Abstract
Salp15, a salivary protein of Ixodes ticks, inhibits the activation of naïve CD4 T cells. Treatment with Salp15 results in the inhibition of early signaling events and the production of the autocrine growth factor, interleukin-2. The fate of the CD4 T cells activated in the presence of Salp15 or its long-term effects are, however, unknown. We now show that Salp15 binding to CD4 is persistent and induces a long-lasting immunomodulatory effect. The activity of Salp15 results in sustained diminished cross-antigenic antibody production even after interruption of the treatment with the protein. Transcriptionally, the salivary protein provokes an acute effect that includes known activation markers, such as Il2 or Cd44, and that fades over time. The long-term effects exerted by Salp15 do not involve the induction of either anergy traits nor increased populations of regulatory T cells. Similarly, the treatment with Salp15 does not result in B cell anergy or the generation of myeloid suppressor cells. However, Salp15 induces the increased expression of the ectoenzyme, CD73, in regulatory T cells and increased production of adenosine. Our study provides a profound characterization of the immunomodulatory activity of Salp15 and suggests that its long-term effects are due to the specific regulation of CD73.
Introduction
Tick saliva is composed of a cocktail of pharmacologically active biomolecules that modulate among other host responses, the activation of CD4 T cells1–5. One of the best-studied components of tick saliva is the protein Salp15 from Ixodes scapularis. This protein inhibits the activation of naïve CD4 T cells, blocking early T cell signaling pathways, and thereby, the production of the autocrine growth factor IL-26–10. Salp15 specifically binds to the first domain (D1) of the CD4 co-receptor causing a conformational change that impedes its interaction with the Src kinase, Lck and its activation11. Due to the specificity of Salp15 for CD4 and its capacity to inhibit the activation of CD4 T cells, the use of this tick saliva protein has been suggested for the treatment of immune diseases. In addition, our group has demonstrated the ability of Salp15 to inhibit the development of CD4 T cell-mediated immune responses in vivo upon challenge with different antigens7, 10, 12. Moreover, Salp15 prevents the development of experimental asthma in a mouse model7. However, whether the inhibition of CD4 T cells by Salp15 has long-term effects on future encounters with specific or unrelated antigens is currently unknown.
A common complication associated with allogeneic hematopoietic stem cell transplantation (HSCT) is the appearance of Graft-versus-host disease (GvHD). GvHD appears when donor T cells recognize as foreign the recipient antigens, including both human leukocyte (HLA) and minor histocompatibility antigens. Allogeneic HSCTs are used both in treatments of malignant disease and in ordinary transplants. GvHD appears in 50% of the transplants and causes death in 15% of the cases13. Despite its effectiveness, the induction of immunosuppression after HSCT can produce undesirable effects. These include the inhibition of graft-versus-tumor effector cells (GvT) and the appearance of infections and neoplasms13–17. Other treatments used for the mitigation or elimination of this disease are ineffective and unspecific. In fact, pre-transplantation chemotherapy and radiotherapy treatments (conditioning) applied in these cases for the elimination of the cancer cells and the establishment of the transplanted cells can result in nonspecific inflammatory events, helping create the necessary conditions for the activation of donor T cells18. Although several murine models of transplantation exist19, 20, none recapitulates in full the pathology observed in human transplantation. The transplant model of pure strains into F1 offspring does not require previous conditioning and results in mild episodes of acute GvHD followed by a period of chronic disease characterized by the production of autoantibodies21.
Because Salp15 is able to inhibit early T cell signaling events, we hypothesized that the protein could preclude the activation of CD4 T cells and induce a long-term unresponsive or anergic after the exposure to the salivary protein. Our results show that Salp15 is able to change the transcriptional program of CD4 T cells during activation that nevertheless fades over time and does not result in increased populations of anergic or regulatory T cells. However, the protein induces the upregulation of the ectoenzyme, CD73 on the surface of Tregs, inducing increased production of the immunosuppressive molecule adenosine. Overall, the activity of Salp15 is evident in a long-term transplantation murine model and prevents the deposition of immune complexes in the kidney, a hallmark of murine chronic GvHD21.
Results
The effect of Salp15 on activating CD4 T cells is long-lasting
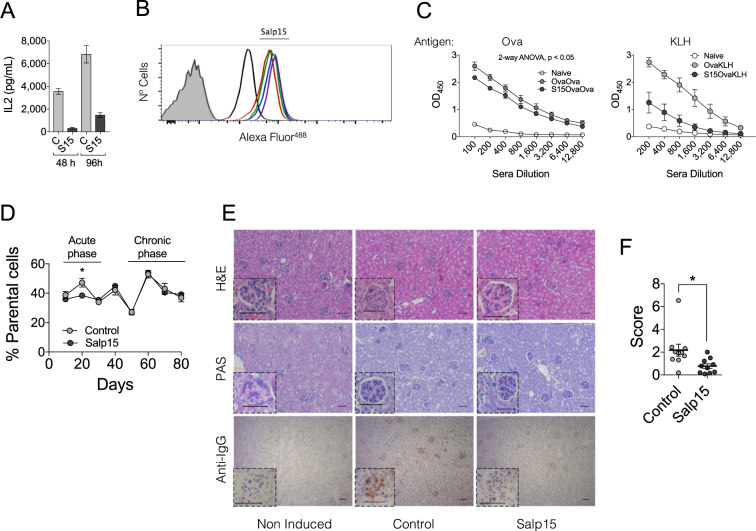
In order to determine whether the effect of Salp15 on the activation of CD4 T cells is sustained, we activated purified splenic CD4 T cells in the presence of the salivary protein for 2 days, followed by their extensive washing and re-stimulation for 2 more days. The production of IL-2 was significantly reduced at both time points, including after 4 days of activation when Salp15 was no longer present (Fig. 1A). The longer-term effect of Salp15 could be due to its persistent binding to the surface of CD4 T cells. Thus, we determined the binding of Alexa Fluor488-labeled Salp15 as well as the inactive control (Salp15ΔP11) by flow cytometry. Although both Salp15 and Salp15ΔP11 bound to purified CD4 T cells, the deletion of the C-terminal peptide, P11, resulted in decreased binding (Fig. 1B) in agreement with its reported lack of activity22. Importantly, binding of Salp15 to CD4 T cells was detectable for up to 72 h (Fig. 1B), indicating a persistent ability of this protein to remain attached to CD4.
Figure 1.
