Abstract
We have constructed a stable cell line, human embryonal kidney 293M+, containing a lacZ reporter gene controlled by an in vitro methylated hormone-responsive enhancer. Methylation of the enhancer–promoter abolishes lacZ expression controlled by ponasterone A (an analogue of ecdysone). Ponasterone A-induced expression is restored by the short-chain fatty acids valeric > butyric > propionic > acetic acid, but not by the histone deacetylase inhibitors trichostatin A and suberoylanilide hydroxamic acid (SAHA). lacZ expression is restored to levels approaching that from an unmethylated counterpart. Incubation with short-chain fatty acids alone does not promote demethylation of the lacZ promoter, however, some demethylation (30%) is observed when transcription is triggered by addition of ponasterone A. Similar levels of hyperacetylated histones H3 and H4 were observed in cells treated with short-chain fatty acids, trichostatin A or SAHA. In vivo DNase I footprinting indicates a more open chromatin structure at the promoter region for butyric acid-treated cells. A synergistic effect in reversing the methylation-mediated repression of the lacZ gene is obtained by combined treatments with the normally ineffective compounds trichostatin A and the short-chain fatty acid caproic acid. Our results suggest the existence of an alternative silencing mechanism to histone deacetylation in executing methylation-directed gene silencing.
INTRODUCTION
Cytosine methylation at the dinucleotide CpG is an important regulator of gene expression in higher eukaryotes, where it is responsible for the transcriptional inactivation of methylated genes. It is essential for normal development in mammals and also plays a crucial role in processes like imprinting and X chromosome inactivation (1).
While methylation can directly affect gene expression by either preventing the assembly of transcription complexes or the binding of necessary transcription factors to promoter sequence elements containing methylated cytosines (2–4), this mode of action is believed to be of minor overall significance. A more general mechanism by which DNA methylation can give rise to transcriptional silencing is via the involvement of methylated DNA-binding proteins (MDBP) which, as the name suggests, bind specifically to methylated DNA and recruit accessory factors which are ultimately responsible for transcriptional silencing. This is illustrated by the example of MeCP2, which is capable of specifically binding methylated DNA at an 85 amino acid domain termed the methyl-CpG binding domain (MBD) (5). Once tethered in position, MeCP2 recruits, via a separate transcriptional repressor domain (TRD), the co-repressor mSin3A, which is a member of a large repressor complex containing histone deacetylases (HDAC) 1 and 2 (6,7). Localised histone deacetylase activity in the vicinity of the original methylation signal leads to the formation of transcriptionally inert heterochromatin resulting in long-term gene silencing.
Although this model adequately explains the role of the histone deacetylases in executing methylation-directed silencing, several observations point to the existence of an alternative mechanism that is histone deacetylase independent. First, both MeCP2 and the related MBD2 protein have been shown to be able to directly repress transcription without the mediation of histone deacetylases (8–10). Secondly, transcriptional repression of methylated reporter genes are in many cases not reversed or only partially relieved by trichostatin A, a potent and highly specific HDAC inhibitor (6,11,12). These results are consistent with the observation that aberrantly methylated endogenous genes like FMR1 are refractory to any transcriptional reactivation by trichostatin A (13–15). Indeed, the expression of only a surprisingly low number of genes (2%) are altered in cultured human cells exposed to trichostatin A (16).
We have constructed a stable transgenic cell line containing a lacZ reporter gene coupled with a hormone-inducible promoter. In the methylated form, the promoter is unable to induce lacZ expression and is insensitive to reactivation by trichostatin A and the chemically similar compound suberoylanilide hydroxamic acid (SAHA). In contrast, the short-chain fatty acids propionic, butyric and valeric acid are capable of overcoming methylation-mediated repression and restore lacZ expression to levels approaching that from an unmethylated promoter.
MATERIALS AND METHODS
Stable transgenic cell lines
The ecdysone-inducible mammalian expression system (further information on www.invitrogen.com/) was used for the construction of stably transfected cell lines. EcR293, a derivative of human embryonal kidney 293 cells, was used as the parent cell line for all constructs. This cell line expresses the functional ecdysone receptor which is required for steroid hormone-inducible reporter gene expression. For the expression system, we used the pIND/lacZ plasmid, which contains the lacZ structural gene under the regulation of a Drosophila minimal heat shock promoter coupled to an ecdysone-responsive enhancer (EcRE) element (Fig. 4). When introduced into EcR293 cells and in the presence of the ecdysone analogue ponasterone A the ecdysone receptor binds to the EcRE to trigger lacZ expression (17).
