SUMMARY
The epithelial FGF9 subfamily specifically binds and activates the mesenchymal ‘c’ splice isoform of FGF receptor 1–3 to regulate organogenesis and tissue homeostasis. The unique N and C-termini of FGF9 subfamily ligands mediate a reversible homodimerization that occludes major receptor binding sites within the ligand core region. Here we provide compelling X-ray crystallographic, biophysical and biochemical data showing that homodimerization controls receptor binding specificity of FGF9 subfamily by keeping the concentration of active FGF9 monomers at a level, which is sufficient for a normal FGFR ‘c’ isoform binding/signaling, but is insufficient for an illegitimate FGFR ‘b’ isoform binding/signaling. We show that deletion of the N-terminus or alanine substitutions in the C-terminus of FGF9 skews the delicate ligand equilibrium towards active FGF9 monomers causing off-target binding and activation of FGFR ‘b’ isoforms. Our study is the first to implicate ligand homodimerization in the regulation of ligand-receptor specificity.
Keywords: Fibroblast Growth Factor 9, FGF Receptor 1, Homodimerization, Autoinhibition, Specificity, X-ray crystallography, Surface Plasmon Resonance, multi-angle light scattering, BaF3 cells
eTOC BLURB
Liu et al. show that receptor binding specificity of FGF9 ligand is controlled by a reversible homodimerization, which maintains the concentration of monomeric active ligands below the threshold that can cause illegitimate binding and activation of ‘b’ splice isoforms of FGFR receptor by FGF9.
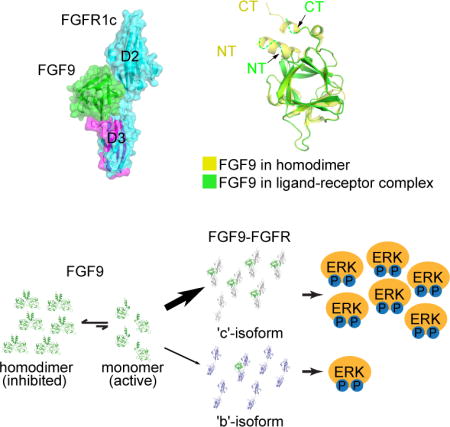
INTRODUCTION
The mammalian FGF family of ligands consists of 18 structurally-related, secreted polypeptides that are divided into six subfamilies: FGF1, FGF4, FGF7, FGF8, FGF9, and FGF19 (Itoh and Ornitz, 2004). The first five subfamilies are retained in the pericellular matrix due to their strong affinity for heparan sulfate (HS) glycosaminoglycans and hence act in a paracrine fashion within tissues, primarily during embryonic development but also throughout adulthood (Thisse and Thisse, 2005; Wills et al., 2008). FGF19 subfamily ligands have poor HS affinity, and hence are able to freely permeate through the extracellular matrix and enter the blood circulation to act in an endocrine fashion (Goetz et al., 2007).
FGFs transduce their signals by binding, dimerizing, and activating cell surface FGF receptors (FGFRs) in HS or Klotho co-receptor dependent fashion (Kurosu et al., 2006; Mohammadi et al., 2005; Schlessinger et al., 2000; Urakawa et al., 2006). FGFRs, which comprise four members in mammals (FGFR1-4), form a distinct subfamily within the superfamily of receptor tyrosine kinases. The prototypical FGFR ectodomain contains three immunoglobulin-like domains termed D1, D2, and D3, of which the D2 and D3 domains and the linker between them are sufficient for FGF ligand binding (Plotnikov et al., 1999). Ligand-receptor binding specificity is an important mechanism in the regulation of FGF signaling and is achieved by temporal and spatial expression of FGFs and FGFRs, and by primary sequence differences among ligands and receptors (Beenken and Mohammadi, 2009; Olsen et al., 2006). Sequence diversity is further elaborated by alternative splicing of certain FGFs and FGFR1-3. The second half of the D3 domain in FGFR1-3 is encoded by two mutually exclusive exons, termed ‘b’ and ‘c’ which are spliced in a tissue-specific fashion. The ‘b’ splice isoforms (FGFR1b-FGFR3b) are primarily expressed in the epithelium whereas the ‘c’ isoforms (FGFR1c-FGFR3c) are restricted to the mesenchyme. Indeed, the FGF-FGFR binding specificity is principally partitioned along this tissue-specific alternative splicing event. This reciprocal expression pattern of cognate ligands and receptors establishes a bidirectional signaling loop between mesenchymal and epithelial compartments of organs. Derangements in this signaling loop can give rise to human skeletal disorders and cancer (Oldridge et al., 1999; Pollock et al., 2007).
A wealth of literature has demonstrated that the FGF7 subfamily comprising FGF3, FGF7, FGF10 and FGF22, which are secreted by the mesenchyme, activate epithelial FGFR2b to mediate critical processes in the development of organs such as the lung, inner ear, kidney, and glands including prostate, mammary, salivary, and lacrimal glands (Cancilla et al., 1999; Jaskoll et al., 2005; Lin et al., 2007; Pirvola et al., 2004; Weaver et al., 2003). Conversely, gene knock-out studies in mice and cell-based studies have shown that FGF9 signaling constitutes the epithelial-to-mesenchymal arm of the bidirectional signaling loop required for morphogenesis of the lung, gonads, cecum, small intestine, and inner ear (Geske et al., 2008; Jameson et al., 2003; Pirvola et al., 2004; Weaver et al., 2003; Zhang et al., 2006b). Previously, we solved the crystal structures of FGF10, a mesenchymal ligand, in complex with its epithelial receptor, FGFR2b, and of FGF8b, an epithelial ligand, in complex with its mesenchymal receptor, FGFR2c (Olsen et al., 2006; Yeh et al., 2003). Analysis of these two structures has revealed that both FGF10 and FGF8b rely primarily on their divergent N-termini to attain specificity for their respective cognate FGFRs. The binding specificity of FGF10 including its other three subfamily members, for FGFR2b can be traced to the hydrogen bonding between a conserved N-terminal aspartic acid in this subfamily and a serine unique to the βC′-βE loop of the FGFR2b isoform. Additionally, the β1 strand in the FGF7 subfamily is extended N-terminally, which introduces steric clashes and electrostatic repulsion with the βC′-βE loop of FGFR ‘c’ isoforms further accounting for the tight specificity of FGF10, and its subfamily members for FGFR2b. Likewise, the specificity of FGF8 subfamily ligands for the FGFR ‘c’ isoforms and FGFR4 can be accounted for by hydrophobic contacts between a conserved N-terminal phenylalanine in this subfamily and a hydrophobic groove in the receptor D3 domain that is present only in the FGFR ‘c’ isoforms and in FGFR4. However, the molecular basis by which the epithelial FGF9 specifically binds and activates the mesenchymal FGFR ‘c’ isoforms is currently unknown. Based on our structural data on the receptor binding specificity of FGF7 and FGF8 subfamilies, we anticipated that the divergent N-termini of FGF9 subfamily would also directly engage the alternatively spliced regions in FGFRs to mediate receptor-binding specificity of the subfamily.
Unique among FGFs, FGF9 subfamily members namely FGF9, FGF16, and FGF20, undergo a reversible homodimerization mediated by their exclusive N- and C-termini as well as by their conserved β-trefoil core. Because most of the core and N-terminal residues that partake in ligand homodimerization are also predicted to mediate FGFR binding, homodimerization suppresses the ability of the FGF9 subfamily to bind and activate cognate FGFRs. Indeed, the strong propensity of FGF9 subfamily to form receptor-binding incapable ‘autoinhibited’ homodimers has made structural studies of FGF9-FGFR complexes technically challenging. To understand the molecular basis by which epithelial FGF9 subfamily ligands selectively activate mesenchymal FGFR ‘c’ isoforms, we engineered a ‘monomerized’ FGF9 and solved its structure in complex with a high affinity FGFR1c harboring a P252R pathogenic mutation. Unexpectedly, analysis of this structure reveals that the FGF9 subfamily deviates from FGF7 and FGF8 subfamilies in that, the N-terminus of FGF9 does not contribute to FGFR binding specificity by directly engaging in divergent contacts with the alternatively spliced regions in the receptor D3 domain. Instead, the FGF9-N-terminus regulates specificity in a negative fashion by promoting homodimerization thereby keeping the concentration of receptor-binding-capable ‘active’ FGF9 monomers too low to grant an illegitimate FGF9-FGFRb binding and signaling. However, the concentration of ligand monomers is still sufficient for a single specific contact between the core of the ligand and the alternatively spliced βF-βG loop in the D3 domain of FGFRc isoforms, to prevail and determine the binding specificity for FGFR ‘c’ isoforms. In support of our model, we show that disruption of this autoinhibitory dimer either by introducing triple point mutations into the C-terminus or deletion of the N-terminus elevates the pool of FGF9 monomers sufficiently high so as to cause off-target binding and activation of ‘b’ splice isoforms of FGFRs. The FGF9-FGFR1c structure also provides the molecular bases for the dramatic increase in binding affinity of FGF9 subfamily members towards FGFR1-3 carrying the Pro->Arg (P252R) pathogenic mutation (Ibrahimi et al., 2004b), and for the loss-of-function effects of the S99D FGF9 mutation found in multiple synostoses syndrome (SYNS) patients (Wu et al., 2009), and of N143T FGF9 substitution found in mice with Elbow knee synostosis (Eks) (Harada et al., 2009).