The effect of Salp15 on CD4 T cells is long-lasting. (A) IL-2 production by purified splenic CD4 T cells activated with anti-CD3/CD28 for 2 days in the presence of 50 μg/ml of Salp15, washed and re-stimulated under the same conditions for another 48 h in the absence of the immunosuppressive protein. The results represent the average ± SE of one experiment in triplicate and are representative of 3 performed. (B) Binding of Salp15 or the C-terminal deletion mutant, Salp15ΔP11 to purified CD4 T cells. The proteins were labeled with Alexa Fluor488 and tested for their binding for different time periods. The binding was assessed by flow cytometry. The shaded histogram represents unlabeled cells. The black histogram represents binding of Salp15ΔP11. Color histograms represent binding of Salp15 at different time points (12, 24, 48 and 72 h). (C) Antibody titers specific for ovalbumin (left panel) and KLH (right panel) in mice immunized with the antigens as described in Methods and treated with 50 μg of Salp15 (S15OvaOva or S15OvaKLH) or left untreated (OvaOva or OvaKLH). Non-immunized mice served as controls (Naive) (D) Percentage of parental (H2b) cells in the blood of transplanted CB6F1 mice (H2b,d) over a period of 80 days. The mice were either treated with 50 μg of Salp15 or Salp15ΔP11 (Control) (E) Histological features of the kidneys of CB6F1 mice transplanted with B6 splenocytes after 80 days. Kidney sections were stained with H&E (top panels), periodic acid schiff (PAS) staining (middle panels) and anti-mouse IgG immune complex deposition (lower panels). The scale bars represent 50 μm. (F) Assessment of IgG immune complex deposition scores in the kidneys of the transplanted mice by analysis of 5 different micrographs with the FriDA software package and averaged per section. The experiments in vivo were performed with groups of 5 mice and performed at least twice.
In order to assess whether Salp15 could also exert long-term effects in vivo, we performed an immunization regime against ovalbumin and the unrelated protein, keyhole limpet hemocyanin (KLH). Groups of mice were immunized with ovalbumin in aluminum hydroxide in the absence or presence of Salp15 (day 0). Seven days later, the mice were boosted with ovalbumin under the same conditions. At day 14, the mice in each group were subdivided and immunized with ovalbumin or KLH in the absence of Salp15. All the mice were sacrificed at day 21. Sera were then analyzed for the presence of ovalbumin- and KLH-specific IgG levels. As expected, ovalbumin-specific IgG levels were significantly lower in mice that had received Salp15 in the first 2 immunizations (Fig. 1C). In order to establish whether the effect of Salp15 was circumscribed to ovalbumin, we also determined sera IgG levels against KLH. Mice that had not been immunized with this antigen did not show KLH-specific IgG in the sera. Notably, the levels of KLH-specific IgG were high in those mice receiving the antigen, and significantly lower in those that had received two previous doses of Salp15, but not at the time of immunization with KLH (Fig. 1C). These data show that the effect of Salp15 lasts beyond the treatment and can affect the response to unrelated antigens, such as KLH.
To further address the long-term effect associated with the treatment with Salp15, we utilized a murine model of GvHD resulting from the adoptive transfer of spleen cells from B6 into unconditioned CB6 F1 mice21. In this model of transplantation, the proliferation of drafted cells can be monitored in the blood and results in two different phases of the disease: an acute phase in which the transplanted cells proliferate, followed by a chronic phase predominantly characterized by symptomatology similar to autoimmune disease, including the deposition of IgG immune complex in the kidney21. CB6 F1 mice were transplanted 60 × 106 spleen cells and divided into two groups. One of the groups received intraperitoneal injections of Salp15 every other day for 10 days, while the control group received the same dose of Salp15ΔP11. Control animals showed a peak of parental cells at day 20 relative to the transplant, followed by a decline and another increase around 60 days post-transplant (Fig. 1D). In contrast, the treatment with Salp15 during the first 10 days post transplantation resulted in a significantly reduced level of parental cells in the blood (Fig. 1D). However, no effect was detected during the chronic phase of the disease. We also analyzed the potential effect of the treatment with Salp15 on the pathology associated with this model at day 80 after transplantation of the parental splenocytes. CB6 F1 mice transplanted with B6 splenocytes showed some disorganization in the glomerular basement membrane and tubular brush border of the kidney upon transplantation that was not affected by the treatment (Fig. 1E). However, immune complex deposition that was readily detected in the control-treated mice was significantly reduced in the animals that had been treated with Salp15 (Fig. 1E,F). Overall, our results show that Salp15 has short-term and long-term effects during the development of immune responses that specifically affect the expansion of CD4 T cells and the production of antibodies.
Identification of transcriptional traits in activating CD4 T cells treated with Salp15
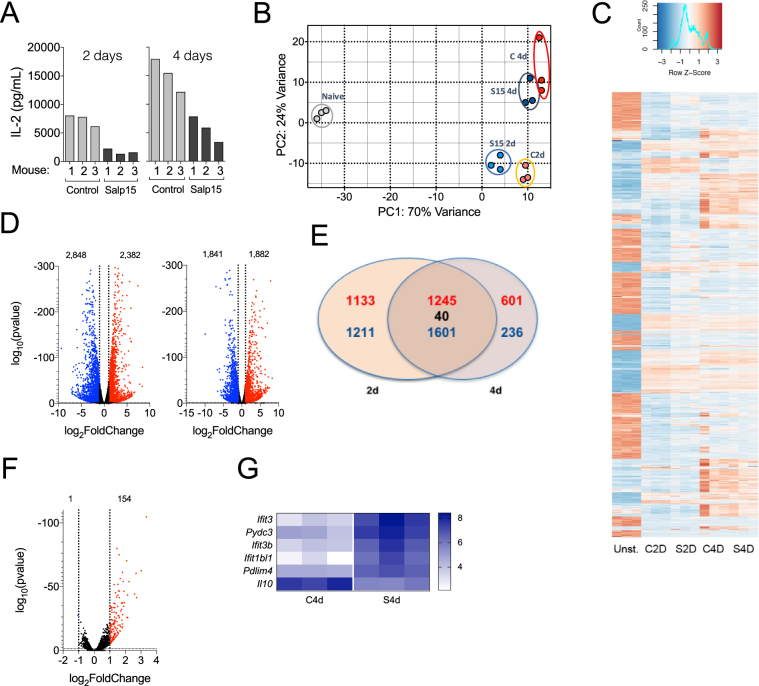
We then sought to determine the transcriptional signature of activated CD4 T cells for 48 and 96 hours with plate-bound anti-CD3 and soluble anti-CD28 in the presence of Salp15 or its control, Salp15ΔP11 (Fig. 2A). To allow the survival of the CD4 T cells throughout the activation process, a low dose of Salp15 (25 μg/ml) was used. This dose reduced the activation of CD4 T cells (Fig. 2A) without significant cell death at 4 days post-activation (data not shown). Principal component analysis (PCA) showed a distinct pattern of gene expression in control activated cells over the analyzed period of time (Fig. 2B). The presence of Salp15 resulted in variations in the PCA at 2 days of activation, while the differences faded when the transcriptome was analyzed after 4 days of activation (Fig. 2B). These differences were also noticeable when the 1000 most regulated genes were analyzed, with maximal differences between Salp15-treated and control activated CD4 T cells at 2 days and more discrete differences when analyzed at 4 days of activation (Fig. 2C). The activation of CD4 T cells under control conditions revealed 2382 genes upregulated and 2848 genes downregulated at 2 days of activation (Fig. 2D), while 1882 genes were upregulated and 1841 genes downregulated after 4 days of activation, using a cut-off value of 1 log2fold change and an adjusted p-value < 0.05 (Fig. 2D). Of these genes, 1245 were upregulated at both 2d and 4d of activation, 1601 were downregulated at both time points and 40 were regulated in opposite directions (Fig. 2E). Using the same cut-off values, we found 154 genes upregulated as a consequence of the presence of Salp15 during activation at 2d while only 1 gene was downregulated (Fig. 2F). Notably, the number of genes regulated at 4d of activation in the presence of Salp15 was dramatically reduced to 5 genes upregulated and just 1 (Il10) downregulated (Fig. 2G). Selected transcriptional changes were validated as shown in Supplementary Fig. 1.