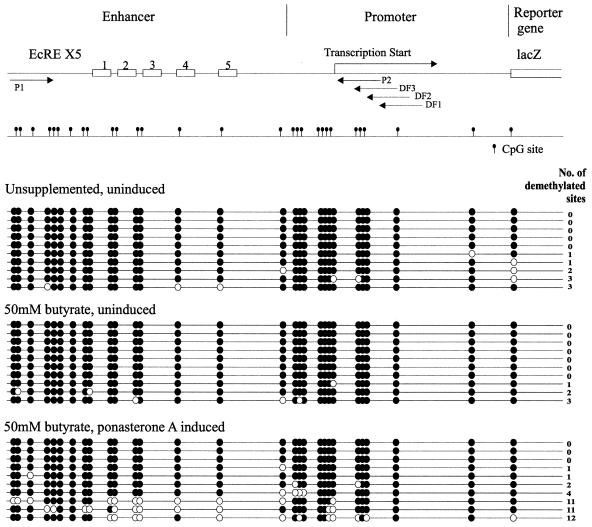
Figure 4.
Bisulphite genomic sequencing of the lacZ enhancer–promoter region from 293M+ cells. There are 29 CpG sites within the ∼500 bp enhancer–promoter region. Methylated CpG sites are shown as filled circles and unmethylated sites as open circles. The EcRE, which is repeated five times, is shown schematically as open boxes.
For the construction of cell lines containing the lacZ gene under control of a methylated enhancer–promoter, the entire enhancer–promoter fragment was excised from the pIND/lacZ plasmid and in vitro methylated with M.SssI methyltransferase. The methylated fragment was reinserted upstream of the lacZ gene and transfected into EcR293. Stable transfectants were obtained after neomycin selection (400 µg/ml). Cells were grown in Dulbecco’s medium, 10% fetal calf serum, 2 mM l-glutamine under 5% CO2 at 37°C.
lacZ gene induction
lacZ expression was induced with 10 µM ponasterone A. The period of induction was 48 h, with a fresh medium change at 24 h. The optimal concentrations for the histone deacetylase inhibitors employed were empirically determined to give the greatest lacZ induction with the lowest cell mortality. The structurally similar compounds trichostatin A (Sigma) and SAHA (Calbiochem) were used at 330 nM and 1.25 µM, respectively. The short-chain fatty acids utilised were acetic, propionic, butyric, isobutyric, valeric and caproic acid. Stock solutions were made up in PBS and adjusted to pH 7. Concentrations employed in this study were 75, 50, 50, 20, 20 and 25 mM, respectively.
lacZ reporter gene assays
Quantitative determination of lacZ expression was obtained by assaying β-galactosidase activity from cell lysates in a colourimetric reaction with ortho-nitrophenyl-β-d-galactopyranoside (ONPG) as substrate (β-galactosidase assay kit; Invitrogen). In situ staining of cells for lacZ expression was performed using X-gal.
Chromatin immunoprecipitation (CHIP)
CHIP was performed as recommended by the supplier of the anti-acetyl-H3 and anti-acetyl-H4 antibodies (Upstate Biotechnologies). These antibodies were generated against chemically synthesised peptides corresponding to amino acids 2–19 of Tetrahymena thermophila histone H4 and residues 1–21 of T.thermophila histone H3, respectively. Immunoprecipitated DNA was obtained from cells subjected to varying treatments and 25 ng of DNA was used per PCR amplification. Semi-quantitative PCR of the lacZ promoter region was performed with primers P1 (5′-GGG AGA TCT CGG CCG CAT ATT AAG TGC ATT GTT) and P2 (5′-GTT TAT TTG CTT AGC TTT CGC TTA GCG ACG TGT T) at 94°C for 30 s, 65°C for 30 s and 72°C for 30 s for 35 cycles. As a control, the promoter of the housekeeping gene HPRT was amplified.
Bisulfite genomic sequencing
Bisulfite modification of genomic DNA was performed as previously described (18). The modified DNA was amplified with primers spanning the entire lacZ enhancer–promoter region, 5′-GAA TGT ATT TAG AAA AAT AAA TAA ATA GGG and 5′-ATA ATA AAA ACC CCC CAT AAT AAA CTT AAA at 94°C for 30 s, 54°C for 30 s and 72°C for 30 s for 35 cycles. The amplified product was cloned into Escherichia coli and individual clones sequenced to determine the original methylation status at the region.
Histone purification and acid–urea gel electrophoresis
Histone extraction was performed as described (19) with some modifications. Briefly, cells were homogenised with 10 strokes of a glass–glass homogeniser and the nuclear pellet sedimented through a sucrose cushion. The nuclear pellet was acid extracted twice with ice-cold 0.4 M HCl and the supernatant was acetone precipitated overnight at –20°C. The pellet was collected by centrifugation and re-extracted with freshly prepared 25% trichloroacetic acid. The pellet was washed three times with fresh acetone/0.1 M HCl (10:1) and washed a further three times with acetone. The pellet was dried and resuspended in water.
Histones were resolved on an acid–urea gel system. Aliquots of 20 µg of purified histones were loaded on 15% polyacrylamide acid–urea gels and run for 16 h at 150 V. The bands were visualised by Coomassie staining.