RESULTS AND DISCUSSION
Engineering of a stabilized FGF9-FGFR1c complex tractable for X-ray structure determination
We have previously used sedimentation equilibrium ultracentrifugation analysis to derive a KD of 680 nM for FGF9 homodimerization (Plotnikov et al., 2001). In a later study, we also showed that FGF20, another FGF9 subfamily ligand, also homodimerizes with a KD of about 100 nM implying that homodimerization is a general property of the entire FGF9 subfamily (Kalinina et al., 2009). Moreover, we have previously performed comprehensive SPR spectroscopy experiments to characterize binding interactions of all FGFs including FGF9 with all seven FGFR isoforms including FGFR1c and FGFR1b (Ibrahimi et al., 2004b). According to these published data, FGF9 binds FGFR1c with a KD of 890 nM but it does not recognize FGFR1b isoform. Because the dissociation constants for autoinhibitory homodimerization and the FGF9-FGFR1c interaction are comparable, the FGF9-FGFR1c complex is intrinsically weak and hence not tractable by X-ray crystallography. To overcome this obstacle, we decided to engineer monomeric FGF9 variants. Based on our previous FGF-FGFR complex structures, both the unique N-terminus and the conserved core region of FGF9 are predicted to mediate binding of FGF9 to its cognate FGFRs. In contrast, C-termini of FGFs have not been implicated in FGFR binding in any of the previous crystal structures. In light of these structural considerations, we chose to monomerize FGF9 by introducing triple alanine substitutions namely, Asp-195, Ile-204, and Leu-205 into its C-terminal region (FGF9mut). As shown in Figure 1A, these three C-terminal residues mediate key contacts at the ligand dimer interface (Kalinina et al., 2009; Plotnikov et al., 2001). Previously we used size exclusion chromatography to show that the single D195A substitution in the FGF9 C-terminal tail significantly diminishes the ability of FGF9 to form autoinhibited dimers. We then used SPR spectroscopy to demonstrate that this mutated, monomerized FGF9 variant binds FGFR1c, a main cognate receptor of FGF9, with a KD value that is similar to that of the wild type FGF9-FGFR1c interaction (Kalinina et al., 2009). It should be emphasized that the lack of an effect of ligand monomerization on FGFR1c binding affinity is expected because monomerizing mutations simply increase the available pool of monomeric ligands that are capable of receptor binding but do not create additional contacts between FGF9 and FGFR1c (Ibrahimi et al., 2004b). To extend and improve upon these data, we used multi-angle light scattering (MALS) analysis to compare the masses of the engineered FGF9 triple mutant (FGF9mut) and wild type FGF9 (FGF9WT) in solution. As shown in Figure 1B, consistent with our previous data, FGF9WT migrated as a dimer with measured MW of 45.6 kD. By contrast, FGF9mut eluted as a single species with MW of 25.3 kD indicative of being predominantly a monomer. Consistent with our previous SPR data on the D195A FGF9 mutant, injection of 1 μM of predominantly dimeric FGF9WT and monomerized FGF9mut onto biosensor chips containing immobilized wild type FGFR1c ectodomain (FGFR1cecto/WT) shows that FGF9mut produces greater response units indicating that, compared to FGF9WT, it forms more complexes with the immobilized ectodomain (Figure 1C). Consistent with the SPR data, the monomerized FGF9mut was found to be more efficacious than the predominantly dimeric FGF9WT in binding and activating full length, wild type FGFR1c isoform (FGFR1cWT) that was expressed on the surface of BaF3 cells (Figure 1E).
Figure 1. Engineering of a stabilized FGF9-FGFR1c complex by employing the Pfeiffer syndrome gain-of-function FGFR1c mutation and ‘monomerizing’ FGF9 mutations.
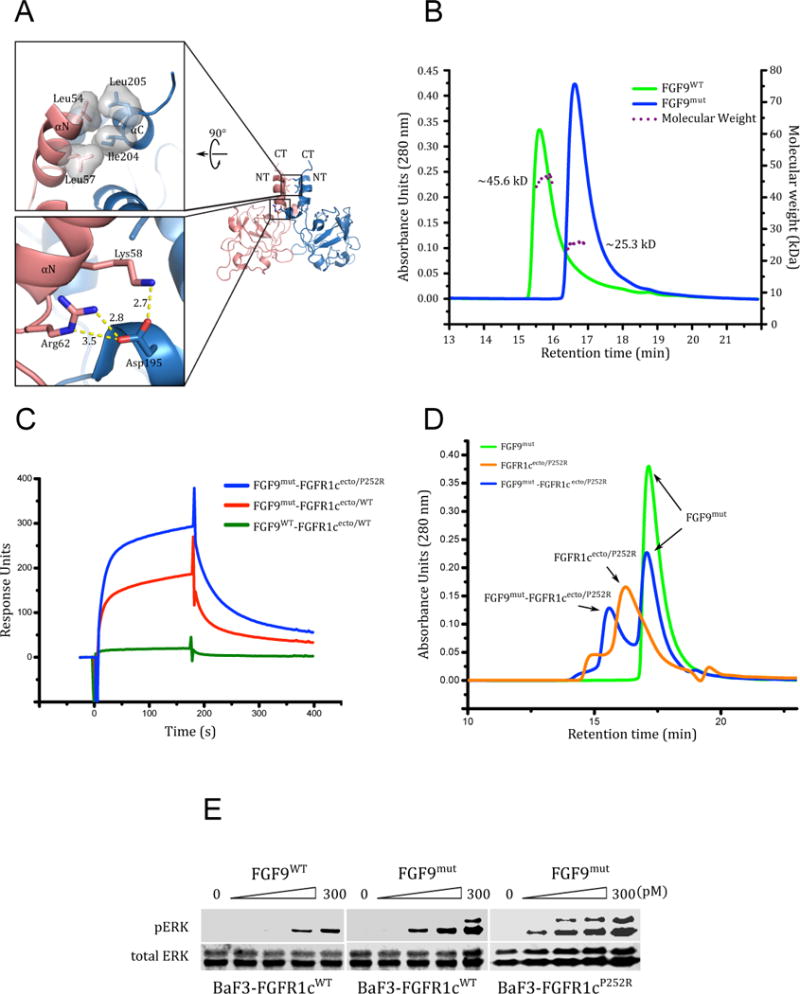
(A) Cartoon representation of the FGF9 homodimer (PDB ID: 1IHK) (Plotnikov et al., 2001) with monomers colored pink and light blue, respectively. Upper and lower boxes show close-up views of the hydrophobic and hydrogen bonding interactions at the dimer interface portion involving the unique N- and C-terminal helices that flank the β-trefoil core of FGF9 subfamily ligands. NT and CT denote the N- and C-termini of the monomers. (B) The D195A, I204A, and L205A triple mutation at the C-terminus of FGF9 render FGF9 monomeric. Purified samples of wild type (FGF9WT) and FGF9 triple mutant (FGF9mut) at 50 μM were analyzed by SEC-MALS. Solid and dashed lines denote UV absorption unit (AU) and molecular weight (MW), respectively. The theoretical MW of an FGF9 monomer is 20 kDa. (C) The P252R Pfeiffer syndrome gain-of-function FGFR1c mutation and ‘monomerizing’ FGF9 mutations enhance FGF9-FGFR1c binding. An overlay of SPR sensorgrams obtained by injecting FGFR1cecto/WT and FGFR1cecto/P252R over CM5 chips containing immobilized FGF9WT and FGF9mut. (D) Analysis of a 1.1:1 mixture of FGF9mut and FGFR1cecto/P252R on a TSKgel SuperSW3000 size-exclusion column mounted on a Waters Breeze HPLC system. The UV traces of FGF9mut alone, FGFR1cecto/P252R alone, and FGF9mut-FGFR1cecto/P252R complex are colored green, orange, and blue, respectively. (E) Detection by immunoblotting of ERK phosphorylation in total cell lysates from BaF3-FGFR1cWT and BaF3-FGFR1cP252R cells stimulated with 0–300 pM FGF9WT or FGF9mut and 5 μg/mL heparin. Bottom blots: Detection of total ERK protein as a sample loading control.
Further stabilization of the FGF9-FGFR1c complex was achieved by employing an FGFR1c ectodomain harboring the P252R mutation (FGFR1cecto/P252R), a gain-of-function mutation that causes the Pfeiffer craniosynostosis syndrome (Muenke et al., 1994). Using X-ray crystallography and SPR spectroscopy, we previously demonstrated that the Pro->Arg pathogenic mutations introduce additional, non-specific hydrogen bonds between the mutated FGFR and the backbone atoms of FGFs that manifest as a generalized (non-specific) increase in binding affinities of mutated FGFR for all FGFs (both normal and illegitimate ligands) (Beenken et al., 2012; Ibrahimi et al., 2001; Ibrahimi et al., 2004a; Ibrahimi et al., 2004b; Olsen et al., 2006). However, these mutations do not switch the binding specificity of the mutated FGFRs; i.e the mutated FGFRs retain their natural ligand binding specificity but simply gain some affinity towards non-cognate/illegitimate ligands (Ibrahimi et al., 2004b). In the case of FGF9, the enhancement in binding affinity is particularly robust (over 20-fold) as measured previously by SPR spectroscopy implicating this ligand as a candidate FGF in the etiology of craniofacial disorders (Ibrahimi et al., 2004b). Indeed, FGF9mut formed a more stable complex with the mutated FGFR1cecto/P252R ectodomain than with the wild type FGFR1c ectodomain (FGFR1cWT) as judged by SPR binding analysis and size-exclusion chromatography (Figure 1C, 1D). To validate these in vitro binding data, we generated a BaF3 cell line ectopically expressing FGFR1c harboring the P252R mutation (FGFR1cP252R). BaF3-FGFR1cWT and BaF3-FGFR1cP252R were exposed to FGF9WT, and receptor activation was assessed by western blotting of cell lysates with Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibodies. As shown in Figure 1E, FGFR1cP252R responded by eliciting much stronger signal than FGFR1cWT thus corroborating the in vitro data that FGF9 forms a more stable/tighter complex with FGFR1cP252R than with FGFR1cWT.
Having confirmed the stability and functionality of the FGF9mut-FGFR1cP252R in vitro and in living cells, we then proceeded with crystallizing the complex. The complex crystallized in the monoclinic space group C2 with a single 1:1 complex in the asymmetric unit. The crystal structure of the complex was solved by molecular replacement and has been refined to 2.6 Å resolution with Rwork and Rfree of 18.26% and 22.82%, respectively (Table 1). The final model consists of a 1:1 FGF9-FGFR1c complex, 58 water molecules, and 3 sulfate ions. The N-terminal residues 35 to 59 of FGF9 and the βC-βC′ loop (residues 298 to 305) in the D3 domain of FGFR1c are disordered in the structure.
Table 1.
X-ray Data Collection and Refinement Statistics
| Construct | FGF9-FGFR1c |
|---|---|
| Data Collection | |
| X-ray wavelength | 0.97900 |
| Space group | P212121 |
| Unit Cell Dimensions | |
| a, b, c (Å) | 42.159,86.793,136.857 |
| α, β, γ (°) | 90.00,90.00,90.00 |
| Resolution (Å) | 50–2.50(2.54–2.50)a |
| No. measured reflections | 114188 |
| No. unique reflections | 18063(867) |
| Data redundancy | 6.3(6.1) |
| Data completeness (%) | 99.4(97.6) |
| Rsym (%)b | 8.7(37.8) |
| I/sig | 29.8(4.0) |
|
| |
| Refinement | |
| Resolution (Å) | 29.5–2.50(2.57–2.50) |
| No. unique reflections | 18002(1330) |
| No. Reflections (Rfree)c | 1799(133) |
| Rwork/Rfree | 18.26/22.82(21.82/29.73) |
| No. atoms | |
| Protein | 2764 |
| Ligand/Ion | 15 |
| Solvent | 58 |
| R.m.s. deviations | |
| Bond length (Å) | 0.009 |
| Bond angle (°) | 1.208 |
| Average Bfactors (Å2) | |
| Protein | 52.25 |
| Ligand/Ion | 126.63 |
| Solvent | 44.04 |
| Ramachandran Plot | |
| Outliers (%) | 0.00 |
| Allowed (%) | 4.46 |
| Favored (%) | 95.54 |
| Rotamer outliers (%) | 1.68 |
| No. C-Beta Deviations | 0 |
| All-Atom Clashscore | 5.27 |
|
| |
| PDB ID | 5W59 |
Values in parenthesis are for the highest resolution shell.