Figure 2.
Transcriptional traits of CD4 T cells activated in the presence of Salp15. (A) IL-2 production by CD4 T cells used for the transcriptomic analysis. Each bar represents one of the 3 mice used. The amount of Salp15 used was 25 μg/ml. Control cells were treated with Salp15ΔP11 (Control). (B) Principal component analysis showing the grouping of the different assay conditions according to their transcriptome. Non-activated cells (Naive, grey); Salp15ΔP11-treated at 2 days (C 2d, orange) or 4 days of activation (C 4d, red); Salp15-treated at 2 days (S15 2d, light blue) or 4 days (S15 4d, dark blue). (C) Heatmap corresponding to the 1000 most regulated genes over the conditions analyzed by RNAseq. (D) Volcano plots showing the genes upregulated (red) or downregulated (blue) by activation with anti-CD3 and CD-28 at 2 (left) and 4 days (right) of stimulation. (E) Venn diagram showing the number of genes regulated at both 2 and 4 days of activation in the absence of Salp15. (F) Volcano plot showing the number of genes differentially regulated during the activation of CD4 T cells in the presence of Salp15 or Salp15ΔP11 (control) after 2 days of stimulation. (G) Heatmap of the genes regulated in the presence of Salp15 at 4 days of activation.
Salp15 affects CD4 T cell genes early during the activation process
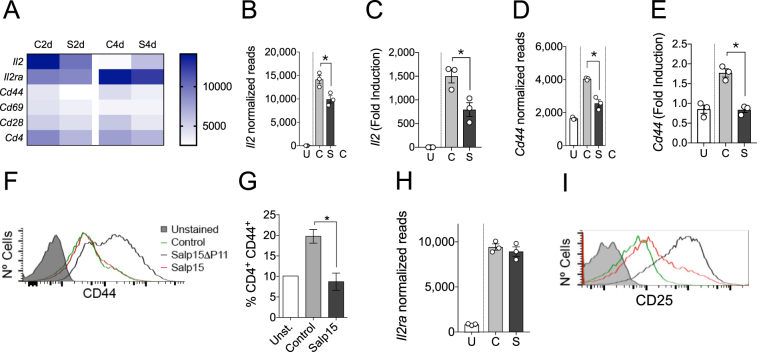
The activation of CD4 T cells produced the expected profile, involving genes such as Il2, Cd44 or IL2ra (Fig. 3A). Gene ontology analysis of Biological Processes (GOBP) revealed that the most over-represented groups included genes related to leukocyte cell-cell adhesion and aggregation or T cell activation, among other immune-related processes (Supplementary Fig. 2). As expected, the presence of Salp15 induced a reduction of Il2 gene expression and the production of IL-2 at 2 days of activation (Fig. 3A,B,C; see also Fig. 2A). Furthermore, the activation marker CD44 was significantly reduced both at the gene expression level (Fig. 3A,D,E), by surface analysis of the protein by flow cytometry in in vitro activated CD4 T cells (Fig. 3F), and in vivo in the spleens of ovalbumin-immunized mice at day 7 post-immunization (Fig. 3G). Although, the effect of Salp15 on Il2ra expression was not evident at this concentration (25 μg/ml, Fig. 3H), the analysis of CD25 on the surface of 2-day activated CD4 T cells revealed decreased levels of the protein in the presence of this dose of Salp15 (Fig. 3I).
Figure 3.
Salp15 affects the activation of CD4 T cells. (A) Heatmap of genes involved in the activation of CD4 T cells at 2 and 4 days of stimulation with anti-CD3/CD28 in the absence or presence of Salp15. (B) Normalized reads corresponding to the Il2 gene at 2 days of activation in the presence of Salp15 (S) or Salp15ΔP11 (C). U- Unstimulated cells. (C) Transcriptional levels of Il2 at 2 days of activation in the presence of Salp15 (S) or Salp15ΔP11 (C) of Salp15 determined by qRT-PCR. Normalized reads of Cd44 (D) and transcriptional levels by qRT-PCR (E) in CD4 T cells at 2 days of activation, as before. (F) Surface CD44 levels on CD4 T cells activated for 2 days in the presence of Salp15 or Salp15ΔP11. Control: Unstimulated cells. The grey histogram represents an unstained control. (G) Percentage of CD4 T cells expressing CD44 in the spleen of mice immunized with ovalbumin and treated with Salp15 compared to control treated, immunized mice (Control) and non immunized mice (Unst.). (H) Normalized reads of Il2ra in CD4 T cells at 2 days of activation, as before. (F) Surface CD25 levels on CD4 T cells activated for 2 days in the presence of Salp15 or Salp15ΔP11. The color legend is as indicated in (F). All the experiments were performed with 25 μg/ml of Salp15 or Salp15ΔP11 (control).
Salp15 induces the expression of 5′-ectonucleotidase (CD73) in regulatory T cells
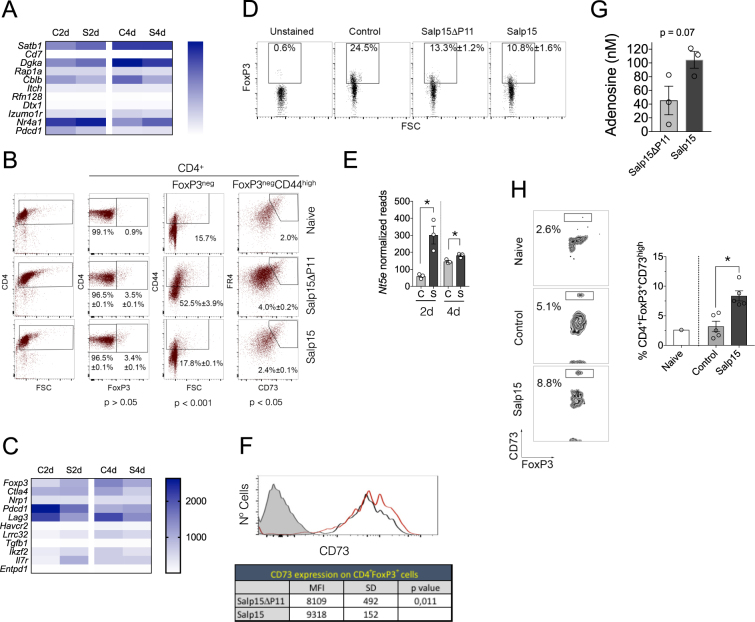
The repressed activation of CD4 T cells in the presence of Salp15 could result in the induction of anergy. We therefore analyzed the expression levels of genes associated with this phenomenon in CD4 T cells, including Satb1, Cd7, Rap1a, Itch, Rnf128, Dtx1, Izumo1r, Cblb, Dgka 23, Nr4a1 or Pdcd1 24. Salp15 induced small and inconsistent changes in these genes (Fig. 4A, Table 1), suggesting that this protein does not induce anergy in CD4 T cells. In order to confirm these results, we analyzed by flow cytometry markers of anergy in CD4 T cells that were activated in vitro in the presence of Salp15. The percentage of CD4+FoxP3−CD44highCD73highFR4high [ 24 cells increased upon CD4 T cell activation but remained as low as in naive cells in the presence of Salp15 (Fig. 4B). Furthermore, the analysis of anergic CD4 T cells in mice either immunized with ovalbumin (Table 2) or induced GvHD (Table 3) showed no effect on this population as a consequence of the treatment with Salp15. Similarly, the treatment with Salp15 did not result in the generation of a significant anergic B cell population or increased the pool of monocytic myeloid suppressor cells (Table 2). These data confirm that Salp15 act as an immunosuppressor on CD4 T cells that depends on its interaction with CD4 and that does not induce a long-term anergic state in T or B cells, nor the generation of myeloid suppressor cells.
