Abstract
Introduction
Charcot–Marie–Tooth disease type 1C (CMT1C) is a rare, dominantly inherited neuropathy caused by mutations in the lipopolysaccharide-induced tumor necrosis factor (LITAF) or small integral membrane protein of the lysosome/late endosome (SIMPLE) gene.
Methods
We present a case series comprised of 10 patients in whom CMT1C is caused by a Gly112Ser substitution in the encoded protein. We focus on clinical presentation, electrodiagnostic analyses, and our findings in the context of previously described cases.
Results
The Gly112Ser mutation causing CMT1C is a mild form of CMT, as patients walked on time, had less weakness than those with Charcot–Marie–Tooth disease type 1A (CMT1A), had a CMT neuropathy score (CMTNS) indicative of mild disease, and had faster ulnar and median motor nerve conduction velocities compared to those with CMT1A.
Conclusion
The G112S mutation in LITAF seems to be clinically indistinguishable from a mild presentation of CMT1A. Muscle Nerve, 2017
Charcot–Marie–Tooth disease type 1C (CMT1C) is a rare, dominantly inherited neuropathy caused by mutations in the lipopolysaccharide-induced tumor necrosis factor (LITAF) gene, also known as small integral membrane protein of the lysosome/late endosome (SIMPLE).[1–3] Given the rarity of CMT1C, we looked at our patients with the Gly112Ser (G112S) mutation and the specific clinical and electrodiagnostic phenotype associated with this mutation.
Previous reports have not focused on the G112S mutation in clinical and electrodiagnostic detail. Clinically, CMT1C has been reported to have minor symptoms with no risk of being wheelchair-bound.[4, 5] A previous report of the G112S mutation resulted in 1 patient with minimal weakness and another with severe weakness and pseudoradicular symptoms.[6] Electrodiagnostically, patients with CMT1C have been reported previously to have slowed nerve conduction velocities (<33 m/s),[4, 6] but some studies have reported conduction blocks,[7] temporal dispersion,[8] or even an axonal CMT pattern.[6]
Herein we evaluate 10 patients with the G112S mutation and compare their electrodiagnostic and clinical features to those with the most common inherited demyelinating neuropathy, CMT1A (Charcot–Marie–Tooth disease type 1A).
METHODS
Patients and Chart Review
This study was approved by the institutional review board at the University of Iowa and informed consent was obtained from each subject. We performed a retrospective analysis of data from both electrodiagnostic testing (EDx) and clinical assessment for 10 patients previously diagnosed with CMT1C caused by the c.334G>A (p.Gly112Ser) mutation in the LITAF gene, which has been reported to be a pathogenic missense mutation.[6] Patients were evaluated by highly experienced neurologists in a CMT clinic, at either Wayne State University or the University of Iowa. Patients were diagnosed with CMT1C if they had either of the following: (1) a positive genetic test; or (2) a first- or second-degree relative with a positive genetic test and a CMT phenotype. Clinical and electrophysiological features of a patient with this mutation reported in this study were described previously.[6] In all patients except patient 2 an autosomal-dominant pattern of inheritance was determined. Patient 2 had a spontaneous de-novo mutation. In addition, for all but patient 2, the mother had a normal pregnancy and delivery. In the case of patient 2, the mother used amphetamines, methamphetamines, marijuana, and alcohol during her pregnancy.
