Abstract
While most DNA polymerases discriminate against ribonucleotide triphosphate (rNTP) incorporation very effectively, the Family X member DNA polymerase μ (Pol μ) incorporates rNTPs almost as efficiently as deoxyribonucleotides. To gain insight into how this occurs, here we have used X-ray crystallography to describe the structures of pre- and post-catalytic complexes of Pol μ with a ribonucleotide bound at the active site. These structures reveal that Pol μ binds and incorporates a rNTP with normal active site geometry and no distortion of the DNA substrate or nucleotide. Moreover, a comparison of rNTP incorporation kinetics by wildtype and mutant Pol μ indicates that rNTP accommodation involves synergistic interactions with multiple active site residues not found in polymerases with greater discrimination. Together, the results are consistent with the hypothesis that rNTP incorporation by Pol μ is advantageous in gap-filling synthesis during DNA double strand break repair by nonhomologous end joining, particularly in nonreplicating cells containing very low deoxyribonucleotide concentrations.
The primary function of DNA polymerases is to replicate and maintain genomic information. Replicative polymerases play a pivotal role in this endeavor by copying the genetic material efficiently and faithfully (reviewed in (1)). Polymerase fidelity is largely upheld by encouraging binding and incorporation of correct nucleotides, while discouraging binding and incorporation of incorrect nucleotides. Nucleotide discrimination has two distinct facets: selection of both the correct base for pairing with a template strand, and selection of the correct sugar containing only a 3′-OH (deoxyribonucleotides, dNTPs) versus those containing a hydroxyl at both the 2′ and 3′ positions (ribonucleotides, rNTPs). Despite the high fidelity exhibited by replicative polymerases, ribonucleotides are incorporated into the genome during replication (2–4), likely due to the high relative cellular concentration of ribo- versus deoxyribonucleotides (5). Ribonucleotides can also be inserted by repair polymerases, presumably with higher incidence than exhibited by replicative polymerases (6,7) (Supplementary Figure S1A). The presence of these ribonucleotides can ultimately lead to genome instability, commonly due to DNA strand breaks resulting from the increased likelihood of hydrolytic backbone cleavage near 2′-hydroxyls (8).
Recent published reports indicate that ribonucleotide incorporation may not always be deleterious. It has been suggested that the presence of rNMPs in DNA may serve in a signaling capacity, by initiating mating type switching in Schizosaccharomyces pombe (9,10), or by dictating strand discrimination during mismatch repair (11,12). Additionally, it has been suggested that repair of DNA double strand breaks (DSBs) during nonhomologous end joining (NHEJ) may be effectively accomplished by insertion of ribonucleotides by repair polymerases (13–15), during both mammalian NHEJ as well as during bacterial and archaeal NHEJ (16–18). In mammals, the candidate enzymes for ribonucleotide insertion during NHEJ are Family X polymerases Mu (Pol μ) (13,19) and terminal deoxynucleotidyl transferase (TdT) (20). While Pol μ functions in general NHEJ and is widely expressed in a variety of tissues (21–23), the expression of TdT is spatially and temporally limited to developing pre-B and pre-T lymphocytes (24). TdT is a primarily template-independent polymerase whose function is to generate sequence heterogeneity for immunoglobulin gene maturation during V(D)J recombination (24). Recent studies have also shown that TdT can exhibit template-dependent behavior (25). Pol μ functions specifically in immunoglobulin kappa light chain rearrangements in V(D)J recombination (22,26), and more broadly in repair of DSBs by NHEJ in non-lymphocyte cells (27–29), but does so in a largely template-driven fashion.
The precise mechanism by which Pol μ and TdT incorporate ribonucleotides with low discrimination (<11-fold) (13,19,30), while their Family X siblings Pols β and λ exclude them 102–104-fold more efficiently is currently unclear (31–33) (Supplementary Figure S1A). It is believed that Pol μ and TdT owe their nucleotide preferences to the loss of sequence conservation of the ‘steric gate’ residues that endow Pols β and λ with their considerably higher degree of substrate specificity (31–33) (Supplementary Figure S1B). In Pols β and λ, the aromatic residues (Tyr271 and Phe272 in Pol β, Tyr505 and Phe506 in Pol λ) comprising the ‘YF motif’ contribute to their ability to exclude 2′-OH-containing nucleotides from the active site, thus reducing the likelihood of their incorporation during DNA repair. For Pol μ and TdT, this motif is not conserved, and the corresponding residues are glycine and tryptophan (Gly433 and Trp434 in Pol μ, Gly448 and Trp449 in TdT; henceforth referred to as the ‘GW motif’). The first residue of each pair has been found to play the stronger role in ribonucleotide discrimination, as loss of Tyr271 in Pol β results in a 10-fold reduction of sugar discrimination (32), and replacement of Gly433 with tyrosine in Pol μ increased discrimination by an average of 5-fold (19). Though mutational studies have indicated that the individual side chains contribute to ribonucleotide discrimination, recent structural information for Pols β and λ suggests that the critical interaction for rNTP exclusion is likely mediated by the backbone carbonyl of the ‘steric gate’ residues, rather than the side chains (32,33). Given the structural similarity between the Family X polymerases (34), differences in amino acid sequence surrounding the incoming ribonucleotide could affect efficiency of incorporation.
To gain a better understanding of ribonucleotide discrimination by the Family X polymerases, we present X-ray crystal structures of human Pol μ, in complex with a single-strand break gapped DNA during distinct steps of the catalytic cycle. These structures represent the first evidence that Pol μ binds and incorporates correctly paired incoming ribonucleotides without distortion of catalytically competent active site geometry. A panel of mutants identified multiple sites that, when altered, increased selectivity for deoxyribose sugars, yet the improved discrimination never approached that observed for related Family X polymerases β and λ. Our work argues that ribonucleotide accommodation by Pol μ is a product of a synergistic system within the active site—which cannot be solely attributed to a single ‘steric gate’—and is consistent with suggestions that this relaxed sugar selectivity is beneficial in physiological contexts.
MATERIALS AND METHODS
Expression and purification of human Pol μ constructs
Sequences encoding full-length (Met1-Ala494) or the truncated catalytic domain (Pro132-Ala494) of human Pol μ were cloned into the pGEXM vector (35), using the NotI and SalI restriction sites. For crystallization of the catalytic domain, Loop2 residues Pro398-Pro410 were deleted using site-directed mutagenesis and a glycine residue (labeled Gly410 in these structures) was added in order to fuse the ends of β-strands 4 and 5 (construct henceforth referred to as hPol μΔ2). Full-length wildtype or Δ2 human Pol μ constructs were expressed in Rosetta2 (DE3) cells, as previously described (35). Individual amino acid substitutions were also generated using site-directed mutagenesis, and were expressed and purified in similar fashion to the wildtype proteins, lacking only the ion exchange step.
Crystallization of the hPol μΔ2 pre-catalytic complex with a 1-nt gapped DNA substrate and incoming ribonucleotide
The following DNA oligonucleotides were used to create the 1-nt gapped DNA complex: template (5′-CGGCATACG-3′), upstream primer (5′-CGTA-3′), and a 5′-phosphorylated downstream primer (5′-pGCCG-3′), and were prepared for crystallization as previously described (35). The annealed DNA was then mixed in a 4:1 molar ratio, added to concentrated hPol μΔ2 (∼10 mg/mL), and incubated on ice at 4°C for one hour. Details of the crystallization conditions for each structure are included in Supplementary Table S1. Crystals were grown using either the sitting drop or hanging drop vapor diffusion techniques (36), then transferred to a cryoprotectant solution containing 100 mM HEPES pH 7.5, 20% (w/v) PEG 4000, 10 mM MgCl2, 50 mM NaCl, 5% (v/v) glycerol, 15% (v/v) ethylene glycol. Nucleotide soaks were carried out in cryoprotectant solution, as indicated in Supplementary Table S1. After soaking, the crystals were then flash frozen in liquid nitrogen and placed into a stream of nitrogen gas cooled to –173°C for data collection.
