Abstract
The methylcytosine-binding domain 2 (MBD2) protein recruits the nucleosome remodeling and deacetylase complex (NuRD) to methylated DNA to modify chromatin and regulate transcription. Importantly, MBD2 functions within CpG islands that contain 100s to 1000s of potential binding sites. Since NuRD physically rearranges nucleosomes, the dynamic mobility of this complex is directly related to function. In these studies, we use NMR and single-molecule atomic force microscopy and fluorescence imaging to study DNA binding dynamics of MBD2. Single-molecule fluorescence tracking on DNA tightropes containing regions with CpG-rich and CpG-free regions reveals that MBD2 carries out unbiased 1D diffusion on CpG-rich DNA but subdiffusion on CpG-free DNA. In contrast, the protein stably and statically binds to methylated CpG (mCpG) regions. The intrinsically disordered region (IDR) on MBD2 both reduces exchange between mCpG sites along the DNA as well as the dissociation from DNA, acting like an anchor that restricts the dynamic mobility of the MBD domain. Unexpectedly, MBD2 binding to methylated CpGs induces DNA bending that is augmented by the IDR region of the protein. These results suggest that MBD2 targets NuRD to unmethylated or methylated CpG islands where its distinct dynamic binding modes help maintain open or closed chromatin, respectively.
INTRODUCTION
DNA methylation contributes to gene regulation, defines cellular differentiation and genome organization, and therefore plays a diverse role in normal growth and development as well as pathogenesis (1,2). Approximately 1% of nucleotides and 75% of CpG dinucleotides are methylcytosines (3). However, regions of increased CpG content (CpG islands) typically remain unmethylated and often occur in gene promoters (4,5). The methylcytosine-binding domain (MBD) family of proteins selectively bind to methylated CpG sites and modify transcription of the associated gene (6,7). MBD2, a founding member of this family, is a core component of the nucleosome remodeling and deacetylase (NuRD) complex (8). MBD2 localizes the NuRD complex to CpG sites, particularly methylated CpG islands (9,10), and contributes to chromatin remodeling and silencing of the associated gene. The MBD2 protein itself contains three distinct regions, a methylcytosine binding domain (MBD), an intrinsically disordered region (IDR), and a coiled-coil domain. Importantly for the studies reported here, we previously found that the MBD2-IDR both increases binding affinity for DNA and recruits core components of the histone deacetylase sub-complex in NuRD (11).
Unlike most transcription factors, MBD2 functions within the context of CpG islands that contain 100s to 1000s of individual binding sites (5,12). In addition, the NuRD complex contains a chromatin remodeling component that moves nucleosomes along the DNA. Hence, how MBD2 exchanges between binding sites is vital for understanding how methylation-dependent genetic silencing works at a molecular level. Over the past few years, we have been studying how different members of the MBD family bind and distribute on DNA using several NMR-based techniques, including paramagnetic relaxation enhancement (PRE) (13), residual dipolar coupling measurements (14), chemical shift analyses and NZ-exchange spectroscopy (14,15). These studies have shown that intra- and intermolecular exchange rates vary between the different MBDs and correlate with their distinct functions. For example, we found that MBD3, a paralogue of MBD2 with low DNA binding affinity, retains an ability to recognize CpG sites and preferentially localize, albeit very weakly, to methylated CpGs. However, MBD3 rapidly exchanges between mCpG specific and non-specific binding modes consistent with the very weak preference for mCpG and a dynamic distribution along the DNA within CpG islands (14). In contrast, MBD2 binds selectively and almost exclusively to a single mCpG when present (14). This observation correlates with whole genome studies that show both MBD2 and MBD3 localize to unmethylated CpG islands while MBD2 more exclusively localizes to methylated CpG islands with silencing of the associated gene (9,10,16,17). In a separate study, we measured intra- and intermolecular exchange between mCpGs by MBD4, an MBD that contains a DNA glycosylase domain involved in mCpG/TpG mismatch repair (15). We found that MBD4 exchanges between methylated sites more rapidly than MBD2. This difference likely reflects a lower binding selectivity for mCpG over CpG and the biological role of MBD4 to rapidly scan methylated CpG regions where mCpG/TpG mismatches occur.
Together, these studies have shown that DNA binding dynamics likely contribute to the specific function of individual MBD proteins. However, the size limitations imposed by NMR restricted the previous studies to relatively small (10–20 base pairs) fragments of DNA. Recently, single-molecule fluorescence techniques have been developed to study transcription factors moving along DNA (18,19). These approaches open the door to studying the dynamic behavior of transcription factors on much larger DNA substrates. In the studies reported here, we correlate our findings on small oligonucleotides with structure and dynamic behavior on much larger, and hence more biologically relevant, fragments of DNA observed using single-molecule fluorescence and atomic force microscopy. These studies lead to several novel observations that help build a model of MBD2 function. We found an unexpected difference in dynamic behavior between CpG-rich and CpG-free regions, in that MBD2 diffuses more rapidly and extensively on CpG-rich sequences than those that lack CpGs (CpG-free). In contrast, MBD2 binds statically or with slow exchange rates when binding to methylated CpG-rich regions. In addition, examining MBD2-DNA complexes by atomic force microscopy shows that binding to CpG-rich sequences leads to marked DNA bending that is further augmented by the IDR when DNA is methylated. Together these studies lead to a model in which MBD2 allows for rapid diffusion and nucleosome remodeling by NuRD within or through CpG rich regions, yet greatly restricts movement and nucleosome remodeling when these islands are methylated. Therefore, the dynamic DNA binding behavior of MBD2 correlates with the biological observation that unmethylated CpG islands are associated with open and transcriptionally active promoters while methylated CpG islands are closed and silenced.
MATERIALS AND METHODS
Protein expression and purification
The chicken MBD2MBD (amino acids 1–71) and human MBD2MBD+IDR (amino acids 150–214) were expressed and purified as previously reported (11,13). Chicken MBD2MBD+IDR, which likewise includes an additional 46 residues (amino acids 1–117), was cloned into the previously described pET32a based vector (13), and further modified to replace the thrombin cleave site with a TEV protease cleavage site. The resulting thioredoxin fusion construct was transformed into BL21(DE3) Escherichia coli cells grown at 37°C until an OD600 of 0.7 and induced with 1 mM IPTG. Bacteria were pelleted and lysed in 20 mM Tris pH 8, 1 M NaCl with sonication. Proteins were further purified via HisTrapFF (GE Life Sciences) and cleaved using either thrombin (MBD2MBD) or TEV protease (MBD2MBD+IDR). Cleaved proteins were further purified over a Resource-S (GE Life Sciences) ion exchange column and Superdex-75 (GE Life Sciences) using an ÄKTA pure FPLC system (GE Life Sciences). Proteins for NMR analysis were either 2H,15N,13C or 2H,15N labeled. For fluorescence polarization assays the thioredoxin fused protein was purified via HisTrapFF and Superdex-75. For single-molecule experiments MBD2MBD+IDR and MBD2MBD were cloned into pET28a with N-terminal His6 tags and expressed and purified in a similar manner.
Fluorescence polarization
Protein samples were buffer exchanged into FP buffer [10 mM HEPES pH 7.5, 50 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA, 2 mM DTT and 5% Tween 20]. A 6-FAM labeled 17-bp dsDNA containing a central methylated CpG site (1xmCpG) (14), was annealed and purified by ion exchange chromatography on a Resource Q column (GE Life Sciences). Serial dilutions of the MBD2 protein were added to 10 nM DNA and polarization was measured on a CLARIOstar microplate reader (BMG Labtech) as described previously (11).
NMR spectroscopy
Uniformly H, 15N,13C labeled MBD2MBD+IDR at 0.5 mM bound to methylated DNA was buffer exchanged into 10 mM NaPO4 pH 6.5, 1 mM dithiothreitol, and 10% 2H2O. TROSY based version of standard double and triple resonance experiments (HNCOCACB, HNCA, HNCO, HNCACO, HNCACB, 15N-NOESY-HSQC and 15N-HSQC) were collected on a 700 MHz Bruker Avance III spectrometer. Residual dipolar couplings were collected on a 2H,13C,15N labeled sample aligned with 12 mg/ml pf1 bacteriophage (20,21). All data was processed using NMRPipe (22) and analyzed in CcpNMR (23,24).
Nz-exchange spectroscopy
In previous studies, we established that MBD2 shows distinct chemical shifts for several residues when bound to either of two dsDNA with a centrally located mCpG. The two sequences differ in the base pairs immediately surrounding the mCpG, which inverts the orientation of bound MBD2MBD (WT: 5′-GGAT(mC)GGCTCCTC-3′; INV: 5′-GGAC(mC)GACTC-3′) (13,15). A 350 μM sample of 2H,15N labeled MBD2MBD was incubated with 275 μM WT DNA and 275 μM INV DNA (yielding 175 μM of each complex with 100 μM excess of each DNA). Samples with 200 and 300 μM excess DNA were collected by adding a 1.25 mM mixture of WT and INV DNA. For each titration point a series of 2D-15N TROSY Nz-exchange spectra were obtained with the following delays: 11.9, 14.3, 16.8, 21.8, 29.3, 36.8, 49.3, 61.8, 118.9 and 211.8 ms. Two samples of MBD2MBD+IDR were prepared: one with 370 μM of MBD2MBD+IDR and 285 μM of each DNA (yielding 185 μM of each complex with 100 μM excess of each DNA) and a sample with 370 μM protein with 740 μM of each DNA (185 μM of each complex with 555 μM excess of each DNA). 2D-15N TROSY Nz-exchange spectra (25) were collected and processes as described previously (15).
