-like breast cancers in which PTEN and HOXA9 levels are lowest, thereby leading to predict that high miR-18a expression predicts poor prognosis in patients with luminal breast cancers [13]. Schwentner and colleagues mentioned the gene expression is affected by DNA methylation induced by external mechanical forces or TME, and may also affect the expression of miRNA through the DNA methylation pathway. They further found that miR-17–92 connects a node to TGF /BMP pathway, resulting in the activation of the stemness regulatory transcriptional repressors ID1 and ID3, in Ewing Sarcoma [44]. Reversely, concomitant inhibition of miR-133a and miR-696 accelerates differentiation of muscle stem cells, elevated the metabolic coactivator PGC-1α, and increases the contractile force in 3D engineered human skeletal muscle bundles; however, the underlined mechanism remains to be elucidated [45]. Tension force-induced bone formation in periodontal ligament cells shows 818 mRNAs and 32 miRNAs including core microRNAs (miR-195-5p, miR-424-5p, miR-1297, miR-3607-5p, miR-145-5p, miR-4328, and miR-224-5p) responsible for tension force-induced bone formation [46]. All of these suggest that miRNAs mediate trans-mechanical transduction of forces to gene expression in a tissue specific manner. Given the nature of tissue specific miRNAs, some proposed to design therapeutic cytotoxic genes specific for small interfering RNA (siRNA) and micro RNA (miRNA)] with co-regulated LHRH receptors, which are selective overexpression on human tumors compared to normal tissues [47].
WHAT MECHANISMS OF miRNA-MEDIATED GENE EXPRESSION AFFECT TISSUE ELASTICITY IN REGULATION OF CSC GROWTH?
Cancer stem cells release cytokines/chemokines, small membranous extracellular vesicles (EVs), and miRNAs into their microenvironments and into the circulatory system [48]. Not much is known about the mechanism by which miRNAs regulate gene expression in cancer progression as a response to microenvironmental cues. Evidence shows that miRNAs play a variety of roles in development and biological processes and they have been reported to be important in some cancers [49]. They were initially thought to silence gene expression only via translation repression, with the implication that the transcripts are not degraded during repression, but recent studies have determined that miRNAs may primarily silence gene expression by destabilizing and degrading mRNA transcripts [50], leading to reduced protein output [51]. Silencing of gene expression by miRNAs can be tissue- and cell-specific [52]. miRNAs that positively regulate gene expression could act as small activating mRNAs (samRNAs) [53]. Such microRNA regulons function in a feedback loop, responding to the TME in cancer cells [54] and being modulated by crosstalk between cancer cells and the surrounding microenvironment. This crosstalk evolves with tumor formation, metastasis and refractoriness to therapy, the last of which is an area that is actively under investigation.
Increased matrix stiffness in human and mouse tissue induces miR-18a to reduce levels of the tumor suppressor phosphatase and tensin homolog (PTEN) and to decrease levels of homeobox A9 (HOXA9), thereby driving tumor progression through integrin activation of β-catenin and MYC [43]. Clinical evidence shows that upregulation of miR-18a is associated with downregulation of PTEN and HOXA9. Thus, tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. MiR-18a, PTEN, and HOXA9 are used as biomarkers to predict prognosis in patients.
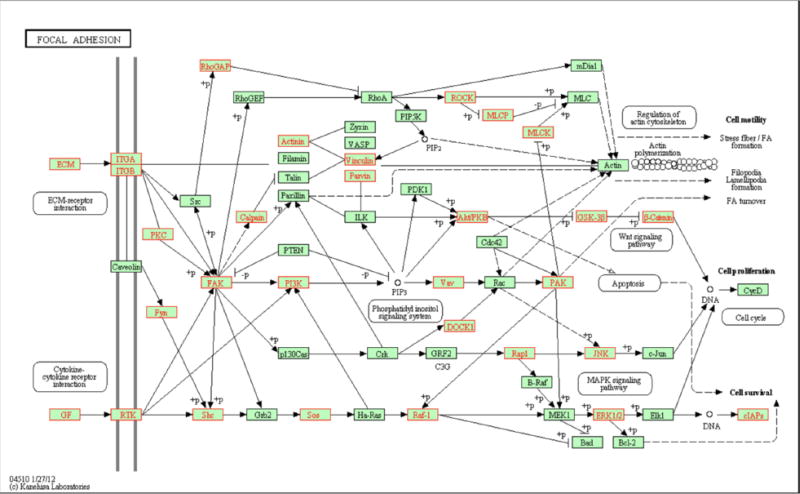
All of these miRNA studies prompted us to hypothesize that tissue mechanics-driven changes in miRNA expression can lead to global genomic-scale changes in cancer cells. We began testing this hypothesis by examining the effects of the ECM on gene expression in tumor cells, in order to uncover possible mechanisms by which tumor cells grown on soft polyacrylamide (PAA, ∼0.1 kPa) hydrogel plates downregulate gene expression compared with cells grown on standard polystyrene (PS, ∼100 kPa) Petri dish plates (Fig. 2) [14]. As extracellular matrix (ECM) consists of extracellular molecules secreted by cells, ECM in multicellular organisms evolves multicellular structures, cell adhesion, and cell-to-cell communication. As CSCs cross-talk with tissue elasticity through focal adhesion pathway as shows previously [10], we focused on this pathway (Fig. 8) [14] and we show here on Fig. (2), which illustrates that functional gene enrichment of the KEGG focal adhesion pathway, which regulates ECM. Differentially regulated genes are mapped onto a KEGG pathway (hsa04510) that is found to be enriched between the culture conditions. Genes highlighted red indicate probes downregulated on PAA (FC < −2). Our results suggest that tissue elasticity, a key ECM microenvironmental factor, plays a role in miRNA expression and thereby regulates tumor gene expression as well as tumor growth. A whole-genome microarray was used to measure transcriptional expression and identify processes that are affected by the difference in the elasticity of the surface medium. The preferential downregulation of transcripts was identified when cells were grown on PAA. We argue that the silencing of these transcripts was caused by miRNAs. A change in the physiological behavior of cells and their response to cytokines occurred due to the change in tissue elasticity. We identified specifically that AKT signaling pathway is regulated by tissue elasticity through analysis of our microarray data. We then investigated AKT-regulated physiologically relevant signaling in cancer cells by gelatin zymography, which was used to measure cell invasiveness. We showed the downregulation of several ECM transcripts in tissue grown on PAA [14]. These studies may lead to the identification of novel miRNA prognostic biomarkers. We conclude by synthesizing our findings with a review of the literature and postulate a mechanism for elasticity-driven gene regulation.
Fig 2. Functional gene enrichment of the KEGG focal adhesion pathway.
Differentially regulated genes are mapped onto the KEGG pathway (hsa04510), which is found to be enriched between the culture conditions. Genes highlighted red indicate probes downregulated on PAA (FC < −2). No probes highlighted were upregulated on PAA (FC > 2) within this pathway (Adopted from Vu, 2015 [14]) (doi:10.1371/journal.pone.0120336.g008).
In their elucidation of how cells communicate within the TME, Lee and colleagues showed that MSC-derived exosomes, which carry mRNAs and miRNAs, mediate cell-to-cell communication within the TME to suppress angiogenesis by transferring anti-angiogenic molecules [55]. We set out to examine processes that involve interactions between cell receptors and the ECM because intuitively these proteins would likely be most affected by a change in tissue matrix elasticity [14]. Based on this criterion, we identified genes involved in the KEGG focal adhesion pathway enriched through transcriptome profiles (Fig. 2). Among the genes downregulated on PAA was EGFR, an oncogene known to have mutated splice variants. We examined the genomic regions that the probes complement and found that some of the probes mapped to intron regions and were downregulated on PAA. Upon further analysis, many ECM and cell receptor genes were identified to have a greater abundance of introns present when grown on PS but highly downregulated when grown on the softer PAA hydrogel. The downregulation of ECM and cell receptor transcripts on PAA may reflect processes that occur on softer ECM, and the finding of introns that are downregulated on PAA may be a physiological mechanism that cells use to regulate gene expression under natural cellular conditions. During the process of preparing total RNA for hybridization to the gene chip, mature mRNAs with a polyA tail are selected for by an oligo dT primer during cDNA synthesis. The process selects for mature mRNAs that have introns spliced out of the transcript. The expression of transcripts containing introns suggests that they are generated from the activation of alternative polyadenylation sites within introns rather than intronretention splicing events that would be subject to nonsense-mediated decay (NMD), but further studies would be needed to show their relative contributions.