In vivo DNase I footprinting
In vivo DNase I footprinting of the lacZ promoter was performed as described (20). Cells from a single cell suspension were permeabilised in a buffer containing 0.2% NP40 and subjected to DNase I digestion in increasing concentrations (0, 40, 80 and 120 Kunitz U/ml) for 5 min at 20°C. The reaction was quenched and genomic DNA extracted. Aliquots of 3 µg of DNA were used for first strand synthesis with T7 polymerase using primer DF1 (5′-AAT TGA TTC ACT TTA ACT TGC ACT) (Fig. 4). A linker (LP1, 5′-GCG GTG ACC CGG GAG ATC TGA ATT C, annealed to LP2, 5′-GAA TTC AGA TC) was added to the 3′-end of the first strand extension with T4 ligase. Exponential amplification of the region of interest was performed with primers LP1 and DF2 (5′-TCT TTA ACT TGC ACT TTA CTG CAG ATT) at 94°C for 30 s, 58°C for 30 s and 72°C for 30 s for 45 cycles. The PCR products were precipitated and the in vivo footprint obtained by linear amplification with radioactively labelled primer DF3 (5′-ACT TTA CTG CAG ATT GTT TAG CTT G) at 94°C for 30 s, 58°C for 30 s and 72°C for 30 s for 20 cycles. Half the amplified reaction was run on an 8% sequencing gel and the footprint visualised by autoradiography.
RESULTS
Construction of stable cell lines containing an unmethylated and in vitro methylated promoter for the lacZ reporter gene
EcR293 cells were transfected with linearised pIND/lacZ plasmid and stable integrants obtained by selection with neomycin. Inducible lacZ expression with ponasterone A was confirmed by histochemical staining with X-gal (Fig. 1) and one cell line, designated 293M–, was retained as a positive non-methylated control for further studies.
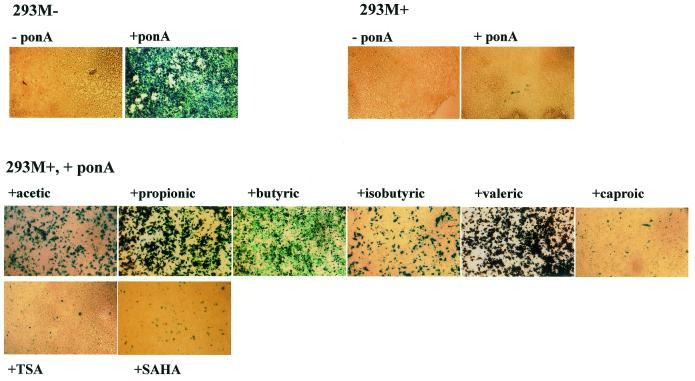
Figure 1.
In situ X-gal staining for detection of lacZ expression. Cells were induced with 10 µM ponasterone A with or without the appropriate supplement for 48 h before staining. Short-chain fatty acids range in size from acetic (two-carbon chain) through propionic (three-carbon), butyric (four-carbon), isobutyric (four-carbon, branched chain) and valeric (five-carbon) to caproic (six-carbon) acid. Concentrations of short-chain fatty acids, trichostatin A and SAHA employed were acetic acid (75 mM), propionic acid (50 mM), butyric acid (50 mM), isobutyric acid (20 mM), valeric acid (20 mM), caproic acid (25 mM), trichostatin A (330 nM) and SAHA (1.25 µM).
For the construction of cell lines containing the lacZ gene under the control of a methylated promoter, EcR293 cells were transfected with pIND/lacZ bearing an in vitro methylated lacZ promoter fragment (Materials and Methods) and selected with neomycin for stably transfected cell lines. Twenty neomycin-resistant clones were chosen and retention of the in vitro methylation pattern was checked by Southern hybridisation using the methylation-sensitive restriction enzymes HpaII and HhaI (both with two sites in the promoter region). The pattern of digestion of the promoter fragments revealed that a majority of the cell lines had lost the in vitro methylated pattern (data not shown). However, two clones remained methylated at these sites and were confirmed to have retained methylation at all 29 CpG sites within the enhancer–promoter region by bisulphite sequencing. One clone, designated 293M+, was retained for further study. Hybridisation with a lacZ-specific probe revealed a single site of insertion in both 293M– and 293M+ (data not shown).
Responses to ponasterone A induction of reporter gene expression
As anticipated, addition of the inducing agent ponasterone A elicits high levels of lacZ expression from the unmethylated lacZ promoter in 293M–. However, no lacZ expression was observed under similar conditions for the methylated 293M+ (Fig. 1). The loss of inducible lacZ expression was confirmed as being due to hypermethylation of the promoter as it could be reversed by exposure to the DNA demethylating agent 5-azacytidine prior to induction with ponasterone A (21).