Structure solution and refinement
Data collected on a rotating Cu anode or at APS (Supplementary Table S2) were indexed, integrated, and scaled using either HKL3000 or HKL2000, respectively (37,38). The 1-nt gapped pre-catalytic ternary complex with a nonhydrolyzable incoming deoxyribonucleotide (PDB ID code: 4M04 (35)) was used as the starting model for refinement for all structures, and the same Rfree test reflections were used to avoid potential model bias. All structures were refined using iterative cycles of manual model building and refinement in COOT (39,40) and Phenix (41). Data refinement statistics are listed in Supplementary Table S2. Ramachandran statistics were generated using MolProbity (42).
Steady-state kinetics for single-nucleotide incorporation
DNA substrates for steady-state measurements of single nucleotide incorporation were prepared by hybridizing a 5′-end Cyanine3-labeled 14-nucleotide primer (5′-Cy3-GTCAGACTGACGTA-3′, where the underlined nucleotide on the primer terminus was either a dA or an rA, depending on the reaction in question) and a 14-nucleotide 5′ downstream primer (5′-pGCCGGACGACGGAG-3′) to a 29-mer template (5′-CTCCGTCGTCCGGCATACGTCAGTCTGAC-3′, where the underlined nucleotide is the templating base) to create a 1-nt gapped substrate. Reaction mixtures (20 μl) contained 50 mM Tris, pH 7.5, 1 mM dithiothreitol, 4% (v/v) glycerol, 0.1 mg/ml bovine serum albumin, 5 mM MgCl2, 100 nM DNA, and full-length enzyme (Supplementary Table S3). Reactions were initiated by adding dTTP or UTP at varying concentrations. The reaction mixtures were incubated at 37°C for 3–4 min. After adding an equal volume of loading dye (99% (v/v) formamide, 5 mM EDTA, 0.1% (w/v) xylene cyanol, and 0.1% (w/v) bromophenol blue), products were resolved on a 16% denaturing polyacrylamide gel, imaged using a Typhoon 9400 (GE Healthcare), and quantified using ImageQuant TL software. The data were fit to the Michaelis-Menten equation using nonlinear regression from KaleidaGraph software version 3.6 (Synergy Software, http://www.synergy.com).
Nonhomologous end-joining assays
The human Ku and XRCC4-Ligase IV complex proteins, as well as the GCG3’ substrate, were prepared as described previously (43). End joining reactions were carried out in a buffer containing: 25 mM Tris (pH 7.5), 1 mM DTT, 150 mM KCl, 4% glycerol, 50 μg/ml bovine serum albumin, 0.1 mM EDTA and 9% polyethyelene glycol (MW: 8000 kDa). Ligations were carried out by adding (final concentrations) 10 nM Ku, 20 nM XRCC4-LigaseIV complex and 0.5 nM Pol μ, to a mixture containing 2 nM DNA substrate, 2 mM Mg2+, 100 ng supercoiled DNA and 100 μM CTP or dCTP. The reactions were carried out at 37°C for 10 min, stopped by addition of EDTA and SDS, and either purified by a Minelute cartridge (Qiagen) or extracted once with a 1:1 mixture of phenol and chloroform. RNase HII (New England Biolabs) digestion of joined products was carried out overnight as per the manufacturer's instructions. Ligation products were resolved using a 5% native PAGE gel and visualized using a Typhoon Imager for qualitative comparisons. End joining products were quantified using qPCR, with primers specific for substrate and product.
RESULTS
Structural exploration of sugar selectivity by Pol μ
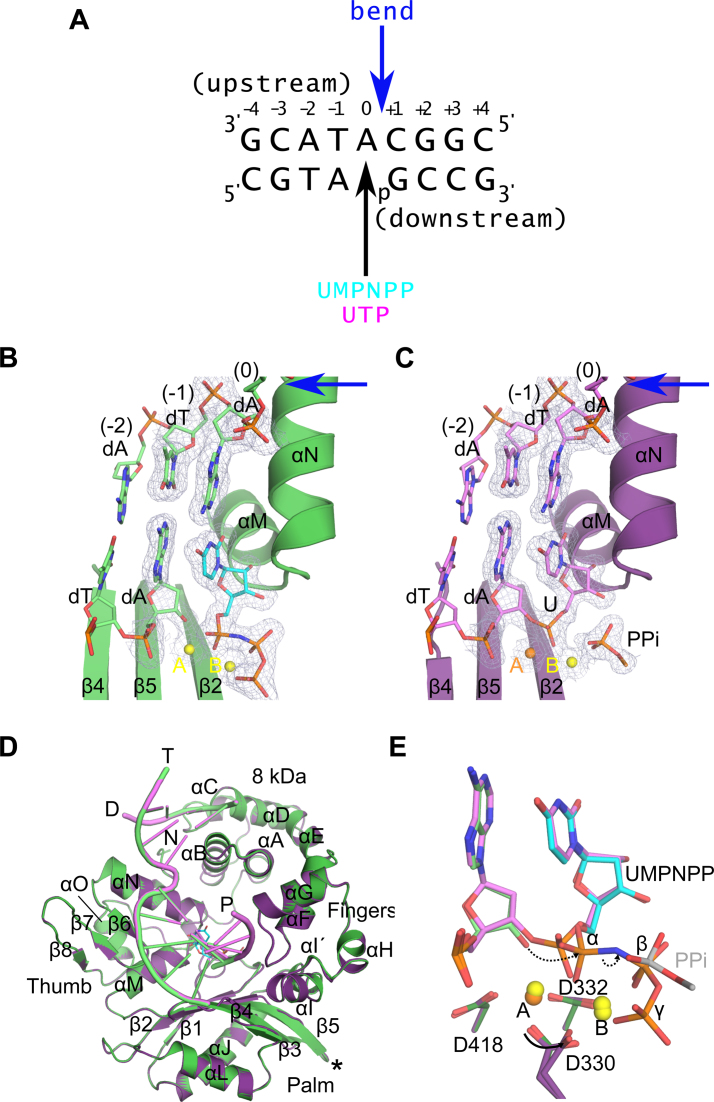
In order to understand the underlying structural mechanism of ribonucleotide accommodation by Pol μ, we have determined X-ray crystal structures of a single-nucleotide gap DNA substrate, with incoming ribonucleotides (Figure 1 and Supplementary Table S2). A truncation variant of the catalytic domain of human Pol μ (Pro132-Ala494) lacking the flexible ‘Loop2’ region between β-strands 4 and 5 (Pro398-Pro410, fused with a single glycine residue) was used for these structural studies. This loop deletion variant, known as hPol μΔ2, displays an enzymatic activity profile indistinguishable from the wildtype, but has a greater proclivity for crystallization (35). To achieve optimal conditions for comparison of ribo- versus deoxyribonucleotide incorporation, hPol μΔ2 was crystallized as a pre-catalytic ternary complex with a 1-nt gapped DNA substrate (Figure 1A), with an incoming nonhydrolyzable deoxyribonucleotide (dUMPNPP), the structure of which has already been reported (PDB ID code 4M04 (35)). This nucleotide was then removed via soaking, and rebound with a nonhydrolyzable ribonucleotide, UMPNPP (PDB ID code 5TWP, Figure 1B). A post-catalytic nicked complex was then obtained by soaking the same dUMPNPP-containing crystals with UTP, followed by a back-soak with pyrophosphate to aid in visualization of the leaving group (PDB ID code 5TWQ, Figure 1C).
Figure 1.
Structural evaluation of ribonucleotide incorporation by wildtype hPol μΔ2. Wildtype hPol μΔ2 was crystallized in complex with a 1-nt gapped DNA substrate and an incoming dUMPNPP (A). A pre-catalytic ternary complex (B, green) was obtained by soaking with nonhydrolyzable UMPNPP (cyan), and a post-catalytic nicked complex (C, purple) was obtained by soaking with hydrolyzable UTP. Structures of the active sites of each complex are shown, with the protein drawn in cartoon and the DNA substrate in stick. Protein secondary structural elements are marked, with β-strands numbered, and α-helices labeled alphabetically. Magnesium (yellow) and manganese (orange) ions are drawn as spheres. 2Fo – Fc electron density (contoured at 1σ) is shown for the DNA substrate near the catalytic center of each complex. The location of the 90° bend in the DNA template strand is indicated by a blue arrow. The partially disordered pyrophosphate leaving group is shown in stick. (D) Global superpositions of the pre- (green) and post-catalytic (purple) complexes. The location of the Loop2 truncation is shown as a black asterisk. (E) Detailed comparison of the wildtype hPol μΔ2 active center in the pre- (protein in dark green; primer terminus in light green; UMPNPP in cyan), and post-catalytic (protein in purple; DNA in lavender) complexes. Dashed arrows indicate movement of electrons during progression of the reaction. All structural figures were generated using PyMOL (Schrödinger).