Figure 4.
Salp15 does not induce increased anergic or Treg numbers but augments the levels of CD73 in FoxP3+ cells. (A) Heatmap representing genes associated with anergy in CD4 T cells according to the RNAseq analysis in the presence of Salp15 (S) or Salp15ΔP11 (C) at 2 and 4 days of activation. (B) Percentage of FoxP3+ and anergic CD4 T cells upon their exposure to 25 μg/ml of Salp15 or Salp15ΔP11 for 2 days. The average ± SE of triplicates is indicated. The p values (Student´s t test) correspond to the comparison between the Salp15ΔP11 and Salp15 groups. The data presented is representative of 2 independent experiments with similar results. (C) Heatmap corresponding to genes associated with regulatory T cells according to the RNAseq analysis in the presence of Salp15 (S) or Salp15ΔP11 (C) at 2 and 4 days of activation. (D) Percentage of FoxP3-positive CD4 T cells upon exposure to 25 μg/ml of Salp15 or Salp15ΔP11 in vitro for 2 days during activation. The values correspond to the average ± SE of 5 mice per group. No significant differences were detected between the Salp15ΔP11 and Salp15 groups. The data is representative of 2 independent experiments. (E) Normalized reads corresponding to the expression levels of Nt5e at 2 and 4 days of activation in vitro in the presence of Salp15 (S) or Salp15ΔP11 (C). (F) Surface expression levels of CD73 on CD4+FoxP3+ T cells (top) activated for 2 days in the presence of 25 μg/ml of Salp15 (red histogram) or Salp15ΔP11 (black histogram). The shaded histogram represents an unstained control. The table shows the average mean fluorescence intensity (MFI) ± standard deviation (SD) of 5 mice per group. Differences of the means were analyzed by the Student´s T-test. (G) Average adenosine levels in the supernatants of CD4 T cells activated in the presence of 25 μg/ml of Salp15 or Salp15ΔP11 for 2 days. The results represent 3 independent mice. (H) Increased percentage of CD4+FoxP3+CD73high cells in the blood of CB6F1 mice transplanted with B6 splenocytes at day 50 post induction. The histogram on the right represents the average ± SE of 5 control (Salp15ΔP11) and 5 Salp15-treated mice. *p < 0.05.
Table 1.
Differential expression of genes associated with CD4 T cell anergy and regulatory T cells.
| Gene | S2D vs. C2D | S4D vs. C4D | ||
|---|---|---|---|---|
| Log2FI | Adj. p-value | Log2FI | Adj. p-value | |
| Anergy | ||||
| Satb1 | 0,304 | 0,012 | 0,002 | 0,997 |
| Cd7 | 0,750 | 0,001 | 0,166 | NA |
| Rap1a | 0,030 | 0,894 | −0,223 | 0,116 |
| Itch | 0,087 | 0,613 | −0,065 | 0,816 |
| Rnf128 | −0,077 | 0,858 | −0,321 | 0,186 |
| Dtx1 | −0,039 | 0,908 | 0,047 | 0,921 |
| Izumo1r | −0,090 | 0,573 | −0,251 | 0,094 |
| Cblb | −0,403 | 8,40E-06 | −0,656 | 3,74E-07 |
| Dgka | 0,282 | 0,001 | −0,215 | 0,114 |
| Nr4a1 | 0,148 | 0,189 | 0,426 | 1,1E-04 |
| Pdcd1 | −0,568 | 3,9E-11 | −0,144 | 0,612 |
| Regulatory T cells | ||||
| Foxp3 | 0,815 | 7,85E-09 | −0,352 | 0,095 |
| Ctla4 | 0,109 | 0,535 | −0,691 | 1,19E-07 |
| Nrp1 | 0,048 | 0,859 | 0,015 | 0,975 |
| Pdcd1 | −0,568 | 3,90E-11 | 0,144 | 0,612 |
| Lag3 | −0,721 | 1,81E-11 | −0,540 | 5,44E-07 |
| Havcr2 | −0,166 | 0,668 | −0,177 | 0,620 |
| Lrrc32 | 0,527 | 0,044 | −0,268 | 0,345 |
| Tgfb1 | −0,148 | 0,197 | −0,164 | 0,473 |
| Ikzf2 | 0,438 | 0,117 | −0,558 | 0,00027 |
| Il7r | 1,504 | 6,21E-15 | −0,040 | 0,914 |
| Entpd1 | 0,402 | 0,163 | −0,004 | 0,995 |
| Type 1 Regulatory T cells | ||||
| Il10 | −0,236 | 0,492 | −1,069 | 4,01E-10 |
| Eomes | 0,136 | 0,477 | −0,193 | 0,351 |
| Il2rb | 0,171 | 0,367 | −0,069 | 0,808 |
| Itga4 | 0,536 | 0,027 | −0,260 | 0,178 |
| Itgb7 | −0,578 | 1,78E-06 | 0,047 | 0,878 |
| Ly6c1 | 0,200 | 0,233 | −0,190 | 0,511 |
| Tigit | −0,387 | 0,001 | −0,500 | 0,001 |
Table 2.
Cell populations in blood and spleen of mice immunized with ovalbumin (O) and treated with Salp15 (S) or the control protein (Δ).
| Blood B cell populations* | Control | ΔO | SO | ||
|---|---|---|---|---|---|
| Average | SD | Average | SD | ||
| B cells (B220+) | 45,10 | 43,50 | 1,87 | 40,07 | 4,43 |
| B220+CD69+ | 28,50 | 12,41 | 4,15 | 11,50 | 4,76 |
| Anergic B cells (B220+CD93+IgM+CD23+) | 3,85 | 2,28 | 0,40 | 1,80 | 0,76 |
| Mature B cells (B220+CD93−CD23+) | 27,87 | 28,77 | 0,56 | 27,80 | 1,50 |
| Spleen populations* | Control | ΔOO | SOO | ||
| Average | SD | Average | SD | ||
| CD4+ | 11,60 | 11,02 | 2,31 | 10,77 | 2,08 |
| CD8+ | 7,84 | 6,10 | 0,98 | 6,38 | 0,87 |
| CD4+CD25+ | 12,90 | 8,47 | 1,33 | 5,60 | 2,34 |
| CD4+CD44+ | 28,10 | 7,48 | 2,43 | 7,55 | 4,21 |
| Anergic T cells (CD4+CD44highCD73highFR4high) | 3,60 | 0,92 | 0,35 | 0,86 | 0,39 |
| Tregs (CD4+NRP1+) | 2,02 | 1,54 | 0,20 | 1,46 | 0,16 |
| Macrophages (F4/80+GR-1−) | 2,15 | 2,08 | 0,09 | 2,44 | 1,30 |
| Neutrophils (F4/80−GR-1+) | 1,30 | 1,52 | 0,21 | 2,73 | 0,71 |
| Monocytic myeloid suppressor cells (CD11bhighGR-1highLy6C+) | 1,01 | 0,56 | 0,24 | 0,82 | 0,36 |
| Inflammatory macrophages (F4/80+GR-1intLy6Chigh) | 1,97 | 2,42 | 0,38 | 2,52 | 0,64 |
| n = 5 | n = 5 | ||||
All comparisons between ΔO vs. SO, p > 0.05. *The gating strategies are shown in Supplementary Fig. 3.