DNA substrates for single-molecule imaging
To generate a DNA substrate incorporating a known target sequence for MBD2 (26), we subcloned a portion (3837 base pairs) of the death associated protein kinase 1 (DAPK1) promoter (chromosome 9, bases 87497573 to 87501409) which includes CpG-rich (4689 base pair) and CpG-poor (2150 base pairs) regions. This sequence was then further sub-divided into clones that contain only the CpG-rich region (CpG-rich: 4705 bp) or rich plus poor regions to generate the CpG-rich-poor DNA (6839 bp) within the pGEM backbone (Supplementary Figure S2). For a CpG-free DNA substrate, the pCpGfree-vitroNmcs plasmid (5488 base pairs) was purchased from InvivoGen that is devoid of any CpG dinucleotides. The CpG-rich region from the DAPK1 promoter was then sub-cloned into the ScaI/NcoI restriction sites to generate a 1697 base pair CpG-rich region within a 5466 CpG-free backbone (CpG-free-rich DNA: 7163 bp).
For DNA methylation, purified plasmids were treated with CpG Methyltransferase (M.SssI) and S-Adenosine methionine (SAM) as a cofactor at 37°C for overnight. The methylation status of the DNA was confirmed by restriction digestion using HpaII. A total of 42 HpaII sites are present in both the CpG-rich and CpG-rich-poor substrates. Among these, 30 sites are in the CpG-rich region on the CpG-rich and CpG-rich-poor DNA substrates. To generate longer DNA substrates for the DNA tightrope assay, linearized DNA substrates were ligated using the Quick Ligation™ Kit (New England BioLabs) at room temperature for overnight. Ligated DNA samples were then purified using phenol-chloroform extraction.
Protein-QD conjugation
Streptavidin-conjugated quantum dots (SAv-QDs-655) were purchased from Invitrogen. For QD labeling of N-terminal His6-tagged MBD2 (His-MBD2MBD and His-MBD2MBD+IDR), 1 μl of SAv-QDs-655 (1 μM) was incubated with 1 μl of the multivalent chelator tris-nitrilotriacetic acid (BTtris-NTA, 2 μM) for 20 min at room temperature (27). The His-MBD2 protein (1 μl of 1 μM) was then added to the SAv-QD-NTA solution and incubated for an additional 20 min at room temperature. All samples were diluted 100X before being introduced into the flow cell in the imaging buffer (50 mM HEPES pH 7.5, 100 mM NaCl, and 1 mM MgCl2).
Fluorescence imaging and analysis
The oblique angle total internal reflection microscopy based particle tracking of QD-labeled proteins on DNA tightropes was described previously (28). Briefly, an inverted microscope (Nikon Ti-E) and a 100× objective (APO TIRF, Nikon) was used in the experiments. Red (655 nm) QD-protein complexes were excited at 488 nm by a solid-state laser (Sapphire DPSS). The signal was split into two channels using a dichroic mirror (T605LPXR, Chroma) and an emission filter (ET655/40nm, Chroma). Flow cells were assembled as methods demonstrated in preceding studies (28). Furthermore, we immobilized poly-l-lysine (2.5 mg/ml, M.W. >300 kDa, Wako Chemicals) treated silica beads onto a cover slip surface with PEGlyation, and then introduced ligated DNA substrates into the flow cell with a syringe pump at a flow rate of 300 μl/min. All images were taken using an EMCCD (iXon DU897, Andor Technology) at a time resolution of 50 ms/frame.
The mean square displacement (MSD) as a function of time interval is given by:
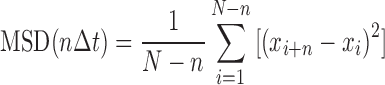 |
(1) |
where N is the total number of frames in the trajectory, n is the number of frames for different time intervals, Δt is the time between frames and xi is the position of the protein-QD in the frame i. The 1D diffusion constant (D) and diffusion exponent (alpha factor) were analyzed by a custom routine developed in LabView Software based on the following equation (29):
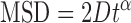 |
(2) |
A protein was categorized as being mobile if the diffusion constant was greater than 1 × 10−5 μm2/s and the R2 value from data fitting using Equation (2) was >0.8. The diffusion range was analyzed using a custom code written using MATLAB.
AFM imaging and image analysis
All DNA and protein samples were pre-incubated for 20 min at room temperature and diluted 10-fold in 1× AFM buffer [25 mM NaOAc, 25 mM HEPES–KOH (pH 7.5) and 10 mM Mg(OAc)2] before being deposited onto a freshly cleaved mica (SPI Supply). The samples were then washed with MilliQ water and dried with nitrogen gas. The final concentration of DNA substrates on mica was ∼0.5 ng/μl and of protein was 30 nM. All images were obtained in the AC mode using a MFP-3D-Bio AFM (Asylum Research). Cantilevers (PPP-FMR, Nanosensors) with spring constants at ∼2.8 N/m were used. The images were collected at the scan size of 3 μm × 3 μm, scan rate of 1–2 Hz, and resolution of 512 × 512 pixels. The DNA bending angle was analyzed using either the Asylum software or a custom MATLAB code, which provide similar results. The MATLAB code determines the center of the DNA-bound protein by performing a 2D Gaussian fit in the vicinity of the highest point of the complex, followed by a sampling of the height along a circle around that point. The angle between the intersection points of DNA and sampling circle was determined by local Gaussian fitting in the vicinity of the two height maxima.
RESULTS
The IDR of MBD2 influences DNA binding affinity
We previously found that a short, positively charged region just C-terminal to the human MBD2 (MBD) augments binding affinity for methylated DNA by 10–100-fold (11). In the current work, we studied the chicken orthologue (96%, 94%, and 83% identical to human MBD2MBD, MBD2MBD+IDR, and MBD2b, respectively) for which we have a high-resolution solution structure and NMR assignments (13). Of note, the human MBD2a isoform includes a long unstructured N-terminal glycine–arginine repeat region that likely increases DNA binding affinity and unique binding interactions (30). Yet, the MBD2b isoform can compact chromatin and functionally replace MBD2a (10). Fluorescence polarization binding analysis shows that the homologous region (IDR) increases binding affinity of the chicken MBD2 by ∼12-fold (Supplementary Figure S1A). In addition, we measured residual dipolar couplings (RDC) for MBD2MBD+IDR bound to methylated DNA and fit them to the known structure of the isolated domain (13). The overall quality of fit (Q factor = 47%) and correlation between measured and predicted structure (r = 0.85) indicate that the IDR does not significantly alter the fold of the MBD (Supplementary Figure S1B and C). As we demonstrated previously, though, both the lack of chemical shift dispersion and RDC measurements indicate that the IDR does not adopt a regular structure when bound to DNA.
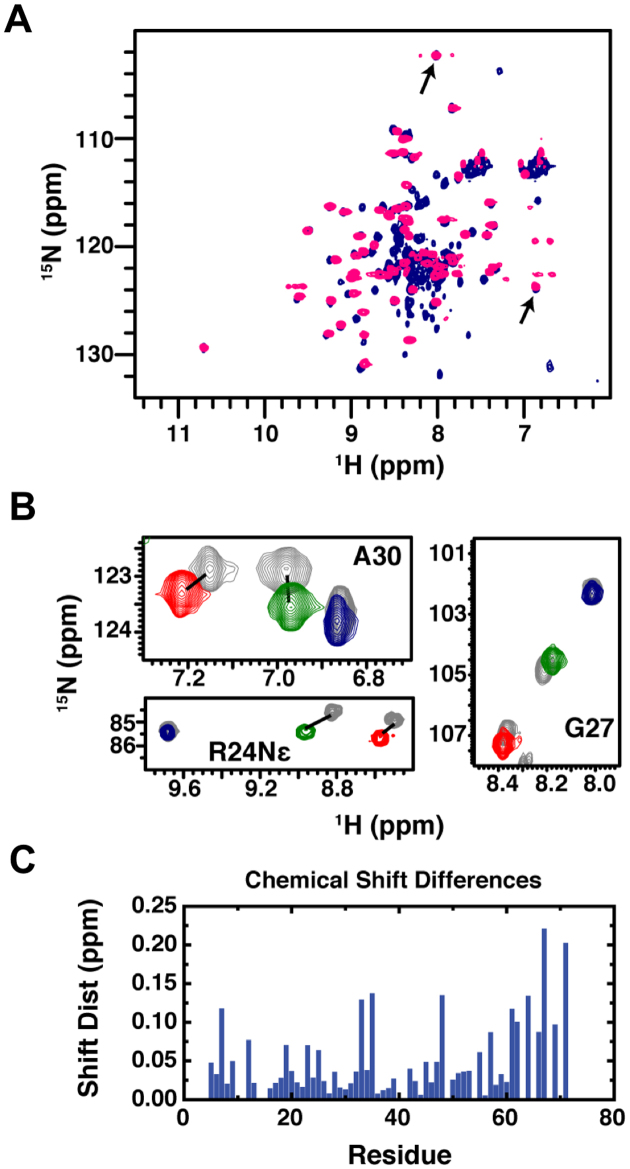
In previous studies of MBD proteins, we have consistently found that the 1H–15N chemical shifts for backbone (G27 and A30) and side chain (R24) resonances of three highly conserved residues reflect the distribution between methylation specific and non-specific binding modes (14,15,31). Hence, the location of these resonances reflects the amount of time spent bound to the methylated site. Comparing the chemical shifts for these reporter residues between MBD2MBD+IDR and MBD2MBD (Figure 1A and B) shows that the IDR does not alter the relative distribution of MBD2 between binding modes. Likewise, the largest chemical shift differences (Figure 1C) map to the C-terminal portion of the MBD, just prior to the additional amino acid sequence unique to the MBD2MBD+IDR clone. Therefore, the increase in binding affinity provided by the IDR does not change the structural recognition of mCpG or methylation selectivity of the protein.