Rsym = Σ|I− < I >|/ΣI, where I is the observed intensity of a reflection, and <I> is the average intensity of all the symmetry related reflections.
Number of reflections randomly excluded from the refinement for Rfree calculation.
Overall structure of the FGF9mut-FGFR1cecto/P252R complex
As anticipated based on the previous FGF-FGFR crystal structures, FGF9 binds FGFR1c at the junction between the D2 and D3 domains making extensive contacts with both domains and the intervening short D2-D3 linker region (Figure 2A). There are no significant structural changes in the core region between the receptor-bound and the free FGF9 molecules (Figure 2B). However, the entire N-terminus of the receptor-bound FGF9 molecule including the αN helix is disordered (Figure 2B). The interface between FGF9 and D2 of FGFR1c preserves most of the conserved contacts seen in previous FGF-FGFR structures. Notably, at the heart of this interface, Tyr-67 of FGF9 packs against Ala-167 of FGFR1c whilst also making two hydrogen bonds with backbone atoms of D2 (Figure 2C, upper panel). Likewise, the hydrophobic contacts between Tyr-145 and Leu-188 of FGF9 and D2 are also conserved features of all the FGF-FGFR complexes studied to date (Figure 2C, upper panel). The hydrogen bonding between Arg-190 of FGF9 and Asp-246 of D2 at the periphery of the interface resonates that observed in the FGF8b-FGFR2c structure (Figure 2C, lower panel) (Olsen et al., 2006). The interface between FGF9 and the receptor D2-D3 linker contains three hydrogen bonds between Arg-250 of FGFR1c and Trp-144 and Asn-146 of FGF9 that are observed universally in all FGF-FGFR complex structures (Figure 2D, upper panel). Unique to the FGF9-FGFR1c complex, however, Phe-140 (in β8) and Trp-144 (in β9) of FGF9 interact with Arg-250 in the receptor D2-D3 linker through π-cation and hydrophobic contacts, respectively (Figure 2D, lower panel). These two aromatic residues are exclusively conserved in the FGF9 subfamily whereas ligands from other FGF subfamilies typically have aliphatic residues at these two locations (Figure S1A). Hence, the FGF9-FGFR1c structure indicates that FGF9 draws significantly greater binding affinity through interactions with the D2-D3 linker than other FGFs typically do (Figure 2D). The interface between FGF9 and D3 of FGFR1c also features the predicted conserved hydrogen bonds between Glu-138 (in β8) and Arg-62 (in β1) of FGF9 with the backbone of the βB′-βC loop in the ‘unspliced’ constant region of the D3 domain (Figure 2D). Reminiscent of several other FGF-FGFR complexes, the ligand binding results in formation of a cleft in the receptor D3 domain between the alternatively spliced βC′-βE loop and the constant βB′-βC loop. These conserved FGF-FGFR contacts provide general (non-specific) binding affinity and impose upon the receptor, the characteristic elongated conformation akin to previous FGF-FGFR complex structures (Figure 2A). The overall topology of the FGF9-FGFR1c complex is closest to that of the FGF2-FGFR1c complex (RMSD 1.114 Å) and is most distant from that of the FGF10-FGFR2b complex (RMSD 2.199 Å).
Figure 2. Structural features of the FGF9mut-FGFR1cecto/252R complex.
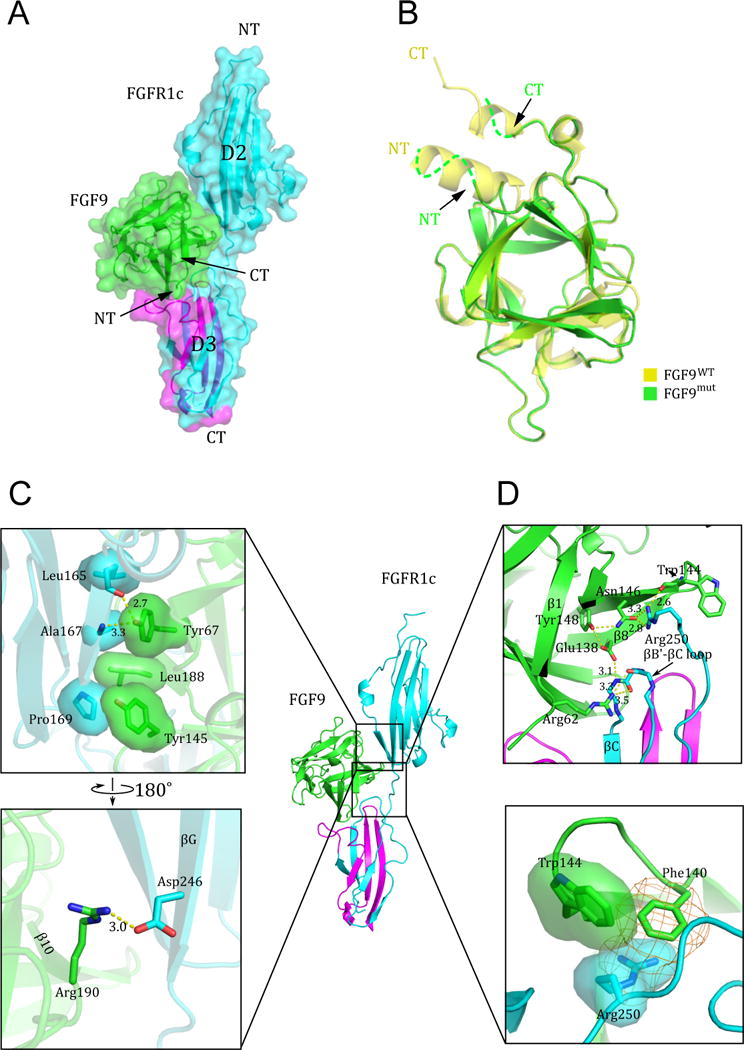
(A) The FGF9mut-FGFR1cecto/P252R complex structure in both cartoon and surface representations in traditional “front view” orientation. The receptor is colored cyan except for its alternatively spliced region in D3, which is colored magenta. The ligand (in green) engages D2, D3, and D2-D3 linker of the receptor. (B) Superimposition of of free (unbound) FGF9 (PDB ID: 1IHK) (Plotnikov et al., 2001) onto the receptor-bound FGF9 shows disordering of the FGF9 N-terminus upon receptor binding. NT and CT denote N- and C-termini of FGF9 and FGFR1c. Arrows point to the N- and C-terminus of receptor-bound FGF9 to emphasize their disordering in the receptor-bound state. (C) The interface between FGF9 and FGFR1c D2 domain. Hydrophobic interactions between Tyr-67, Tyr-145, and Leu188 of FGF9 and Leu-165, Ala-167, and Phe-169 in D2 of receptor (upper box) are highly conserved, whereas the hydrogen bonding contact between Arg-190 of FGF9 and Asp-246 of FGFR1c (lower box) is seen only in a subset of FGF-FGFR complexes. (D) The interface between FGF9 and FGFR1c D2-D3 linker and the constant (unspliced) region of D3. The upper box shows the conserved network of hydrogen bonds between Trp-144 and Asn-146 FGF9 and Arg-250 of the receptor D2-D3 linker, and between Glu-138 (in β8) and Arg-62 (in β1) of FGF9 with the backbone of the βB′-βC loop in the ‘unspliced’ constant region of the D3 domain. The π-cation (orange mesh) and hydrophobic interactions (transparent surface) between Phe-140 and Trp-144 of FGF9 and the side chain of Arg-250 are unique to the FGF9 subfamily (lower box). See also Figure S1.
The core region of FGF9 plays a primary role in the determination of FGF9-FGFR1c binding specificity
We have previously determined the crystal structures of eight distinct FGF-FGFR complexes containing FGF1, FGF2, FGF8, and FGF10 as bound ligands (Belov and Mohammadi, 2013). Through analysis of these structures, we have previously shown that FGF-FGFR binding specificity is principally dictated by contacts between the highly divergent N-termini of FGFs and the alternatively spliced regions in D3 of FGFRs in particular the βC′-βE loop and βF-βG strands (Belov and Mohammadi, 2013; Olsen et al., 2006; Yeh et al., 2003). The structural data have also shown that secondary specificity is mediated by contacts made between the less divergent elements in FGF core homology region namely β4, and β8 strands and β4-β5 loop and the alternatively spliced βC′-βE and βF-βG loops of the FGFR D3 domain. Hence, by analogy we anticipated that FGF9 would also attain its FGFR isoform selectivity by primarily relying on its divergent N-terminus to make specific contacts with the alternatively spliced regions in D3. Unexpectedly, in the FGF9-FGFR1c structure, the entire N-terminal region (residues 35–59) including the αN helix is disordered implying that it does not directly participate in receptor binding (Figure 2B). Indeed, modeling studies show that the presence of a rigid αN helix is incompatible with FGFR1c binding as it directly clashes with the receptor βC′-βE loop (Figure 3A). Instead, detailed analysis of the FGF9-FGFR1c complex structure reveals that the FGF9 core region assumes a primary role in determining FGF9-FGFR specificity. In particular, the interface between the β8 strand of FGF9 and the βF-βG loop of FGFR1c D3 domain appear to play a dominant role in providing receptor-binding specificity of FGF9. Here, Ser-346 in the βF-βG loop of receptor D3 makes water-mediated hydrogen bonds with Glu-138 and Phe-140 of FGF9 (Figure 3B, right panel). Modeling studies show that replacement of Ser-346 by a tyrosine or phenylalanine, the corresponding residues in the ‘b’ splice isoforms and in FGFR4 (Figure 3C), would severely reduce receptor binding affinity of FGF9. Specifically, three out of four possible rotameric positions for a tyrosine would cause major steric clashes with FGF9 besides the loss of hydrogen bonding contacts. The fourth rotamer position would not create steric conflict, but it would still result in the loss of water-mediated hydrogen bonding contacts with FGF9 (Figure S1C). The receptor-binding specificity of FGF9 is further biased towards FGFR1c and FGFR2c due to the hydrophobic contacts between the FGF9 core and Val-316 at the upper ridge of the receptor D3 cleft (Figure 3B, left panel). This valine is conserved in FGFR2c but is replaced by an alanine in FGFR3c (Figure 3C), which should weaken this hydrophobic interaction and hence lower FGF9-FGFR3c affinity.