Alternative polyadenylation of introns in coding sequences avoids NMD (nonsense-mediated decay) surveillance because the termination codons within the introns would be downstream from the 3’ most exon-exon junction of the transcript that is recognized during NMD. The exon junction complex is deposited 20–24 base pairs upstream from the exon-exon junction on the mRNA after splicing [56]. NMD is triggered when a premature termination codon is recognized by the ribosome before the exon junction complex [57]. The probability of a finding a termination codon within a randomly generated intron sequence would be 3/64 codons or once every ∼21.33 codons. The likelihood of finding at least one nonsense codon within an intron sequence is greater than 80% for intron sequences of a length of 100 nucleotides and approaches 100% for longer intron sequences [58]. The location of transcripts containing introns suggests the introns are at least several hundred codons in length, and would be likely to contain at least one stop codon. The polyadenylation mechanism is coupled to alternative splicing and to the transcriptional machinery, such that the splicing of an alternative splice site can determine if a polyadenylation site is cleaved, and the strength of a polyadenylation site can affect the usage of an alternative splice site [59, 60]. Alternative polyadenylation of introns has been associated with the activation of weak 5’ splice sites that allow the recognition of the alternative polyadenylation sites within introns [61]. Polyadenylation of introns creates a composite terminal exon with an alternative 3’ UTR which alters binding sites of regulatory protein and miRNA binding sites. Though reports vary, the average length of an intron is 3–4 times longer than the 3’ UTR length in humans [62, 63], which raises the likelihood that alternative polyadenylation sites within introns having an alternative 3’ UTR that would be more susceptible to downregulation by miRNA as was detected when the cells were grown on PAA. Alternative polyadenylation suggests modifications in RNA binding between the two conditions, and was among the processes that were identified to be functionally enriched in our study. RNA binding of proteins affects the selection of the 5’ and 3’ splice sites which are important in alternative splicing and polyadenylation.
We then searched for introns of downregulated genes on other ECM and receptor genes. Probes complimentary to introns were found in mRNA transcripts of primitive neuroectodermal tumor (PNET) cells grown on polystyrene (PS) but were highly downregulated when grown on PAA [14]. This was found to be true of both non-coding and coding genes. This pattern of expression points to activation of alternative polyadenylation of sites within introns of coding genes of PNET cells grown on PS. If the introns detected were formed by the intron retention mode of alternative splicing and flanked by exons, the transcript would likely contain a nonsense codon within its reading frame and promote NMD of the transcript. For non-coding genes the interpretation is less clear. For example, MEG3 is an imprinted non-coding gene containing several probes covering several introns. This gene was expressed on PS, suggesting selection by either retained introns or alternative poly (A) sites, since retained introns of non-coding genes are not subject to NMD. But since the introns were highly downregulated on PAA, NMD could not have caused the degradation of the transcripts in the non-coding mRNA. Non-coding RNAs are unique, in that they do not have a start codon or coding region and in humans are not subject to NMD (nonsense-mediated decay) surveillance, which requires the translation of the transcript to activate it. Intron retention introduces stop codons into the transcript. These stop codons activate NMD in coding regions. We observed intron sequences of coding sequences that are present on PS but absent on PAA, which are not likely to be formed from the retention of introns because of NMD acting on the intron sequences to degrade them. Alternative polyadenylation within introns of coding sequences would be more likely because it avoids NMD. Non-coding RNA [64] has been less well studied than coding regions [65], and the intron and exon regions [66] for many have not been very well characterized to determine the extent of alternative polyadenylation [67] or other types of 3’ end processing [68].
It is possible that the PNET (primitive neuroectodermal tumor) cells have a mechanism in place that avoids NMD (nonsense-mediated decay) of transcripts. NMD has been shown to be inhibited in a stressed TME that includes hypoxia, amino acid starvation, and reactive oxygen species [69]. The differences between the PAA and PS environments may be enough that the cells recognize PS as a stressed environment and inhibit NMD, but on the softer PAA hydrogel, which resembles the elasticity of their natural brain tissue environment, they behave as non-stressed and NMD becomes active. This could be one explanation for why intron sequences were found on cells grown on PS but not on PAA. Silencing of transcripts by NMD or miRNA is not mutually exclusive and both processes may be active at the same time. Examination of the non-coding gene, MEG3, suggests that alternative polyadenylation is active but further studies are required to show if NMD is contributing to these observations. It is clear that miRNA silences mRNA transcripts in our study, but we do not know if it is the only mechanism that degrades mRNA.
The alternative polyadenylation transcripts within intron regions are able to bypass NMD surveillance, which better support our results on PS for genes with a coding sequence. The loss of the alternatively polyadenylated transcripts on PAA can be explained by the increased miRNA bound to target sites on PAA. Introns are longer on average than the 3’ UTR: in humans, introns were reported to have a mean length of 3749 base pairs [63] compared to the 3’ UTR mean length of 988 base pairs [62]. Alternative polyadenylation could provide a longer 3’ UTR binding region and alternative binding sites that would be different from the wild type and would more likely be downregulated on PAA. The mechanisms that cause the activation of alternative polyadenylation sites in introns may also function in coding regions but this would require further studies to confirm. The alternative binding sites generated by alternative polyadenylation would help explain tissue-specific gene expression.
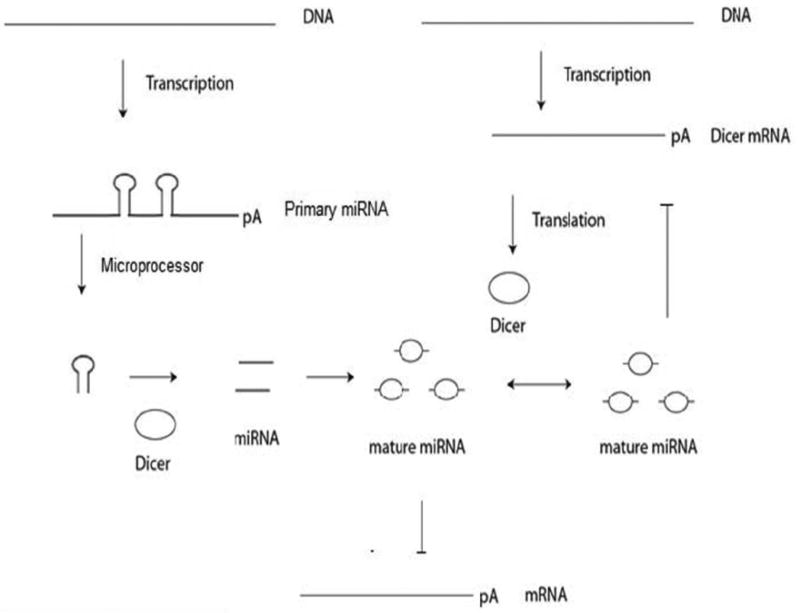
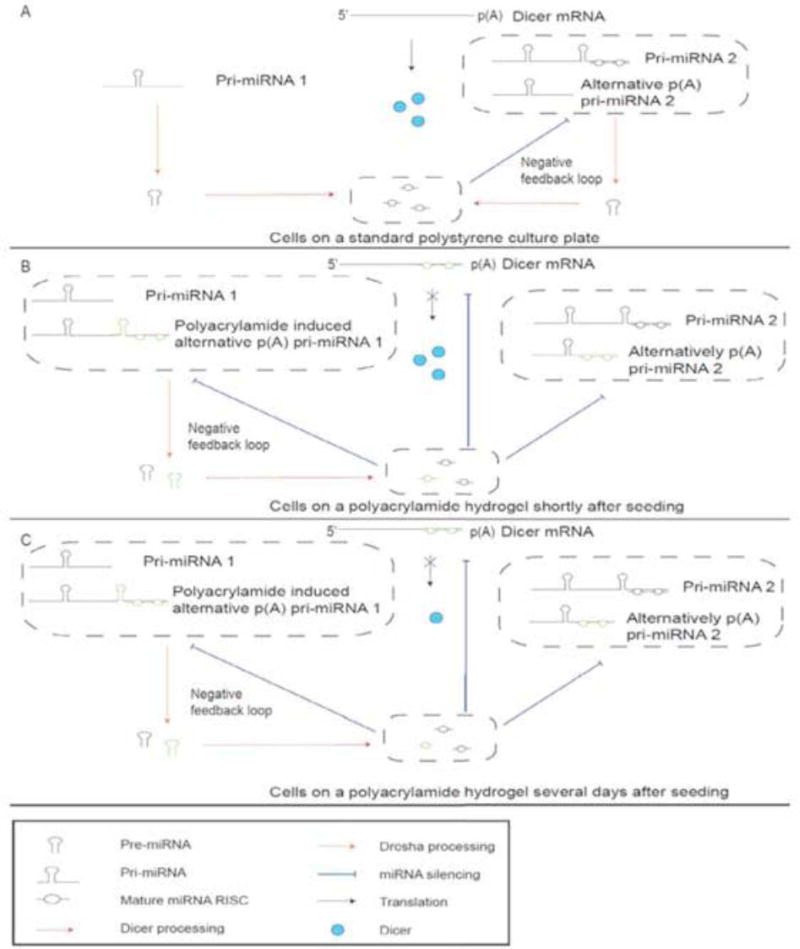