Structural superposition of the pre- and post-catalytic complexes with a 1-nt gapped DNA substrate indicates that these structures are nearly indistinguishable (RMSD of 0.091Å over 293 Cα atoms, Figure 1D). In both structures, the protein and DNA are observed in a ‘closed’ conformation. Direct interactions with the template strand are few, and are mediated primarily by the palm and thumb subdomains. The upstream primer is tightly held by the fingers subdomain, while the downstream primer containing the 5′-phosphate is bound by the 8 kDa subdomain. In the pre-catalytic ternary complex, the incoming nonhydrolyzable UMPNPP is bound in the active site, correctly pairing with the templating adenine base (Figure 1B). The ribonucleotide is easily accommodated in the active site, with no distortion of protein side chains. Two divalent magnesium ions occupy the catalytic (metal A) and nucleotide binding (metal B) sites. The primer terminal nucleotide is observed with a C3′-endo sugar pucker that places the 3′-OH in a catalytically relevant position, 3.5 Å from the α-phosphate of the incoming nucleotide (Figure 1E). Nucleotide incorporation occurs in crystallo upon soaking with UTP, with minimal structural changes resulting from catalysis. There is clear electron density in the post-catalytic complex for the newly formed phosphodiester bond, accompanied by a concomitant loss of contiguous density between the α- and β-phosphates (Figure 1C). As a consequence of bond formation, the α-phosphate undergoes stereochemical inversion, which results in a 1.2 Å movement of the phosphorous atom toward the primer terminal base (Figure 1E). The bound PPi is partially disordered, and is present in a slightly different conformation than observed in the pre-catalytic complex. The metal A and B sites remain occupied, but the metal A site is now filled by a manganese ion (likely a contaminant from the pyrophosphate stock used in this experiment, whose identity was assigned based on coordination distance, geometry and the presence of anomalous scattering, Supplementary Figure S2A). A partial rotation of the side chain of Asp330 is also observed after incorporation. Detailed analysis of the active site before and after catalysis suggests that the reaction mechanism proceeds in a similar fashion to that of deoxyribonucleotide incorporation (Figure 1E) (35).
Analysis of interactions stabilizing binding of the incoming ribonucleotide
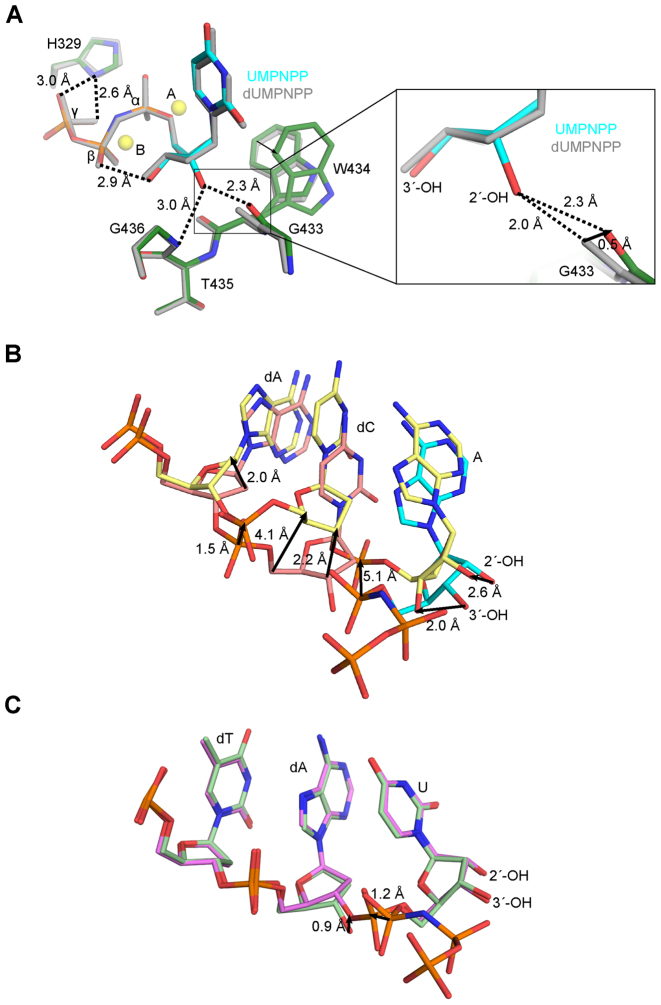
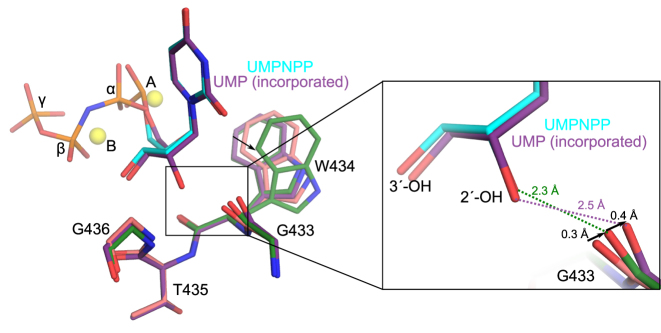
In order to understand its lack of discrimination against ribonucleotides, the hPol μΔ2 1-nt gapped pre-catalytic ternary complex with bound UMPNPP was compared with previously reported structures containing a bound deoxyribonucleotide (35). The crystals were grown and soaked under identical conditions—differing only in the identities of the incoming nucleotides—to provide an unbiased microenvironment for comparison. It should be noted that the efficient exchange of a correctly paired deoxyribonucleotide with a ribonucleotide is consistent with Pol μ’s weak sugar selectivity (13,19,30). In the 1-nt gapped pre-catalytic ternary complexes, a notable difference in the conformation of Trp434 is observed between the two structures. With a bound dUMPNPP, Trp434 adopts only a single conformation (Figure 2A, left), which likely stabilizes the position of the primer terminal sugar via π–CH interactions. However, when the UMPNPP is bound, Trp434 displays considerable mobility, which is apparent in its weakened electron density and higher relative B-factors (Supplementary Figure S2B). Trp434 is modeled with two alternate conformations. The higher occupancy conformation is nearly identical to that observed in the dUMPNPP-bound structure, differing only by a slight rotation around the Cβ-Cγ bond. The lower occupancy conformation assumes a different rotamer, where the planar aromatic group is rotated drastically back toward the backbone between Gly433 and Trp434. This rotation appears to be conformationally coupled with a rotation of Val420, which is located proximal to the Trp434 side chain. The increased mobility of the Trp434 side chain appears to cause a slight decrease in the positional stability of the primer terminal nucleotide. This decrease is reflected in the occupancy of the primer terminus, which refined to 0.88. The density for the mobile conformation is too weak to accurately model.
Figure 2.
Analysis of interactions involving the 2′-OH on the incoming ribonucleotide. (A) Superposition of pre-catalytic ternary complexes of wildtype hPol μΔ2 with incoming deoxy- (dUMPNPP, gray) or ribonucleotide (protein in dark green; UMPNPP in cyan). Magnesium ions (yellow) from the ribonucleotide bound structure are shown for orientation. Zoomed in view (right) of putative hydrogen bonding interactions (dashed lines) with the 2′-OH. (B) Superposition of the Pol λ Loop1 deletion variant pre- (pink, PDB ID code 3UPQ (33); AMPNPP in cyan) and post- catalytic (yellow, PDB ID code 3UQ0 (33)) complexes. (C) Superposition of the hPol μΔ2 pre- (light green) and post-catalytic (lavender) complexes. Direction and calculated distances for movement of paired atoms are shown as black arrows.
The 2′-OH of the UMPNPP makes only two direct interactions with the protein. First, a putative hydrogen bond might exist between the 2′-OH and the backbone amide of Gly436, though the geometry of this interaction is suboptimal (Figure 2A, left). A second hydrogen bond, between the 2′-OH and the backbone carbonyl of Gly433, is also observed. A potentially key difference between the ribo- versus deoxyribonucleotide bound structures is visible in the disposition of the backbone carbonyl of Gly433 in the presence of the 2′-OH (Figure 2A, right). If the carbonyl maintained the position observed in the dUMPNPP-bound structure (35), the 2′-OH would present a steric clash. Upon binding of the 2′-OH on the UMPNPP, this carbonyl shifts by 0.5 Å, increasing the distance from the hydroxyl to 2.3 Å. A similar short-distance hydrogen bond in Pol λ was proposed to be a potentially high energy interaction between the 2′-OH and the Tyr505 backbone carbonyl (33).