Figure 1.
Comparison of NMR chemical shifts between MBD2MBD+IDR and MBD2MBD bound to DNA. (A) An overlay of 2D 15N-HSQC spectra for MBD2MBD+IDR (navy) and MBD2MBD (pink) bound to mCpG DNA reveals very similar chemical shifts for most amide resonances including those for reporter resonances G27 and A30 (arrows). (B) Expanded regions from 2D 15N-HSQC spectra for MBD2MBD+IDR bound to DNA without CpG dinucleotides (red), with 3 CpG dinucleotides (green) and 1 mCpG dinucleotide (navy) are shown for reporter residues A30, R24 Nε, and G27. For comparison, the same regions are shown for MBD2MBD (gray) bound to the same DNA with equivalent resonances connected by lines (if they do not overlap). (C) The chemical shift distances between MBD2MBD+IDR and MBD2MBD backbone amide resonances shows relatively small changes for most residues, with some of the larger differences immediately preceding the additional residues in MBD2MBD+IDR.
MBD2MBD+IDR and MBD2MBD carry out unbiased 1D diffusion on CpG-rich DNA and subdiffusion on CpG-free DNA
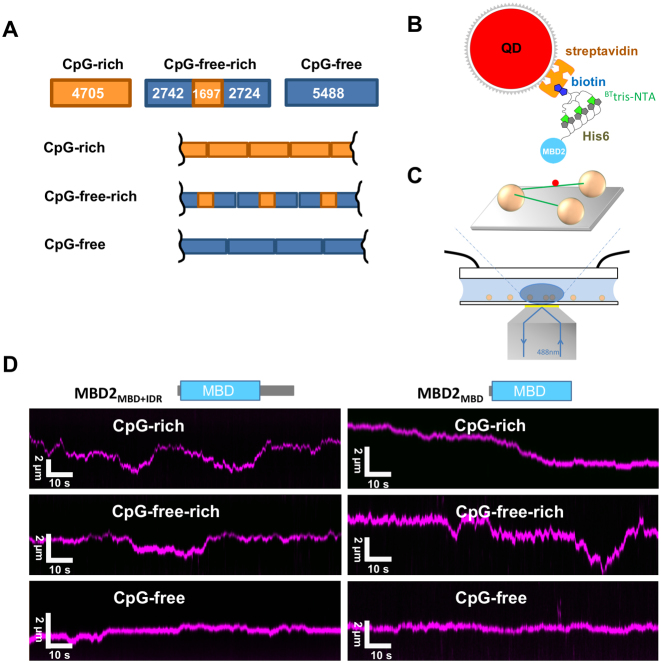
Dynamic movements of proteins on DNA, such as 1D sliding, jumping, and hopping are essential for a protein to find its targeted sequences or structures on DNA (18,32–36). Furthermore, the MBD2–NuRD complex rearranges nucleosomes within the context of large CpG islands and modifies higher order chromatin organization (37,38). Therefore, to further understand how MBD2 dynamically distributes on much larger DNA substrates and the role of the IDR, we applied oblique angle total internal reflection fluorescence (OAF) microscopy imaging of quantum dot (QD)-labeled proteins (28,39–41). We tested MBD2 with (MBD2MBD+IDR) or without (MBD2MBD) a portion of the IDR on DNA tightropes with or without CpG sequences (Materials and Methods, Figure 2A). The CpG-rich and CpG-poor regions contain 6.5–9.5 and 0.6 CpG sites per 100 bp DNA, respectively (Supplementary Figure S2). The DNA substrates used for the DNA tightrope assay were generated by ligating linear DNA fragments containing unmethylated CpG-rich, CpG-free plus rich (CpG-free-rich), CpG-free (CpG-free), or CpG-rich plus poor (CpG-rich-poor) sequences (Figure 2 and Supplementary Figure S2). Both fluorescence imaging of YOYO1-stained ligated DNA and AFM imaging of unstained ligated DNA demonstrated the formation of longer DNA fragments after ligation (Supplementary Figure S3 A,B). We conjugated His-tagged MBD2MBD+IDR and MBD2MBD to streptavidin-coated quantum dots (SAv-QDs) through the multivalent chelator tris-nitrilotriacetic acid (BTtris-NTA) linker (27) (Figure 2B). The three Ni-NTA moieties on the circular scaffold of the tris-NTA adaptor bind the His-tag with subnanomolar affinity (27,42). AFM and fluorescence imaging established that SAv-QDs without MBD2 did not have significant binding affinity for DNA. Hydrodynamic flow was used to stretch DNA and suspend ligated DNA strands between poly-L-lysine coated silica microspheres at an elongation of ∼90% of the DNA contour length (Figure 2C) (28). Following formation of the DNA tightropes, either QD-labeled MBD2MBD+IDR or MBD2MBD was introduced into the flow cell. The binding of MBD2-QDs molecules to DNA tightropes was long lived, with ∼99% of the MBD2MBD+IDR and MBD2MBD binding on the DNA tightropes throughout the observation window (2 min, N = 257 for MBD2MBD+IDR and N = 254 for MBD2MBD). Analysis of MBD2 on DNA tightropes revealed two populations (Table 1): static and mobile molecules. Overall, on the unmethylated DNA tightropes, the majority of MBD2MBD+IDR and MBD2MBD molecules are mobile (>65%, Table 1).
Figure 2.
MBD2MBD+IDR and MBD2MBD carry out 1D diffusion on tightropes with or without unmethylated-CpG sequences. (A) Cartoon drawing of three ligated DNA substrates for the DNA tightrope assay: CpG-rich, CpG-free-rich and CpG-free. The numbers in the boxes show the length of the DNA. (B) Schematic representation of QD-conjugated MBD2MBD+IDR and MBD2MBD using BTtris-NTA as the linker between the His6-tag on the MBD2 proteins and streptavidin coated-QD. (C) Schematic drawing of the DNA tightrope assay depicting a QD-labeled protein (red dot) on the DNA tightrope (green lines) stretched between silica beads (golden spheres) immobilized on a passivated glass surface. (D) Representative kymographs of red (655 nm) QD-labeled MBD2MBD+IDR (left panel) and MBD2MBD (right panel) on three different DNA tightropes.
Table 1. Proportion of static MBD2 on unmethylated- and methylated-DNA tightropes.
| DNA | MBD2MBD+IDR | MBD2MBD | |||
|---|---|---|---|---|---|
| Percentage (%) | N | Percentage (%) | N | ||
| Unmethylated | CpG-rich | 32.1%±4.9% | 74 | 33.8%±5.2% | 91 |
| CpG-free-rich | 22.7%±2.3% | 127 | 22.9%±1.2% | 95 | |
| CpG-free | 10.6%±3.9% | 69 | 15.7%±3.7% | 56 | |
| Methylated | CpG-rich | 91.2%±2.3% | 284 | 93.6%±2.2% | 173 |
| CpG-free-rich | 94.8%±1.8% | 234 | 95.5%±1.3% | 272 | |
| CpG-free | N.A. | N.A. | |||
The values represent mean ± SEM from two to three experiments for each data set.