Four women and 6 men were included in the study. The mean age at examination was 35 (range 5–72) years. EDx data were available for 7 of the 10 patients studied, and data from a complete neurological exam were available for 9 of the 10 patients. Motor nerve (MNCS) and sensory nerve conduction study (SNCS) testing of the median, ulnar, fibular, and sural nerves had been performed using conventional methods (Nicolet Viking, Synergy EMG, or Nihon-Kohden system).[9] A block in motor conduction was defined as a ≥50% reduction in the amplitude of the proximal negative (vs. distal negative) peak compound muscle action potential (CMAP), if the distal negative peak CMAP is ≥20% of the normal lower limit.[10]
Calculation of Neuropathy Score
A CMT neuropathy score (CMTNS) was calculated using a modified scale that had been validated previously as a measure of disability caused by CMT.[6] Using this scale, level-of-impairment scores were classified as follows: 0–10 = mild impairment; 11–20 = moderate impairment; and ≥21 = significant impairment.[6] The CMTNS has 9 scored components: 3 are symptoms; 4 are signs observed by a clinician; and 2 are EDx data. Each measurement is scored from 0–4, for a possible total of 36 points. For those patients who did not have EDx data, the CMTNS was out of 28 points, and thus a weighted average was used to generate an average CMTNS score.
RESULTS
Clinical Symptoms
A complete list of clinical symptoms for all patients is listed in Table 1. All patients walked on time (at or before 18 months) and the first symptoms included being the slowest runner, walking on toes, or having a foot deformity. Motor and sensory symptoms in the lower extremities were observed in 60% of patients. All patients had foot deformities. The average CMTNS was 7.5, range 2–11 (0 = normal, 36 = maximum abnormal score).[6] All patients complained of cramps and pain in feet or legs. Fifty percent of patients complained of exertional calf pain, which was worse after exercise or at the end of the day.
Table 1.
Clinical symptoms of CMT1C G112S
| Clinical symptoms | Number of patients with CMT1C (%) (n = 10) |
|---|---|
| 1. a | |
| Only 1 patient wore a foot orthotic. | |
| Childhood onset of presentation | 8 (80%) (in the other 2 patients, age of onset was 30 and 60 years, respectively) |
| Walked on time (≤18 months) | 10 (100%) |
| Ethnicity | French, German, Welsh, Russian, Polish, Slavic |
| First symptom | |
| Slowest runner | 3 (30%) |
| Toe walker | 4 (40%) |
| Foot deformity | 3 (30%) |
| Motor symptoms | |
| Lower limbs (trip, catch toes, shoe inserts, slap feet, ankle–foot orthotica) | 9 (90%) |
| Upper limbs (difficulty with buttons) | 1 (10%) |
| Foot surgery | 3 (30%) |
| Cramps and pain | 10 (100%) |
| Sensory symptoms: | |
| Lower limbs (reduction or loss of sensation) | 6 (60%) |
| CMTNS | 7.5 of 36 (range 2–11) |
Clinical Signs
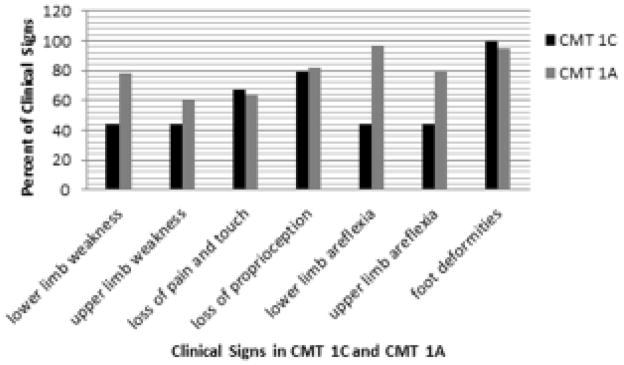
In patients with the CMT1C G112S mutation, sensory findings and foot deformities were similar to those reported for CMT1A, but muscle weakness and areflexia occurred less frequently (Fig. 1).
Figure 1.
Clinical signs. Comparison of CMT1C Gly112Ser mutation with CMT1A. Histogram shows percent of clinical signs in patients with CMT1C (n = 9) and CMT1A (n = 119).[17]
MNCS in CMT1C
No conduction block or temporal dispersion was seen in patients with the CMT1C G112S mutation. Velocity of motor conduction ranged from 19.7 to 38.5 m/s (Table 2). When compared with the EDx in those with CMT1A, the CMT1C motor nerve conduction studies had a higher CMAP, faster conduction velocities, and shorter distal motor latencies.