Figure 3. Molecular determinants of FGF9-FGFR1c binding specificity.
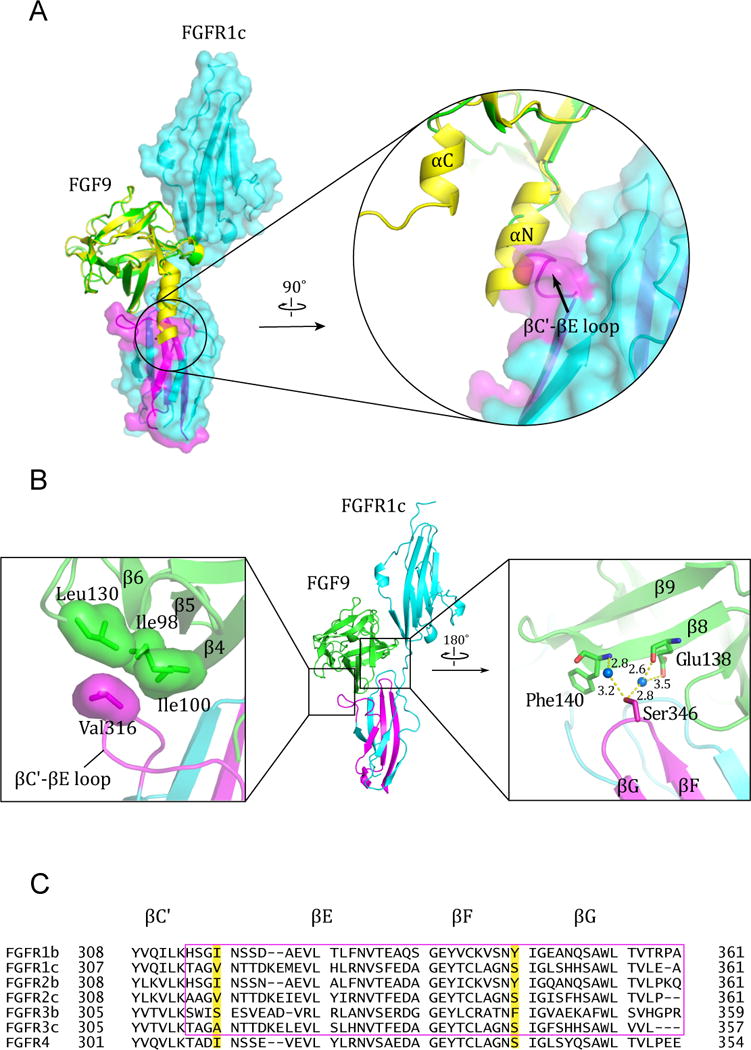
(A) Superimposition of free (unbound) FGF9 (PDB ID: 1IHK) (Plotnikov et al., 2001) onto the FGF9 in the FGF9mut-FGFR1cecto/P252R structure. Note the steric clash between the the N-terminal helix αN of free FGF9 and the βC′-βE loop of the receptor. (B) Specific contacts at the interface between FGF9 core region and the alternatively spliced βC′-βE and βF-βG loops in FGFR1c D3. Hydrophobic interactions between Ile-98, Ile-100, and Leu-130 of FGF9 and Val-316 in the βC′-βE loop of receptor D3 (left box) account for the higher affinity of FGF9 for FGFR1c and FGFR2c isoforms relative to FGFR3c. Shown in the right box are the water-mediated hydrogen bonds between Ser-346 from the βF-βG loop of receptor D3 and Glu-138 and Phe-140 from the β8 strand in the FGF9 β-trefoil core. Blue balls represent water molecules. Hydrogen bonds are represented as dashed yellow lines and their distances are given. (C) Structure based sequence alignment of the second half the D3 of the seven principal FGFR isoforms. β-strands are denoted as arrows above the sequences and the strands that pair with one another in upper and lower β-sheets of the Ig fold of D3 are indicated with the same color. Val-316 and Ser-346 on the FGFR1c sequence and their corresponding residues in other receptors are highlighted with yellow color. The alternative spliced regions of the receptors are boxed with a magenta rectangle. See also Figure S1.
Functional validation of the mode of FGF9-FGFR specificity observed in the FGF9-FGFR1c crystal structure
To validate the structurally deduced mode of FGF9-FGFR binding specificity, we tested the effects of mutating Ser-346 to tyrosine and Val-316 to alanine on the affinity of FGFR1c for FGF9. The binding interactions of FGFR1cecto/WT and the mutated FGFR1c ectodomains (FGFR1cecto/S346Y and FGFR1cecto/V316A) with FGF9 were studied using SPR spectroscopy (Figure 4A–C). Consistent with the structural data, FGF9 bound poorly to both mutant FGFR1c ectodomains, and FGFR1cecto/S346Y mutant showed a greater loss in FGF9 binding than the FGFR1cecto/V316A mutant. Next, the impacts of these two mutations on FGF9-FGFR1c signaling were investigated in the context of BaF3 cell lines engineered to ectopically express FGFR1cS346Y and FGFR1cV316A mutants. In parallel, we also studied the effects of the receptor mutations on the ability of FGF1 to activate FGFR1c in order to further assess the specific contribution of these two receptor residues in FGF9 recognition. Both receptor mutations significantly diminished the ability of FGF9 to activate FGFR1c in BaF3 cells as measured by MAPK activation (Figure 4D). In contrast, the V316A substitution had nearly no effect on FGF1-FGFR1c signaling, whereas the S346Y mutation actually enhanced the ability of FGF1 to activate FGFR1c. Importantly, the enhanced signaling by the mutant in response to FGF1 is expected because FGF1 has an arginine in the position corresponding to Gln-139 of FGF9 (Figure S1A), which enables FGF1 to engage in π-cation interaction with the mutated Tyr-346 reminiscent of that seen in the FGF1-FGFR2b structure (Beenken et al., 2012) (Figure S1B). Taken together the differential importance of these two receptor residues in mediating interactions of FGFR1c with FGF9 and FGF1 supports our structurally deduced mode of FGF9-FGFR1c binding specificity.
Figure 4. SPR and cell-based experiments confirm the importance of Val-316 and Ser-346 of FGFR1c in contributing to FGF9-FGFR1c specificity.
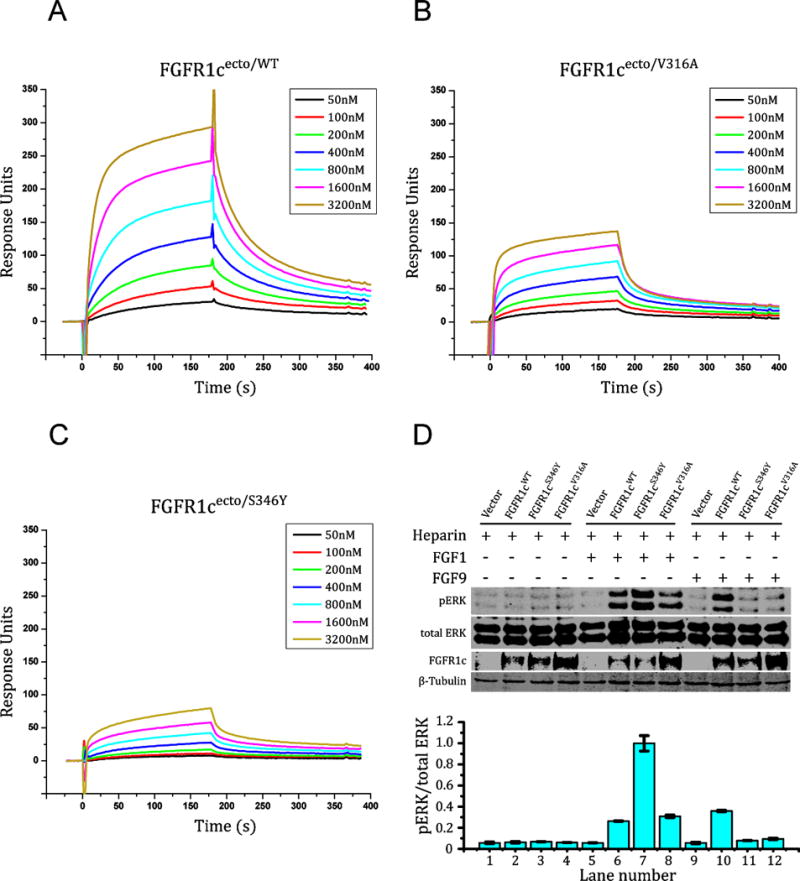
(A–C) Overlays of SPR sensorgrams illustrating FGF9mut interaction with the ectodomains of FGFR1cWT, FGFR1cV316A, and FGFR1cS345Y. FGF9mut was immobilized on an SPR biosensor chip, and increasing concentrations of FGFR1cecto/WT, FGFR1cecto/S346Y, or FGFR1cecto/V316A were injected over the chip. (D) Comparison of the activity of FGF9WT on BaF3 cell lines ectopically expressing FGFR1cWT, FGFR1cV316A, or FGFR1cS345Y. Soluble lysates from untreated cells or cells treated with 2 nM of FGF9 or FGF1 (control) in the presence of heparin were analyzed by westernblotting with pERK1/2 antibodies. To control for similar levels of FGFR1c expression, cell lysates were blotted with anti-FGFR1c antibodies directed against the C-terminal tail of FGFR1c. To control for equal sample loading, the membranes were developed with anti-β tubulin antibodies. pERK signals were normalized to total ERK. Error bars represent standard deviations. See also Figure S1.