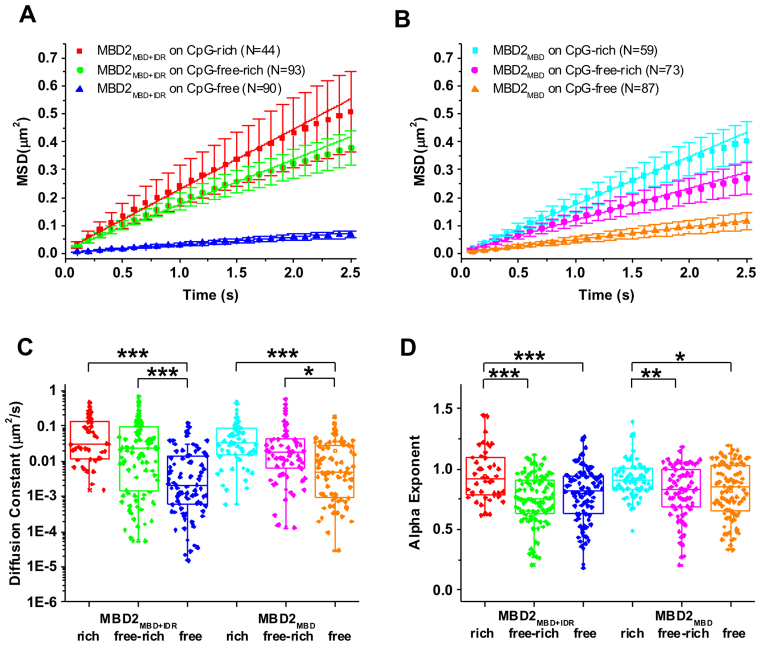
To obtain diffusion constants for mobile MBD2MBD+IDR and MBD2MBD on DNA tightropes, the position of MBD2MBD+IDR- and MBD2MBD-QDs on DNA was tracked by Gaussian fitting to kymographs (particle position versus time plots, Figure 2D) (39,41). Diffusion constants and diffusive exponents (alpha factors) were obtained from fitting the MSD versus time plots (Figure 3A and B). An alpha exponent of 1 indicates an unbiased random walk and a value <1 indicates subdiffusion (29). At the same protein concentrations (10 nM in the flow cell), the diffusion constants of MBD2MBD+IDR and MBD2MBD on CpG-rich and CpG-free-rich substrates were indistinguishable (Figure 3C and Table 2). However, the diffusion constants displayed by MBD2MBD+IDR and MBD2MBD on the CpG-free DNA tightrope were significantly slower compared to those on DNA tightropes containing CpG sequences (CpG-rich and CpG-free-rich) (Figure 3C).
Figure 3.
Diffusion constant, alpha exponent of MBD2MBD+IDR and MBD2MBD on three types of DNA substrates. Plots of average values of MSD versus time for MBD2MBD+IDR (A) and MBD2MBD (B) on CpG-rich, CpG-free-rich and CpG-free DNA tightropes. The error bars represent SEM. (C and D) Diffusion constants (C) and alpha exponents (D) of MBD2MBD+IDR and MBD2MBD on CpG-rich, CpG-free-rich, and CpG-free DNA tightropes (Table 2). ***P < 0.0005; **P < 0.005; *P < 0.5.
Table 2. Diffusion of MBD2MBD+IDR and MBD2MBD on three DNA substrates.
| DNA | MBD2MBD+IDR | MBD2MBD | ||||
|---|---|---|---|---|---|---|
| D (μm2/s) | α exponent | N | D (μm2/s) | α exponent | N | |
| CpG-rich | 0.089±0.016 | 0.95±0.03 | 44 | 0.078±0.012 | 0.92±0.02 | 59 |
| CpG-free-rich | 0.071±0.013 | 0.75±0.02 | 93 | 0.057±0.013 | 0.81±0.03 | 73 |
| CpG-free | 0.014±0.003 | 0.79±0.02 | 90 | 0.019±0.003 | 0.82±0.03 | 88 |
The values represent mean ± SD from 2 to 5 experiments for each data set.
In addition, on the CpG-rich tightropes MBD2MBD+IDR and MBD2MBD displayed similar alpha exponents that are close to 1 (0.95 ± 0.03 and 0.92 ± 0.02, respectively), indicating unbiased 1D diffusion on DNA (Figure 3D, Table 2). Surprisingly, inclusion of CpG-free sequences in the DNA tightropes significantly (P < 0.05) reduced the alpha exponents for both MBD2MBD+IDR and MBD2MBD (Figure 3D, Table 2), indicating subdiffusive motion. Overall, both MBD2MBD+IDR and MBD2MBD display significantly (P<0.005) narrower diffusion ranges on the CpG-free DNA tightropes compared to all the other DNA tightropes containing CpG-rich sequences (CpG-rich and CpG-free-rich, Supplementary Figure S4). In summary, fluorescence imaging shows that the IDR does not significantly modify the 1D diffusion of MBD2 on DNA. However, both MBD2MBD+IDR and MBD2MBD show unbiased diffusion on CpG-rich DNA while subdiffusive motion on CpG-free sequences.
Both MBD2 MBD and MBD2MBD+IDR become static on methylated DNA
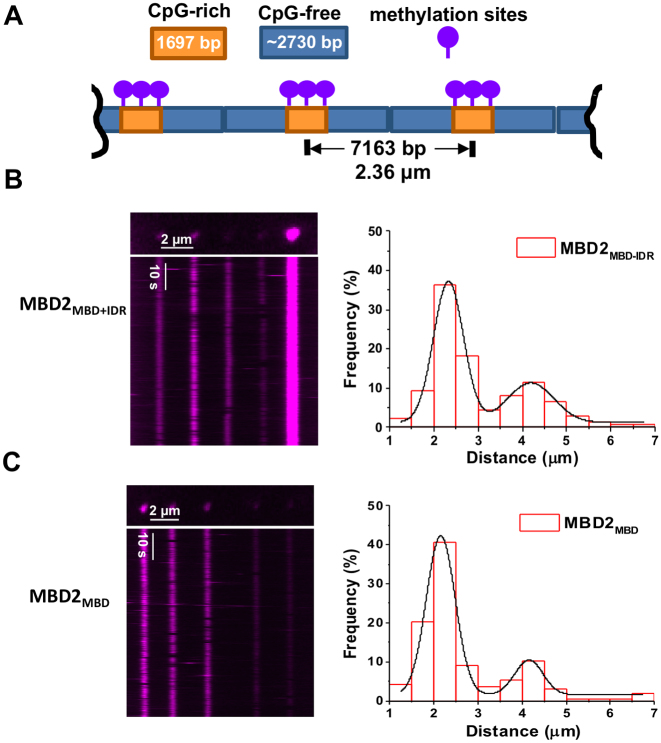
To evaluate how DNA methylation affects the dynamics of MBD2 on DNA, we imaged QD-labeled MBD2MBD+IDR and MBD2MBD on the CpG-free-rich (Figure 4), CpG-rich, and CpG-rich-poor (Supplementary Figures S5 and S6) DNA tightropes after methylation. Linear DNA substrates were methylated before ligation using CpG Methyltransferase (M.SssI) with S-Adenosine methionine (SAM) as a cofactor (Supplementary Figure S3C). The methylation status of the linear DNA substrates was confirmed by inhibition of digestion by HpaII restriction enzyme (Supplementary Figure S3C). When methylated, the CpG-rich, CpG-free-rich, and CpG-poor-rich DNA contain 42, 30 and 42 HpaII restriction sites, respectively. Importantly, lack of methylation of just one site permits cleavage of the vector by the enzyme. Hence, absence of digestion indicates a high level of methylation (43). We then ligated the methylated DNA and used hydrodynamic flow to form DNA tightropes between silica beads inside the flow cell. AFM imaging showed that QD-labeled MBD2MBD+IDR retains binding specificity for mCpG-rich region on the linear mCpG-free-rich DNA (Supplementary Figure S5). As compared to unmethylated DNA, the binding events for both MBD2MBD+IDR and MBD2MBD increased dramatically on methylated DNA tightropes (Supplementary Figure S6A,B). Combining the data for all three DNA substrates (CpG-rich, CpG-rich-poor, and CpG-free-rich), the density of both MBD2MBD+IDR and MBD2MBD complexes increased from 1.5/40 kb on unmethylated DNA to 4/40 kb on methylated DNA (Supplementary Figure S6C and D). Similar to what was observed on the unmethylated DNA, the binding of MBD2-QDs molecules to methylated DNA was long lived, with over 99% of the MBD2MBD+IDR and MBD2MBD binding on the DNA tightropes throughout the observation window (2 min, N = 518 for MBD2MBD+IDR and N = 445 for MBD2MBD). However, in contrast to the unmethylated DNA tightropes, the majority (∼91.2% to 95.5%) of the MBD2MBD+IDR and MBD2MBD bound statically to the methylated DNA tightropes (Figure 4, Supplementary Figure S6, and Table 1). Furthermore, the distance between adjacent proteins on the methylated CpG-free-rich DNA tightropes are Gaussian distributed with peaks centered at 2.33 (± 0.38) μm and 4.19 (± 0.65) μm for MBD2MBD+IDR, and 2.15 (± 0.39) μm and 4.15 (± 0.37) μm for MBD2MBD, respectively (Figure 4B and C). These spacing match the distances between methylated CpG-rich regions on DNA tightropes (Figure 4A). Taken together, fluorescence imaging of MBD2MBD+IDR and MBD2MBD on DNA tightropes establishes that MBD2 recognizes methylated CpG sites through stable and static binding.