Table 2.
Motor nerve conduction findings in patients with CMT1C Gly112Ser LITAF/SIMPLE mutation and in those with CMT1A
| Normal | CMT1C (n = 7) | CMT1A[17] (n = 119) | |
|---|---|---|---|
| 1. Data expressed as mean ± standard deviation (range). CMAP, compound muscle action potential (motor, millivolts); DML, distal motor latency; MNCV, motor nerve conduction velocity; amplitudes represent baseline to peak values; TA, tibialis anterior. | |||
| 2. a | |||
| Fibular motor recording from the tibialis anterior (TA) was not performed. | |||
| Median nerve | No response = 0 | No response = 4 | |
| DML (ms) | <4.5 | 6.1 ± 1.3 (4.6–7.9) | 9.8 ± 2.6 (4.6–22.3) |
| CMAP (mV) | |||
| Distal | 4.9 ± 1.4 (3.6–7.5) | 2.4 ± 1.9 (0.1–9.5) | |
| Proximal | >4 | 4.2 (2.7–5.8) | |
| MNCV (m/s) | >48 | 27.9 ± 6.4 (20–38) | 20.2 ± 4.9 (7–33) |
| Ulnar nerve | No response = 0 | No response = 0 | |
| DML (ms) | <3.5 | 4.3 ± 0.81 (2.9–5.4) | 8 ± 3.1 (4.5–16.4) |
| CMAP (mV) | |||
| Distal | >6 | 5.5 ± 1.93 (2.9–5.4) | 2.4 ± 1.6 (0.1–9.5) |
| Proximal | >6 | 4.0 (0.99–7.5) | |
| MNCV (m/s) | >49 | 28 ± 7.2 (20.3–38.5) | 17.3 ± 5.1 (6–30) |
| Fibular nerve/EDB | No response = 1 | No response = 24 | |
| DML (ms) | <5.5 | 9.5 ± 3.1 (5.8–13.3) | 11.9 ± 3.4 (6–19.4) |
| CMAP (mV) | |||
| Distal | >3 | 1.9 ± 5.7 (0.27–4) | 0.9 ± 1.2 (0.07–4) |
| Proximal | >3 | 1.4 (0.2–2.97) | |
| MNCV (m/s) | >41 | 20 ± 0.058 (19.7–20) | 17 ± 4.6 (5–24) |
| Fibular nerve, TA | No response = 0 | Not availablea | |
| DML (ms) | <6.7 | 4.6 ± 0.86 (3.8–6) | Not availablea |
| CMAP (mV) | Not availablea | ||
| Distal | >5 | 3.7 ± 0.11 (3.2–4.5) | |
| Proximal | >5 | 3.4 (2.7–4.4) | |
| MNCV (m/s) | >44 | 26 ± 2.83 (19.7–33) | |
SNCS in CMT1C
The sural sensory nerve action potential (SNAP) was absent in all patients tested, and the median and ulnar SNAPs were absent in 43% of patients tested. Sensory nerve conduction velocities ranged from 20 to 41 m/s (Table 3).
Table 3.
Sensory nerve conduction findings in 7 patients with the Gly112Ser LITAF/SIMPLE mutation
| Normal | Mean | Range | No response | |
|---|---|---|---|---|
| 1. Amplitudes represent baseline to peak values. SNAP, sensory nerve action potential (sensory, microvolts); DSL, distal sensory latency; SNCV, sensory nerve conduction velocity. | ||||
| Median nerve | ||||
| DSL (ms) | <3.5 | 5.1 | 3.5–6.6 | 3 |
| SNAP (μV) | >5 | 9.9 | 6.9–13 | |
| SNCV (m/s) | >45 | 36.5 | 32–41 | |
| Ulnar nerve | ||||
| DSL (ms) | <3.5 | 5 | 2.6–7.1 | 3 |
| SNAP (μV) | >18 | 7.9 | 7.3–8.8 | |
| SNCV (m/s) | >45 | 31 | 20–40 | |
| Sural nerve | 7 | |||
DISCUSSION
CMT1C is a rare hereditary demyelinating neuropathy with few data regarding clinical outcomes for various disease-causing mutations. Herein we have analyzed 10 patients with the G112S mutation in the LITAF gene.