Taking our observations together with the literature on this topic, we postulate a model to incorporate Dicer negative feedback of miRNA and alternative polyadenylation for gene regulation (Fig. 3). Our model is supported by the previous study which found that miRNA production may be regulated by a negative feedback loop by a miRNA, let-7, that is able to down-regulate Dicer expression when it is at higher levels but is up-regulated when let-7 is reduced [70]. Indeed, Dicer may be subject to negative feedback regulation by a wider range of miRNAs in a spatiotemporal fashion when tumor cells are placed in different physical environments, such as PS or PAA. In our model (Fig. 3A), cells grown on PS produce miRNAs which are able to bind to their own primary miRNA or other primary miRNAs to form a negative feedback loop that regulates the production of miRNAs. When cells are initially seeded on PAA (Fig. 3B), PAA causes the induction and production of a set of alternative p(A) pri-miRNAs, increasing miRNA levels. The increased miRNA levels act to silence Dicer mRNA, other mRNAs, and pri-mRNAs that have target miRNA binding sites on their 3’ UTR. The miRNAs induced by PAA bind to other primary miRNAs to continue the negative feedback regulation of miRNA production. Silencing of Dicer mRNA does not immediately lower levels of Dicer protein (Fig. 3B), but over time, the degradation of Dicer proteins lower miRNA production (Fig. 3C). When miRNA production lowers, less miRNA binds to Dicer mRNA, which allows more Dicer protein to be produced. This in turn increases the amount of miRNA that can bind to Dicer mRNA. The negative feedback mechanism continues until equilibrium of Dicer and miRNA expression is reached. The regulation of Dicer mRNA by miRNA represents a second delayed negative feedback regulation of Dicer protein, Dicer mRNA, and miRNA. Alternative polyadenylation within introns present in cultures grown on PS and downregulated on PAA were likely only a subset of the possible polyadenylation events. There likely exists a set of alternative polyadenylated transcripts within introns and exons that avoid miRNA silencing when grown on PAA but has not been highlighted by the observed differential expression between the culture conditions. These alternative polyadenylated transcripts would likely have a shorter 3’ UTR that would provide less miRNA binding sites or have less miRNA binding sites that correspond to miRNA that is abundant, both of which would avoid silencing by miRNA.
Fig 3. Dicer negative feedback regulation by miRNA.
Primary miRNA is processed by Dicer protein, producing mature miRNA. The mature miRNA is able to bind to the 3’ UTR of Dicer mRNA that degrades the Dicer transcript. The downregulation of Dicer mRNA does not immediately lower Dicer protein levels. The level of Dicer protein is gradually reduced over time, which lowers the amount of miRNA produced, and as a result less miRNA is available to silence Dicer mRNA.
In support of our hypothesis, miR-1258 as a candidate micro RNA may directly target HPSE (Heparanase) and suppress BMBC (brain metastatic breast cancer) [71], which indicates miR-1258 directly regulate heparanase activity to break down TME-elasticity scaffold in breast tissue to metastasize to the brain. The levels of miR-1258 inversely correlate with heparanase expression, enzymatic activity, and cancer cell metastatic propensities, being lowest in highly aggressive BMBC cell variants compared with either nontumorigenic or nonmetastatic human mammary epithelial cells. MicroRNA mechanisms are linked to brain-metastatic breast cancer through heparanase control, and they offer a strong rationale to develop heparanase-based therapeutics for treatment of cancer patients with brain metastases [71].
However, clinical validation is needed for this assessment. About half of miRNA genes are located within introns of coding regions and the other half are within intergenic non-coding regions [24]. They are usually transcribed by RNA polymerase II to generate a primary miRNA (primiRNA) transcript that is capped and polyadenylated [26]. Primary miRNAs can contain multiple stem loop secondary structures that represent precursor miRNAs (pre-miRNAs), which are cleaved co-transcriptionally within the nucleus by the Microprocessor complex [25]. The pre-miRNA is exported to the cytoplasm where the RNA-induced silencing complex (RISC) is then able to select and associate to one of the strands to form a mature miRNA that is able to bind imperfectly to complementary sequences on the 3’ UTR of mRNA to downregulate gene expression [24]. In addition to regulating mRNA transcripts, primary miRNA transcripts, like all mRNAs, also contain a 3’ UTR with potential miRNA binding sites that can be targeted by miRNAs originating from other primary miRNA transcripts as well as the same primary miRNAs. These situations result in negative and auto-negative feedback loops, respectively, that influence the expression of miRNA. This feedback leads to potential cross-talk and represents a source of regulation between different miRNAs and the processes that they govern. One reported example of miRNA regulation is the negative feedback relationship between of DICER1 and an miRNA, let-7, which is able to downregulate DICER1 mRNA by binding to its 3’ UTR, but as DICER1 protein is reduced, in turn DICER1 leads to reduce production of let-7 allowing DICER1 transcripts to translate and, process new mature miRNA [70].
Another mechanism by which microRNAs are expressed and regulated may be through the alternative splicing and alternative polyadenylation of primary miRNA transcripts. Cells utilize alternative splicing and alternative polyadenylation as a mechanism to adapt to their extracellular environment and respond to external signals and cues to produce isoforms of mRNAs and proteins. When this principle is applied to primary miRNA transcripts, changes in a cell’s environment may result in changes which affect the miRNAs that are processed and change the dynamics of how they are regulated. A primary miRNA transcript may contain multiple splice sites and alternative polyadenylation sites. The specific miRNA processed from the transcript is likely dependent on the cell’s response to its extracellular environment; the quantity may change as the cell migrates to a different environment. The alternative splicing and polyadenylation of primary miRNA can affect the identity of miRNA that is able to bind to the 3’ UTR of these transcripts by creating alternative 3’ UTR regions. In addition, the new 3’ UTR may be longer or shorter, which would correlate with a greater or smaller number of potential miRNA binding sites, respectively. A longer 3’ UTR would have a higher chance of being repressed by existing miRNA, while a shorter 3’ UTR would be a mechanism for the primary miRNA transcript to bypass negative regulation by existing miRNA and begin processing new miRNA products which can later bind and downregulate other mRNA transcripts. These regulatory mechanisms, taken together, help mediate the behavior and response of the cell to its environment.