Figure 4.
Both MBD2MBD+IDR and MBD2MBD become static at the methylated CpG-rich regions on DNA tightropes. (A) A cartoon drawing of the ligated DNA substrates containing alternating mCpG-rich and CpG-free sequences (mCpG-free-rich) for the DNA tightrope assay. The DNA is methylated using CpG Methyltransferase (M.SssI) and S-Adenosine methionine (SAM) as a cofactor. The expected center-to-center distance between two nearest neighbor CpG-rich regions on DNA tightropes is 2.1 μm, assuming 90% extension of the DNA contour length. QD-labeled MBD2MBD+IDR (B) and MBD2MBD (C) on the ligated mCpG-free-rich DNA tightropes. Left panels: Fluorescence images (top) and corresponding kymographs (bottom). Right panels: histograms of the distance between two adjacent MBD2MBD+IDR (B) and MBD2MBD (C) on mCpG-free-rich DNA tightropes. The solid lines represent double Gaussian fits to the data (R2 > 0.98) with peaks centered at 2.33 (±0.38) μm and 4.19 (±0.65) μm for MBD2MBD+IDR (N = 138), and 2.15 (±0.39) μm and 4.15 (±0.37) μm for MBD2MBD (N = 167), respectively.
Intra- and intermolecular exchange by MBD2
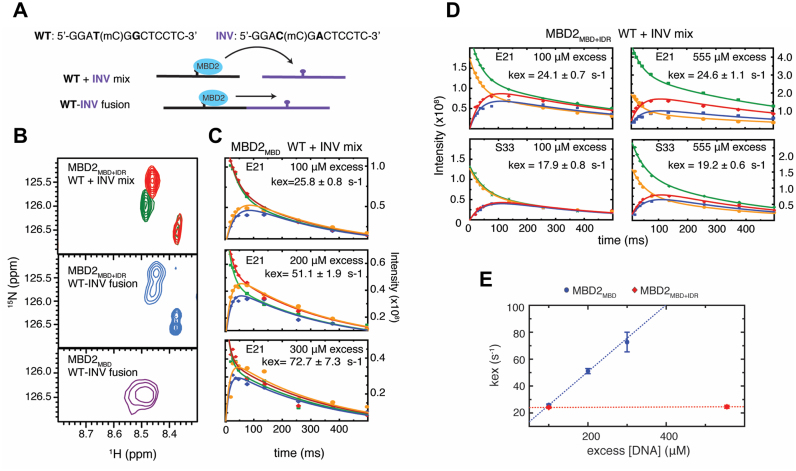
While the single-molecule studies clearly demonstrate that methylation greatly restricts dynamic mobility of MBD2 on DNA, the spatial resolution (∼16 nm) of fluorescence imaging limits localization accuracy of the MBD2 protein to ∼50 base pairs of DNA (28), which contains ∼5 CpG dinucleotides based on the density of the CpG sites (Supplementary Figure S2). In previous studies, we found that the MBD can exchange relatively rapidly between closely-spaced mCpG sites (10–20 base pair separation) on the same DNA (intramolecular exchange, lifetimes <1 ms) and between different molecules of DNA (intermolecular exchange, lifetimes of 20–30 ms) (15). Hence, to test whether the IDR modifies local exchange between individual methylated sites, we compared intra- and intermolecular exchange for MBD2MBD+IDR and MBD2MBD. Several resonances in the 2D 15N-HSQC spectrum of MBD2 show different chemical shifts when bound to closely related 10 base pair oligonucleotides that differ in sequence immediately adjacent to the central mCpG dinucleotide (WT and INV, Figure 5A and B). When MBD2MBD+IDR is bound to a 20-base pair fusion of these two sequences, with the mCpGs separated by only 10 bases, the 2D 15N-HSQC spectrum contains two separate peaks consistent with slow exchange between the binding sites (Figure 5B middle panel). In contrast, when MBD2MBD is bound to the same sequence, the spectrum contains only a single broadened peak at the average position between the two resonances (Figure 5B bottom panel). Hence, the IDR decreases the rate of intramolecular exchange from fast to slow exchange on the NMR timescale.
Figure 5.
The MBD2 IDR influences both intramolecular between mCpG sites and intermolecular exchange between different DNA molecules. (A) Sequence of WT and INV DNA with a central methylated CpG. The two base pairs that differ between the two sequences (bold) lead to unique chemical shifts in the bound state. MBD2 was bound to a mixture or fusion of the WT and INV sequences for inter- and intra-molecular exchange rate measurements, respectively. (B) An overlay of 2D 15N-HSQC spectra for MBD2MBD+IDR bound to a mixture of WT and INV DNA (top panel), MBD2MBD+IDR and MBD2MBD bound to the fusion of WT and INV DNA (middle and bottom panels, respectively). (C) NZ exchange curves for MBD2MBD residue E21 bound to a mixture of WT and INV DNA with 100 μM, 200 μM, and 300 μM excess free DNA. (D) NZ exchange curves for MBD2MBD+IDR residues E21 and S33 bound to 100 μM and 555 μM excess free DNA. (E) Plot of the measured intermolecular exchange rates versus excess DNA concentration.
To determine whether the IDR influences intermolecular exchange, we used Nz exchange spectroscopy to measure the intermolecular exchange rate between the WT and INV methylated CpG sequences, as we have described previously (15). We followed the auto and exchange cross peaks as a function of NZ delay, and fit the intensities to four coupled equations describing molecular exchange in the slow limit (44). We measured very similar exchange rates for both MBD2MBD+IDR and MBD2MBD with 100 μM excess DNA (Figure 5C and D). However, increasing the excess DNA concentration enhanced the exchange rates for MBD2MBD but not for MBD2MBD+IDR (Figure 5C–E). Without the IDR, the strong free DNA concentration dependence for MBD2MBD indicates that intermolecular exchange is rate limited by the DNA re-binding on-rate or intersegment transfer (45,46). In contrast, in the presence of the IDR, the lack of free DNA concentration dependence indicates that DNA dissociation is rate limiting for MBD2MBD+IDR (45,46). Therefore, the IDR both reduces exchange between mCpG sites along the DNA as well as dissociation from DNA, acting like an anchor that restricts dynamic mobility of the MBD domain.
MBD2 bends DNA and IDR augments the bending upon binding to mCpG
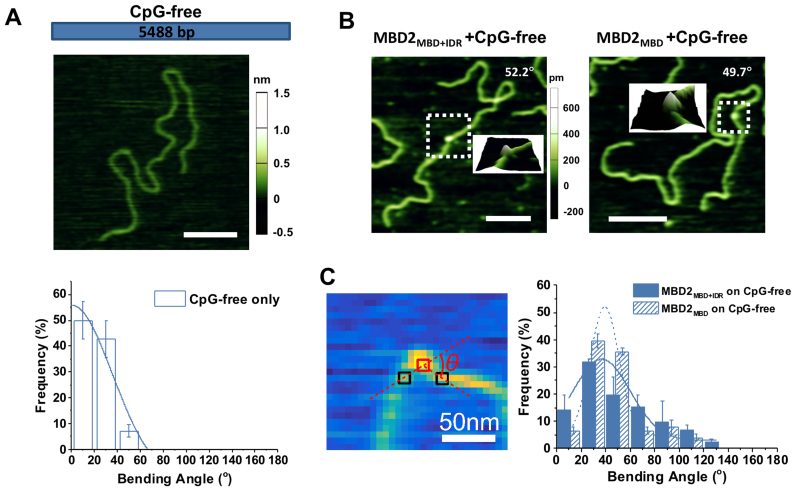
In order to evaluate whether MBD2 with or without the IDR affects the DNA conformation upon binding, we applied AFM imaging in air to visualize the MBD2MBD+IDR- and MBD2MBD-DNA complexes on linear CpG-free DNA (Figure 6), as well as unmethylated and methylated CpG-rich DNA (Figure 7). Due to its small size, it is challenging to unambiguously identify MBD2MBD+IDR (138 amino acids) and MBD2MBD (91 amino acids) on DNA in topographical AFM images. To increase the size of MBD2MBD+IDR and MBD2MBD and facilitate visualization, the thioredoxin (TRX) tag was not removed from the N-termini of the proteins. Importantly, the TRX tag does not significantly modify methylated DNA binding affinity (Supplementary Figure S1). Representative images of CpG-free only (Figure 6A), as well as TRX-MBD2MBD+IDR and TRX-MBD2MBD on linear CpG-free DNA substrates (Figure 6B) show examples of DNA only and the protein–DNA complexes. The heights of TRX-MBD2MBD+IDR and TRX-MBD2MBD on DNA (0.49 ± 0.06 nm and 0.45 ± 0.06 nm, respectively) were significantly (P < 0.00001) taller than that of dsDNA itself (0.29 ± 0.04 nm) allowing us to unambiguously identify the protein-DNA complexes (Figure 6B). A striking feature from these images is the DNA bending induced by both proteins on CpG-free (Figure 6), as well as CpG-rich, and mCpG-rich DNA (Figure 7). As compared to DNA alone, MBD2MBD+IDR and MBD2MBD induced a small degree of DNA bending on CpG-free DNA (36.7°±27.6° and 39.2°±13.7°, respectively, Table 3). The DNA bending angles induced by MBD2MBD+IDR and MBD2MBD on CpG-rich DNA (66.4° ± 32.4° and 68.8°± 30.7°, respectively) were larger than observed for CpG-free, but still comparable between the two proteins (Table 3). In contrast, MBD2MBD+IDR induced significantly (P < 1 × 10−6) larger bending angles on methylated CpG-rich (103.5° ± 23.1°) DNA, while MBD2MBD did not (69.0°± 32.6°, Table 3).