Previous reports of CMT1C have suggested that this condition results from conduction block, temporal dispersion, or axonal neuropathy.[6, 7] However, our study did not reveal either conduction blocks or temporal dispersion in any of the patients expressing the CMT1C-associated G112S form of LITAF, suggesting that this protein causes a uniformly demyelinating form of CMT.
The patients in our study more frequently displayed lower more than upper extremity symptoms, and the lower extremity problems were predominantly structural, involving mainly eversion of the foot. Although all of the patients had abnormal foot structure and 90% complained of problems with their feet, only 44% had weakness in the lower extremities (Table 1). In addition, only 30% had undergone foot surgery. Consistent with this finding, over half of the patients had preserved reflexes (Tables 1 and 2) and did not exhibit as much weakness as their CMT1A counterparts on physical examination (Table 1). We also observed a lower CMTNS in patients with CMT1C G112S (7.5 of 36, on average); this is milder than the average CMTNS for patients with CMT1A (~13–14).[11, 12]
Fifty percent of our patients exhibited exertional calf pain; this is not typical of the usual foot pain seen in the majority of CMT1A patients.[13]
Consistent with previous reports, we observed some variability in the age at which symptoms became apparent.[2, 4, 7] Although in the majority of patients the symptoms began in childhood, in 2 patients the symptoms did not emerge until later in life; 1 presented in his fourth decade and the other in his sixth decade. Therefore, the G112S mutation causing CMT1C does not appear to have a consistent age at which symptoms become apparent.
Previous reports have suggested that nerve conduction studies of the lower limbs are more frequently abnormal in those with CMT1C.[7] Previous reports have also identified patients with CMT1C with a few reported intermediate nerve conduction velocities and conduction block.[7, 14] In our study of those with the G112S mutation, the sural SNAP was absent in all patients, whereas the ulnar CMAPs and SNAPs were affected in only a subset, consistent with the lower extremities being more frequently involved electrodiagnostically. Conduction block was not noted in our patients. Also, in our study, only 2 nerve conduction velocities in the intermediate range were observed: 38 m/s and 39 m/s. However, it should be noted that conduction velocity values for a single nerve are not sufficient to classify CMT as intermediate. The term “intermediate” reflects the type of CMT and takes into account axonal and demyelinating pathology; it does not reflect an nerve conduction velocity (NCV) value.[14] EDx abnormality in CMT1C due to the G112S mutation is found in the lower extremities more than the upper extremities. Findings are in the demyelinating, not intermediate, range with no evidence of conduction block. Although the NCVs are in the demyelinating range for CMT1C, they are slightly faster in the upper extremities compared with the lower extremities and have a higher CMAP amplitude (Table 2), suggesting that the motor nerve conduction studies are milder in those with CMT1C compared to those with CMT1A.