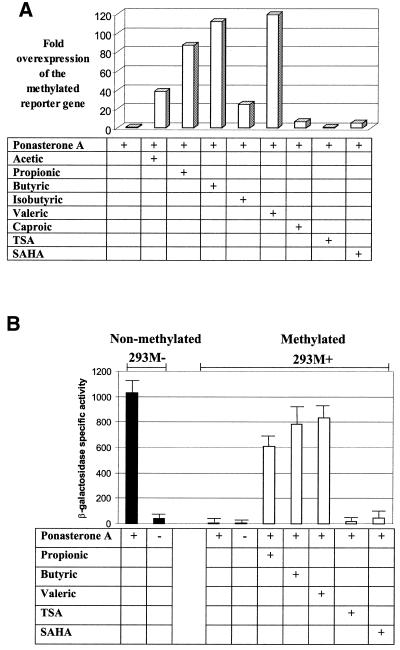
To see if methylation-mediated repression of the lacZ promoter could be overcome, we induced 293M+ cells with ponasterone A in the presence of known histone deacetylase inhibitors. Two general classes of compounds were used, the hybrid polar compounds trichostatin A and SAHA, which are highly specific histone deacetylase inhibitors and are active at nano- and micromolar concentrations (22), and the short-chain fatty acids, which are less specific, invoke a wide spectrum of cellular responses and are active histone deacetylase inhibitors at millimolar levels. As seen in Figure 1, induction in the presence of propionic, butyric and valeric acid resulted in a dramatic regain of inducible lacZ expression. Lower levels of expression were observed with acetic, isobutyric and caproic acid. In contrast, only weak relief from repression was obtained with trichostatin A and SAHA. The release from methylation-mediated repression, where occurring, appears to be a feature of all cells undergoing a particular treatment and not a sub-population of cells, as we observed almost all cells staining blue following X-gal staining (Fig. 1). Quantitative determination of β-galactosidase activity in expressing cells showed that the most potent treatments (with propionic, butyric and valeric acid) restored expression to levels comparable to that from an unmethylated promoter (Fig. 2), indicating near total release from methylation-mediated repression.
Figure 2.
(A) Recovery of inducible lacZ expression in 293M+ cells following exposure to short-chain fatty acids. Quantitative determination of lacZ expression was obtained by β-galactosidase activity assays. Each bar represents the mean value of at least three independent measurements normalised to that of the unsupplemented control (bar 1, value set as 1×) and expressed as fold overexpression. (B) Comparison of raw β-galactosidase assay values for selected treatments. β-Galactosidase specific activity is defined as nmol ONPG hydrolysed/min incubation at 37°C/mg protein of cell lysate. All data points represent the means ± SD of at least three independent measurements.
No lacZ expression was observed on addition of any of these compounds in the absence of ponasterone A (data not shown).
Kinetics of induced lacZ expression from methylated and non-methylated lacZ promoters
Cultures of 293M– and 293M+ were induced with ponasterone A and cells were harvested at regular intervals and assayed for β-galactosidase activity (Fig. 3A). lacZ expression is rapidly induced in 293M– (curve a), reaching significant levels by 12 h post-induction, with a constant rate of increase until the final time point (48 h). In contrast, only very low expression is observed in 293M+ (curve b), with no significant increase above the baseline expression level throughout the entire period of determination. Induction of 293M+ in conjunction with butyric acid (curve c) shows a lag period of response to ponasterone A induction with no measurable β-galactosidase activity at 12 h post-induction. After 24 h, there is a steep rate of increase in β-galactosidase activity until it reaches comparable levels to that from 293M–.
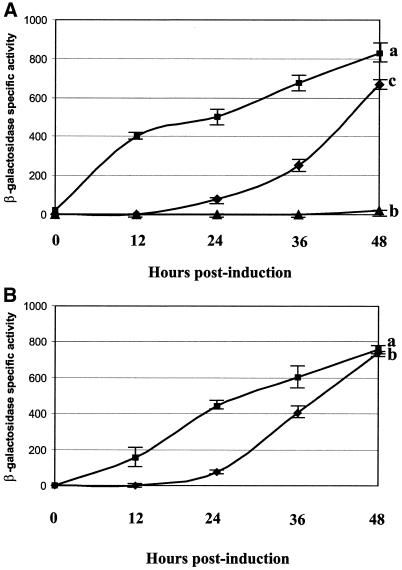
Figure 3.
(A) Increase in lacZ expression following addition of ponasterone A (0 h) as measured by β-galactosidase activity. Curve a is 293M– cells induced with ponasterone A, curve b is 293M+ cells induced with ponasterone A and curve c is 293M+ cells induced with ponasterone A in the presence of butyric acid. Each experiment was repeated three times. (B) Reversibility of the effects of butyric acid on lacZ induction in 293M+ cells. Curve a, cells cultured for 24 h in medium containing butyric acid before induction with ponasterone A (0 h); curve b, cells cultured in butyric acid-containing medium for 24 h. This was replaced with normal growth medium for a further 24 h before induction with ponasterone A and butyric acid (0 h). Results are from three independent experiments.