Table 3.
Cell populations (average ± SD) in blood upon B6 splenocyte transplant into CB6 F1 mice.
| Blood cell populations | Day 20 | Day 30 | Day 40 | Day 60 | Day 80 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control | Salp15 | Control | Salp15 | Control | Salp15 | Control | Salp15 | Control | Salp15 | |
| CD4+ T cells | 26.1 ± 0.7 | 26.5 ± 0.5 | 33.5 ± 1.6 | 30.0 ± 1.2 | 34.9 ± 2.2 | 38.4 ± 2.7 | 30.9 ± 1.6 | 28.5 ± 1.8 | 21.0 ± 1.8 | 25.2 ± 2.7 |
| CD8+ T cells | 12.8 ± 1.2 | 13.8 ± 0.4 | 17.6 ± 1.0 | 18.0 ± 0.9 | 18.5 ± 1.1 | 17.4 ± 0.8 | 15.6 ± 0.8 | 16.2 ± 0.4 | 15.9 ± 1.4 | 18.4 ± 1.6 |
| Tregs (CD4+FoxP3+) | 7.1 ± 0.4 | 6.2 ± 0.3 | 7.3 ± 0.3 | 7.1 ± 0.2 | 4.7 ± 0.4 | 3.9 ± 0.7 | 6.8 ± 0.4 | 7.2 ± 0.5 | 7.7 ± 0.6 | 7.7 ± 1.2 |
| Anergic T cells (CD4+CD44highCD73highFR4high) | 6.2 ± 1.8 | 3.9 ± 0.9 | 20.0 ± 2.7 | 16.5 ± 1.9 | 13.4 ± 1.1 | 12.3 ± 1.1 | 4.4 ± 0.9 | 5.4 ± 0.5 | 7.0 ± 1.4 | 5.9 ± 1.0 |
All comparisons between Control vs. Salp15, p > 0.05. n = 5 mice per group.
The transcriptomic data also helped us elucidate whether activated CD4 T cells in the presence of Salp15 acquired specific markers of regulatory T cells, such as Foxp3, Ctla4, Nrp1, Pdcd1, Lag3, Havcr2, Lrrc32, Tgfb1, Ikzf2, Il7r or Entpd1 25, 26. No major differences were found between controls and CD4 T cells activated in the presence of Salp15 (Fig. 4C, Table 1). Moreover, the analysis of FoxP3-positive cells after 4 days of activation in vitro did not show changes in the percentage of Tregs associated with the presence of Salp15 (Fig. 4B). The effect of Salp15 on the pool of Tregs in vivo was also negligible in mice that had been immunized with ovalbumin (Fig. 4D). Furthermore, the analysis of FoxP3-positive CD4 T cells in the peripheral blood of CB6 F1 mice transplanted with B6 splenocytes did not show differences associated with the treatment with Salp15 throughout the life of the experiment (Table 3). Similar results were found when we analyzed the expression of Nrp1 on CD4 T cells, another marker of regulatory T cells27 (Table 2). The analysis of type 1 regulatory T (Tr1) cells markers, including Il10, Eomes, Il2rb, Itga4, Itgb7, Ly6c1 or Tigit 28, showed that Salp15 does not induce the generation of these cells, although Il10 expression levels were significantly reduced in the presence of the salivary protein at 4 days of activation (Table 1). These results demonstrate that the treatment with Salp15 does not result in the generation of a population of regulatory T cells that could account for long-term immunomodulatory effects.
We then addressed whether the treatment with Salp15 would impact the activity rather than the size of the Treg population. The ectoenzyme CD73 is expressed by Tregs and mediates the production of adenosine, an immunosuppressive molecule on T cells29–31. We observed that expression levels of Nt5e (which encodes CD73) were increased upon the treatment of CD4 T cells with Salp15 (Fig. 4E). Therefore, we determined the expression levels of CD73 on the surface of FoxP3-positive cells. We found that the activation of CD4 T cells in vitro in the presence of Salp15 resulted in a significant increase in the surface expression of CD73 (Fig. 4F). Furthermore, the levels of adenosine increased upon the treatment of activating CD4 T cells with Salp15 (Fig. 4G). Importantly, we observed an increase in a population of CD4+FoxP3+CD73high in the blood of mice that have been induced GvHD at day 50 post transplant (Fig. 4H). Overall, our results show that Salp15 is able to induce long-term effects on activating CD4 T cells that involve, at least in part, the increased expression and activity of CD73 on regulatory T cells.
Discussion
The tick salivary protein Salp15 inhibits early CD4 T cell signaling events and, in consequence, their activation. The activity of Salp15 on CD4 T cells is well-characterized9, 10, 24. Both in vitro and in vivo, this protein is able to prevent a full activation program on these immune cells6. The fate of CD4 T cells affected by the activity of Salp15 is not known. They could become permanently unable to respond to antigen or, alternatively, revert to a state in which they are amenable to become activated when antigen is present again. Here, we show that the presence of Salp15 during the activation of CD4 T cells results in long-term effects that affect their encounter with new antigens. Using transcriptomics, two models of CD4 T cell activation in vivo and the polyclonal activation of these cells in vitro, we show that whereas Salp15 does not affect the generation of anergic CD4 T cells or Tregs, it induces the increased expression of the ectoenzyme, CD73, in Foxp3+ regulatory T cells. This effect results in an elevated production of adenosine, a known immunomosuppressive molecule produced by Tregs32, 33.
Treatment with Tregs has been proposed as an appropriate therapy for GvHD and other immune disorders34, 35. In fact, several biological agents, including CTLA4 fusion proteins or anti-TNF antibodies34, have the ability to either induce an increase in number or the activity of Tregs. Treg function involves CD73 activity and other mechanisms of action36, 37. Indeed, the importance of CD73 activity has been studied in a murine model of GvDH, demonstrating that the ectonuclease helps control the disease30. The conversion of 5´-AMP to adenosine mediated by CD73 in Tregs has been broadly described38. The anti-inflammatory effect of adenosine present in the pericellular microenvironment results in the suppression of proliferation of effector CD4 T cells and the reduction of cytokine production39. Indeed, we found that the levels of this molecule were increased in in vitro assays of CD4 T cell activation in the presence of Salp15. The capacity of Salp15 to increase Nt5e transcription and the upregulation of CD73 on the surface of FoxP3+ T cells can therefore help explain the long-lasting effects elicited by Salp15. Future studies will determine whether the absence of CD73 on Tregs can indeed suppress the long-term immunomodulatory effect exerted by the salivary protein.