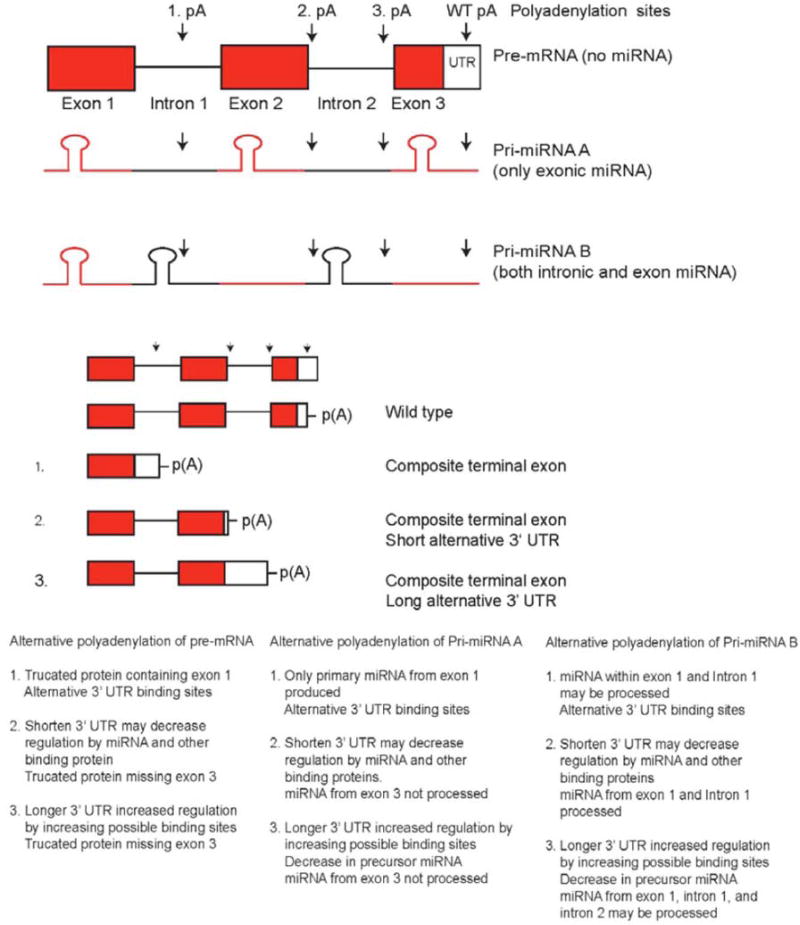
We expanded the idea of alternative polyadenylation of mRNA to pri-miRNA sequences, which can also be spliced and polyadenylated, and created a model where alternative polyadenylation of pri-miRNA can regulate miRNA levels (Fig. 4). About half of all miRNA are within intron regions of coding regions and the other half are within the intron and exon regions of non-coding RNA, as shown in genome-scale analyses [24]. Alternative polyadenylation of these sites can create alternative 3’ UTR that have different binding sites, which are recognized by particular tissue-specific miRNAs. Alternative polyadenylation that results in a shortened 3’ UTR would have fewer targets for miRNA whereas a lengthened 3’ UTR would have more possible targets to be regulated by miRNA. In our model, alternative polyadenylation of pri-miRNA within introns can act to prevent miRNA from being processed. Our model predicts that mRNA from PNET cells grown on PS are alternatively polyadenylated within introns of coding sequences, but when the cells are grown on PAA, newly processed miRNA that is induced by the PAA growth conditions binds to the 3’ UTR and degrades the mRNA more rapidly. The newly processed miRNA is dependent on the polyadenylation site of pri-miRNA. The increase in miRNA predicts that when cells are grown on PAA, polyadenylation occurs further downstream in primiRNA, which would result in more miRNA being produced. Pri-miRNA can also be downregulated by other miRNAs or by the miRNA that they code for, which can act as a negative feedback mechanism to downregulate their production of miRNA and affect the regulation of other mRNAs and miRNAs, with a cascading effect. The opposite may also occur, where alternative polyadenylation isoforms that avoid downregulation by miRNA are produced.
Fig 4. Model of miRNA regulation by alternative polyadenylation sites located within introns.
Alternative polyadenylation causes a change in possible miRNA binding sites and truncation of mRNA. Polyadenylation that results in a shortened 3’-UTR generally lowers regulation by miRNA, while a lengthened 3’-UTR increases regulation by miRNA. Alternative polyadenylation of pri-miRNA affects the ability of miRNA to be produced by preventing miRNA stem loop regions from being processed. Alternative polyadenylation sites within the 3’ UTR of a transcript can have shorter or longer 3’ UTRs. Alternative polyadenylation within intron sequences creates a transcript with an extended coding region that ends at the first stop codon within an intron and also creates a 3’ UTR. The alternative polyadenylation within an exon does not create a 3’-UTR region. (Adopted from Vu, 2015 [14] (doi:10.1371/journal.pone.0120336.g013)).
All Figs. (1–4) help us to understand Fig. (5), which illustrates three modes of action (mechanisms). We expanded this model (Fig. 4) to incorporate Dicer negative feedback of miRNA and alternative polyadenylation (Fig. 5). Figure 5, as newly conceived, with three models of tissue specific negative feedback and spatiotemporal regulation of Dicer and miRNA by alternative polyadenylation, may shed new light into miRNA mediated regulation of cancer tissue elasticity. It was previously reported that miRNA production may be regulated by a negative feedback loop involving let-7, a miRNA that is able to downregulate Dicer expression in human cancer cell lines, showing association of the tightly regulated, equilibrated state of Dicer and various miRNAs with cell growth and cell cycle phases
Fig 5. Model of tissue-specific negative feedback regulation of Dicer and miRNA by alternative polyadenylation.
(A) Cells on PS plates have microRNAs produced from pri-miRNAs that are able to silence other pri-miRNAs in a negative feedback loop.
(B) Cells seeded on PAA can induce the polyadenylation of transcripts containing miRNAs (green) that can silence Dicer mRNAs and other pri-miRNAs.
(C) Over time, the degradation of Dicer mRNA results in less Dicer protein. This constitutes a second delayed negative feedback mechanism, in which increases in miRNA levels decrease the amount of Dicer mRNA but do not result in an immediate reduction of Dicer protein. As Dicer proteins are degraded, miRNA production will begin to fall, which reduces miRNAs silencing of Dicer mRNA until an equilibrium level is reached.
To explain how alternative polyadenylation sites utilized in cultures grown on PS are downgraded in cultures grown on PAA, we show that on PAA, alternative polyadenylation of introns can induce a new set of miRNAs not present on PS. Some alternative polyadenylation sites that were originally present on PS would be silenced by the newly synthesized miRNA. Synthesis of new miRNA would also cause miRNA levels to rise and increase binding to 3’ UTR sites of Dicer and to the newly synthesized pri-miRNA, which would also be silenced if it had the same target sites on its 3’ UTR. The negative feedback of Dicer would prevent the continued production of miRNAs that are not negatively regulated by specific miRNAs. Over time, the change in miRNA levels due to their being degraded and not replaced would result in a change in the characteristics of the cell.
The softer PAA surface represents a cellular microenvironment that may be closer to native cell conditions, while the PS condition represents a microenvironment that is closer to that of stiffer tissue. The negative feedback mechanisms that control miRNA and Dicer may also control the expression of genes on different types of tissue. Cells grown on bone, muscle, and brain tissue may have a set of tissue-specific miRNAs that are induced in their microenvironment and regulated by Dicer and miRNA negative feedback mechanisms.
The process of tumor metastasis, the migration of cancer cells and stem cells from one tissue type to another, and the ability of a cell to adapt to its new environment are dependent on the ability of cells to recognize their ECM environment and adapt to it in order to survive, grow, and replicate. This study suggests that the mechanism that allows cells to adapt to their environment involves miRNA regulation of gene expression which is influenced by the elasticity of surface environment. When cells adapt to their environment, the cells’ behavior and responses to external stimuli are also changed. Global miRNA-driven changes in gene regulation represent a strategy by which tumors adapt to their local TME.
Strategies affecting the expression of specific miRNAs can control certain cancer subclones to become or remain dormant and ensure the success of a “watch-and-wait” approach to manage cancer. For example, miR-135b and miR-146b target the calcium-sensing receptor (CaSR) and reduce its expression in colorectal tumors, reducing the antiproliferative and prodifferentiating actions of calcium - CaSR mediates the antitumorigenic effects of calcium against colorectal cancer (CRC) [72].
CLINICAL RELEVANCE AND IMPLICATIONS
Reports show strategies to regulate miRNAs to suppress cancer progression. Some chemotherapy can regulate miRNA to modulate tumor growth, such as integrative miRNA/mRNA regulatory network mediates temozolomide resistance in glioblastoma [73]. Some reports advocate nutritional intervention as part of the “watch-and-wait” approach, because diverse dietary bioactive components, e.g., butyrate, folate, retinoids, curcumin, do exert their biological or clinical effects [74], in part, through modulation of miRNA expression [75]. It is important to test these agents at the levels of let-7 on the PS vs PAA hydrogel model. Nutrient management care of cancer is clinically relevant because such an experimental model may imply that nutrient management care is a part of the “watch-and-wait” practice. Similarly, alternative splicing may regulate gene expression at both mRNA and miRNA levels. It is plausible that epigenetics capture the true nature, the dynamic, complex, and transient nature, of cancer. Unlike genetics, epigenetics is currently difficult to monitor and to target. However, of all the epigenetic factors, miRNA could be one of the more useful and practical targets for the diagnosis, prognosis, and therapy of cancer. This realization is derived from the failure of single-mutation-target clinical trials, which implies the need for new comprehensive approaches to cancer, leading to take the TME into account. One aspect of miRNAs that is currently neglected in clinical diagnoses and cancer research is that miRNA expression can rapidly change when a cell changes from one environment to another, and one dimension of this change can be measured by taking into account differences in tissue elasticity. When a cancer stem cell migrates to another region of the body or spreads to a surrounding tissue, there may be a rapid change in miRNA expression once the cell establishes itself in its new location. Current protocols typically sample and profile tissues once the cells are established and a new tumor develops. The miRNA expression profiles of the original tumor and metastases would, of course, differ. But during the time frame when a cancer stem cell establishes a new tumor, the cell has to adapt and change according to its extracellular environment.