The lag period appears to be due to some form of butyric acid-induced remodelling of the chromatin, as preculturing 293M+ in the presence of butyric acid for 24 h yielded an immediate response to stimulation by ponasterone A (Fig. 3B, curve a). The effects of butyric acid can be reversed by replacement of butyric acid-containing medium with normal medium for 24 h prior to ponasterone A stimulation. In this case a lag period is again observed before a rise in lacZ expression levels (Fig. 3B, curve b). Upon removal of butyric acid and ponasterone A, lacZ expression in 293M+ drops within 8 days to undetectable levels. The effects appear to be fully reversible, as further culturing and transcriptional stimulation of previously stimulated cells show an essentially similar response to that in Figure 3A (data not shown).
Bisulphite genomic sequencing of the lacZ promoter
To establish whether the activation of lacZ expression is concomitant with or is due to DNA demethylation, we performed bisulphite genomic sequencing across the promoter region. Sequencing of clones derived from 293M+ cells grown in unsupplemented medium confirmed the initial hypermethylated state of the promoter (Fig. 4). No changes in methylation were observed for cells grown in the presence of butyric acid alone. However, the combination of butyric acid and ponasterone A, which gives high levels of lacZ expression, also results in extensive demethylation in 30% of the clones sequenced. A similar result was observed for cells grown in valeric acid and ponasterone A (demethylation in 20% of clones; data not shown). As seen, butyric acid per se is insufficient to induce demethylation at the promoter. Demethylation is only observed under conditions where active transcription is proceeding through the promoter. Taken together, these data suggest that demethylation is not responsible for the regain of promoter activity. Furthermore, in situ staining reveals almost 100% of cells staining positively for β-galactosidase activity whereas the proportion of cells undergoing demethylation for the lacZ promoter is far lower. In conclusion, the demethylation observed in the present case is most likely a consequence rather than a cause of lacZ transcription.
Histone deacetylation and chromatin remodelling at the lacZ promoter
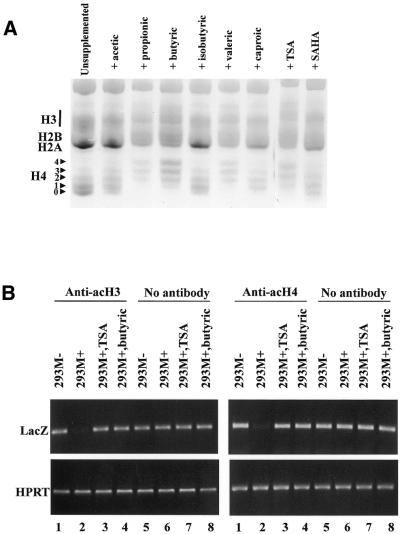
To determine if the divergent responses to short-chain fatty acids and hybrid polar compounds are due to the relative effectiveness of these compounds in invoking histone hyperacetylation, we extracted histones from 293M+ grown in medium supplemented with the various compounds. Purified histones were resolved on acid–urea gels which display the extent of acetylation on the core histones H3 and H4. Figure 5A reveals that the various short-chain fatty acids differ in their ability to induce histone hyperacetylation. In general, there appears to be a good correlation within the short-chain fatty acids between histone hyperacetylation capability and the ability to reverse transcriptional repression. For example, propionic, butyric and valeric acid give a strong relief of repression (Figs 1 and 2) and are at the same time potent histone hyperacetylating agents. On the other hand, isobutyric and caproic acid, which give a very weak relief of repression, also yield less acetylation of histone H4 (Fig. 5). This correlation, however, is not maintained in the case of trichostatin A and SAHA. While trichostatin A was capable of extensive histone hyperacetylation, SAHA yielded levels of hyperacetylated histones that were only marginally greater than that of untreated cells. In either case, both compounds are ineffective at reversing methylation-directed silencing (Figs 1 and 2).
Figure 5.
(A) Acid–urea gel electrophoresis of histones extracted from 293M+ cells grown for 14 h in the presence of the various compounds. The positions of acetylated forms of histone H4 are indicated corresponding to the number of modifications. (B) Semi-quantitative PCR of immunoprecipitated DNA from 293M+ and 293M– cells pulled down with antibodies against acetylated histones H3 and H4. The no antibody control was performed using genomic DNA extracted from an aliquot of total chromatin taken prior to addition of the antibody. As seen, the lacZ promoter is originally under-represented in 293M+ DNA precipitated by these antibodies but is enriched following culture in either butyric acid or trichostatin A. The constitutively active housekeeping gene HPRT was used as a positive control.