The N-terminus of FGF9 regulates receptor binding specificity of FGF9 by controlling the availability of receptor binding sites in the FGF9 core region
According to the crystal structure, the N-terminus of FGF9 does not make specific contacts with FGFR1c to dictate receptor binding specificity of FGF9. However, since FGF9 N-terminus plays a major role in the formation of autoinhibited FGF9 homodimers, we hypothesized that the FGF9 N-terminus may indirectly modulate its receptor binding specificity by controlling the pool of receptor-binding capable ‘active’ FGF9 monomers. This hypothesis was particularly worth exploring in light of the limited number of specific contacts between FGF9 core and FGFR seen in the crystal structure (Figure 3B). To this end, guided by the current crystal structure of the FGF9-FGFR1c complex, we further truncated the N-terminus of our FGF9 construct to residue Gly-59 (FGF9ΔNT), the first ordered residue of FGF9 in the FGF9 mut-FGFR1cecto/P252R crystal structure. According to the FGF9-FGFR1c structure, these disordered N-terminal residues (Asp-35 to Lys-58) are dispensable for receptor binding. However, as seen in Figure 1A, residues Leu-54, Leu-57, and Lys-58 from this N-terminal region play key roles in the formation of the autoinhibited FGF9 homodimer. Indeed, analysis of the truncated FGF9ΔNT by SEC-MALS showed that it elutes as a single peak with a calculated mass of 20.3 kD consistent with being a monomer (Figure 5A). We then treated BaF3-FGFR1cP252R and BaF3-FGFR1bP252R cells side-by-side with the dimeric FGF9WT and the two monomerized FGF9 ligands (FGF9ΔNT and FGF9mut) and analyzed receptor activation by immunoblotting the cell lysates with Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibodies. In agreement with the reported specificity of FGF9 for FGFR ‘c’ isoforms (Zhang et al., 2006a), FGF9WT treatment led to MAPK phosphorylation in the BaF3-FGFR1cP252R but failed to induce MAPK phosphorylation in the BaF3-FGFR1bP252R cells (Figure 5B). Remarkably, however, both monomerized FGF9mut and FGF9ΔNT acquired substantial ability to induce MAPK phosphorylation in BaF3-FGFR1bP252R cells (Figure 5B–F). Consistent with their monomeric state, both FGF9mut and FGF9ΔNT also exhibited greater potencies than FGF9WT in inducing MAPK phosphorylation in BaF3-FGFR1cP252R cell line (Figure 5B, Figure 1E). These data show that interference with homodimer formation by manipulating either the N or C-terminus confers upon the mutated monomeric FGF9 molecules the ability to illegitimately bind and activate ‘b’ isoform of FGFR1. To test the generality of this mechanism, we also compared the abilities of FGF9WT, and FGF9mut to activate ‘b’ and ‘c’ splice isoforms of FGFR2 that were ectopically expressed on the surface of BaF3 cell lines. Consistent with the known specificity of FGF9 for FGFR ‘c’ isoforms, FGF9WT treatment led to robust MAPK phosphorylation in the BaF3-FGFR2c cell line but not in BaF3-FGFR2b cells. By contrast, FGF9mut acquired significant ability to induce MAPK phosphorylation in BaF3-FGFR2b cells (Figure 5C). These data establish the generality of homodimerization as a regulatory mechanism for FGF9 to discriminate between ‘b’ and ‘c’ isoforms of FGFRs.
Figure 5. The divergent N-termini of FGF9 subfamily ligands indirectly govern FGFR binding pecificity by controlling the availability of active FGF9 monomers.
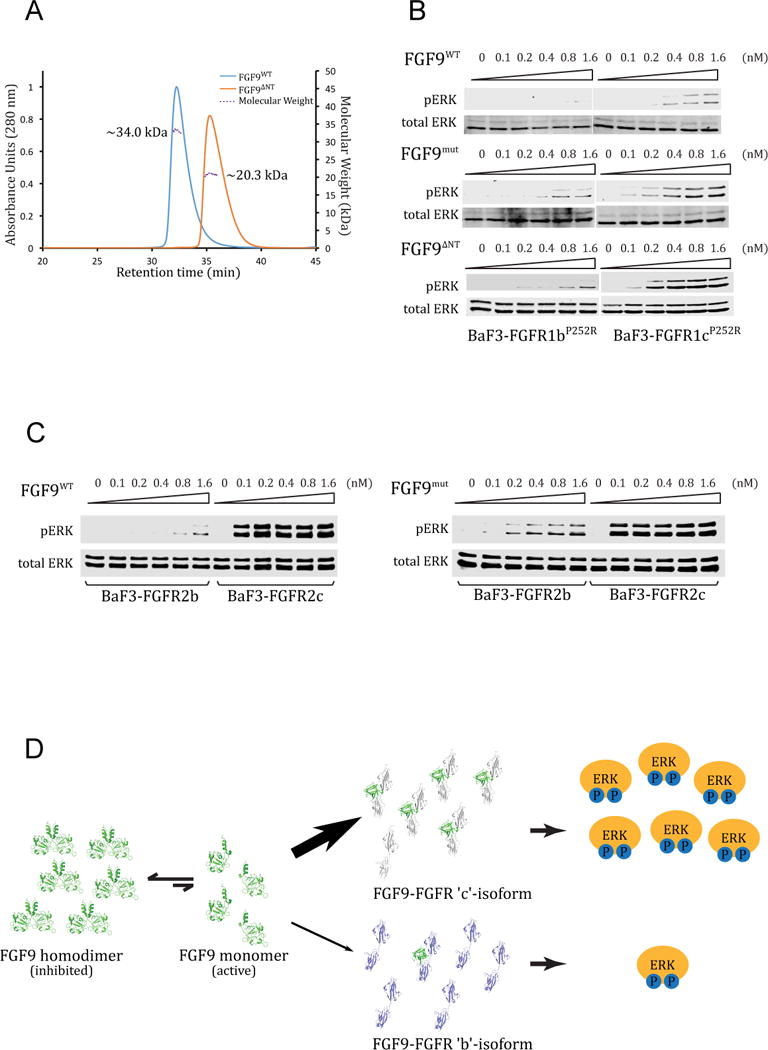
(A) SEC-MALS analysis of FGF9WT and its N-terminally truncated derivative FGF9ΔNT. Solid curves represent the UV absorbance trace of each ligand eluted from Superdex 200 10/300 GL column. Dashed lines represent the molecular weight of each component measured by MALS. Note that the theoretical MW of FGF9ΔNT is 17.7 kDa. (B) Detection by immunoblotting of ERK phosphorylation in total cell lysates from BaF3 cells expressing either FGFR1bP252R (BaF3-FGFR1bP252R) or FGFR1cP252R (BaF3-FGFR1cP252R) that were stimulated with a gradient concentration ranging from 0.1 to 1.6 nM of FGF9WT, FGF9mut, or FGF9ΔNT. (C) Comparison of the activities of FGF9WT and FGF9mut on BaF3 cell lines expressing FGFR2b (BaF3-FGFR2bWT) or FGFR2c (BaF3-FGFR2cWT). Soluble lysates from untreated cells and cells stimulated with indicated doses of each ligand were analyzed by westernblotting with pERK1/2 antibodies. Bottom blots in panels B and C: Detection of total ERK protein as a sample loading control. (D) Illustration of the working model by which FGF9 homodimerization controls the receptor binding specificity of FGF9. FGF9 exits in an equilibrium between the ‘autoinhibited’ receptor-binding incompetent FGF9 dimers and receptor-binding capable ‘active’ monomers. The concentration of active monomers is enough to activate FGFR ‘c’ isoforms but is insufficient to cause off-target/illegitimate activation of FGFR ‘b’ isoforms. See also Figure S2.
Based on these data we can conclude that, in stark contrast to FGF7 and FGF8 subfamilies, the FGF9 N-terminus regulates receptor binding specificity in a negative fashion by promoting homodimerization, which suppresses the concentration of active monomers to a level that is below the threshold capable of causing illegitimate binding to FGFR ‘b’ isoforms. However, the population of the active monomers is still sufficient for the limited specific binding between the FGF9 core and the FGFRc D3 to prevail and lead to FGF9-FGFRc complex formation and signaling (Figure 5D). Deletion of the FGF9 N-terminus relieves this autoinhibition increasing the concentration of monomeric FGF9 ligands to a sufficiently high level that can cross the ligand specificity barrier set by alternative splicing and activate FGFR ‘b’ isoforms.
Structural basis for the gain-of-function by the P252R FGFR1 Pfeiffer syndrome mutation, and loss-of-function effects of the S99N FGF9 mutation identified in multiple synostoses syndrome (SYNS) patients and of the N143T FGF9 substitution found in Elbow knee synostosis mice (Eks)
Analysis of the FGF9-FGFR1c structure also provides an explanation for why the P252R mutation confers upon FGFR1c a drastic gain in FGF9 binding vis-a-vis other FGFs (Ibrahimi et al., 2004b). At the interface between FGF9 and the receptor D2-D3 linker, the guanidinium group of the pathogenic Arg-252 residue makes conserved hydrogen bonds with backbone carbonyl oxygens of FGF9, as observed in previous structures of FGF ligands bound to FGFR harboring this linker mutation (Figure 6A). However, the mutated Arg-252 residue also engages in favorable π-cation contacts with Phe-140 and Trp-144 of FGF9 (Figure 6A). As mentioned earlier, Phe-140 and Trp-144 are unique to FGF9 subfamily ligands (Figure S1A), which explains the robust increase in binding of these ligands by the pathogenic Pro->Arg mutation in FGFR1c, FGFR2c and FGFR3c.
Figure 6. Structural insights into mechanisms of action of the P252R FGFR1c Pfeiffer syndrome mutation, the S99N FGF9 mutation in multiple synostoses syndrome (SYNS) patients, and the N143T FGF9 substitution in mice with Elbow knee synostosis (Eks).