A hydrogen bond is observed between the side chain of His329 and the γ-phosphate oxygens of the incoming nucleotide, distal from the 2′-OH (Figure 2A). His329 has been shown to be indispensable for gap-filling during NHEJ of noncomplementary DNA double-strand breaks and template independent synthesis by Pol μ, but is dispensible for gap-filling during single-strand break repair (45). In these contexts, the histidine side chain is thought to stabilize the binding of an incoming nucleotide, by ‘bridging’ the triphosphate and the primer terminal phosphate. It has been hypothesized that for incoming ribonucleotides, stabilizing the triphosphate position using multiple hydrogen bonding interactions may serve to overcome unfavorable interactions involving the 2′-OH (33). This is consistent with His329 being conserved only in Pol μ and TdT, rather than in Pols β or λ, which do not incorporate ribonucleotides as effectively.
Though interactions with the triphosphate may aid in initial binding of an incoming ribonucleotide despite the unfavorable interaction between the 2′-OH and the nearby backbone carbonyl, this hydrogen bonding network is usually lost during dissociation of the pyrophosphate leaving group after incorporation (33). It has been hypothesized that losing these stabilizing interactions results in build-up of repulsive strain in the polymerase active site, making the energetically unstable binding mode of the 2′-OH untenable in the post-catalytic conformation. Consistent with this hypothesis, the crystal structure of a Loop1 deletion variant of Pol λ post-catalytic nicked complex reveals that the 2′-OH on the newly incorporated nucleotide is expelled from its position in the pre-catalytic ternary complex, and the phosphate backbone of the preceding nucleotide becomes grossly distorted (Figure 2B) (33). By comparison, structural superposition of the pre- and post-catalytic 1-nt gapped complexes for Pol μ reveals no such distortion (Figure 2C). The only movement observed in the Pol μ post-catalytic nicked complex is a slight upward shift of the C3 and O3 atoms (∼0.9 Å). This shift is common to both deoxy- and ribonucleotide incorporation, and appears to be characteristic of bond formation by Pol μ (35,44).
Biochemical characterization of ribonucleotide incorporation by Pol μ
Steady-state kinetic parameters were determined for wildtype Pol μ, using a sequence and structural context identical to that used in the crystallization experiments. These experiments show that Pol μ exhibits only a 2.7-fold preference of incorporating dTTP versus UTP in this sequence context (Table 1). Previous studies have suggested that the presence of a ribonucleotide on the primer terminus functions as a weak chain terminator for this enzyme (13). The activity of Pol μ was therefore examined in the presence of a ribonucleotide-terminated primer. There was no significant difference in catalytic efficiency for deoxribonucleotide extension from a rMNP-terminated primer, but there was a slight decrease in insertion of a ribonucleotide in the same context (Table 1).
Table 1. Kinetic characterization of nucleotide incorporation by human wildtype Pol μ, in the context of a 1-nt gapped DNA substrate.
| Primer terminus | Nucleotide | K m (μM) | k cat (s−1) | k cat/Km (s−1·μM−1) |
|---|---|---|---|---|
| dA | dTTP (4) | 4.0 ± 0.90 | 0.020 ± 0.0050 | 5.60 × 10−3 ± 2.2 × 10−3 |
| dA | UTP (6) | 21.0 ± 3.0 | 0.04 ± 0.017 | 2.1 × 10−3 ± 0.65 × 10−3 |
| rA | dTTP (4) | 3.7 ± 0.70 | 0.017 ± 0.0050 | 4.40 × 10−3 ± 1.0 × 10−3 |
| rA | UTP (3) | 23.6 ± 5.80 | 0.015 ± 0.0007 | 0.65 ± 0.17 × 10−3 |
Values represent the mean ± standard deviation for each calculation, with values in parenthesis indicating the number of independent measurements per sample.
We next endeavored to modulate the ribonucleotide discrimination of Pol μ, using a structure-guided site-directed mutagenesis approach. Because Pols β and λ contain the ‘YF motif’ and exhibit greater sugar selectivity (31–33), a strategy involving mutation of the ‘GW motif’ in Pol μ to mimic the ‘YF motif‘ (Supplementary Figure S1B) would be logical. These variations, however, have been previously generated in Pol μ (19), and revealed that the W434F mutation yielded a protein with similar enzymatic activity as compared to wildype, that was still capable of incorporating ribonucleotides. Substitution of Gly433 with tyrosine greatly reduced insertion of ribonucleotides (3- to 18-fold better discrimination than wildtype, depending on the identity of the incoming rNTP), but had a severely deleterious effect on overall catalytic efficiency of dNTP incorporation as well. From a structural standpoint, these results are unsurprising, since the presence of the tyrosine side chain might produce a slight steric clash with the nearby Ser458, and a more severe clash with the large, aromatic side chain of Trp434. The G433Y/W434F double mutant had almost no detectable enzymatic activity (19). Therefore, we decided to take a more conservative mutagenesis approach, substituting Gly433 with either alanine (as was used to decrease ribonucleotide discrimination by Pols β and λ (32,46)) or serine. Increasing the size of the side chain could, hypothetically, decrease the backbone flexibility in this region, and limit the mobility of the carbonyl interacting with the 2′-OH. The side chain of Trp434 was reduced in size to either a single ring (histidine) or alanine. His329 was substituted with alanine, to remove hydrogen bonds with the γ-phosphate of the incoming ribonucleotide. These mutations were generated in full-length Pol μ, and kinetic parameters were determined in a steady-state enzymatic activity assay, using a 1-nt gapped single-strand break DNA substrate of identical sequence context to that used in the structural studies (Table 2).
Table 2. Comparison of ribonucleotide discrimination in wildtype and mutant Pol μ proteins.
| Enzyme | xNTP | K m (μM) | k cat (s−1) | k cat/Km (s−1·μM−1) | Discrimination |
|---|---|---|---|---|---|
| Pol μ wt | dTTP (4) | 4.0 ± 0.9 | 0.02 ± 0.005 | 5.6 × 10−3 ± 2.2 × 10−3 | |
| UTP (6) | 21 ± 3.0 | 0.04 ± 0.012 | 2.1 × 10−3 ± 0.65 × 10−3 | 2.7 | |
| Pol μ H329A | dTTP (5) | 7.5 ± 1.98 | 0.017 ± 0.005 | 2.2 × 10−3 ± 4.23 × 10−4 | |
| UTP (5) | 65.8 ± 10.8 | 0.005 ± 0.003 | 8.1 × 10−5 ± 3.34 × 10−5 | 27.0 | |
| Pol μ G433A | dTTP (3) | 3.1 ± 0.28 | 0.018 ± 0.006 | 5.8 × 10−3 ± 2.3 × 10−3 | |
| UTP (3) | 29 ± 2.8 | 0.022 ± 0.008 | 0.76 × 10−3 ± 0.23 × 10−3 | 7.6 | |
| Pol μ G433S | dTTP (5) | 6.0 ± 1.4 | 0.026 ± 0.005 | 4.5 × 10−3 ± 1.6 × 10−3 | |
| UTP (5) | 72.7 ± 22.9 | 0.027 ± 0.014 | 0.41 × 10−3 ± 0.28 × 10−3 | 11.0 | |
| Pol μ W434A | dTTP (6) | 9.4 ± 1.13 | 0.032 ± 0.013 | 3.7 × 10−3 ± 1.68 × 10−3 | |
| UTP (3) | 79.1 ± 8.20 | 0.012 ± 0.003 | 1.6 × 10−4 ± 0.56 × 10−4 | 23.0 | |
| Pol μ W434H | dTTP (4) | 6.7 ± 1.62 | 0.020 ± 0.006 | 3.1 × 10−3 ± 1.37 × 10−3 | |
| UTP (3) | 61.1 ± 8.20 | 0.021 ± 0.006 | 3.5 × 10−4 ± 0.88 × 10−4 | 8.8 |
Values represent the mean ± standard deviation for each calculation, with values in parenthesis indicating the number of independent measurements per sample.