The utility of circulating tumour cells and plasma microRNA profiling is being recognized in first-line screening for cancer diagnosis and monitoring of cancer progression as indicated in multiple clinical trials across cancer types (Table 1). A keyword search of https://clinicaltrials.gov [accessed December 16, 2016] for “miRNA” AND “cancer” results in 158 ongoing clinical trials. The trials that take miRNA into consideration are investigating esophageal adenocarcinoma (ClinicalTrials.gov Identifier: NCT02812680), Breast Cancer (Newly Diagnosed Breast Cancer; Recurrent Breast Cancer) (ClinicalTrials.gov Identifier: NCT01722851), lung cancer (ClinicalTrials.gov Identifier: NCT02247453), the Identification of Bevacizumab response predictors in metastatic breast cancer (ClinicalTrials.gov Identifier: NCT01598285), neoadjuvant or adjuvant treatment for locally advanced & inflammatory breast cancer (ClinicalTrials.gov Identifier: NCT01231386), prostate cancer (ClinicalTrials.gov Identifier: NCT01220427), colon cancer (ClinicalTrials.gov Identifier: NCT02466113), and epithelial ovarian cancer (ClinicalTrials.gov Identifier: NCT02758652). Interestingly, some clinical data support our predicted model (Fig. 5), showing predictable and prognostically valuable expression levels of the microRNA processing enzymes Dicer and Drosha in epithelial skin cancer (ClinicalTrials.gov Identifier: NCT00849914). It is very interesting to see that NCT02869399 tackles on the clinical opportunity of tertiary prevention of head and neck cancer with a dietary intervention (DietINT). Clinical relevance of miRNA can be described with the following categories.
Table 1.
Clinical trials evolving in miRNAs in cancer treatment.
| ClinicalTrials.gov Identifier | Cancer Type | Study type/Clinical Trial Phase |
|---|---|---|
| NCT02812680 | Esophageal adenocarcinoma (Circulating Tumour Cells and Plasma microRNA) | Observational |
| NCT01722851 | Newly Diagnosed Breast Cancer; Recurrent Breast Cancer | Observational |
| NCT02247453 | Lung cancer | Observational |
| NCT01598285 | Identification of Bevacizumab response predictors in metastatic breast cancer | Observational |
| NCT01231386 | Neoadjuvant or adjuvant treatment for locally advanced & inflammatory breast cancer | Observational |
| NCT01220427 | Prostate cancer | Observational |
| NCT02466113 | Colon cancer | Observational |
| NCT02758652 | Molecular Mechanisms Leading to Chemoresistance in Epithelial Ovarian Cancer | Observational |
| NCT00849914 | microRNA processing enzymes Dicer and Drosha in epithelial skin cancer | Interventional |
| NCT01829971 | Liver cancer: A Multicenter Phase I Study of MRX34, miRNA miR-RX34 Liposomal Injection | Phase I |
| NCT02065908 | Circulating MicroRNA as Biomarker of Cardiotoxicity in Breast Cancer | Observational |
| NCT01541800 | Circulating miRNAs as Disease Markers in Pediatric Leukemia, Lymphoma, CNS tumor. | Observational |
| NCT02366494 | Micro RNAs to Predict Response to Androgen Deprivation Therapy | Observational |
| NCT02635087 | microRNAs Tool for Stratifying Stage II Colon Cancer | Observational |
| NCT02656589 | Breast cancer: microRNA of HER2-Positive Patient Treated With Herceptin | Observational |
| NCT00806650 | Anti-IMP3 Autoantibody and MicroRNA Signature Blood Tests in Finding Metastasis in Patients With Localized or Metastatic Kidney Cancer | Interventional |
| NCT01119573 | Biomarkers in Tissue Samples From Patients With Stage I or Stage III Endometrial Cancer | Observational |
| NCT02869399 | Tertiary Prevention of Head and Neck Cancer With a Dietary Intervention (DietINT) | Observational |
| NCT02471469 | Personalizing Enzalutamide Therapy by Understanding the Relation Between Tumor mRNAs, miRNAs and Treatment Response (ILUMINATE) | Observational |
| NCT02634502 | Radiofrequency Ablation Combined With S-1 for Pancreatic Cancer With Liver Metastasis | Interventional |
| NCT02964351 | micro RNA Profiles Identification in Adeno Carcinoma Prostate Cancer | Observational |
A keyword search of https://clinicaltrials.gov for “miRNA” AND “cancer” results in 158 ongoing clinical trials.
MicroRNAs for Diagnosis
Tumors are unpredictable tissues comprised of diverse cells, including stromal cells, fibroblasts, invulnerable cells and mesenchymal undifferentiated stem cells, as well as non-cell segments (physical scaffold), neoplastic cells notwithstanding. There is increasing evidence to support the notion that these non-neoplastic cell segments bolster disease initiation, movement and metastasis, and that their removal or reconstruction can hinder tumor development. Our comprehension of various parts of the tumor stroma in propelling disease has been enhanced by the utilization of platform- and framework-based 3D frameworks initially created for regenerative medicine. Furthermore, tranquilize conveyance frameworks (e.g., disintegration studies) produced using engineered and common biomaterials convey medications to kill stromal cells or reinvent the microenvironment for tumor restraint. Additionally, drug delivery systems made from synthetic and natural biomaterials deliver drugs to kill stromal cells or reprogram the microenvironment for tumor inhibition [76]. These help investigate the effect of 3D tumor models in expanding our comprehension of tumorigenesis to tumor microenvironment.
Many researchers look towards miRNAs as diagnostic tools and molecular markers to characterize diseases. For example, the miRNA expression profile of a tumor may reveal to physicians a specific tumor type and can be used to predict the aggressiveness of a tumor and the probability of the tumor spreading. Eventually we may estimate survival rates based on the miRNA profile of a patient and manage risk and treatment based on these assessments. MicroRNA profiling is already used to separate several grades of medulloblastoma tumors and more aggressive tumors can be identified based on expression of their miRNA [77].
MicroRNA profiling can also be used to help diagnose and prevent the misdiagnoses of patients. CNS-PNETs, for example, are difficult to diagnose because of the lack of genetic and immunohistochemical markers, but recent evidence has shown that miRNAs within the C19MC cluster may be used as a diagnostic tool to help characterize these tumors [49].
MicroRNAs as Drug Targets
MicroRNA profiling can aid in the discovery of biomarkers for other diseases and guide physicians in deciding the best course of treatment to serve patients. For example, if a tumor is not likely to spread or cause any side effects, it may be best to avoid the risks associated with surgery (even biopsy) as surgery breaks down the physical scaffold boundary, a protection frontier of immune defense for non-malignant normal tissues. However, if such miRNAs leaked out of TME, targeting such messenger miRNAs should be in place to block the uncontrolled signaling of miRNAs. Indeed, “some traditionally ‘undruggable’ molecules can be targeted via their miRNA gene regulators, enabling the treatment of diseases that, at present, seem impossible to cure”[78]. Some chemically modified miRNA-targeting antisense oligonucleotides equipped with in vivo local delivery strategies are on the clinical development of miRNA-targeting therapeutics [79]. Such miRNAs specific blockade of miRNAs signaling can serve as cancer subclonal switching board management [80], effectively keeping certain cancer subclones in the sleep stage (dormant state) so that we can exercise “watch-and-wait” approach to cancer. This concept is directly supported by Liu and colleagues’ report showing that while miR-34a, a p53 target, underexpressed in CD44+ prostate cancer cells, enforced expression of miR-34a in CD44+ prostate cancer cells inhibits clonogenic expansion, tumor regeneration, and metastasis [81, 82]. Chang and colleagues show that by acting as a tumor-suppressor, microRNA-7 (miR-7)-mediated KLF4/PI3K/Akt/p21 pathway is critical for prostate cancer stem-like cells (PCSCs) stemness - A negative correlation between miR-7 expression and prostate tumor progression implicates its potential application for prostate cancer therapy [83]. Discovery of functional master regulator (MR) proteins necessary to maintain tumour cell state shed new light on breaking up the autoregulated modules (termed tumour checkpoints) by therapeutically targeting dysregulated post-translational modifications [84]. The related miRNAs are emerging as optimal biomarkers as they can regulate protein expression, a new way to tackle tumors rather than genes. Indeed, their study of 2,600 patients revealed only 6.4% of druggable targets of mutations and for the vast majority of patients; their MR-associated protein activity can sustain a tumor, which is a likely viable drug target.