Although trichostatin A and butyric acid appear equally effective at histone hyperacetylation on a global level, the possibility remains that local changes in chromatin conformation at the lacZ promoter are responsible for the divergent results, with the short-chain fatty acids instituting a more permissive chromatin conformation for inducible expression than the hybrid polar compounds. To address this question, we performed chromatin immunoprecipitation assays on the lacZ promoter with antibodies specific against acetylated histones H3 and H4. As shown in Figure 5B, the lacZ promoter is poorly represented in 293M+ DNA co-precipitated with these antibodies (tracks 2). However, DNA from the same cell line grown in the presence of either butyric acid or TSA is now enriched for this region in the DNA fraction pulled down by these antibodies (tracks 3 and 4). Corresponding controls of the same treatments with non-immunoprecipitated DNA show equal representation of the lacZ promoter in all samples (tracks 5–8). Within the limit of precision afforded by these assays, we cannot distinguish any difference between butyric acid and trichostatin A treatments on the levels of hyperacetylated histones H3 and H4 associated with the lacZ promoter.
In vivo DNase I footprinting of the lacZ promoter
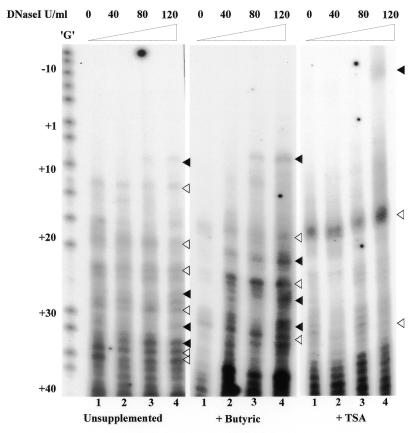
In order to determine other forms of chromatin remodelling besides histone hyperacetylation, we performed in vivo DNase I footprinting at the lacZ promoter for 293M+ cells. Permeabilised cells were incubated with increasing concentrations of DNase I. Lanes 1 of Figure 6 (0 U DNase I) display nicking due to endogenous nuclease activity. Lanes 2–4 of the three panels show the effect of increasing concentrations of DNase I. A comparison between lanes 2–4 and their corresponding controls in lanes 1 clearly shows that butyric acid has a marked effect on the accessibility of chromatin to DNase I. A comparison of the left and the middle panels shows that the cleavage sites are at roughly the same nucleotide positions. In cells treated with trichostatin A, only one major band at nucleotide position +20 is observed. Incubation with increasing concentrations of DNase I resulted in only a slight increase in nuclease sensitivity at this band. Taken together these results suggest that despite apparently similar levels of histone hyperacetylation, butyric acid creates a more open chromatin structure than trichostatin A.
Figure 6.
In vivo DNase I footprinting of the lacZ promoter in 293M+ cells. Hypersensitive sites cleaved by endogenous nucleases are indicated by open arrows. Filled arrows indicate hypersensitive sites revealed by DNase I cleavage. The positions of the sites are shown relative to the transcription start site (+1).
Synergism between trichostatin A and caproic acid in reversing silencing
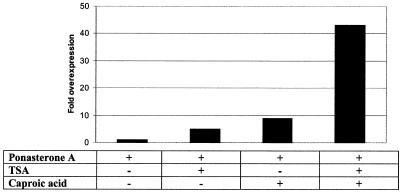
In order to determine if the effect obtained with the short-chain fatty acids is due to the action of an alternative silencing mechanism to histone deacetylation, we induced 293M+ cells with ponasterone A in the presence of both trichostatin A and caproic acid, either singly or combined. As previously shown, caproic acid is the only member among the short-chain fatty acids tested which yields poor reversal of methylation-directed silencing (Fig. 2). In addition, it is also a poor histone deacetylase inhibitor (Fig. 5). However, trichostatin A, in spite of being a potent histone deacetylase inhibitor, is incapable of lifting repression of the reporter gene. It remains possible that successful reversal of silencing requires two distinct activities, suppression of an alternative silencing mechanism and histone hyperacetylation-associated chromatin remodelling through inhibition of histone deacetylases to give a permissive chromatin structure for efficient transcription. Figure 7 displays the values for lacZ expression obtained under the different treatments. There is a clear cooperation between trichostatin A and caproic acid, which interact to provide partial relief from silencing. It should be noted that the level of lacZ expression achieved is only ∼10% of that obtained with butyric acid. No additive effect on lacZ transcription is observed on addition of trichostatin A in combination with butyric acid, possibly because butyric acid is a potent histone deacetylase inhibitor in its own right (data not shown).
Figure 7.
Interaction between trichostatin A and caproic acid in reversing methylation-directed silencing in 293M+ cells. Induction of lacZ expression was performed for 48 h. Overexpression is visualised by normalising the mean value for β-galactosidase activity from treated cell extracts to that of the unsupplemented control (bar 1, lacZ expression level defined as 1×). All data points represent the means of at least six independent experiments.