Whereas the wildtype Pol μ displayed only 2.7-fold preference for deoxyribonucleotides, the G433A and G433S mutants improved discrimination against ribonucleotides incrementally, directly correlated with increasing side chain size (7.6- and 11-fold preference for deoxyribonucleotides, respectively) (Table 2). For G433S, this effect was largely achieved by a reduction in apparent binding affinity for the incoming ribonucleotide (72.7 μM), as compared to the deoxyribonucleotide (21 μM). Interestingly, G433A exhibited a similarly low binding affinity for UTP (29 μM), as compared to the wildtype. Both Gly433 mutants displayed a slight decrease in catalytic rate for ribonucleotide incorporation. Therefore, improved ribonucleotide discrimination was successfully achieved, without a significant decrease in catalytic efficiency for deoxyribonucleotide incorporation. With respect to Trp434, modulation of ribonucleotide discrimination was inversely correlated with side chain size. The alanine substitution yielded substantially higher discrimination (23-fold), while the effect of the histidine substitution were more moderate (8.8-fold) (Table 2). Similar to the G433A/S mutants, the increased discrimination by the Trp434 mutants was largely due to a reduction in apparent binding affinity for the incoming UTP (79.1 μM and 61.1 μM for W434A and W434H, respectively). The increase in ribonucleotide discrimination comes at a cost, however, because these mutations also decrease the catalytic efficiency of deoxyribonucleotide incorporation. Surprisingly, despite its distance from the 2′-OH, substitution of His329 with alanine had the greatest effect on ribonucleotide discrimination (27-fold). The H329A mutant exhibited reduced binding affinity for the incoming ribonucleotide, to a similar extent as for the Gly433 and Trp434 mutants, but also an 8-fold decrease in the catalytic rate for the reaction. It should be noted that, in all cases, the 1-nt single-strand break was filled using a single nucleotide, regardless of the identity of the sugar moiety involved. Neither the wildtype Pol μ, or any of the ribonucleotide discrimination mutants displayed any evidence of strand displacement or multiple incorporations, which is consistent with the observation that Pol μ fills small (1- or 2-nt) gaps in vivo using a single incorporation event (44).
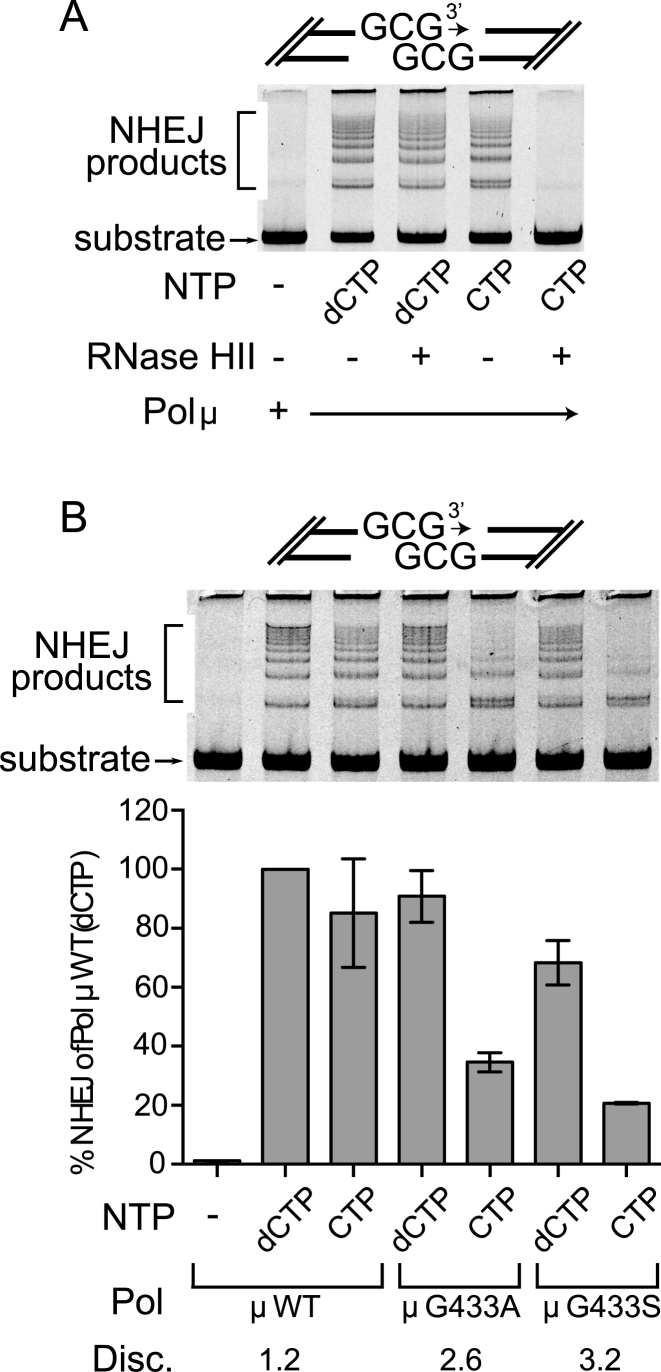
We next assessed the relevance of ribonucleotide incorporation in the context of DSB repair by NHEJ, using an in vitro reconstitution assay. The DSB substrate for this assay was comprised of ends that align to form a 1-nt gap that must be filled using complementary dCTP or CTP before end-joining can occur (Figure 3A). End joining is apparent upon addition of Ku, XRCC4-ligase IV complex, wildtype full-length Pol μ, and either dCTP or CTP. Joining efficiency was similar when using dCTP when compared with CTP, and the incorporation of cytidine ribonucleotide in joined product was verified with RNAseHII digestion (Figure 3A). Using CTP, Pol μ G433A and G433S mutants supported less joining, compared with wild type Pol μ using CTP, and a slight decrease when dCTP was used (Figure 3B). This is consistent with these mutants’ increased sugar discrimination (Table 2). Trp434 and His329 mutations have been previously reported to cause a severely deleterious effect on the ability to perform nonhomologous end-joining (15,45), so these mutants were not tested in this study.
Figure 3.
Comparison of nucleotide discrimination by Pol μ in NHEJ. (A) In vitro NHEJ concatamerization assay for full-length wildtype Pol μ, using either dCTP or CTP, as indicated. Following completion of NHEJ, dCTP- or CTP-incorporated reaction mixtures were treated with RNase HII, which cleaves where ribonucleotides have been incorporated into DNA (52). (B) Comparison of nucleotide discrimination by wildtype or mutant Pol μ in the same in vitro NHEJ concatamerization assay used above (top), lacking only the RNase HII treatment step. Extent of end-joining by each protein variant was quantitated by real-time PCR (bottom). All values represent calculated averages of end-joining (normalized to the activity of wildtype Pol μ with dCTP incorporation) ± the standard deviation for each experiment (Supplementary Table S4). Each assay was performed in triplicate.