Figure 6.
Bending angles induced by MBD2MBD+IDR and MBD2MBD on the linear CpG-free DNA substrate. (A) Cartoon drawing of the linear CpG-free DNA substrate (top panel), an AFM image of the linear CpG-free DNA (middle panel), and statistical analysis of the intrinsic DNA bending along the CpG-free DNA substrate (bottom panel). The solid line represent Gaussian fit to the data with the peak centered at 0°. (B) Representative AFM images of MBD2MBD+IDR (left panel) and MBD2MBD (right panel) on the CpG-free DNA substrate. Scale bars are at 200 nm. The inserts show the 3D surface plots of the zoomed (boxed) regions. (C) Statistical analysis of the DNA bending angles induced by MBD2MBD+IDR and MBD2MBD binding on the linear CpG-free DNA. The left panel shows an example of using the custom Matlab code to measure the DNA bending angle (red square: center of DNA-bound MBD2; black squares: DNA identified through sampling along a circle around the protein; θ: protein-mediated DNA bending angle. The solid and dotted lines in the right panel are Gaussian fits to the data (R2 > 0.90) with peaks centered at 36.7° (±27.6°) for MBD2MBD+IDR (N = 91) and 39.2° (±13.7°) for MBD2MBD (N = 77) (Table 3).
Figure 7.
MBD2MBD+IDR but not MBD2MBD mediates additional DNA bending upon binding to mCpG-rich DNA. (A) Cartoon drawing of the linear unmethylated and methylated CpG-rich DNA substrates. (B) Representative AFM images of CpG-rich and mCpG-rich DNA only. (C) Statistical analysis of the intrinsic DNA bending along the CpG-rich and mCpG-rich DNA substrates. The solid lines represent Gaussian fits to the data with the peaks centered at 0°. (D) Representative AFM images of MBD2MBD+IDR (top panels) and MBD2MBD (bottom panels) binding to linear unmethylated (left panels) and methylated (right panels) CpG-rich DNA. Scale bars are at 200 nm. The inserts show the 3D surface plots of the zoomed (boxed) regions. (E) DNA bending angles upon binding of MBD2MBD+IDR (left panel) and MBD2MBD (right panel) to unmethylated (orange bars) and methylated (purple bars) CpG-rich DNA. The lines represent Gaussian fits to the data (R2 > 0.95). The error bars represent SD. The mean and SEM of the bending angles are reported in Table 3.
Table 3. Summary of DNA bending angles induced by MBD2 binding.
| DNA | MBD2MBD+IDR | MBD2MBD | |||
|---|---|---|---|---|---|
| Bending angle (°) | N | Bending angle (°) | N | ||
| Unmethylated | CpG-free | 36.7°±27.6° | 91 | 39.2°±13.7° | 77 |
| CpG-rich | 66.4°±32.4° | 167 | 68.8°±30.7° | 102 | |
| CpG free region | 42.5°±26.3° | 113 | 48.9°±20.1° | 57 | |
| CpG rich region | 74.1°±35.7° | 76 | 69.9°±36.1° | 56 | |
| Methylated | CpG-free | N.A. | N.A. | ||
| CpG-rich | 103.5°±23.1° | 162 | 69.0°±32.6° | 182 | |
| CpG free region | 42.1°±38.1° | 105 | 41.4°±23.3° | 181 | |
| CpG rich region | 102.2°±41.3° | 73 | 62.6°±27.6° | 114 | |
The values represent mean ± SD from two to three experiments for each data set. The significance values regarding the difference among the data sets are reported in Supplementary Figure S8.
To rule out the possibility that sample variation contributed to these differences in DNA bending, we analyzed MBD2MBD+IDR- and MBD2MBD-induced DNA bending angles on the CpG-free and CpG-rich regions present in the same linear DNA (unmethylated and methylated CpG-free-rich DNA substrates, Supplementary Figures S7 and S8). Even though we could not differentiate the two DNA ends from each other, the central location of the CpG-rich region on the linear DNA substrate (38% to 62% measured from either end) allowed us to assign the MBD2 binding to either the CpG-free or CpG-rich regions (Supplementary Figures S7C and S8C). Analysis of the binding position of MBD2MBD+IDR and MBD2MBD on the unmethylated and methylated CpG-free-rich DNA showed that both MBD2MBD+IDR and MBD2MBD preferentially bound to the CpG-rich region (Supplementary Figures S7C and S8C). Bending angle analysis on the CpG-free-rich DNA substrates confirmed the results from using DNA substrates of CpG-free (Figure 6) and CpG-rich only sequences (Figure 7). On the linear unmethylated CpG-free-rich DNA, both MBD2MBD+IDR and MBD2MBD induced additional bending on the CpG-rich region as compared to at the CpG-free region (Supplementary Figure S7). Importantly, while both MBD2MBD+IDR and MBD2MBD induced a similar degree of slight bending at the CpG-free region, upon binding to the methylated CpGs, MBD2MBD+IDR mediated additional DNA bending compared to MBD2MBD (Supplementary Figure S8D, Table 3). In summary, while the IDR region does not further modify the DNA bending by MBD2 at the CpG-free region or unmethylated CpG, it facilitates additional DNA bending upon recognition of the methylated CpG DNA (Supplementary Figure S9).
While MBD2MBD does not bend CpG-free DNA to the same extent as mCpG-rich DNA, it is unclear whether bending requires increased CpG content. Therefore, we isolated the CpG-poor region (0.6 CpG per 100 bp) and measured bending by MBD2MBD+IDR for both unmethylated and methylated DNA. Analysis of the bending angles (Supplementary Figure S10A,B) shows that MBD2MBD+IDR bends CpG-poor DNA (56.3° ± 24.7°), but somewhat less than CpG-rich DNA (66.4° ± 32.4o); and that methylation increases the bending (72.4° ± 60.7°), but not to the same extent as for CpG-rich DNA (103.5° ± 23.1°). This result indicates that the average of MBD2-mediated DNA bending depends on CpG density and methylation status.
Finally, given that we have high-resolution structural analysis, we chose to study the chicken MBD2 orthologue. While the human MBD2b and chicken MBD2 orthologues are highly homologous (94% identity for MBD2MBD+IDR), we sought to confirm that the human MBD2MBD+IDR can bend methylated DNA as well. As expected, AFM of human MBD2MBD+IDR shows very similar bending to that of the chicken orthologue when bound to bound to CpG-free (51.5° ± 29.4°), CpG-rich (67.7° ± 27.2°) and mCpG-rich (96.8° ± 13.4°) DNA (Supplementary Figure S10C and D). Hence, the behavior we have characterized applies to the human orthologue as well.
DISCUSSION
The current model of MBD2 function postulates that the protein selectively targets methylated CpG islands, recruiting the NuRD co-regulatory complex to silence gene expression through histone deacetylase and nucleosome remodeling activities. However, genomic localization and gene expression studies have indicated that while MBD2-NuRD predominately localizes to methylated CpG islands, it can be found at a subset of unmethylated CpG islands associated with active promoters (9,10,16,17). Interpretation of these results is further complicated by the low affinity MBD3 paralogue that likewise recruits NuRD complex in a mutually exclusive manner (30). These observations have led us to question whether MBD2-NuRD functions at both unmethylated and methylated promoters, contributing to either nucleosome mobilization or stabilization depending on the distinct dynamic DNA binding modes of the MBD2.