Our results show that the binding of Salp15 to CD4 persists along the activation period in vitro. However, the changes induced by Salp15 at the transcriptional level fade over time. Furthermore, Salp15 does not affect CD4 T cell differentiation in the absence of polarizing cytokines, including the induction of Th1, Th2, Th17 or Treg gene markers (Suppl. Fig. 4A). We cannot exclude that under polarizing conditions, Salp15 may affect CD4 T cell differentiation. In fact, in the presence of IL-6, the inhibition of IL-2 production during CD4 T cell activation can lead to their differentiation towards a Th17 phenotype22. Nevertheless, the restimulation of splenocytes of ovalbumin and KLH immunized mice resulted in similar levels of IL-4, while IFNγ was not detected in the restimulation supernatants (Suppl. Fig. 4B), arguing against an effect of Salp15 on Th1 or Th2 differentiation under these conditions. Our analysis also show that Salp15 does not seem to exert a direct effect on other cellular types, including the generation of anergic B cells or myeloid-derived suppressor cells (MDSCs). Nevertheless, in both in vivo models, the treatment with Salp15 results in an indirect effect on the ability of B cells to produce antigen-specific antibodies, as we have previously demonstrated10.
In summary, we show that the tick salivary immunosuppressor Salp15 is able to induce the sustained repression of CD4 T cell activation that involves the direct effect on these immune cells during the activation period and the increased expression of CD73 on regulatory T cells. These data support the notion that Salp15 is able to maintain its immunomodulatory action through the induction of increased Treg activity, leading to long-term effects in two in vivo models, including a pre-clinically relevant murine model of graft versus host disease.
Methods
Protein purification and labeling
Salp15 and an inactive deletion mutant lacking the last 20 aminoacids (Salp15ΔP11) were purified from Drosophila S2 cells, as described10. Protein labeling was performed using the Alexa Fluor® 488 Protein Labeling Kit (Thermo Fisher Scientific, Eugene, OR), following the manufacturer’s instructions.
Cell purification and activation
CD4 T cells were purified from the spleens of C57BL/6 mice by negative selection using a CD4 T cell isolation Kit (Miltenyi Biotec, Bergisch Gladbach, GE) according to the manufacturer’s instructions. Purified CD4 T cells at the indicated concentrations were activated with 5 μg/ml of plate-bound anti-CD3ε and 1 μg/ml of soluble anti-CD28 (BD Biosciences, San Diego, CA) in the presence of the indicated concentration of Salp15 or Salp15ΔP11. Cells were incubated at 37 °C in TexMACS Medium (Miltenyi Biotec).
Flow cytometry
Blood was extracted from the saphenous vein in the presence of EDTA and depleted of erythrocytes by hypotonic lysis. Whole splenocytes were isolated from immunized or GvHD mice by mechanical disruption followed by lysis of erythrocytes. The cells (106/ml) were incubated with Fc Block (anti-CD16/CD32; BD BioSciences) and labelled with fluorochrome-labeled antibodies against CD4, CD8, B220, CD69, Ly6C, F4/80, GR-1, CD25, CD11b, CD44, CD73, FR4, NRP1, Foxp3, CD93, IgM, CD23 (Miltenyi Biotec). To detect Salp15 binding to CD4 T cells, splenic purified CD4 T cells were labeled with Salp15-Alexa Fluor488 or Salp15ΔP11-Alexa Fluor488.
Immunizations
Groups of C57Bl/6 mice were immunized subcutaneously with 50 μg of ovalbumin in aluminum hydroxide (50% Hydrogel, Invivogene. Tolouse, France) containing 50 mg of Salp15 or PBS (conbtrol). The mice were boosted under the same conditions 7 days later. At day 14, each group of mice was subdivided into 2 groups and immunized with either 50 μg of ovalbumin or keyhole lympet hemocyanin (KLH). The mice were sacrificed 7 days later and the spleens and sera were analyzed for cellular composition and antigen-specific immunoglobulin titers, respectively.
Graft versus host disease murine model
Splenocytes were extracted from 8-week old C57BL/6 (H-2b) mice and 60 × 106/mouse were injected intraperitoneally into CB6F1 (H-2b,d) mice (Envigo, Gannat, France). The mice were treated with 50 μg of Salp15 by intraperitoneal injection starting the day of cell transfer and every other day until day 10. Blood was extracted from day 10 to day 80 at 10-days intervals. Erythrocytes were removed by hypotonic lysis and the cells were analyzed by flow cytometry. At sacrifice, kidneys were processed for histochemical evaluation.
Renal deposited IgG detection, PAS and HE
Kidneys were fixed in 10% neutral buffered formalin, dehydrated, embedded in paraffin and cut into 5 μm thick sections. For histopathology, sections were hydrated and stained with hematoxylin – eosin (HE) or periodic acid-Schiff (PAS) according to standard protocols. For immunohistochemical analysis, tissue sections were subjected to antigen retrieval using protease K for 20 min at 37 °C. After blocking, sections were incubated with primary antibody overnight. The slides were then sequentially incubated with DAB chromogen for 5 min, counterstained with Mayer’s hematoxylin and mounted for microscopy. Goat Anti-Mouse IgG-HRP Light chain specific (Jackson ImmunoResearch Laboratories) was used at 1/250 concentration as the primary antibody. Photographs were taken with an Axioimager A1 microscope and analyzed with Frida software40.
RNAseq
Purified CD4 T cells from three mice were activated independently with 5 μg/ml of plate-bound anti-CD3 and 1 μg/ml of soluble anti-CD28 (BD Bioscience) in the presence of 25 μg/ml of Salp15 or Salp15ΔP11 (control). Cells were incubated at 37 °C in TexMACS Medium (Miltenyi Biotec) for 48 and 96 h. RNA extraction was performed using the PureLink RNA Micro Scale Kit (Thermo Fisher Scientific) according to the manufacturerʼs protocol. The quantity and quality of the RNAs were evaluated using the Qubit RNA Assay Kit (Invitrogen, Eugene, OR) and RNA Nano Chips in a 2100 Bioanalyzer (Agilent Technologies, Waldbronn, GE), respectively. Libraries for sequencing were prepared using the TruSeq RNA Sample Preparation Kit v2 (Illumina Inc, San Diego, CA) following the protocol provided by the manufacturer. Single-read, 50 nt sequencing of pooled libraries was carried out in a HiScanSQ platform (Illumina Inc.).
The quality control of the sequenced samples was performed with FASTQC software (www.bioinformatics.babraham.ac.uk/projects/fastq). Reads were mapped against the mouse (mm10) reference genome by using the program Tophat41 to account for spliced junctions. The resulting BAM alignment files for the samples were the input for the Differential Expression (DE) analysis, carried out by DESeq. 242, to detect differentially expressed genes among the different conditions. GO enrichment was tested using the ClusterProfiler43 Bioconductor package and the Panther Database44. Transcriptomics data were also analyzed using QIAGENʼs Ingenuity Pathway Analysis (IPA, QIAGEN, Red Wood city, CA).