A previous report of an analysis of patients with the c.430G>A p.Val144Met mutation in LITAF demonstrated that this mutation results in clinical features similar to those reported in our study.[2] Although conduction blocks were not reported, the age of onset was variable, and patients displayed a demyelinating neuropathy. As in our study of the G112S mutation, patients with the Val144Met variant had normal tendon stretch reflexes, were more affected in the lower extremities, and reported foot pain in both mother and son.[2] Thus, our study brings to light another mutation in LITAF that results in a milder form of CMT. In addition, a study involving 38 patients with 4 missense mutations (p.Ala111Gly, p.Gly112Ser, p.Pro135Ser, p.Pro135Thr) included several adults who presented with minor symptoms, and none of the 38 patients were wheelchair-bound.[4] The report of a family with the p.Pro135Ser mutation suggested variable age of onset, variable clinical presentation (from mild symptoms of sensory loss, bilateral plantar ulcers, and no muscle weakness to more significant gait difficulties and distal weakness), and the presence of a demyelinating neuropathy with conduction blocks without excessive temporal dispersion.[7]
It is suspected that CMT1C resembles CMT1A because of the possible function of LITAF, a mutation that could potentially result in an excess of peripheral myelin protein-22 (PMP-22). LITAF is highly expressed in peripheral nerves and Schwann cells.[15] The G112S mutation in LITAF near the transmembrane domain has been suggested to result in mislocalization of the LITAF protein to the mitochondria rather than to the endosome or lysosome.[16] It has been hypothesized that PMP-22 could be one of the substrates of the LITAF pathway in protein degradation.[6] Further studies characterizing mutations that target other regions of LITAF and analyses of potential differences in their clinical phenotypes are expected to be informative with regard to understanding the cellular and molecular mechanisms that underlie CMT.
In conclusion, we have studied a group of patients with the G112S mutant form of LITAF. Our findings demonstrate that patients with this mutation have a degree of sensory loss and foot deformity similar to those observed in patients with CMT1A, but muscle weakness and areflexia are milder. The motor nerve conduction studies are also milder. Thus, the G112S-causing mutation in LITAF results in a mild form of a demyelinating CMT that is almost clinically indistinguishable from mild CMT1A. One distinguishing trend seen in half of our patients with the G112S mutation is exertional calf pain that is not typically seen in those with CTM1A; this trend would be of interest to assess in future cases of CMT1C with the G112S mutation.
Acknowledgments
This work was supported by a grant from the National Institute of Neurological Disorders and Stroke (to M.E.S.) and the Office of Rare Diseases (U54NS065712 to M.E.S.), Muscular Dystrophy Association (to M.E.S., and a clinical research training grant to N.U.J.), and the Charcot-Marie-Tooth Association (to M.E.S.).
References
- 1.Potulska-Chromik A, Sinkiewicz-Darol E, Kostera-Pruszczyk A, Drac H, Kabzinska D, Zakrzewska-Pniewska B, et al. Charcot-Marie-Tooth type 1C disease coexisting with progressive multiple sclerosis: a study of an overlapping syndrome. Folia Neuropathol. 2012;50:369–374. doi: 10.5114/fn.2012.32366. [DOI] [PubMed] [Google Scholar]
- 2.Gerding WM, Koetting J, Epplen JT, Neusch C. Hereditary motor and sensory neuropathy caused by a novel mutation in LITAF. Neuromuscul Disord. 2009;19:701–703. doi: 10.1016/j.nmd.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Street VA, Bennett CL, Goldy JD, Shirk AJ, Kleopa KA, Tempel BL, et al. Mutation of a putative protein degradation gene LITAF/SIMPLE in Charcot-Marie-Tooth disease 1C. Neurology. 2003;60:22–26. doi: 10.1212/wnl.60.1.22. [DOI] [PubMed] [Google Scholar]