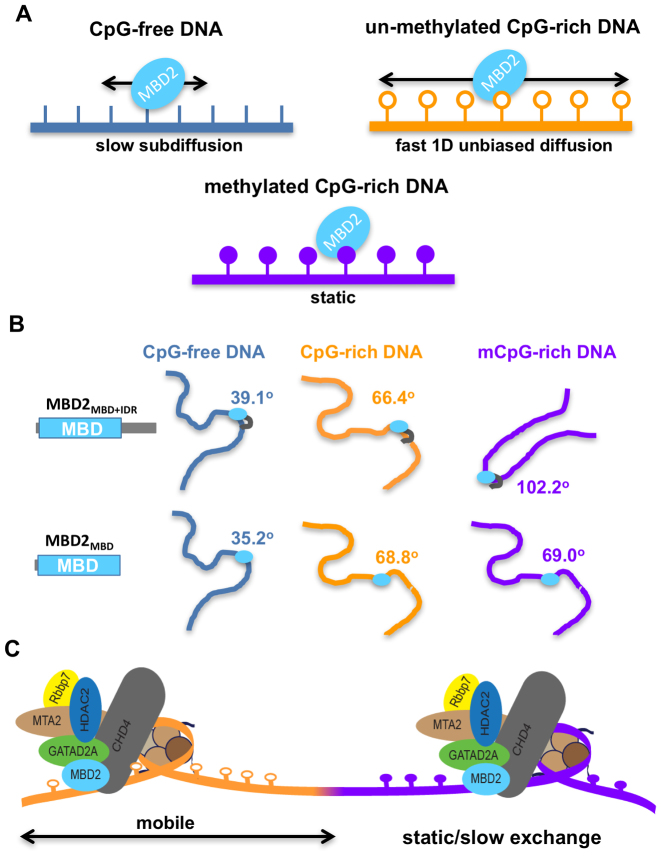
In the studies reported here, we have found that the isolated MBD2MBD shows at least three distinct DNA binding modes: (i) relatively slow 1D subdiffusion on CpG-free regions without significant DNA bending, (ii) relatively rapid 1D unbiased diffusion on unmethylated CpG-rich regions with intermediate DNA bending, (iii) stable binding on methylated CpG-rich regions associated with marked DNA bending (Figure 8A and B). Furthermore, results from tracking of MBD2 on DNA tightropes strongly support that MBD2 carries out curvilinear motion during rapid diffusion along the DNA, as has been described for other transcription factors (47–49). If the hydrodynamic radius of the red QD (655 nm) is assumed to be 11.5 nm (50) and the radius of gyration of free MBD2 is 1.28 nm based on the solution structure (13), then the expected upper limit for diffusion constants for a single red QD labeled MBD2 sliding on DNA is 21.1 μm2/s. Assuming MBD2 rotates around DNA helix, the expected upper limits for diffusion constants based on the modified version of the Stokes-Einstein relation is 0.03 μm2/s (51), which is close to what was observed experimentally (Table 2). The associated IDR does not significantly change the 1D diffusion dynamics of MBD2, which is consistent with the notion that IDR does not change the structural recognition of mCpG or methylation selectivity supported by results from NMR experiments reported in the current and previous studies (11).
Figure 8.
CpG and methylation-dependent DNA binding models of MBD2. (A) The difference in MBD2 DNA binding dynamics at CpG-free, CpG-rich and mCpG regions. MBD2 diffuses more rapidly and extensively on CpG-rich sequences than those that lack CpGs (CpG-free). MBD2 binds statically or with slower exchange rates when binding to mCpG-rich regions. (B) MBD2 recognizes CpG and mCpG through DNA bending. MBD2 induces sharper DNA bending at CpG-rich regions compared to at CpG-free, which is further augmented by IDR when binding to methylated CpG-rich sequences. The summary of the MBD2-induced DNA bending angles on different DNA substrates is presented in Table 3 and Supplementary Figure S9. (C) Proposed model for the dynamics of MBD2 in the context of the NuRD complex. MBD2 enables the NuRD complex to freely mobilize nucleosomes within unmethylated CpG islands, while impeding mobility on densely methylated CpG DNA.
Once MBD2 enters densely methylated regions, however, diffusion is greatly restricted, manifested as stable DNA binding in fluorescence imaging (spatial resolution 16 nm) and slower exchange rates on methylated CpGs. The stable binding of both MBD2MBD and MBD2MBD+IDR on DNA tightropes containing methylated CpGs are consistent with previous observations. Binding analyses (6,13,52,53) and quantum mechanics calculations (54) have established that methylation contributes to a favorable change in free energy for MBD2 binding to DNA. In addition, computational studies suggest that methylation leads to subtle structural rearrangements within the DNA and MBD binding sites (54). These structural rearrangements and the free energy difference are consistent with the chemical shift changes we observe between methylation specific and non-specific binding modes and likely contribute to the marked stabilization of MBD2 when bound to methylated CpG regions as observed by single-molecule fluorescence. However, unexpectedly, this study uncovered a new role for the IDR in recognition of methylated DNA. Combining results from NMR and AFM experiments strongly supports that the IDR serves as an additional anchor by reducing exchange between CpG sites and dissociation from DNA leading to stable binding over a methylated CpG region, as well as introducing additional bending at methylated CpG sites.
Therefore, these data support a model of MBD2MBD function in which the dynamic mobility of this domain regulates whether MBD2-NuRD efficiently rearranges nucleosomes along an unmethylated CpG island or fixes nucleosomes in place within methylated CpG islands (Figure 8C). This difference suggests that MBD2-NuRD will tend to move nucleosomes away from unmethylated and towards methylated regions and allow for more dynamic mobility within unmethylated regions. Furthermore, AFM analyses clearly shows that MBD2 bends methylated DNA, an observation that has not been previously reported. Whether this bending occurs in the context of the full MBD2-NuRD complex and nucleosome laden chromatin is yet to be determined. Nonetheless, this observation raises the possibility that MBD2 itself contributes to DNA compaction.
Taken together, these results potentially shed light on the enigmatic function of MBD2-NuRD in cells. The established model that MBD2 solely targets NuRD to methylated CpG islands and silences the associated genes based on previous biochemical and biophysical characterizations does not fit with recent whole genome localization studies and gene expression analyses (9,16,55). Instead, the cellular function of MBD2 appears to be context dependent, such that MBD2-NuRD can be associated with either gene activation or silencing, although predominantly the latter. If, however, one considers that the function of MBD2 reflects dynamic mobility on DNA as opposed to an on-off binding event, then the function is perhaps less enigmatic. Given both the relatively high affinity for non-methylated DNA as well as the presence of additional chromatin and DNA interacting domains within NuRD, then it is unlikely that the complex spends time dissociated from DNA. Instead, the complex most likely dynamically spreads across chromatin depending on the relative binding affinity and mobility of its constituent parts. Hence, MBD2 should allow the NuRD complex to freely mobilize nucleosomes within unmethylated CpG islands, while impeding mobility on densely methylated DNA. This model implies that MBD2-NuRD would accumulate both at densely methylated CpG islands as well as at the junction between unmethylated and methylated CpG rich regions. We suggest that the latter has been observed in ChIP analyses which revealed a peak for MBD2 localization at the shoulder of an unmethylated CpG island (i.e. Figure 5 B in Manefera et al. (16)). In fact, MBD2-NuRD accumulation at the edges of CpG islands helps explain the strong correlation between gene expression and differential methylation of CpG shores, the regions immediately adjacent to islands (56,57).