Structural characterization of the hPol μΔ2 ribonucleotide discrimination mutants
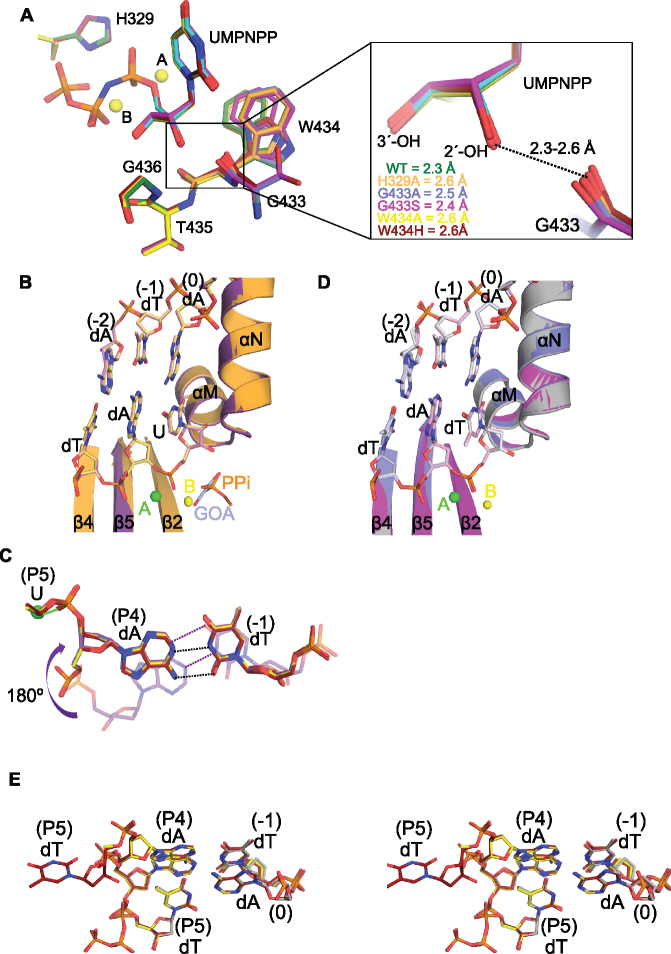
To better understand how Pol μ active site mutations may affect ribonucleotide discrimination, structures of all of the hPol μΔ2 mutants were determined in complex with the same DNA substrate used for the wildtype protein (Supplementary Tables S1 and S2). Despite the decreased binding affinity of each mutant for the incoming ribonucleotide (Table 2), the UMPNPP is clearly bound in the active site, with catalytically competent geometry indistinguishable from that of the wildtype enzyme (RMSD of 0.15–0.21 Å over 325 Cα atoms) (Figure 4A). Both glycine substitutions are easily accommodated in the active site and cause no distortion in surrounding residues. In the G433S ternary complex, the serine side chain is observed in two alternate conformations, neither of which would clash with Trp434. As in the wildtype ternary complex, the Trp434 side chain is highly mobile in the G433S structure. Conversely, in the G433A ternary complex, the side chain is observed in the sharply rotated conformation likely incapable of stabilizing the position of the primer terminal nucleotide. All of the Gly433 and Trp434 mutants exhibit some primer terminal instability (occupancies refined to 0.79, 0.72, 0.82 and 0.88 for G433A, G433S, W434A and W434H, respectively). There is no indication of this instability for the H329A mutant. In this structure, the incoming nucleotide triphosphate is easily accommodated in the canonical position, even in the absence of the His329 side chain, and displays no increase in B-factors that might be indicative of mobility. Though there are no distortions of active site geometry, it should be noted that distances associated with the short-range hydrogen bond between the 2′-OH and the Gly433 carbonyl vary in the mutants, but all are longer (2.4–2.6 Å) than is observed in the wildtype enzyme (2.3 Å) (Figure 4A, right).
Figure 4.
Structural characterization of Pol μ ribonucleotide discrimination mutants. (A) Structural superpositions of the active site residues in the pre-catalytic ternary complexes of hPol μΔ2 wildtype (green, UMPNPP in cyan) and ribonucleotide discrimination mutants (H329A in orange, G433A in blue, G433S in magenta, W434A in yellow, W434H in maroon). The two magnesium ions (yellow spheres) from the wildtype structure are shown for orientation. The panel on the right gives a magnified view of the 2′-OH distances to the carbonyl of residue 433. (B) Structural superpositions of the post-catalytic complexes of the hPol μΔ2 wildtype (protein in purple, DNA in lavender) and H329A mutant (protein in orange, DNA in khaki). The partially disordered pyrophosphate leaving group (PPi, orange) from the wildtype structure, and a bound glycolate molecule (GOA, gray) from the H329A structure are modeled in the active site in stick. (C) Stick diagram of the expulsion of the newly incorporated rUMP from the active sites of the G433A/S (G433A in blue, G433S in magenta) and W434A/H (W434A in yellow, W434H in maroon) mutants. The canonical position maintained by the wildtype protein (purple, transparent) is shown for comparison. Putative hydrogen bonding interactions for the wildtype (purple dashes) and mutant (black dashes) base pair are indicated. Coordination of the HhH2 sodium ion (green sphere) by the phosphate between residues P4 and P5 in the mutants is drawn as a green line. (D) Structural superpositions of the post-catalytic complexes of the hPol μΔ2 wildtype (gray), G433A (blue), and G433S (magenta) mutants with incorporated dTMP. E. Stereo diagram of DNA substrate mixtures from the post-catalytic complexes of the hPol μΔ2 wildtype (gray, PDB ID code 4M0A (35)), W434A (yellow), and W434H (maroon) mutants with incoming dTTP/incorporated dTMP.
Post-catalytic structures of each mutant were then determined by soaking binary complex crystals with UTP. H329A incorporated the ribonucleotides with no global structural changes to either the protein or the DNA substrate (Figure 4B) as a result of nucleotide incorporation (RMSD of 0.21 Å over 325 Cα atoms). In contrast to the post-catalytic structures of the wildtype and H329A mutants, all of the Gly433 and Trp434 substitution mutants showed a drastic change in DNA substrate conformation after incorporation. Once the reaction is completed, the newly incorporated nucleotide is expelled from the active site and is largely disordered beyond the phosphodiester bond. Meanwhile, the sugar-base moiety of the primer residue immediately upstream is flipped 180° from the canonical position, which allows it to maintain base pairing with the template base in the -1 position (Figure 4C). The phosphate of the newly incorporated rUMP now coordinates with the metal in the HhH2 site. Due to the relatively high concentration of free nucleotide triphosphate in the soaking solution (5 mM), a new UTP molecule is bound in the active site, mimicking a pre-catalytic complex. Since the primer terminus is no longer in a catalytically relevant conformation, the bound triphosphate is unincorporated.
To determine whether the observed DNA substrate rearrangement was a general characteristic of catalysis, or was due specifically to the presence of a ribonucleotide, post-catalytic structures of the mutants were determined by soaking binary complex crystals with dTTP. Both Gly433 substitution mutants incorporated the deoxyribonucleotide with no subsequent conformational changes in DNA substrate (Figure 4D). In contrast, both Trp434 substitution mutations displayed a roughly equal mixture of reaction products—a population where the newly incorporated dTMP remains in the active site, in a conformation nearly identical to that observed in the wildtype protein (35); and a population that has expelled the newly incorporated dTMP similarly to the observed conformation of the incorporated ribonucleotides, and bound an unincorporated dTTP (Figure 4E). The entire newly incorporated deoxyribonucleotide is visible in the W434H structure, but refines with higher B-factors and relatively weak 2Fo – Fc electron density.
DISCUSSION
Diminished ribonucleotide discrimination is one of the distinguishing features of Family X Pol μ (13,19) (Supplementary Figure S1). Because Pol μ plays a unique role in repair of DNA double-strand breaks containing noncomplementary ends during NHEJ, ribonucleotide incorporation by this enzyme could prove to be a critical consideration. NHEJ is the major repair pathway during the G1 phase of the cell cycle, and in nonreplicating cells (47). At these particular times, the deoxyribonucleotides typically favored by DNA polymerases are at their lowest concentrations, while ribonucleotide levels are correspondingly high (5). Given the tremendous threat that DSBs pose to genomic integrity, the most efficient means of their repair might be through incorporation of ribonucleotides by Pol μ, which could be rapidly repaired afterwards through the ribonucleotide excision repair pathway (reviewed in (48)). To investigate how Pol μ accommodates rNTPs, we used X-ray crystal structures of the catalytic domain of Pol μ, in complex with DNA single-strand break substrates and incoming ribonucleotides (Figure 1 and Supplementary Table S2), to reveal that Pol μ binds and incorporates ribonucleotides, with no distortion of canonical catalytic geometry. Given the drastic difference in ribonucleotide discrimination between Pol μ and Pols β or λ (Supplementary Figure S1), it is perhaps surprising that a more substantial structural distinction between the active sites of these Family X polymerases is not observed.