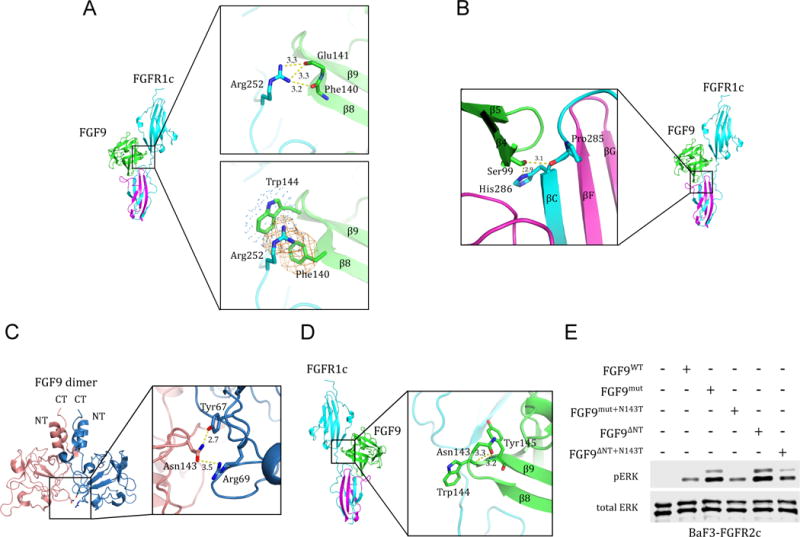
(A) Molecular basis for the robust increase in FGF9 binding affinity of pathogenic FGFR1-3 variants harboring Pro->Arg mutations in their D2-D3 linker region. The upper box shows the conserved hydrogen bonds between the side chain of the mutated Arg-252 residue in the D2-D3 linker of FGFR1c and the backbone carbonyl oxygens of Phe-140, Glu-141, and Glu-142 in FGF9 core region. The lower box depicts the FGF9 subfamily-specific π-cation and hydrophobic interactions (rendered as orange mesh and blue dots, respectively) between the side chain of the mutated Arg-252 residue and Phe-140 and Trp-144 of FGF9. (B) Structural basis for the loss-of-function effects of the S99N FGF9 mutation identified in multiple synostoses syndrome (SYNS) patients. Hydrogen bonds between Ser-99 of FGF9 and D3 of FGFR1c are depicted as dashed yellow lines. (C) The FGF9 N143T mutation found in Elbow knee synostosis (Eks) mice is a loss-of-function mutation. Cartoon presentation of the autoinhibited FGF9 dimer (PDB ID: 1IHK) (Plotnikov et al., 2001) with a close-up view of the dimer interface mediated by Asn-143. Asn-143 on one of the FGF9 molecules (pink) and Tyr-67, Arg-69 on the other FGF9 (blue) molecule are shown as sticks. Hydrogen bonds are depicted by dashed yellow lines and their lengths are given in Å. (D) FGF9-FGFR1c structure model showing the location of Asn-143 of FGF9 in the complex. Dashed yellow lines represent the hydrogen bonds between the side chain of Asn-143 and the backbone amide nitrogen of Trp-144 and Tyr-154 that facilitate formation of β8-β9 turn. (E) Comparison of the activities of FGF9WT, FGF9mut, FGF9ΔNT, and derivatives of FGF9mut and FGF9ΔNT carrying the N143T mutation (FGF9mut+N143T and FGF9ΔNT+N143T). BaF3-FGFR2c cells were stimulated with 500 pM of each ligand and FGFR2c activation was assessed by blotting total cell lysates with phospho-Erk1/2 antibodies. Bottom blot: Detection of total ERK protein as a sample loading control. See also Figure S1.
The FGF9-FGFR1c crystal structure also provides the molecular basis for the loss-of-function conferred by the S99N mutation in FGF9, which has been identified in 12 patients with multiple synostoses syndrome (SYNS) in a large Chinese family (Wu et al., 2009). In the FGF9-FGFR1c crystal structure, the side chain of Ser-99 of FGF9 makes two short hydrogen bonds with the backbone carbonyl oxygen of Pro-285 and the side chain of His-286 in the βB′-βC loop of receptor D3 (Figure 6B). Mutation of Ser-99 to Asn should cause a major loss in receptor binding both due to the loss of these hydrogen bonds (Figure 6B). Consistent with our structural prediction, a recent study showed that the S99N mutation decreases the binding affinity between FGF9S99N and FGFR, ultimately attenuating FGF9 signal transduction (Tang et al., 2017).
The gain-of-function behavior of our monomerized FGF9ΔNT and FGF9mut stands at odds with the previously published data on the N143T FGF9 substitution found in Elbow knee synostosis (Eks) mice (Harada et al., 2009). N143T mutation also suppresses FGF9 dimerization however, in contrast to our monomeric FGF9ΔNT and FGF9mut variants, FGF9N143T exhibits a loss-of-function as measured by its lower mitogenic activity compared to the wild type FGF9 (Harada et al., 2009). As alluded to earlier, many of the core residues that are involved in the homodimer interface correspond precisely to the same residues that are necessary for FGFR binding. Asn-143, which resides on the β8-β9 hairpin loop within FGF9 β-trefoil core, is a prime example of such FGF9 residues that fulfill such dual roles in both FGF9-FGF9 homodimerization and FGF9-FGFR binding. As shown in Figure 6C, in the crystal structure of FGF9 homodimer, Asn-143 makes two hydrogen bonds with Tyr-67, Arg-69 that promote formation of the autoihibited FGF9 homodimer. On the other hand, analysis of the crystal structure of FGF9-FGFR1c complex shows that Asn-143 plays a critical role in FGF9-FGFR1c binding by facilitating the β8–β9 hairpin formation, which provides key contacts with FGFR1c (Figure 6D). Hence, according to this structural analysis, the N143T mutation should impair the ability of FGF9 to bind and activate FGFR even though it is able to monomerize the ligand. To unambiguously demonstrate that N143T is indeed a loss-of-function mutation, we introduced this mutation into both FGF9mut and FGF9ΔNT mutant background (FGF9mut+N143T and FGF9ΔNT+N143T). We reasoned that if the N143T were a loss-of-function mutation then these compound mutants should incur a loss in bioactivity compared to their parent FGF9mut and FGF9ΔNT mutants. Consistent with our FGF9-FGFRc crystal structure, westernblot analysis of FGFR1c expressing cells treated with FGF9mut+N143T or FGF9ΔNT+N143T ligands showed that the N143T mutation diminishes the activities of both monomerized FGF9mut and FGF9ΔNT mutants (Figure 6E). Hence, the dual involvement of N143 in both ligand homodimerization and FGFR binding is the reason why despite being a monomer, the N143T FGF9 mutant exhibited a loss of function in the cell-based assays (Harada et al., 2009).
FGF-FGFR binding specificity is a key regulatory mechanism in FGF signaling and hence understanding the molecular rules that govern FGF-FGFR specificity is essential for comprehension of this important signaling system in biology and pathophysiology. Previous crystallographic studies of 8 different FGF-FGFR complexes have firmly established that contacts between FGFs and alternatively spliced regions in D3 provide the primary mechanism of achieving specificity and or promiscuity in FGF-FGFR interactions. These studies have revealed two distinct structural modes by which the divergent N-termini of FGFs engage the alternatively spliced βC′-βE loop of FGFR to mediate FGF-FGFR binding specificity. One of the structural modes is used by the FGF1 and FGF7 subfamilies, and another one is FGF8 subfamily specific (Figure S2A, B). However, our FGF9-FGFR1c structure discloses a third mode of FGF-FGFR binding specificity whereby the N-terminus of FGF9 does not partake directly in mediating FGF-FGFR specificity. Instead, it acts indirectly by controlling the availability of residues in the core domain of FGF9 to engage in specific contacts with the βF-βG loop of the alternatively spliced region in FGFR D3.
FGF9 secreted by the epithelium is a key mitogen for lung, kidney, and prostate mesenchyme (Colvin et al., 2001; Giri et al., 1999; Vainio, 2012). Moreover, FGF9 mediates hair follicle regeneration following skin injury (Gay et al., 2013), and also maintains pluripotency of nephron progenitor cells (Barak et al., 2012). Together with retinoic acid, FGF9 determines mammalian germ cell sexual fate commitment (Bowles et al., 2010). FGF9 has also been implicated in oncogenesis (Abdel-Rahman et al., 2008). It is the downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas (Hendrix et al., 2006), and participates in driving osteoblastic bone metastases in prostate cancer (Li et al., 2008). Given the breadth of physiological and pathobiological activities of FGF9, agonists and antagonists of this ligand hold vast translational potential. The structure of the FGF9-FGFR1c complex reported in this study may serve as a template to guide the discovery of both agonists and antagonists for this pleiotropic FGF ligand. The FGF9 agonists have the potential to serve as important therapeutic tools for tissue regeneration, repair, and long-term maintenance of self-renewing nephron progenitors in culture for therapeutic cell-replacement approaches. The antagonists of FGF9 signaling could have potential therapeutic value for treating human cancers.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for reagents should be directed to the Lead Contact, Dr. Moosa Mohammadi (moosa.mohammadi@nyumc.org).
Experimental Model and Subject Details
E. coli BL21(DE3) was used for heterologous expression of FGF9 and FGFR1c. To express the proteins, the bacteria cells were seeded in 1 L Luria Broth medium (RPI, Cat#L24045) and incubated at 37 °C until OD600 reaches 0.8~1.2. In case of FGF9, protein expression was induced with 1 mM IPTG (Sigma-Aldrich, Cat#5820-100GM) at 20°C, and the cells were harvest for protein purification after ~16 hours. For FGFR1c, the cells were induced for protein expression with 1 mM IPTG at 37 °C and were harvested after 4 hours’ incubation.
Method Details
Protein Purification and Crystallization
The cDNA fragments encoding residues 35-208 of the human FGF9 (termed FGF9WT), residues 59–208 of human FGF9 (termed FGF9ΔNT) and residues 141–365 of human FGFR1c (FGFR1c ectodomain; FGFR1cecto) were amplified by polymerase chain reaction and subcloned into the bacterial expression vector pET28a. The QuikChange XL site-directed mutagenesis kit (Agilent Technologies) was used to introduce the D195A/L204A/L205A triple mutations into FGF9WT (termed FGF9mut), and the N143T single mutation into FGF9ΔNT (termed FGF9ΔNT+N143T) and into FGF9mut (termed FGF9mut+N143T), and the single mutations P252R, V316A, and S346Y into FGFR1cecto (FGFR1cecto/P252R, FGFR1cecto/V316A, and FGFR1cecto/Y346Y) for in vitro experiments and into full length FGFR1c for cell-based assays. FGF9WT, FGF9mut, FGF9ΔNT, FGF9mut+N143T and FGF9ΔNT+N143T were expressed in Escherichia coli strain BL21 (DE3) and were precipitated from the soluble cell lysate fraction using 500 mM ammonium sulfate. The precipitated protein is then pelleted down at 18000 rpm (Sorvall RC-5C Plus superspeed centrifuge, Rotor SS-34) and dissolved in HEPES-NaOH pH 7.5 buffer containing 150 mM NaCl buffer. The protein solution is then filtered with 0.45 μm membrane (Corning sterile syringe filter, Part#431220) and loaded onto Source 15S ion exchange column (GE Healthcare Life Sciences, Prod#17094405). FGF9WT and all the mutant proteins are eluted using 100 ml linear NaCl gradient (0 – 1.0 M). The fractions containing FGF9 proteins are concentrated to ~5 mL and applied onto HiLoad 16/600 Superdex 75 prep grade (GE Healthcare Life Sciences, Prod#28989333) size-exclusion chromatography with HEPES-NaOH pH 7.5, 150 mM sodium chloride as running buffer for final purification.