DISCUSSION
Transcriptional silencing of methylated transgenes has been well studied and has received ample experimental confirmation. The great majority of these studies are based on the transient transfection (23–25) of in vitro methylated constructs, although some studies have taken advantage of stable episomal systems (26). We have chosen to investigate methylation-mediated gene silencing and re-expression in a stable cell line because the situation of a stably integrated transgene after chromatin assembly and passage through many cycles of cell division is probably closer to that of an actual endogenous gene. In addition, the ecdysone-inducible reporter gene system offers the unique advantage of extremely low baseline expression and tight regulation of gene activity with no known non-specific interaction in a mammalian cell background (17). Hence, it is possible to study the silencing, transcriptional inactivation and restoration of inducible gene expression in isolation.
We have observed in this system that methylation-mediated silencing can be reversed by high concentrations of selected short-chain fatty acids but not by specific histone deacetylase inhibitors like trichostatin A and SAHA. While previous studies have reported some relief of silencing in methylated transgenes with sodium butyrate, this was accomplished only with partially methylated (∼10% of available CpGs methylated) genes (25) or with only a modest relief of repression from fully methylated genes (26). In our case, we obtained near total reversal of silencing from a fully methylated promoter of high CpG density, indicating that the flooding of cells with certain short-chain fatty acids can fully overcome the effects of methylation-directed gene silencing. Although the concentrations of short-chain fatty acids employed may appear excessively high, this is not without precedent (26) and it has been previously shown that virus-transformed cells, such as 293 cells, are capable of tolerating high doses of sodium butyrate (27).
We have reported elsewhere that overexpression of 5-methylcytosine-DNA glycosylase in 293M+ results in demethylation and restoration of inducible expression at the lacZ promoter (21). This is not to be confused with the partial demethylation that we observe in the present case (Fig. 4), which is most likely due to the short-chain fatty acid-induced regain of transcriptional activity at the promoter. Interestingly, most demethylation occurs in the upstream enhancer region which is required for ecdysone receptor binding to facilitate transcription. This is consistent with observations that the mere act of protein binding at an active promoter can lead to progressive demethylation of the binding site in a stable cell system (28,29).
No differences in histone hyperacetylation potential were observed between the short-chain fatty acids and the hybrid polar compounds (trichostatin A and SAHA) on a nucleus-wide and at a locus-specific level. There is a good correlation among the short-chain fatty acids between histone hyperacetylation capacity and reversal of methylation-mediated repression. However, the equally effective histone hyperacetylating agent trichostatin A is nevertheless incapable of reversing silencing. Trichostatin A and SAHA are highly specific histone deacetylase inhibitors whereas the short-chain fatty acids, notably butyric acid, are capable of evoking a wide range of cellular responses in addition to histone hyperacetylation. In this context, we do observe a difference between butyric acid and trichostatin A treatments on the chromatin structure at the lacZ promoter.
Our results provide evidence for an alternative silencing mechanism to histone deacetylation in executing methylation-directed gene silencing. This mechanism is resistant to reversal by dedicated histone deacetylase inhibitors but can be almost fully reversed with selected short-chain fatty acids. At this stage, we cannot distinguish whether the effects of the short-chain fatty acids are accomplished by blocking a general repression mechanism analogous to histone hyperacetylation or by interference with direct repression exerted by a methylated DNA-binding protein(s). CHIP assays with monoclonal anti-MeCP2 showed no differences in MeCP2 binding at the lacZ promoter between the different treatments (data not shown). However, this does not rule out the possible involvement of a different methylated DNA-binding protein(s). Furthermore, our results suggest that while the formation of transcriptionally competent chromatin may be a necessary prerequisite for transcriptional activity, it is not sufficient by itself to reverse methylation-directed gene repression in this system.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank H. Angliker for DNA sequencing and P. Muller for oligonucleotide synthesis. This work was sponsored by the Novartis Research Foundation.