Replicative DNA polymerases use Van der Waals interactions with large, usually aromatic side chains to systematically exclude binding of ribonucleotides (49) (Supplementary Figure S3), and mutations of the steric gate residues decrease discrimination by >100-fold (reviewed in (50,51)). In contrast, sugar selectivity in the Family X polymerases would appear to be mediated through hydrogen bonding interactions between the 2′-OH of a ribonucleotide and the protein backbone (Figure 2A and Supplementary Figure S3). The increased sugar selectivity of Pols β and λ (32,33), compared to that of Pol μ and TdT, has been largely attributed to the presence of a putatively high-energy, short-range hydrogen bonding interaction between the 2′-OH and the backbone carbonyl of the ‘YF motif’ tyrosine residues (Tyr271 in Pol β, and Tyr505 in Pol λ). The lengths of this hydrogen bond in the rNTP-bound 1-nt gapped pre-catalytic ternary complex structures of Pols β and λ (Supplementary Figure S4) are measured to be 2.5 and 2.4 Å, respectively. For both of these enzymes, the backbone carbonyl shifts slightly (∼0.4 Å) from its position in the deoxyribonucleotide-bound structure upon binding of the rNTP. Intriguingly, this putative hydrogen bonding interaction in the pre-catalytic ternary complex of Pol μ is similarly short, at 2.3 Å (Figure 2A). Since the amino acid side chains are not directly involved in this interaction, it is perhaps unsurprising their substitution would have more subtle effects (8- to 12-fold changes in discrimination (32,46)).
Since the critical interaction with the 2′-OH could create a steric clash with the backbone carbonyl of the first residue of either the ‘YF or ‘GW’ motif, the energetics of that region must be considered. These residues lie on the C-terminal end of α-helix M in the Family X polymerases. Therefore, a partial negative charge from the helical dipole moment in this region could contribute somewhat to stabilization of the short-distance interaction between the 2′-OH and the backbone carbonyl of Gly433 (32). In addition, since glycine residues have decreased restraints on backbone plasticity (42), a glycine at this position in Pol μ and TdT might offer the carbonyl interacting with the 2′-OH a wider range of motion than would be possible for the tyrosine residues observed in Pols β and λ. The proposed mobility of the Gly433 carbonyl in Pol μ is visible from superpositions of the available 1-nt gapped crystal structures, from the binary complex before binding of the incoming ribonucleotide (35); to the UMPNPP-bound pre-catalytic complex; through in crystallo incorporation in the post-catalytic nicked complex. This superposition reveals that the carbonyl moves ∼0.3 Å from its position in the binary complex upon binding of the incoming ribonucleotide, then a further ∼0.4 Å as a result of incorporation (Figure 5). The relative importance of Gly433 backbone flexibility is supported by the fact that substitution of Gly433 with either alanine or serine—which should incrementally increase restraints on movement—shows a correlated increase in ribonucleotide discrimination (Table 2). Although the Gly433 mutants more effectively excluded ribonucleotides from binding in the active site, they display catalytic rates nearly indistinguishable from the wildtype protein, regardless of the type of nucleotide incorporated. Aside from a slightly longer interaction distance from the 2-OH to the carbonyl observed in the structures of the Gly433 substitution mutants (Figure 4A), there are no overt structural explanations for the differences in ribonucleotide discrimination.
Figure 5.
Putative role of Gly433 backbone carbonyl motions for ribonucleotide binding and incorporation. Structural superposition of hPol μΔ2 1-nt gapped binary (pink, PDB ID code 1LZG (35)), pre-catalytic ternary (protein in green; UMPNPP in cyan), and post-catalytic nicked (purple) complexes. Magnesium ions (yellow) from the pre-catalytic structure are shown for orientation. Global view of regions in close proximity to the 2′-OH are shown on the left, with a zoomed-in view (right) of putative hydrogen bonding interactions (dashed lines) with the 2′-OH.
The role of Trp434 in this system is less clear. The tryptophan side chain, being larger than the phenylalanine present in Pols β or λ, reduces extraneous space inside the active site. Having a tighter, more compact active site could more effectively exclude solvent and decrease vibrational mobility of the incoming nucleotide or the divalent metal ions, allowing the catalytically competent geometry to be maintained more easily. The space-filling hypothesis is supported by the observation that an incremental size reduction from a two-ring system to a single ring (W434H), to deletion of the ring system entirely (W434A), correlated well to an increase in ribonucleotide discrimination (Table 2). Improved sugar selectivity is due primarily to reduced binding affinity for the rNTP, as indicated by the increase in Km. A slight improvement in sugar selectivity was also observed when the equivalent residue in Pol β (Phe272) was mutated to alanine (32). The heterogeneity of DNA product conformations in the post-catalytic structures for the Trp434 substitution mutants (Figure 4C and E) suggests that loss of the tryptophan side chain may fundamentally affect nucleotide incorporation, regardless of the identity of the sugar. Decreased stabilization of the primer terminal sugar, through loss of π–CH interactions, provides a possible explanation for the reduced catalytic efficiency in these mutants. There appears to be an additional electrostatic component to the system, since substitution of Trp434 with histidine rather than phenylalanine had a stronger effect on ribonucleotide discrimination (19).
His329, positioned on the opposite side of the incoming nucleotide far from the 2′-OH, has an unanticipated role in ribonucleotide binding and incorporation. H329A, like the Gly433 and Trp434 mutant, exhibited decreased binding affinity for the ribo- versus deoxyribonucleotide. However, loss of the His329 side chain also decreased the rate of the reaction by 8-fold (Table 2), likely due to loss of putative hydrogen bonding interactions with the triphosphate and with the primer terminal phosphate. Pre- and post-catalytic crystal structures of Pol μΔ2 (H329A) revealed no overt structural changes that could contribute to either the reduction in binding affinity or kinetic rate (Figure 4). It is possible that loss of His329 decreases stabilization of the dense negative charge on the pyrophosphate leaving group. Thus, the ability of the H329A mutant to incorporate the incoming ribonucleotide with no subsequent conformational changes (Figure 4B), indicates that its effect is transmitted catalytically, rather than through strain associated with binding of the 2′-OH.
Though previous studies of the Family X polymerases have attributed sugar selectivity primarily to a single residue of the ‘steric gate’ (19,32,46)—namely the tyrosine or glycine of either the ‘YF’ or ‘GW’ motif—our work also suggests a role for the tryptophan residue present in Pol μ. Our work demonstrates that any energetic strain generated by binding and incorporation of the incoming nucleotide's 2′-OH is easily accommodated in the wildtype enzyme. However, any alteration of the immediate microenvironment diminishes the ability of Pol μ to absorb this energetic strain. Interestingly, ribonucleotide discrimination in Pol μ could also be modulated by the side chain of a residue positioned too far away to directly sense the presence of a 2′-OH (i.e. His329). The structural, biochemical, and kinetic data presented in this work reveal that accommodation of ribonucleotide binding and incorporation cannot be solely attributed to a single residue, but rather is a complex system, with multiple residues uniquely contributing to the overall energetics of nucleotide binding and incorporation.