Real-time RT-PCR
RNA was reverse transcribed using M-MLV reverse transcriptase (Thermo Fisher Scientific) and random hexamers. Real-time PCR was then performed using SYBR Green PCR Master Mix (Quanta Biosciences, Beverly, MA) on a QuantStudio 6 real-time PCR System (Thermo Fisher Scientific). Fold induction of the genes was calculated using the 2−ΔΔCt method relative to the reference, previously validated genes, Rpl19 and Actb, as indicated. The primers used are listed in Table 4.
Table 4.
Sequence of the Primers used for RNAseq validation by qRT-PCR.
| Gene | Forward | Reverse | Purpose |
|---|---|---|---|
| IL2 | TGTGCTCCTTGTCAACAGCG | TTTCAATTCTGTGGCCTGCTTG | RNAseq validation |
| Zfp750 | CCCACCTTTGCTGCAGGT | GGGGGCCGTGGAATATAGTG | RNAseq validation |
| Klf2 | GCCTTCGGTCTTTTCGAGGA | AGGCTTCTCACCTGTGTGTG | RNAseq validation |
| Serpine2 | GATCCAAGCGAGGACGGG | GACCTGGATCCCTGTGTTGG | RNAseq validation |
| Cd44 | ATCCTCGTCACGTCCAACAC | GCTTTCTGGGGTGCTCTTCT | RNaseq validation |
| Jchain | GACGACGAAGCGACCATTCT | GCTCTGGGTGGCAGTAACAA | RNAseq validation |
| Amy2a5 | TTCGTTCTGCTGCTTTCCCT | CATTGGGTGGAGAGACCTGC | RNAseq validation |
| Cd7 | ATGCCCAAGACGTACAACAG | CTCCTGCCGGTCTTCAAAGT | RNAseq validation |
| Art2a-ps | GGCTAACCCAGCAGGTGACT | GGCTTCTGTGGATGTTCCCA | RNAseq validation |
| Rpl19 | GACCAAGGAAGCACGAAAGC | CAGGCCGCTATGTACAGACA | Reference, RNAseq validation |
| Actb | GACGATGCTCCCCGGGCTGTATTC | TCTCTTGCTCTGGGCCTCGTCACC | Reference, RNAseq validation |
Determination of adenosine levels
The levels of adenosine were determined in the culture supernatants of activated CD4 T cells using the fluorometric adenosine assay kit (Abnova, Walnut, CA) following the methods provided by the manufacturer.
Statistical analysis
Results are presented as means ± SE, unless otherwise stated. The differences in means between groups were tested using the Studentʼs T-test. Differences in antibody titers were assessed by a 2-way ANOVA. All calculations were made in GraphPad Prism, version 7. A p-value < 0.05 was considered statistically significant. All experiments were performed at least 3 times. In vivo experiments consisted of groups of 5 mice and were performed at least twice.
Ethics statement
All work involving animals was approved by the Institutional Animal Care and Use Committee (IACUC) at CIC bioGUNE and the competent authority (Diputación de Bizkaia). CIC bioGUNE animal facility is accredited by AAALAC Intl. All experiments were performed in accordance with European and Spanish guidelines and regulations.
Data availability
The transcriptomic data are deposited under GEO accession number GSE98700.
Electronic supplementary material
Acknowledgements
Supported by grants from the Department of Education of the Basque Government (PI2013-49 to JA and PI2012-42 to RB). JA is funded by the European Union (Grant Agreement number 602272). AMA and JLL’s work was supported by the Basque Department of Industry, Tourism and Trade (Etortek and Elkartek Programs), the Innovation Technology Department of Bizkaia and the CIBERehd Network. The work of AC is supported by a Ramón y Cajal award, the Basque Department of Industry, Tourism and Trade (Etortek), ISCIII (PI13/00031), FERO VIII Fellowship, the BBVA foundation, MINECO (SAF2016-79381-R) and the European Research Council Starting Grant (336343). CIBERonc was co-funded with FEDER funds. AC-M was funded by a Juan de la Cierva program award and the European Union MSCA program (CIG 660191). RB was funded by MINECO grants BFU2011-25986 and BFU2014-52282-P and the Consolider Program (BFU2014-57703-REDC). FJB was funded by a MINECO grant (CTQ2014-56966-R). D.B. is funded by a MINECO FPI fellowship. We thank the MINECO for the Severo Ochoa Excellence accreditation (SEV-2016-0644).
Author Contributions
Performed the experiments: J.T.C., I.M.M., D.B., M.A.P.I., V.G.J., A.C.M., N.M., A.M., J.M.F., C.S., V.G.C., J.A. Analyzed the data: J.T.C., J.L.L., A.M.A., H.R., J.A. Provided reagents: F.J.B., J.D.S., R.B., A.R., M.L.M.C. Wrote the manuscript: J.T.C., A.C., L.A., H.R., J.A., J.A. designed the study and provided supervision.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-11354-2
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kotal J, et al. Modulation of host immunity by tick saliva. J Proteomics. 2015;128:58–68. doi: 10.1016/j.jprot.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kern A, et al. Tick saliva represses innate immunity and cutaneous inflammation in a murine model of Lyme disease. Vector Borne Zoonotic Dis. 2011;11:1343–1350. doi: 10.1089/vbz.2010.0197. [DOI] [PubMed] [Google Scholar]
- 3.Oliveira CJ, et al. Deconstructing tick saliva: non-protein molecules with potent immunomodulatory properties. J Biol Chem. 2011;286:10960–10969. doi: 10.1074/jbc.M110.205047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francischetti IM, Sa-Nunes A, Mans BJ, Santos IM, Ribeiro JM. The role of saliva in tick feeding. Front Biosci (Landmark Ed) 2009;14:2051–2088. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hovius JW, Levi M, Fikrig E. Salivating for knowledge: potential pharmacological agents in tick saliva. PLoS Med. 2008;5 doi: 10.1371/journal.pmed.0050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juncadella I, Anguita J. The Immunosuppresive Tick Salivary Protein, Salp15. Pathogen-Derived Immunomodulatory Molecules. 2009;666:121–131. doi: 10.1007/978-1-4419-1601-3_10. [DOI] [PubMed] [Google Scholar]
- 7.Paveglio S, et al. The tick salivary protein, Salp15, inhibits the development of experimental asthma. Journal of Immunology. 2007;178:7064–7071. doi: 10.4049/jimmunol.178.11.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juncadella I, Garg R, Ananthnarayanan S, Yengo C, Anguita J. T-cell signaling pathways inhibited by the tick saliva immunosuppressor, Salp15. Fems Immunology and Medical Microbiology. 2007;49:433–438. doi: 10.1111/j.1574-695X.2007.00223.x. [DOI] [PubMed] [Google Scholar]
- 9.Garg R, et al. Cutting edge: CD4 is the receptor for the tick saliva immunosuppressor, Salp15. Journal of Immunology. 2006;177:6579–6583. doi: 10.4049/jimmunol.177.10.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anguita J, et al. Salp15, an Ixodes scapularis salivary protein, inhibits CD4(+) T cell activation. Immunity. 2002;16:849–859. doi: 10.1016/S1074-7613(02)00325-4. [DOI] [PubMed] [Google Scholar]
- 11.Ashish, et al. Conformational rearrangement within the soluble domains of the CD4 receptor is ligand-specific. Journal of Biological Chemistry. 2008;283:2761–2772. doi: 10.1074/jbc.M708325200. [DOI] [PubMed] [Google Scholar]