MicroRNAs for Prognosis
The miRNA profile of spatiotemporal expression integrated with a patient’s gene expression gives clues to how a specific stage of cancer (temporal factor) may manifest in miRNA profiling, which may be used to separate and predict the prognosis of a patient and aid in deciding what types of treatment should be performed [54]. Analyzing the spatiotemporal miRNA profile of a tumor can help understand how cancers progress and provide multiple targets to stop or prevent their progression. Therapeutic strategies may also be designed to target microRNAs as key modulators of the tumor immune response, part of the TME. For example, in breast cancer, miRNAs mediated TGF-β signaling regulates metastasis through modulating invasion-metastasis-related factors, including epithelial-to-mesenchymal transition (EMT) modulators [85], cancer stem cells (CSCs) activators, matrix metalloproteinase (MMP), tissue inhibitors of MMPs (TIMPs) inflammation factors [86], cell adhesion molecules (CAMs), and tumor microenvironment (TME) [87]. Thus, through a clear understanding of the miRNA-mediated TGF-β pathway, it may provide a novel prognostic benchmark and safer therapeutic target to prevent BC metastasis.
“Watch-and-Wait” Approach to Cancer
Cancer and stem cells biology as well as regenerative medicine should be integrated into “watch-and-wait” approach to cancer as these cells are in action during “watch” periods for balancing the bad (cancer) and the good (stem cell regulated regenerative medicine). In the normal human body, mutated cells have always been well controlled and removed, including cancer cells. When cancer cells undergo the infinite proliferation, they are in the process of the formation of an immunosuppressive microenvironment (cancer organ), in which cancer organ can hijack the immune cells by a variety of mechanisms to suppress the immune response. Moreover, some immune cells are converted to cancer cells, helping to form new blood vessels, plundering of resources and promote the transfer of cancer cells to other organs to open up new territories. Immunotherapy with antibodies against the immune regulators CTLA4 and PD-L1/PD-1 conjures up hope against cancer; however, only a subset of patients responded, suggesting immunity is regulated by tumor, host and environmental factors – all of these define a narrow therapeutic window [88] of a given tumor. Chen and Mellman define elements of cancer immunity and the cancer–immune set point, providing the guidelines for “watch-and-wait” approach to cancer [89]. They define anti-cancer immunity into three main phenotypes: the immunedesert phenotype (“non-inflamed tumour micro- environment with few or no CD8-carrying T cells”), the immune– excluded phenotype (“the immune cells do not penetrate the parenchyma of these tumors but instead are retained in the stroma that surrounds nests of tumour cells”) and the inflamed phenotype (“the presence in the tumour parenchyma of both CD4- and CD8-expressing T cells, often accompanied by myeloid cells and monocytic cells; the immune cells are positioned in proximity to the tumour cells”). Well defined specific underlying biological mechanisms for these three cancer-immune phenotypes may shed new light on “watch-and-wait” approach to cancer, in particular for watching the host’s immune response for eradicating the cancer. The biological mechanisms include factors that contribute to the generation of activated T-cell immunity or tolerance, and factors that are related to the normal genetics of an individual, age, and the microbiota, the presence of infection, exposure to sunlight and the intake of immunemodifying drugs. Their frame-work of a cancer-immune set point concept helps define the vast majority of clinical and biomarker data in to “watch-and-wait” bench marks for cancer. Above concept can be incorporated into dietary practice, because miRNA expression in relation to different dietary habits and lifestyle factors has been documented in stool and plasma. The document showing diet and lifestyle change profiling miRNAs (miR-16, miR-21, mir-34a and miR-222) provides spatiotemporal monitoring for “watch-and-wait” approach to cancer [90].
MicroRNAs are good candidates to identify and target during this transition period since they possess regulatory mechanisms that act on mRNA and can serve as early molecular markers of the ability of tumors to become malignant. When we profile a tumor that has already been established, we have to keep in mind that we are only observing one time point and that some early miRNAs may have already been produced and destroyed by regulatory feedback mechanisms. For example, if a tumor is to metastasize to another region of the body, it has to accomplish several steps: break down the extracellular matrix, enter the blood stream, attach to and establish itself in an extracellular environment and maintain that extracellular environment to form a tumor. Each of these steps may require certain miRNAs that are only active for a short period of time until the cell accomplishes its short-term goals. During each transitional phase, there may be miRNA targets they could be inhibited or promoted to stall the progress of tumorigenesis. In the future, developments in single-cell sequencing technology may be one way to observe the expression of miRNA in cancer stem cells and tumor growth.
Another example, miRNA expression profiles may improve identification of specific tumor subtypes, and improve the diagnosis, prognosis, and prediction of cancer and cancer therapeutics. However, because tumor subtypes have already been established in various cancers [91], such as luminal, basal, HER-2-enriched, triple negatives in breast cancer, would miRNA expression profiles improve the current known clinical, histological, and molecular phenotypes and enhance their diagnosis, prognosis, and therapeutics above and beyond? What is the clinical relevance for miRNA profiling that is already known and done on miRNA in pre-clinical models? A key observation that needs more elaboration and elucidation is the finding that TME-scaffold is an impediment and barrier to malignant or CSC mobilization. Importantly, breakdown of TME-scaffold promotes metastasis and mending of such TME-scaffold may justify the rationale of maintenance therapy and a watch-and-wait approach in cancer care. We found TME-scaffold regulates MMP-9 activity related to cancer invasion in PENT CSC [14]; however, there is scarcely literature of malignant or CSCs using catabolic enzymes to break down the TME-scaffold that may facilitate their invasion, migration, and colonization in the metastatic process. Some questions remain to be elucidated as follows: Could target miRNAs by inhibiting catabolic enzymes, inflammatory mechanisms, HDACs be one way to fulfill this goal? Some specific experimental models are needed to test and confirm our hypothesis. More proof-of-concept experiments are required to appreciate the clinical feasibility and validity of the “watch-and-wait” approach to cancer care.
CONCLUSION
Taken together, miRNAs are major players of posttranscriptional gene regulation that function as both tumor suppressors and promoters, depending on their spatiotemporal behavior within cancer cells and the tumor microenvironment, both proximally and distally. Some miRNAs can transform the tumor microenvironment via non-cell-autonomous mechanisms, whereas other miRNAs in neighboring cells stabilize cancer hallmark traits. Understanding the mechanisms of miRNA-mediated regulation of tumor microenvironments can aid the design of therapeutic interventions. Certain stages of cancer should be managed with a “watch-and-wait” approach to balance risks with benefits, in conjunction with stabilizing and regulating the TME through lifestyle and risk-factor management (Fig. 3 in [2]). The endpoint will be if humans need to live with cancer, they need to guard cancer stem cell dormancy [80] for a lifetime.
Ongoing research shows the role of TME elasticity and its influence on cancer stem cells through regulation of microRNA to sense microenvironment, however; it is unclear how specifically these alterations are driving microRNA expression rather than the overall gene expression in cancer stem cells. It is even less clear how the identified pathways can help in dissecting a watchful waiting as oppose to treatment in cancer patients. All of these remain to be elucidated.
Acknowledgments
This study was supported by the Children’s Hospital of Orange County (CHOC) Children’s Foundation, CHOC-UCI/ICTS pilot grants, CHOC Neuroscience Institute, Austin Ford Tribute Fund, W. M. Keck Foundation, Grant R21CA134391 from the National Institutes of Health, and Grant AW 0852720 from the National Science Foundation. We thank Dr. Merri Lynn Casem, Dr. Robert Koch, and Dr. Douglas Eernisse for their critical reading of the manuscript. We thank Maria Minon, MD, Leonard S. Sender, MD, Philip H. Schwartz, PhD; John H. Weiss, MD, PhD; Hong Zhen Yin, MD, Robert A. Koch, PhD for their enthusiasm and support. Dr. Bridget Samuels’s editorial help is acknowledged.