ACCESSION NUMBERS
5TWP, 5TWQ, 5TWR, 5TWS, 5VZ7, 5VZ8, 5VZ9, 5VZA, 5VZB, 5VZC, 5VZD, 5VZE, 5VZF, 5VZG, 5VZH, 5VZI.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Krahn for optimization of the UMPNPP, UTP, and dTTP parameter files for structural refinement, D. Walton for technical consultation, and B. Beard and M. Schellenberg for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Division of Intramural Research of the National Institute of Environmental Health Sciences, National Institutes of Health [1ZIA ES 102645 to L.C.P., Z01 ES065070 to T.A.K.] (in part); American Cancer Society Postdoctoral Fellowship [PF-14-0438-01-DMC to J.M.P.]; National Cancer Institute [CA097096 to D.A.R.]; US Department of Energy, Office of Science, Office of Basic Energy Sciences [W-31-109-Eng-38].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bebenek K., Kunkel T.A.. Functions of DNA polymerases. Adv. Protein Chem. 2004; 69:137–165. [DOI] [PubMed] [Google Scholar]
- 2. Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Lundstrom E.B., Burgers P.M., Johansson E., Chabes A., Kunkel T.A.. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clark A.B., Lujan S.A., Kissling G.E., Kunkel T.A.. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon. DNA Repair (Amst.). 2011; 10:476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clausen A.R., Zhang S., Burgers P.M., Lee M.Y., Kunkel T.A.. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase delta. DNA Repair (Amst.). 2013; 12:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994; 140:1–22. [DOI] [PubMed] [Google Scholar]
- 6. Makarova A.V., Nick McElhinny S.A., Watts B.E., Kunkel T.A., Burgers P.M.. Ribonucleotide incorporation by yeast DNA polymerase zeta. DNA Repair (Amst.). 2014; 18:63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Donigan K.A., Cerritelli S.M., McDonald J.P., Vaisman A., Crouch R.J., Woodgate R.. Unlocking the steric gate of DNA polymerase eta leads to increased genomic instability in Saccharomyces cerevisiae. DNA Repair (Amst.). 2015; 35:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li Y.A., Breaker R.R.. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2΄-hydroxyl group. J. Am. Chem. Soc. 1999; 121:5326–5372. [Google Scholar]
- 9. Vengrova S., Dalgaard J.Z.. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 2006; 7:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sayrac S., Vengrova S., Godfrey E.L., Dalgaard J.Z.. Identification of a novel type of spacer element required for imprinting in fission yeast. PLoS Genet. 2011; 7:e1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghodgaonkar M.M., Lazzaro F., Olivera-Pimentel M., Artola-Boran M., Cejka P., Reijns M.A., Jackson A.P., Plevani P., Muzi-Falconi M., Jiricny J.. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol. Cell. 2013; 50:323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lujan S.A., Williams J.S., Clausen A.R., Clark A.B., Kunkel T.A.. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol. Cell. 2013; 50:437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nick McElhinny S.A., Ramsden D.A.. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 2003; 23:2309–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramsden D.A. Polymerases in nonhomologous end joining: building a bridge over broken chromosomes. Antioxid. Redox Signal. 2011; 14:2509–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin M.J., Garcia-Ortiz M.V., Esteban V., Blanco L.. Ribonucleotides and manganese ions improve non-homologous end joining by human Polmu. Nucleic Acids Res. 2013; 41:2428–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pitcher R.S., Brissett N.C., Picher A.J., Andrade P., Juarez R., Thompson D., Fox G.C., Blanco L., Doherty A.J.. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J. Mol. Biol. 2007; 366:391–405. [DOI] [PubMed] [Google Scholar]
- 17. Zhu H., Shuman S.. Bacterial nonhomologous end joining ligases preferentially seal breaks with a 3′-OH monoribonucleotide. J. Biol. Chem. 2008; 283:8331–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bartlett E.J., Brissett N.C., Doherty A.J.. Ribonucleolytic resection is required for repair of strand displaced nonhomologous end-joining intermediates. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E1984–E1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruiz J.F., Juarez R., Garcia-Diaz M., Terrados G., Picher A.J., Gonzalez-Barrera S., Fernandez de Henestrosa A.R., Blanco L.. Lack of sugar discrimination by human Pol mu requires a single glycine residue. Nucleic Acids Res. 2003; 31:4441–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boule J.B., Rougeon F., Papanicolaou C.. Terminal deoxynucleotidyl transferase indiscriminately incorporates ribonucleotides and deoxyribonucleotides. J. Biol. Chem. 2001; 276:31388–31393. [DOI] [PubMed] [Google Scholar]
- 21. Aoufouchi S., Flatter E., Dahan A., Faili A., Bertocci B., Storck S., Delbos F., Cocea L., Gupta N., Weill J.C. et al. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. 2000; 28:3684–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bertocci B., De Smet A., Berek C., Weill J.C., Reynaud C.A.. Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity. 2003; 19:203–211. [DOI] [PubMed] [Google Scholar]
- 23. Dominguez O., Ruiz J.F., Lain de Lera T., Garcia-Diaz M., Gonzalez M.A., Kirchhoff T., Martinez A.C., Bernad A., Blanco L.. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000; 19:1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gilfillan S., Dierich A., Lemeur M., Benoist C., Mathis D.. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993; 261:1175–1178. [DOI] [PubMed] [Google Scholar]
- 25. Loc’h J., Rosario S., Delarue M.. Structural basis for a new templated activity by terminal deoxynucleotidyl transferase: implications for V(D)J recombination. Structure. 2016; 24:1452–1463. [DOI] [PubMed] [Google Scholar]
- 26. Bertocci B., De Smet A., Weill J.C., Reynaud C.A.. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006; 25:31–41. [DOI] [PubMed] [Google Scholar]
- 27. Capp J.P., Boudsocq F., Besnard A.G., Lopez B.S., Cazaux C., Hoffmann J.S., Canitrot Y.. Involvement of DNA polymerase mu in the repair of a specific subset of DNA double-strand breaks in mammalian cells. Nucleic Acids Res. 2007; 35:3551–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chayot R., Danckaert A., Montagne B., Ricchetti M.. Lack of DNA polymerase mu affects the kinetics of DNA double-strand break repair and impacts on cellular senescence. DNA Repair (Amst.). 2010; 9:1187–1199. [DOI] [PubMed] [Google Scholar]
- 29. Mahajan K.N., Nick McElhinny S.A., Mitchell B.S., Ramsden D.A.. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol. Cell. Biol. 2002; 22:5194–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roettger M.P., Fiala K.A., Sompalli S., Dong Y., Suo Z.. Pre-steady-state kinetic studies of the fidelity of human DNA polymerase mu. Biochemistry. 2004; 43:13827–13838. [DOI] [PubMed] [Google Scholar]
- 31. Cavanaugh N.A., Beard W.A., Wilson S.H.. DNA polymerase beta ribonucleotide discrimination: insertion, misinsertion, extension, and coding. J. Biol. Chem. 2010; 285:24457–24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cavanaugh N.A., Beard W.A., Batra V.K., Perera L., Pedersen L.G., Wilson S.H.. Molecular insights into DNA polymerase deterrents for ribonucleotide insertion. J. Biol. Chem. 2011; 286:31650–31660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gosavi R.A., Moon A.F., Kunkel T.A., Pedersen L.C., Bebenek K.. The catalytic cycle for ribonucleotide incorporation by human DNA Pol lambda. Nucleic Acids Res. 2012; 40:7518–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moon A.F., Garcia-Diaz M., Batra V.K., Beard W.A., Bebenek K., Kunkel T.A., Wilson S.H., Pedersen L.C.. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair (Amst.). 2007; 6:1709–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moon A.F., Pryor J.M., Ramsden D.A., Kunkel T.A., Bebenek K., Pedersen L.C.. Sustained active site rigidity during synthesis by human DNA polymerase mu. Nat. Struct. Mol. Biol. 2014; 21:253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chayen N.E. Comparative studies of protein crystallization by vapour-diffusion and microbatch techniques. Acta Crystallogr. D Biol. Crystallogr. 1998; 54:8–15. [DOI] [PubMed] [Google Scholar]
- 37. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M.. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006; 62:859–866. [DOI] [PubMed] [Google Scholar]
- 38. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 39. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 40. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lovell S.C., Davis I.W., Arendall W.B. 3rd, de Bakker P.I., Word J.M., Prisant M.G., Richardson J.S., Richardson D.C.. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003; 50:437–450. [DOI] [PubMed] [Google Scholar]
- 43. Nick McElhinny S.A., Snowden C.M., McCarville J., Ramsden D.A.. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell Biol. 2000; 20:2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moon A.F., Gosavi R.A., Kunkel T.A., Pedersen L.C., Bebenek K.. Creative template-dependent synthesis by human polymerase mu. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4530–E4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moon A.F., Garcia-Diaz M., Bebenek K., Davis B.J., Zhong X., Ramsden D.A., Kunkel T.A., Pedersen L.C.. Structural insight into the substrate specificity of DNA Polymerase mu. Nat. Struct. Mol. Biol. 2007; 14:45–53. [DOI] [PubMed] [Google Scholar]
- 46. Brown J.A., Fiala K.A., Fowler J.D., Sherrer S.M., Newmister S.A., Duym W.W., Suo Z.. A novel mechanism of sugar selection utilized by a human X-family DNA polymerase. J. Mol. Biol. 2010; 395:282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takata M., Sasaki M.S., Sonoda E., Morrison C., Hashimoto M., Utsumi H., Yamaguchi-Iwai Y., Shinohara A., Takeda S.. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998; 17:5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Williams J.S., Lujan S.A., Kunkel T.A.. Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat. Rev. Mol. Cell Biol. 2016; 17:350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brown J.A., Suo Z.. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011; 50:1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang W., Wu E.Y., Hellinga H.W., Beese L.S.. Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides. J. Biol. Chem. 2012; 287:28215–28226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ordonez H., Uson M.L., Shuman S.. Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation. Nucleic Acids Res. 2014; 42:11056–11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rydberg B., Game J.. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:16654–16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.