Human FGFR1cecto/WT, FGFR1cecto/P252R, FGFR1cecto/V316A, and FGFR1cecto/Y346Y were expressed in E. coli BL21 DE3 cells. Inclusion bodies enriched in misfolded insoluble FGFR1c protein were dissolved in 6 M guanidinium hydrochloride and were refolded by dialysis for 2 days at 4°C against following buffers: buffer A (25 mM Tris pH 8.2, 150 mM NaCl, 7.5% glycerol), buffer B (25 mM Tris pH 8.2, 100 mM NaCl, 5% glycerol), and buffer C (25 mM Tris pH 8.2, 50 mM NaCl, 5% glycerol). Dialysis against each buffer was for minimally 12 hours. Correctly folded FGFR1c proteins were captured on a 5 ml heparin affinity HiTrap column (GE Healthcare Life Sciences, Prod#17-0407-03) and eluted with a 100 ml linear NaCl gradient (0 – 2.0 M). Fractions containing FGFR1c proteins were pooled, concentrated and applied onto a HiLoad 16/600 Superdex 75 prep grade sizing column. FGFR1c proteins were eluted isocratically in 25 mM HEPES-NaOH pH 7.5 buffer containing 1 M NaCl.
Purified ligand and receptor proteins were then mixed at a molar ratio of 1.1:1, and the mixture was injected onto a HiLoad 16/60 Superdex 200 prep grade (GE Healthcare Life Sciences, Prod#28989335) size-exclusion column in buffer consisting of 25 mM HEPES-NaOH pH 7.5, 150 mM NaCl. Fractions containing the ligand-receptor complex were pooled and concentrated to ~3 mg/ml. The purity of the complex was estimated to be greater than 95% based on SDS-PAGE analysis. The FGF9mut-FGFR1cP252R complex was crystallized at 18 °C in 100 mM Tris pH 8.0, 8% (w/v) PEG 20,000, 0.3 M sodium chloride, and 40 mM L-proline. SDS-PAGE analysis confirmed that both FGF9 (residues 35 to 208) and FGFR1c (residues 141 to 365) proteins used for crystallization are fully contained in the crystals (data not shown).
X-ray Diffraction Data Collection and Structure Determination
Crystals were soaked briefly in cryoprotectants containing mother liquor and up to 25% glycerol and flash frozen in liquid nitrogen. A single crystal was used to collect X-ray diffraction data at the NSLS beamline X4C. Data were indexed, integrated, and scaled using HKL2000. The FGF9mut-FGFR1cecto/P252R complex structure was solved by molecular replacement using the program Phaser in the Phenix Suite with the coordinates of FGF1-FGFR1c (PDB ID 3OJV) (Beenken et al., 2012) serving as the search model. Iterative model building and refinements were carried out using Coot (Emsley and Cowtan, 2004) and Phenix.refine (Adams et al., 2010), respectively. The structural representations were made using PyMOL (Schrodinger, 2015).
Surface Plasmon Resonance (SPR) Spectroscopy
Surface Plasmon Resonance (SPR) experiments were performed on a Biacore 2000 instrument (GE Healthcare Life Sciences). FGF9 proteins were covalently bound to a CM5 biosensor chip (GE Healthcare Life Sciences, Prod#BR100012) by amine coupling as follows: FGF9WT and FGF9mut were diluted to about 5 μg/ml using 10 mM sodium acetate pH 4.5 buffer, and were injected over a chemically activated CM5 chip surface at a flow rate of 5 μl/min. HBS-EP (10mM HEPES-NaOH pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% [vol/vol] polysorbate 20) was used as running buffer. Each FGF9 protein was coupled to a density corresponding to about 1,800 SPR response units (RUs). To control for nonspecific binding, FGF homologous factor 1B (FHF1B), which shares structural homology with FGFs but does not bind to FGFRs (Olsen et al., 2003), was covalently coupled onto the control flow channel of the chip. A range of concentrations of FGFR1cecto/P252R from 50 nM to 3,200 nM was prepared with HBS-EP buffer, and passed over the chip surface at a flow rate of 50 μl/min. The association and disassociation phases were 180 seconds each, with return to baseline achieved between each injection cycle. The binding data were processed and analyzed using BIAevaluation software.
Size Exclusion Chromatography–Multi-Angle Light Scattering (SEC-MALS)
The SEC-MALS instrument setup consisted of a Waters Breeze 2 HPLC system (Waters), a miniDAWN-TREOS 18-angle static light scattering detector with built-in 658.0-nm wavelength laser (Wyatt Technology Corp.), and an Optilab rEX refractive index detector (Wyatt Technology Corp.). Either a TSKgel SuperSW3000 column (for FGF9WT, FGF9mut, FGFR1cecto/P252R, and FGF9mut-FGFR1cecto/P252R complex; TOSOH Bioscience LLC, Prod#21485) or a Superdex 200 10/300 GL column (for FGF9WT and FGF9ΔNT; GE Healthcare life sciences, Prod#17517501) was placed in-line between the HPLC pump (Waters 1525) and the HPLC UV (Waters 2998 Photodiode Array), laser light scattering, and refractive index detectors. Light scattering and refractive index detectors were calibrated following the manufacturer’s guidelines. The refractive index increment (dn/dc) in which n is the refractive index and c is the concentration of the mixture of DDM and CHS in 20 mM Tris-HCl pH 8.0 buffer containing 300 mM NaCl, was determined offline using an Optilab T-rEX refractive index detector. Monomeric bovine serum albumin (#23210, Thermo Scientific, USA) was used as part of routine data quality control.
60 ml or more of 25 mM HEPES pH 7.5 buffer containing 150 mM NaCl were passed through the system at a flow rate of 0.5 mL/min to equilibrate the Superdex 200 10/300 GL column and establish stable baselines for light scattering and refractive index detectors. Wild type and mutate ligand and receptor ectodomain proteins (FGF9WT, FGF9mut, FGF9ΔNT, FGFR1cecto/P252R) were diluted to 50 μM using sodium phosphate buffer pH 6.7, 150 mM sodium sulfate. Mixtures of wild type FGFR1c-FGF9 and their mutants were prepared by adding FGFR1c to FGF9 at a molar ratio of 1:1.1 and concentrating the mixture to 50 μM final concentration. 50 μl of protein samples were injected onto the gel filtration column, and the column eluent was continuously monitored for 280 nm absorbance, laser light scattering, and refractive index. The analyses were performed at ambient temperature. Data were collected every second at the flow rate of 0.1 mL/min for SuperSW3000 column and 0.5 mL/min for Superdex 200 10/300 GL column. Laser light scattering intensity and eluent refractive index (concentration) data were adjusted manually for the volume delay of UV absorbance at 280 nm, and were processed using ASTRA software (Wyatt Technology Corp.). A protein refractive index increment (dn/dc value) of 0.185 ml/g was used for molecular mass calculations.
Lentivirus Generation and Cell Study
HEK293T cells (a gift from Dr. Alka Mansukhani, NYU Langone Medical Center) were maintained in DMEM medium (CORNING, Cat#10-017-CV) supplemented with 10% FBS (CORNING, Ref#35-010-CV), 100 U/ml of Penicillin and 100 μg/ml Streptomycin (Thermo Fisher Scientific, Cat#15140-122). BaF3 cells (a gift from Dr. Sara Byron, TGen), an IL3-dependent hematopoietic pro B cell line, were cultured in RPMI 1640 medium (CORNING, #10-040-CV) supplemented with 10% FBS, 100 U/ml of penicillin, 100 μg/ml streptomycin and 5 ng/ml murine IL-3 (Cell Guidance Systems Ltd, Cat#GFM1-100).
Stable expression of full-length (transmembrane) human FGFR1c and its mutants in BaF3 cells was achieved using lentiviral vectors. To generate lentiviral expression vectors, HEK293T cells were seeded at a density of about 8×105 in 10 cm cell culture dishes and co-transfected by calcium phosphate co-precipitation with 8 μg of lentiviral transfer plasmid encoding wild-type or mutant FGFR1c, 1.6 μg of pMD2.G envelope plasmid, and 2.5 μg of psPAX2 packaging plasmid. Fresh medium was added to the cells for a 3-day period after transfection. Cell culture supernatant containing recombinant lentivirus particles was harvested and used to infect 2×105 BaF3 cells in the presence of polybrene (5 μg/ml; Santa Cruz Biotechnology, Cat#134220). for 48 hours. The infected BaF3 cells were sorted for red fluorescence to isolate the cell clones stably expressing FGFR1cWT, FGFR1cP252R, FGFR1cV316A, or FGFR1cS346Y.
BaF3 cells were starved for 5 hours in FBS/IL-3 free RPMI 1640 medium followed by 10 min stimulation with FGF9WT and its mutated versions or with FGF1 as control in the presence of heparin (final concentration of 5 μg/mL; Fisher Scientific, Cat#BP252450). BaF3 cells overexpressing FGFR2b or FGFR2c were kindly provided by Dr. Pamela Pollock. These two cell lines were stimulated for 10 min with FGF9WT and FGF9mut in the presence of 5 μg/mL heparin. After stimulation, the cells were washed once with phosphate-buffered saline (PBS), and then lysed in radioimmunoprecipitation assay (RIPA) buffer. Protein concentration in the cell lysate was measured with the BCA protein assay reagents (Thermo Scientific, Prod#232284 and #23228). The primary antibodies used for immunoblot analysis were: Cell Signaling Technology® Phospho-p44/42 MAPK XP® mAb (#4370), Santa Cruz Biotechnology® ERK 1 Antibody (K-23) (#sc-153), and Thermo Scientific β Tubulin Antibody (#PA1-41331). The rabbit-source primary antibody used to detect the expression of FGFR1c was generated in our laboratory. The secondary antibody used to expose western results is: Goat anti-Rabbit IRDye 800CW (Li-COR, #926-32211).
Data and Software Availability
The coordinates and structure factors for the human FGF9-FGFR1c structure have been deposited in the RCSB Protein Data Bank under ID code 5W59.
Supplementary Material
Highlights.
Crystal structure of FGF9 in complex with FGFR1c ectodomain at 2.6 Å resolution
Homodimerization autoinhibits the concentration of active FGF9 monomers
FGF9 monomer pool is insufficient for illegitimate FGFR ‘b’ isoform activation
FGF9 core region directly participates in determining receptor binding specificity
Acknowledgments
This work was supported by the DE13686 grant (awarded to M.M.) from the National Institute for Dental & Craniofacial Research. The authors thank Drs. Regina Goetz, and Artur Belov for critically reading the manuscript and providing thoughtful suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTION
Y.L., J.M. and M.M. designed and performed the experiments, analyzed data, and wrote the paper. L.S., A.B. and A.E. contributed to experimental design, data analysis, and manuscript revision.