Footnotes
AUTHOR CONTRIBUTIONS
Conceived and designed the experiments: SCL. Performed the experiments: SCL, LTV, and JFZ. Analyzed the data: SCL, LTV, WGL, and MHK. Contributed reagents/materials/analysis tools: SCL, JFZ, WGL, and MHK. Wrote the paper: LTV SCL. Revised the paper: SCL LTV JJL, JFZ, ZL, BAD, WGL, and MHK.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Li SC, Tachiki LM, Kabeer MH, Dethlefs BA, Anthony MJ, Loudon WG. Cancer genomic research at the crossroads: realizing the changing genetic landscape as intratumoral spatial and temporal heterogeneity becomes a confounding factor. Cancer Cell Int. 2014;14(1):115. doi: 10.1186/s12935-014-0115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chevalier T, Mueller M, Mougiakakos D. Analysis of dendritic cell subpopulations in follicular lymphoma with respect to the tumor immune microenvironment. Leuk Lymphoma. 2016;57(9):I. doi: 10.3109/10428194.2015.1135432. [DOI] [PubMed] [Google Scholar]
- 4.Fan S, Hansen M, Lo Y, Tishkoff S. Going global by adapting local: A review of recent human adaptation. Science. 2016;354(6308):54–59. doi: 10.1126/science.aaf5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watt FM, Hogan BL. Out of Eden: stem cells and their niches. Science. 2000;287(5457):1427–30. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 6.Evans J, Essex A, Xin H, et al. Registered report: Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Elife. 2015:4. doi: 10.7554/eLife.07301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vermeulen L, De Sousa Heijden EMF, van der M, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12(5):468–76. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 8.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 9.Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144(1):92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 11.Mazzio EA, Soliman KF. Basic concepts of epigenetics: impact of environmental signals on gene expression. Epigenetics. 2012;7(2):119–30. doi: 10.4161/epi.7.2.18764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yosef NAR. Writ large: Genomic dissection of the effect of cellular environment on immune response. Science. 2016;354(6308):64–8. doi: 10.1126/science.aaf5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lokody I. Microenvironment: Tumour-promoting tissue mechanics. Nat Rev Cancer. 2014;14(5):296. doi: 10.1038/nrc3727. [DOI] [PubMed] [Google Scholar]
- 14.Vu LT, Keschrumrus V, Zhang X, et al. Tissue elasticity regulated tumor gene expression: implication for diagnostic biomarkers of primitive neuroectodermal tumor. PloS One. 2015;10(3):e0120336. doi: 10.1371/journal.pone.0120336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romero D. Immunotherapy: CAR T cells ready to go mainstream. Nat Rev Clin Oncol. 2016;13(7):396–7. doi: 10.1038/nrclinonc.2016.99. [DOI] [PubMed] [Google Scholar]
- 16.Onea AS, Jazirehi AR. CD19 chimeric antigen receptor (CD19 CAR)-redirected adoptive T-cell immunotherapy for the treatment of relapsed or refractory B-cell Non-Hodgkin’s Lymphomas. Am J Cancer Res. 2016;6(2):403–24. [PMC free article] [PubMed] [Google Scholar]
- 17.Riedmann EM. Agenus’ brain cancer vaccine doubles survival rate in GBM patients. Hum Vaccin Immunother. 2014;10(8):2139–40. doi: 10.4161/hv.36804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson B. Therapeutic vaccine for brain cancer succeeds using a unique approach. Biotechnol Healthc. 2011;8(3):31–2. [PMC free article] [PubMed] [Google Scholar]
- 19.Spira A, Disis ML, Schiller JT, et al. Leveraging premalignant biology for immune-based cancer prevention. Proc Natl Acad Sci USA. 2016 doi: 10.1073/pnas.1608077113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goel A, Maroules C, Mitchell G, et al. Ethnic Difference in Proximal Aortic Stiffness: An Observation From the Dallas Heart Study JACC: Cardiovascular Imaging. JACC Cardiovasc Imaging. 2017;10(1):54–61. doi: 10.1016/j.jcmg.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 21.Tajik A, Zhang Y, Wei F, et al. Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater. 2016;15(12):1287–96. doi: 10.1038/nmat4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meinke P, Nguyen TD, Wehnert MS. The LINC complex and human disease. Biochem Soc Trans. 2011;39(6):1693–7. doi: 10.1042/BST20110658. [DOI] [PubMed] [Google Scholar]
- 23.Kaminska K, Szczylik C, Bielecka ZF, et al. The role of the cell-cell interactions in cancer progression. J Cell Mol Med. 2015;19(2):283–96. doi: 10.1111/jcmm.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci USA. 2007;104(45):17719–24. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15(9):902–9. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jahagirdar D, Purohit S, Jain A, Sharma NK. Export of microRNAs: A Bridge between Breast Carcinoma and Their Neighboring Cells. Front Oncol. 2016;6:147. doi: 10.3389/fonc.2016.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci. 2013;14(7):14240–69. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su MJ, Aldawsari H, Amiji M. Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of micrornas 155 and 125b2 transfection using nanoparticle delivery systems. Sci Rep. 2016;6:30110. doi: 10.1038/srep30110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Nie J, Mei Q, Han WD. MicroRNAs: Novel immunotherapeutic targets in colorectal carcinoma. World J Gastroenterol. 2016;22(23):5317–31. doi: 10.3748/wjg.v22.i23.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shalaby T, Fiaschetti G, Baumgartner M, Grotzer MA. MicroRNA signatures as biomarkers and therapeutic target for CNS embryonal tumors: the pros and the cons. Int J Mol Sci. 2014;15(11):21554–86. doi: 10.3390/ijms151121554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicoloso MS, Calin GA. MicroRNA involvement in brain tumors: from bench to bedside. Brain Pathol. 2008;18(1):122–9. doi: 10.1111/j.1750-3639.2007.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 34.Serguienko A, Grad I, Wennerstrom AB, et al. Metabolic reprogramming of metastatic breast cancer and melanoma by let-7a microRNA. Oncotarget. 2015;6(4):2451–65. doi: 10.18632/oncotarget.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 36.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11(12):849–64. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi YJ, Lin CP, Risso D, et al. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science. 2017;355(6325) doi: 10.1126/science.aag1927. pii: eaag1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostenfeld MS, Jensen SG, Jeppesen DK, et al. miRNA profiling of circulating EpCAM(+) extracellular vesicles: promising biomarkers of colorectal cancer. J Extracell Vesicles. 2016;5:31488. doi: 10.3402/jev.v5.31488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eyking A, Reis H, Frank M, Gerken G, Schmid KW, Cario E. MiR-205 and MiR-373 Are Associated with Aggressive Human Mucinous Colorectal Cancer. PloS One. 2016;11(6):e0156871. doi: 10.1371/journal.pone.0156871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frediani JN, Fabbri M. Essential role of miRNAs in orchestrating the biology of the tumor microenvironment. Mol Cancer. 2016;15(1):42. doi: 10.1186/s12943-016-0525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Marzo L, Desantis V, Solimando AG, et al. Microenvironment drug resistance in multiple myeloma: emerging new players. Oncotarget. 2016;7(37):60698–711. doi: 10.18632/oncotarget.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moses B, Evans R, Slone W, et al. Bone Marrow Microenvironment Niche Regulates miR-221/222 in Acute Lymphoblastic Leukemia. Mol Cancer Res. 2016;14(10):909–19. doi: 10.1158/1541-7786.MCR-15-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mouw JK, Yui Y, Damiano L, et al. Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat Med. 2014;20(4):360–7. doi: 10.1038/nm.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwentner R, Herrero-Martin D, Kauer MO, et al. The role of miR-17-92 in the miRegulatory landscape of Ewing sarcoma. Oncotarget. 2016 doi: 10.18632/oncotarget.14091. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng CS, Ran L, Bursac N, Kraus WE, Truskey GA. Cell Density and Joint microRNA-133a and microRNA-696 Inhibition Enhance Differentiation and Contractile Function of Engineered Human Skeletal Muscle Tissues. Tissue Eng Part A. 2016;22(7–8):573–83. doi: 10.1089/ten.tea.2015.0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang M, Lin H, Luo M, Wang J, Han G. Integrated miRNA and mRNA expression profiling of tension force-induced bone formation in periodontal ligament cells. In vitro Cell Dev Biol Anim. 2015;51(8):797–807. doi: 10.1007/s11626-015-9892-0. [DOI] [PubMed] [Google Scholar]