References
- 1.Jaenisch R. (1997) DNA methylation and imprinting: why bother? Trends Genet., 13, 323–329. [DOI] [PubMed] [Google Scholar]
- 2.Comb M. and Goodman,H.M. (1990) CpG methylation inhibits proenkaphalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res., 18, 3975–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iguchi-Ariga S.M.M. and Schaffner,W. (1989) CpG methylation of the cAMP-responsive enhancer promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev., 3, 612–619. [DOI] [PubMed] [Google Scholar]
- 4.Ammerpohl O., Schmitz,A., Steinmuller,L. and Renkawitz,R. (1998) Repression of the mouse M-lysozyme gene involves both hindrance of enhancer factor binding to the methylated enhancer and histone deacetylation. Nucleic Acids Res., 26, 5256–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nan X., Meehan,R.R. and Bird,A. (1993) Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res., 21, 4886–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nan X., Ng,H.-H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenmann,R.N. and Bird,A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 7.Jones P.L., Veenstra,G.J.C., Wade,P.A., Vermaak,D., Kaas,S.U., Landsberger,N., Strouboulis,J. and Wolffe,A.P. (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet., 19, 187–191. [DOI] [PubMed] [Google Scholar]
- 8.Nan X., Campoy,F.J. and Bird,A. (1997) MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell, 88, 471–481. [DOI] [PubMed] [Google Scholar]
- 9.Ng H.-H., Zhang,Y., Hendrich,B., Johnson,C.A., Turner,B.M., Erdjument-Bromage,H., Tempst,P., Reinberg,D. and Bird,A. (1999) MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nature Genet., 23, 58–61. [DOI] [PubMed] [Google Scholar]
- 10.Kudo S. (1998) Methyl-CpG-binding protein MeCP2 represses Sp1-activated transcription of the human leukosialin gene when the promoter is methylated. Mol. Cell. Biol., 18, 5492–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu F., Thiesen,J. and Stratling,W.H. (2000) Histone deacetylase-independent transcriptional repression by methyl-CpG-binding protein 2. Nucleic Acids Res., 28, 2201–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorincz M.C., Schubeler,D., Goeke,S.C., Walters,M., Groudine,M. and Martin,D.I. (2000) Dynamic analysis of proviral induction and de novo methylation: implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol. Cell. Biol., 20, 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffee B.F., Zhang,F., Warren,S.T. and Reines,D. (1999) Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nature Genet., 22, 98–101. [DOI] [PubMed] [Google Scholar]
- 14.Cameron E.E., Bachman,K.E., Myohanen,S., Herman,J.G. and Baylin,S.B. (1999) Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nature Genet., 21, 103–107. [DOI] [PubMed] [Google Scholar]
- 15.Chiurazzi P., Pomponi,M.G., Pietrobono,R., Bakker,C.E., Neri,G. and Oostra,B.A. (1999) Synergistic effect of histone hyperacetylation and DNA demethylation in the reactivation of the FMR1 gene. Hum. Mol. Genet., 8, 2317–2323. [DOI] [PubMed] [Google Scholar]
- 16.Van Lint C., Emiliani,S. and Verdin,E. (1996) The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr., 5, 245–253. [PMC free article] [PubMed] [Google Scholar]
- 17.No D., Yao,T.P. and Evans,R.M. (1996) Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc. Natl Acad. Sci. USA, 93, 3346–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raizis A.M., Schmitt,F. and Jost,J.-P. (1995) A bisulfite method of 5-methylcytosine mapping that minimizes template degradation. Anal. Biochem., 226, 161–166. [DOI] [PubMed] [Google Scholar]
- 19.Thorne A.W., Cary,P.D. and Crane-Robinson,C. (1998) Extraction and separation of core histone and non-histone chromosomal proteins. In Gould,H. (ed.), Chromatin: A Practical Approach. Oxford University Press, New York, NY.
- 20.Pfeifer G.P., Chen,H.H., Komura,J. and Riggs,A.D. (1999) Chromatin structure analysis by ligation-mediated and terminal transferase-mediated polymerase chain reaction. Methods Enzymol., 304, 548–571. [DOI] [PubMed] [Google Scholar]
- 21.Zhu B., Benjamin,D., Zheng,Y., Angliker,H., Thiry,S., Siegmann,M. and Jost,J.-P. (2001) Overexpression of 5-methylcytosine DNA glycosylase in human embryonic kidney cells EcR293 demethylates the promoter of a hormone-regulated reporter gene. Proc. Natl Acad. Sci. USA, 98, 5031–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richon V.M., Emiliani,S., Verdin,E., Webb,Y., Breslow,R., Rifkind,R.A. and Marks,P.A. (1998) A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc. Natl Acad. Sci. USA, 95, 3003–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bryans M., Kass,S., Seivwright,C. and Adams,R.L.P. (1992) Vector methylation inhibits transcription from the SV40 early promoter. FEBS Lett., 309, 97–102. [DOI] [PubMed] [Google Scholar]
- 24.Kass S.U., Goddard,J.P. and Adams,R.L.P. (1993) Inactive chromatin spreads from a focus of methylation. Mol. Cell. Biol., 13, 7372–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine A., Cantoni,G.L. and Razin,A. (1991) Inhibition of promoter activity by methylation: possible involvement of protein mediators. Proc. Natl Acad. Sci. USA, 88, 6515–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh C.-L. (1994) Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol., 14, 5487–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Haan J.B., Gevers,W. and Parker,M.I. (1986) Effects of sodium butyrate on the synthesis and methylation of DNA in normal cells and their transformed counterparts. Cancer Res., 46, 713–716. [PubMed] [Google Scholar]
- 28.Hsieh C.-L. (1999) Evidence that protein binding specifies sites of DNA demethylation. Mol. Cell. Biol., 19, 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin I.G. and Hsieh,C.-L. (2001) Chromosomal DNA demethylation specified by protein binding. EMBO Rep., 2, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]