References
- Abdel-Rahman WM, Kalinina J, Shoman S, Eissa S, Ollikainen M, Elomaa O, Eliseenkova AV, Butzow R, Mohammadi M, Peltomaki P. Somatic FGF9 mutations in colorectal and endometrial carcinomas associated with membranous beta-catenin. Human mutation. 2008;29:390–397. doi: 10.1002/humu.20653. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak H, Huh SH, Chen S, Jeanpierre C, Martinovic J, Parisot M, Bole-Feysot C, Nitschke P, Salomon R, Antignac C, et al. FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Developmental cell. 2012;22:1191–1207. doi: 10.1016/j.devcel.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken A, Eliseenkova AV, Ibrahimi OA, Olsen SK, Mohammadi M. Plasticity in interactions of fibroblast growth factor 1 (FGF1) N terminus with FGF receptors underlies promiscuity of FGF1. The Journal of biological chemistry. 2012;287:3067–3078. doi: 10.1074/jbc.M111.275891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nature reviews. Drug discovery. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov AA, Mohammadi M. Molecular mechanisms of fibroblast growth factor signaling in physiology and pathology. Cold Spring Harbor perspectives in biology. 2013;5 doi: 10.1101/cshperspect.a015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Feng CW, Spiller C, Davidson TL, Jackson A, Koopman P. FGF9 suppresses meiosis and promotes male germ cell fate in mice. Developmental cell. 2010;19:440–449. doi: 10.1016/j.devcel.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Cancilla B, Ford-Perriss MD, Bertram JF. Expression and localization of fibroblast growth factors and fibroblast growth factor receptors in the developing rat kidney. Kidney international. 1999;56:2025–2039. doi: 10.1046/j.1523-1755.1999.00781.x. [DOI] [PubMed] [Google Scholar]
- Colvin JS, White AC, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gay D, Kwon O, Zhang Z, Spata M, Plikus MV, Holler PD, Ito M, Yang Z, Treffeisen E, Kim CD, et al. Fgf9 from dermal gammadelta T cells induces hair follicle neogenesis after wounding. Nature medicine. 2013;19:916–923. doi: 10.1038/nm.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geske MJ, Zhang X, Patel KK, Ornitz DM, Stappenbeck TS. Fgf9 signaling regulates small intestinal elongation and mesenchymal development. Development. 2008;135:2959–2968. doi: 10.1242/dev.020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri D, Ropiquet F, Ittmann M. FGF9 is an autocrine and paracrine prostatic growth factor expressed by prostatic stromal cells. Journal of cellular physiology. 1999;180:53–60. doi: 10.1002/(SICI)1097-4652(199907)180:1<53::AID-JCP6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Goetz R, Beenken A, Ibrahimi OA, Kalinina J, Olsen SK, Eliseenkova AV, Xu C, Neubert TA, Zhang F, Linhardt RJ, et al. Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Molecular and cellular biology. 2007;27:3417–3428. doi: 10.1128/MCB.02249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M, Murakami H, Okawa A, Okimoto N, Hiraoka S, Nakahara T, Akasaka R, Shiraishi Y, Futatsugi N, Mizutani-Koseki Y, et al. FGF9 monomer-dimer equilibrium regulates extracellular matrix affinity and tissue diffusion. Nature genetics. 2009;41:289–298. doi: 10.1038/ng.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer research. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- Ibrahimi OA, Eliseenkova AV, Plotnikov AN, Yu K, Ornitz DM, Mohammadi M. Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:7182–7187. doi: 10.1073/pnas.121183798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi OA, Zhang F, Eliseenkova AV, Itoh N, Linhardt RJ, Mohammadi M. Biochemical analysis of pathogenic ligand-dependent FGFR2 mutations suggests distinct pathophysiological mechanisms for craniofacial and limb abnormalities. Human molecular genetics. 2004a;13:2313–2324. doi: 10.1093/hmg/ddh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi OA, Zhang F, Eliseenkova AV, Linhardt RJ, Mohammadi M. Proline to arginine mutations in FGF receptors 1 and 3 result in Pfeiffer and Muenke craniosynostosis syndromes through enhancement of FGF binding affinity. Human molecular genetics. 2004b;13:69–78. doi: 10.1093/hmg/ddh011. [DOI] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends in genetics : TIG. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Jameson JL, Achermann JC, Ozisik G, Meeks JJ. Battle of the sexes: new insights into genetic pathways of gonadal development. Transactions of the American Clinical and Climatological Association. 2003;114:51–63. discussion 64–55. [PMC free article] [PubMed] [Google Scholar]
- Jaskoll T, Abichaker G, Witcher D, Sala FG, Bellusci S, Hajihosseini MK, Melnick M. FGF10/FGFR2b signaling plays essential roles during in vivo embryonic submandibular salivary gland morphogenesis. BMC developmental biology. 2005;5:11. doi: 10.1186/1471-213X-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina J, Byron SA, Makarenkova HP, Olsen SK, Eliseenkova AV, Larochelle WJ, Dhanabal M, Blais S, Ornitz DM, Day LA, et al. Homodimerization controls the fibroblast growth factor 9 subfamily’s receptor binding and heparan sulfate-dependent diffusion in the extracellular matrix. Molecular and cellular biology. 2009;29:4663–4678. doi: 10.1128/MCB.01780-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, Baum MG, Schiavi S, Hu MC, Moe OW, et al. Regulation of fibroblast growth factor-23 signaling by klotho. The Journal of biological chemistry. 2006;281:6120–6123. doi: 10.1074/jbc.C500457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZG, Mathew P, Yang J, Starbuck MW, Zurita AJ, Liu J, Sikes C, Multani AS, Efstathiou E, Lopez A, et al. Androgen receptor-negative human prostate cancer cells induce osteogenesis in mice through FGF9-mediated mechanisms. The Journal of clinical investigation. 2008;118:2697–2710. doi: 10.1172/JCI33093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Liu G, Zhang Y, Hu YP, Yu K, Lin C, McKeehan K, Xuan JW, Ornitz DM, Shen MM, et al. Fibroblast growth factor receptor 2 tyrosine kinase is required for prostatic morphogenesis and the acquisition of strict androgen dependency for adult tissue homeostasis. Development. 2007;134:723–734. doi: 10.1242/dev.02765. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Olsen SK, Goetz R. A protein canyon in the FGF-FGF receptor dimer selects from an a la carte menu of heparan sulfate motifs. Current opinion in structural biology. 2005;15:506–516. doi: 10.1016/j.sbi.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Muenke M, Schell U, Hehr A, Robin NH, Losken HW, Schinzel A, Pulleyn LJ, Rutland P, Reardon W, Malcolm S, et al. A common mutation in the fibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nature genetics. 1994;8:269–274. doi: 10.1038/ng1194-269. [DOI] [PubMed] [Google Scholar]
- Oldridge M, Zackai EH, McDonald-McGinn DM, Iseki S, Morriss-Kay GM, Twigg SR, Johnson D, Wall SA, Jiang W, Theda C, et al. De novo alu-element insertions in FGFR2 identify a distinct pathological basis for Apert syndrome. American journal of human genetics. 1999;64:446–461. doi: 10.1086/302245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SK, Garbi M, Zampieri N, Eliseenkova AV, Ornitz DM, Goldfarb M, Mohammadi M. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. The Journal of biological chemistry. 2003;278:34226–34236. doi: 10.1074/jbc.M303183200. [DOI] [PubMed] [Google Scholar]
- Olsen SK, Li JY, Bromleigh C, Eliseenkova AV, Ibrahimi OA, Lao Z, Zhang F, Linhardt RJ, Joyner AL, Mohammadi M. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes & development. 2006;20:185–198. doi: 10.1101/gad.1365406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Zhang X, Mantela J, Ornitz DM, Ylikoski J. Fgf9 signaling regulates inner ear morphogenesis through epithelial-mesenchymal interactions. Developmental biology. 2004;273:350–360. doi: 10.1016/j.ydbio.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Plotnikov AN, Eliseenkova AV, Ibrahimi OA, Shriver Z, Sasisekharan R, Lemmon MA, Mohammadi M. Crystal structure of fibroblast growth factor 9 reveals regions implicated in dimerization and autoinhibition. The Journal of biological chemistry. 2001;276:4322–4329. doi: 10.1074/jbc.M006502200. [DOI] [PubMed] [Google Scholar]
- Plotnikov AN, Schlessinger J, Hubbard SR, Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98:641–650. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- Pollock PM, Gartside MG, Dejeza LC, Powell MA, Mallon MA, Davies H, Mohammadi M, Futreal PA, Stratton MR, Trent JM, et al. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene. 2007;26:7158–7162. doi: 10.1038/sj.onc.1210529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Molecular cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC. The PyMOL Molecular Graphics System. 2015 Version 1.8. [Google Scholar]
- Tang L, Wu X, Zhang H, Lu S, Wu M, Shen C, Chen X, Wang Y, Wang W, Shen Y, et al. A point mutation in Fgf9 impedes joint interzone formation leading to multiple synostoses syndrome. Human molecular genetics. 2017;26:1280–1293. doi: 10.1093/hmg/ddx029. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Developmental biology. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Urakawa I, Yamazaki Y, Shimada T, Iijima K, Hasegawa H, Okawa K, Fujita T, Fukumoto S, Yamashita T. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–774. [Google Scholar]
- Vainio S. How the developing mammalian kidney assembles its thousands of nephrons: Fgfs as stemness signals. Developmental cell. 2012;22:1125–1126. doi: 10.1016/j.devcel.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Weaver M, Batts L, Hogan BL. Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Developmental biology. 2003;258:169–184. doi: 10.1016/s0012-1606(03)00117-9. [DOI] [PubMed] [Google Scholar]
- Wills AA, Kidd AR, 3rd, Lepilina A, Poss KD. Fgfs control homeostatic regeneration in adult zebrafish fins. Development. 2008;135:3063–3070. doi: 10.1242/dev.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XL, Gu MM, Huang L, Liu XS, Zhang HX, Ding XY, Xu JQ, Cui B, Wang L, Lu SY, et al. Multiple synostoses syndrome is due to a missense mutation in exon 2 of FGF9 gene. American journal of human genetics. 2009;85:53–63. doi: 10.1016/j.ajhg.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh BK, Igarashi M, Eliseenkova AV, Plotnikov AN, Sher I, Ron D, Aaronson SA, Mohammadi M. Structural basis by which alternative splicing confers specificity in fibroblast growth factor receptors. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2266–2271. doi: 10.1073/pnas.0436500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. The Journal of biological chemistry. 2006a;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Stappenbeck TS, White AC, Lavine KJ, Gordon JI, Ornitz DM. Reciprocal epithelial-mesenchymal FGF signaling is required for cecal development. Development. 2006b;133:173–180. doi: 10.1242/dev.02175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