- 47.Ghanghoria R, Kesharwani P, Tekade RK, Jain NK. Targeting luteinizing hormone-releasing hormone: A potential therapeutics to treat gynecological and other cancers. J Control Release. 2016 doi: 10.1016/j.jconrel.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 48.Seewaldt V. ECM stiffness paves the way for tumor cells. Nat Med. 2014;20(4):332–3. doi: 10.1038/nm.3523. [DOI] [PubMed] [Google Scholar]
- 49.Li M, Lee KF, Lu Y, et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16(6):533–46. doi: 10.1016/j.ccr.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 51.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103(8):2746–51. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su Q, Li S. Small activating mRNA (samRNA): A hypothesis for a specific positive feedback regulation of gene expression. Bioscience Hypotheses. 2008:44–7. 1/1. [Google Scholar]
- 54.Suzuki HI, Katsura A, Matsuyama H, Miyazono K. MicroRNA regulons in tumor microenvironment. Oncogene. 2015;34(24):3085–94. doi: 10.1038/onc.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee JK, Park SR, Jung BK, et al. Exosomes derived from mesenchymal stem cells suppress angiogenesis by downregulating VEGF expression in breast cancer cells. PloS One. 2013;8(12):e84256. doi: 10.1371/journal.pone.0084256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19(24):6860–9. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 58.Zhang X, Lee J, Chasin LA. The effect of nonsense codons on splicing: a genomic analysis. RNA. 2003;9(6):637–9. doi: 10.1261/rna.5060403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rigo F, Martinson HG. Functional coupling of last-intron splicing and 3’-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol. 2008;28(2):849–62. doi: 10.1128/MCB.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilusz JE, Spector DL. An unexpected ending: noncanonical 3’ end processing mechanisms. RNA. 2010;16(2):259–66. doi: 10.1261/rna.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian B, Pan Z, Lee JY. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007;17(2):156–65. doi: 10.1101/gr.5532707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun M, Hurst LD, Carmichael GG, Chen J. Evidence for a preferential targeting of 3’-UTRs by cis-encoded natural antisense transcripts. Nucleic Acids Res. 2005;33(17):5533–43. doi: 10.1093/nar/gki852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong X, Scofield DG, Lynch M. Intron size, abundance, and distribution within untranslated regions of genes. Mol Biol Evol. 2006;23(12):2392–404. doi: 10.1093/molbev/msl111. [DOI] [PubMed] [Google Scholar]
- 64.Delihas N. Complexity of a small non-protein coding sequence in chromosomal region 22q11.2: presence of specialized DNA secondary structures and RNA exon/intron motifs. BMC Genomics. 2015;16:785. doi: 10.1186/s12864-015-1958-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cenik C, Chua HN, Singh G, et al. A common class of transcripts with 5’-intron depletion, distinct early coding sequence features, and N1-methyladenosine modification. RNA. 2017;23(3):270–283. doi: 10.1261/rna.059105.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patrushev LI, Kovalenko TF. Functions of noncoding sequences in mammalian genomes. Biochemistry (Mosc) 2014;79(13):1442–69. doi: 10.1134/S0006297914130021. [DOI] [PubMed] [Google Scholar]
- 67.Li W, Park JY, Zheng D, Hoque M, Yehia G, Tian B. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol. 2016;14:6. doi: 10.1186/s12915-016-0229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoque M, Ji Z, Zheng D, et al. Analysis of alternative cleavage and polyadenylation by 3’ region extraction and deep sequencing. Nat Methods. 2013;10(2):133–9. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol Cancer Res. 2010;8(3):295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29(11):2073–7. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- 71.Zhang L, Sullivan PS, Goodman JC, Gunaratne PH, Marchetti D. MicroRNA-1258 suppresses breast cancer brain metastasis by targeting heparanase. Cancer Res. 2011;71(3):645–54. doi: 10.1158/0008-5472.CAN-10-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fetahu IS, Tennakoon S, Lines KE, et al. miR-135b- and miR-146b-dependent silencing of calcium-sensing receptor expression in colorectal tumors. Int J Cancer. 2016;138(1):137–45. doi: 10.1002/ijc.29681. [DOI] [PubMed] [Google Scholar]
- 73.Hiddingh L, Raktoe RS, Jeuken J, et al. Identification of temozolomide resistance factors in glioblastoma via integrative miRNA/mRNA regulatory network analysis. Sci Rep. 2014;4:5260. doi: 10.1038/srep05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davis CD, Ross SA. Evidence for dietary regulation of microRNA expression in cancer cells. Nutr Rev. 2008;66(8):477–82. doi: 10.1111/j.1753-4887.2008.00080.x. [DOI] [PubMed] [Google Scholar]
- 75.Davis CD. Vitamin D and cancer: current dilemmas and future research needs. Am J Clin Nutr. 2008;88(2):565S–9S. doi: 10.1093/ajcn/88.2.565S. [DOI] [PubMed] [Google Scholar]
- 76.Adjei IM, Blanka S. Modulation of the tumor microenvironment for cancer treatment: a biomaterials approach. J Funct Biomater. 2015;6(1):81–103. doi: 10.3390/jfb6010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferretti E, De SE, Po A, et al. MicroRNA profiling in human medulloblastoma. IntJCancer. 2009;124(3):568–77. doi: 10.1002/ijc.23948. [DOI] [PubMed] [Google Scholar]
- 78.Schmidt MF. Drug target miRNAs: chances and challenges. Trends Biotechnol. 2014;32(11):578–85. doi: 10.1016/j.tibtech.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 79.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13(8):622–38. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 80.Li SC, Lee KL, Luo J. Control dominating subclones for managing cancer progression and posttreatment recurrence by subclonal switchboard signal: implication for new therapies. Stem Cells Dev. 2012;21(4):503–6. doi: 10.1089/scd.2011.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li J, Lam M Reproducibility Project: Cancer B. Registered report: the microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Elife. 2015;4:e06434. doi: 10.7554/eLife.06434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu C, Kelnar K, Liu B, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17(2):211–5. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang YL, Zhou PJ, Wei L, et al. MicroRNA-7 inhibits the stemness of prostate cancer stem-like cells and tumorigenesis by repressing KLF4/PI3K/Akt/p21 pathway. Oncotarget. 2015;6(27):24017–31. doi: 10.18632/oncotarget.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Califano A, Alvarez MJ. The recurrent architecture of tumour initiation, progression and drug sensitivity. Nat Rev Cancer. 2016 doi: 10.1038/nrc.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li M, Li H, Liu X, Xu D, Wang F. MicroRNA-29b regulates TGF-beta1-mediated epithelial-mesenchymal transition of retinal pigment epithelial cells by targeting AKT2. Exp Cell Res. 2016;345(2):115–24. doi: 10.1016/j.yexcr.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 86.Keklikoglou I, Koerner C, Schmidt C, et al. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-kappaB and TGF-beta signaling pathways. Oncogene. 2012;31(37):4150–63. doi: 10.1038/onc.2011.571. [DOI] [PubMed] [Google Scholar]
- 87.Pollari S, Leivonen SK, Perala M, Fey V, Kakonen SM, Kallioniemi O. Identification of microRNAs inhibiting TGF-beta-induced IL-11 production in bone metastatic breast cancer cells. PloS One. 2012;7(5):e37361. doi: 10.1371/journal.pone.0037361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li SC, Han YP, Dethlefs BA, Loudon WG. Therapeutic Window, a Critical Developmental Stage for Stem Cell Therapies. Curr Stem Cell Res Ther. 2010;5(4):297–303. [PMC free article] [PubMed] [Google Scholar]
- 89.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–30. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 90.Tarallo S, Pardini B, Mancuso G, et al. MicroRNA expression in relation to different dietary habits: a comparison in stool and plasma samples. Mutagenesis. 2014;29(5):385–91. doi: 10.1093/mutage/geu028. [DOI] [PubMed] [Google Scholar]
- 91.Harper KL, Sosa MS, Entenberg D, et al. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature. 2016 doi: 10.1038/nature20609. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]