- 12.Motameni A, et al. Delivery of the immunosuppressive antigen salp15 to antigen-presenting cells by Salmonella enterica serovar typhimurium aroA mutants. Infection and Immunity. 2004;72:3638–3642. doi: 10.1128/IAI.72.6.3638-3642.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blazar BR, Murphy WJ, Abedi M. Advances in graft-versus-host disease biology and therapy. Nat Rev Immunol. 2012;12:443–458. doi: 10.1038/nri3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikulska M, et al. Blood stream infections in allogeneic hematopoietic stem cell transplant recipients: reemergence of Gram-negative rods and increasing antibiotic resistance. Biol Blood Marrow Transplant. 2009;15:47–53. doi: 10.1016/j.bbmt.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 15.Dykewicz CA, et al. Summary of the Guidelines for Preventing Opportunistic Infections among Hematopoietic Stem Cell Transplant Recipients. Clin Infect Dis. 2001;33:139–144. doi: 10.1086/321805. [DOI] [PubMed] [Google Scholar]
- 16.Akhtari M, et al. Therapy-related myeloid neoplasms after autologous hematopoietic stem cell transplantation in lymphoma patients. Cancer Biol Ther. 2013;14:1077–1088. doi: 10.4161/cbt.26342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman DL, Leisenring W, Schwartz JL, Deeg HJ. Second malignant neoplasms following hematopoietic stem cell transplantation. Int J Hematol. 2004;79:229–234. doi: 10.1532/IJH97.03178. [DOI] [PubMed] [Google Scholar]
- 18.Holler E, et al. Inflammatory reactions induced by pretransplant conditioning–an alternative target for modulation of acute GvHD and complications following allogeneic bone marrow transplantation? Leuk Lymphoma. 1997;25:217–224. doi: 10.3109/10428199709114161. [DOI] [PubMed] [Google Scholar]
- 19.Schroeder MA, DiPersio JF. Mouse models of graft-versus-host disease: advances and limitations. Dis Model Mech. 2011;4:318–333. doi: 10.1242/dmm.006668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu YW, Gress RE. Murine models of chronic graft-versus-host disease: insights and unresolved issues. Biol Blood Marrow Transplant. 2008;14:365–378. doi: 10.1016/j.bbmt.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tschetter JR, Mozes E, Shearer GM. Progression from acute to chronic disease in a murine parent-into-F1 model of graft-versus-host disease. J Immunol. 2000;165:5987–5994. doi: 10.4049/jimmunol.165.10.5987. [DOI] [PubMed] [Google Scholar]
- 22.Juncadella I, et al. The tick saliva immunosuppressor, Salp15, contributes to Th17-induced pathology during Experimental Autoimmune Encephalomyelitis. Biochemical and Biophysical Research Communications. 2010;402:105–109. doi: 10.1016/j.bbrc.2010.09.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lechner O, et al. Fingerprints of anergic T cells. Curr Biol. 2001;11:587–595. doi: 10.1016/S0960-9822(01)00160-9. [DOI] [PubMed] [Google Scholar]
- 24.Kalekar LA, et al. CD4(+) T cell anergy prevents autoimmunity and generates regulatory T cell precursors. Nat Immunol. 2016;17:304–314. doi: 10.1038/ni.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaudhary BAA, Samid M, Al-Ramadi BK, Elkord E. Phenotypic alterations, clinical impact and therapeutic potential of regulatory T cells in cancer. Expert Opin Biol Ther. 2014;14:931–945. doi: 10.1517/14712598.2014.900539. [DOI] [PubMed] [Google Scholar]
- 26.Elkord E, Abd Al Samid M, Chaudhary B. Helios, and not FoxP3, is the marker of activated Tregs expressing GARP/LAP. Oncotarget. 2015;6:20026–20036. doi: 10.18632/oncotarget.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruder D, et al. Neuropilin-1: a surface marker of regulatory T cells. Eur J Immunol. 2004;34:623–630. doi: 10.1002/eji.200324799. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, P. et al. Eomesodermin promotes the development of type 1 regulatory T (TR1) cells. Science Immunology2 (2017). [DOI] [PMC free article] [PubMed]
- 29.Whitehill GD, et al. Adenosine Selectively Depletes Alloreactive T Cells to Prevent GVHD While Conserving Immunity to Viruses and Leukemia. Mol Ther. 2016;24:1655–1664. doi: 10.1038/mt.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, et al. Graft-versus-host disease is enhanced by selective CD73 blockade in mice. PLoS One. 2013;8 doi: 10.1371/journal.pone.0058397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrentraut H, et al. CD73+ regulatory T cells contribute to adenosine-mediated resolution of acute lung injury. FASEB J. 2013;27:2207–2219. doi: 10.1096/fj.12-225201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohta A, Sitkovsky M. Role of G-protein-coupled adenosine receptors in downregulation of inflammation and protection from tissue damage. Nature. 2001;414:916–920. doi: 10.1038/414916a. [DOI] [PubMed] [Google Scholar]
- 33.Sitkovsky MV, et al. Physiological control of immune response and inflammatory tissue damage by hypoxia-inducible factors and adenosine A2A receptors. Annu Rev Immunol. 2004;22:657–682. doi: 10.1146/annurev.immunol.22.012703.104731. [DOI] [PubMed] [Google Scholar]
- 34.Fessler J, Felber A, Duftner C, Dejaco C. Therapeutic potential of regulatory T cells in autoimmune disorders. BioDrugs. 2013;27:281–291. doi: 10.1007/s40259-013-0026-5. [DOI] [PubMed] [Google Scholar]
- 35.Trzonkowski P, et al. Treatment of graft-versus-host disease with naturally occurring T regulatory cells. BioDrugs. 2013;27:605–614. doi: 10.1007/s40259-013-0050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalekar LA, Mueller DL. Relationship between CD4 Regulatory T Cells and Anergy In Vivo. The Journal of Immunology. 2017;198:2527–2533. doi: 10.4049/jimmunol.1602031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rueda CM, Jackson CM, Chougnet CA. Regulatory T-Cell-Mediated Suppression of Conventional T-Cells and Dendritic Cells by Different cAMP Intracellular Pathways. Front Immunol. 2016;7 doi: 10.3389/fimmu.2016.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yegutkin GG. Nucleotide- and nucleoside-converting ectoenzymes: Important modulators of purinergic signalling cascade. Biochim Biophys Acta. 2008;1783:673–694. doi: 10.1016/j.bbamcr.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 39.Kobie JJ, et al. T regulatory and primed uncommitted CD4 T cells express CD73, which suppresses effector CD4 T cells by converting 5′-adenosine monophosphate to adenosine. J Immunol. 2006;177:6780–6786. doi: 10.4049/jimmunol.177.10.6780. [DOI] [PubMed] [Google Scholar]
- 40.Gurel B, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu G, Wang LG, Han Y, He Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomic data are deposited under GEO accession number GSE98700.