- 4.Latour P, Gonnaud PM, Ollagnon E, Chan V, Perelman S, Stojkovic T, et al. SIMPLE mutation analysis in dominant demyelinating Charcot-Marie-Tooth disease: three novel mutations. J Periph Nerv Syst. 2006;11:148–155. doi: 10.1111/j.1085-9489.2006.00080.x. [DOI] [PubMed] [Google Scholar]
- 5.Luigetti M, Fabrizi GM, Taioli F, Del Grande A, Lo Monaco M. A novel LITAF/SIMPLE variant within a family with minimal demyelinating Charcot-Marie-Tooth disease. Neurol Sci. 2014;35:2005–2007. doi: 10.1007/s10072-014-1833-2. [DOI] [PubMed] [Google Scholar]
- 6.Saifi GM, Szigeti K, Wiszniewski W, Shy ME, Krajewski K, Hausmanowa-Petrusowicz I, et al. SIMPLE mutations in Charcot-Marie-Tooth disease and the potential role of its protein product in protein degradation. Hum Mutat. 2005;25:372–383. doi: 10.1002/humu.20153. [DOI] [PubMed] [Google Scholar]
- 7.Ciotti P, Luigetti M, Geroldi A, Capponi S, Pazzini I, Gulli R, et al. A novel LITAF/SIMPLE mutation within a family with a demyelinating form of Charcot-Marie-Tooth disease. J Neurol Sci. 2014;343:183–186. doi: 10.1016/j.jns.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 8.Bennett CL, Shirk AJ, Huynh HM, Street VA, Nelis E, van Maldergem L, et al. SIMPLE mutation in demyelinating neuropathy and distribution in sciatic nerve. Ann Neurol. 2004;55:713–720. doi: 10.1002/ana.20094. [DOI] [PubMed] [Google Scholar]
- 9.Jerath NU, Gutmann L, Reddy CG, Shy ME. Charcot–Marie–Tooth disease type 1× in women: electrodiagnostic findings. Muscle Nerve. 2016;54:728–732. doi: 10.1002/mus.25077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joint Task Force of the European Federation of Neurological Societies/Peripheral Nerve Society Guideline on management of chronic inflammatory demyelinating polyradiculoneuropathy: report of a joint task force of the European Federation of Neurological Societies and the Peripheral Nerve Society—First Revision. J Peripher Nerv Syst. 2010;15:1–9. doi: 10.1111/j.1529-8027.2010.00245.x. [DOI] [PubMed] [Google Scholar]
- 11.Shy ME, Chen L, Swan ER, Taube R, Krajewski KM, Herrmann D, et al. Neuropathy progression in Charcot-Marie-Tooth disease type 1A. Neurology. 2008;70:378–383. doi: 10.1212/01.wnl.0000297553.36441.ce. [DOI] [PubMed] [Google Scholar]
- 12.Fridman V, Bundy B, Reilly MM, Pareyson D, Bacon C, Burns J, et al. CMT subtypes and disease burden in patients enrolled in the Inherited Neuropathies Consortium natural history study: a cross-sectional analysis. J Neurol Neurosurg Psychiatry. 2015;86:873–878. doi: 10.1136/jnnp-2014-308826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribiere C, Bernardin M, Sacconi S, Delmont E, Fournier-Mehouas M, Benchortane M, et al. Pain assessment in Charcot-Marie-Tooth (CMT) disease. Ann Phys Rehabil Med. 2012;55:160–173. doi: 10.1016/j.rehab.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson G, Myers S. Intermediate forms of Charcot-Marie-Tooth neuropathy: a review. Neuromol Med. 2006;8:123–130. doi: 10.1385/nmm:8:1-2:123. [DOI] [PubMed] [Google Scholar]
- 15.Lee SM, Olzmann JA, Chin LS, Li L. Mutations associated with Charcot-Marie-Tooth disease cause SIMPLE protein mislocalization and degradation by the proteasome and aggresome-autophagy pathways. J Cell Sci. 2011;124:3319–3331. doi: 10.1242/jcs.087114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacerda AF, Hartjes E, Brunetti CR. LITAF mutations associated with Charcot-Marie-Tooth disease 1C show mislocalization from the late endosome/lysosome to the mitochondria. PloS One. 2014;9:e103454. doi: 10.1371/journal.pone.0103454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birouk N, Gouider R, Le Guern E, Gugenheim M, Tardieu S, Maisonobe T, et al. Charcot-Marie-Tooth disease type 1A with 17p11.2 duplication. Clinical and electrophysiological phenotype study and factors influencing disease severity in 119 cases. Brain. 1997;120:813–823. doi: 10.1093/brain/120.5.813. [DOI] [PubMed] [Google Scholar]