In summary, we have found that the methylcytosine binding domain of MBD2 exhibits distinct dynamic DNA binding mode depending on CpG density and methylation status. These results underscore the importance of understanding how DNA binding domains dynamically associate with DNA and suggest a model for MBD2 function in cells. We propose that relative binding affinity is an inadequate description of DNA binding domain interaction with DNA, especially in the case of the MBD proteins. The different dynamic binding modes are more relevant to the function of MBD proteins in the context of large macromolecular machines. Hence, extending these analyses to the larger NuRD complex will help elucidate the function of this complex in epigenetic transcription regulation, cell differentiation, and development.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the Weninger group at NCSU for technical support, and Changjiang You and Jacob Piehler at Universität Osnabrück for providing the NTA compound.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01-GM107559 to H.W. and R.R., P30-ES025128 Pilot Project to H.W., R01-GM098264 to D.W.]; National Science Foundation [DBI1353897 to R.R.]; North Carolina State University [Research and Innovation Seed Funding to H.W.]. Funding for open access charge: NIH [R01-GM107559 and R01-GM098264].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ambrosi C., Manzo M., Baubec T.. Dynamics and context-dependent roles of DNA methylation. J. Mol. Biol. 2017; 429:1459–1475. [DOI] [PubMed] [Google Scholar]
- 2. Suzuki M.M., Bird A.. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008; 9:465–476. [DOI] [PubMed] [Google Scholar]
- 3. Kanwal R., Gupta S.. Epigenetic modifications in cancer. Clin. Genet. 2012; 81:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deaton A.M., Bird A.. CpG islands and the regulation of transcription. Genes Dev. 2011; 25:1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Illingworth R.S., Bird A.P.. CpG islands–‘a rough guide’. FEBS Lett. 2009; 583:1713–1720. [DOI] [PubMed] [Google Scholar]
- 6. Hendrich B., Bird A.. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998; 18:6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hendrich B., Tweedie S.. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003; 19:269–277. [DOI] [PubMed] [Google Scholar]
- 8. Zhang Y., Ng H.H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D.. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999; 13:1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baubec T., Ivánek R., Lienert F., Schübeler D.. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell. 2013; 153:480–492. [DOI] [PubMed] [Google Scholar]
- 10. Günther K., Rust M., Leers J., Boettger T., Scharfe M., Jarek M., Bartkuhn M., Renkawitz R.. Differential roles for MBD2 and MBD3 at methylated CpG islands, active promoters and binding to exon sequences. Nucleic Acids Res. 2013; 41:3010–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Desai M.A., Webb H.D., Sinanan L.M., Scarsdale J.N., Walavalkar N.M., Ginder G.D., Williams D.C. Jr. An intrinsically disordered region of methyl-CpG binding domain protein 2 (MBD2) recruits the histone deacetylase core of the NuRD complex. Nucleic Acids Res. 2015; 43:3100–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bird A.P. CpG-rich islands and the function of DNA methylation. Nature. 1986; 321:209–213. [DOI] [PubMed] [Google Scholar]
- 13. Scarsdale J.N., Webb H.D., Ginder G.D., Williams D.C. Jr. Solution structure and dynamic analysis of chicken MBD2 methyl binding domain bound to a target-methylated DNA sequence. Nucleic Acids Res. 2011; 39:6741–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cramer J.M., Scarsdale J.N., Walavalkar N.M., Buchwald W.A., Ginder G.D., Williams D.C. Jr. Probing the dynamic distribution of bound states for methylcytosine-binding domains on DNA. J. Biol. Chem. 2014; 289:1294–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Walavalkar N.M., Cramer J.M., Buchwald W.A., Scarsdale J.N., Williams D.C. Jr. Solution structure and intramolecular exchange of methyl-cytosine binding domain protein 4 (MBD4) on DNA suggests a mechanism to scan for mCpG/TpG mismatches. Nucleic Acids Res. 2014; 42:11218–11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Menafra R., Brinkman A.B., Matarese F., Franci G., Bartels S.J.J., Nguyen L., Shimbo T., Wade P.A., Hubner N.C., Stunnenberg H.G.. Genome-wide binding of MBD2 reveals strong preference for highly methylated loci. PLoS ONE. 2014; 9:e99603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shimbo T., Du Y., Grimm S.A., Dhasarathy A., Mav D., Shah R.R., Shi H., Wade P.A.. MBD3 localizes at promoters, gene bodies and enhancers of active genes. PLoS Genet. 2013; 9:e1004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gorman J., Plys A.J., Visnapuu M.-L., Alani E., Greene E.C.. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat. Struct. Mol. Biol. 2010; 17:932–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gorman J., Greene E.C.. Visualizing one-dimensional diffusion of proteins along DNA. Nat. Struct. Mol. Biol. 2008; 15:768–774. [DOI] [PubMed] [Google Scholar]
- 20. Zweckstetter M., Bax A.. Characterization of molecular alignment in aqueous suspensions of Pf1 bacteriophage. J. Biomol. NMR. 2001; 20:365–377. [DOI] [PubMed] [Google Scholar]
- 21. Clore G.M., Starich M.R., Gronenborn A.M.. Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J. Am. Chem. Soc. 1998; 120:10571–10572. [Google Scholar]
- 22. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 23. Skinner S.P., Goult B.T., Fogh R.H., Boucher W., Stevens T.J., Laue E.D., Vuister G.W.. Structure calculation, refinement and validation using CcpNmr Analysis. Acta Crystallogr. D Biol. Crystallogr. 2015; 71:154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D.. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005; 59:687–696. [DOI] [PubMed] [Google Scholar]
- 25. Sahu D., Clore G.M., Iwahara J.. TROSY-based z-exchange spectroscopy: application to the determination of the activation energy for intermolecular protein translocation between specific sites on different DNA molecules. J. Am. Chem. Soc. 2007; 129:13232–13237. [DOI] [PubMed] [Google Scholar]
- 26. Lopez-Serra L., Ballestar E., Fraga M.F., Alaminos M., Setien F., Esteller M.. A profile of methyl-CpG binding domain protein occupancy of hypermethylated promoter CpG islands of tumor suppressor genes in human cancer. Cancer Res. 2006; 66:8342–8346. [DOI] [PubMed] [Google Scholar]
- 27. Reichel A., Schaible D., Furoukh Al N., Cohen M., Schreiber G., Piehler J.. Noncovalent, site-specific biotinylation of histidine-tagged proteins. Anal. Chem. 2007; 79:8590–8600. [DOI] [PubMed] [Google Scholar]
- 28. Lin J., Countryman P., Buncher N., Kaur P., E L., Zhang Y., Gibson G., You C., Watkins S.C., Piehler J. et al. . TRF1 and TRF2 use different mechanisms to find telomeric DNA but share a novel mechanism to search for protein partners at telomeres. Nucleic Acids Res. 2014; 42:2493–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saxton M.J. Single-particle tracking: the distribution of diffusion coefficients. Biophys. J. 1997; 72:1744–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Le Guezennec X., Vermeulen M., Brinkman A.B., Hoeijmakers W.A.M., Cohen A., Lasonder E., Stunnenberg H.G.. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 2006; 26:843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cramer J.M., Pohlmann D., Gomez F., Mark L., Kornegay B., Hall C., Siraliev-Perez E., Walavalkar N.M., Sperlazza M.J., Bilinovich S. et al. . Methylation specific targeting of a chromatin remodeling complex from sponges to humans. Sci. Rep. 2017; 7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Winter R.B., von Hippel P.H.. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The Escherichia coli repressor–operator interaction: equilibrium measurements. Biochemistry. 1981; 20:6948–6960. [DOI] [PubMed] [Google Scholar]
- 33. Berg O.G., Winter R.B., von Hippel P.H.. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981; 20:6929–6948. [DOI] [PubMed] [Google Scholar]
- 34. Winter R.B., Berg O.G., von Hippel P.H.. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor–operator interaction: kinetic measurements and conclusions. Biochemistry. 1981; 20:6961–6977. [DOI] [PubMed] [Google Scholar]
- 35. von Hippel P.H., Berg O.G.. Facilitated target location in biological systems. J. Biol. Chem. 1989; 264:675–678. [PubMed] [Google Scholar]
- 36. Tafvizi A., Mirny L.A., van Oijen A.M.. Dancing on DNA: kinetic aspects of search processes on DNA. ChemPhysChem. 2011; 12:1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Allen H.F., Wade P.A., Kutateladze T.G.. The NuRD architecture. Cell. Mol. Life Sci. 2013; 70:3513–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lai A.Y., Wade P.A.. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer. 2011; 11:588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dunn A.R., Kad N.M., Nelson S.R., Warshaw D.M., Wallace S.S.. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011; 39:7487–7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hughes C.D., Wang H., Ghodke H., Simons M., Towheed A., Peng Y., Van Houten B., Kad N.M.. Real-time single-molecule imaging reveals a direct interaction between UvrC and UvrB on DNA tightropes. Nucleic Acids Res. 2013; 41:4901–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kad N.M., Wang H., Kennedy G.G., Warshaw D.M., Van Houten B.. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single-molecule imaging of quantum-dot-labeled proteins. Mol. Cell. 2010; 37:702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lata S., Piehler J.. Stable and functional immobilization of histidine-tagged proteins via multivalent chelator headgroups on a molecular poly(ethylene glycol) brush. Anal. Chem. 2005; 77:1096–1105. [DOI] [PubMed] [Google Scholar]
- 43. Kaur P., Plochberger B., Costa P., Cope S.M., Vaiana S.M., Lindsay S.. Hydrophobicity of methylated DNA as a possible mechanism for gene silencing. Phys. Biol. 2012; 9:065001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rule G.S., Hitchens T.K.. Fundamentals of protein NMR spectroscopy. 2006; 5:Springer; 1–531. [Google Scholar]
- 45. Iwahara J., Clore G.M.. Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J. Am. Chem. Soc. 2006; 128:404–405. [DOI] [PubMed] [Google Scholar]
- 46. Iwahara J., Zweckstetter M., Clore G.M.. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:15062–15067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blainey P.C., Luo G., Kou S.C., Mangel W.F., Verdine G.L., Bagchi B., Xie X.S.. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009; 16:1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dikić J., Menges C., Clarke S., Kokkinidis M., Pingoud A., Wende W., Desbiolles P.. The rotation-coupled sliding of EcoRV. Nucleic Acids Res. 2012; 40:4064–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marklund E.G., Mahmutovic A., Berg O.G., Hammar P., van der Spoel D., Fange D., Elf J.. Transcription-factor binding and sliding on DNA studied using micro- and macroscopic models. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:19796–19801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Arnspang E.C., Brewer J.R., Lagerholm B.C.. Multi-color single particle tracking with quantum dots. PLoS ONE. 2012; 7:e48521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schurr J.M. The one-dimensional diffusion coefficient of proteins absorbed on DNA hydrodynamic considerations. Biophys. Chem. 1975; 9:413–414. [PubMed] [Google Scholar]
- 52. Hashimoto H., Liu Y., Upadhyay A.K., Chang Y., Howerton S.B., Vertino P.M., Zhang X., Cheng X.. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012; 40:4841–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fraga M.F., Ballestar E., Montoya G., Taysavang P., Wade P.A., Esteller M.. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003; 31:1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zou X., Ma W., Solov’yov I.A., Chipot C., Schulten K.. Recognition of methylated DNA through methyl-CpG binding domain proteins. Nucleic Acids Res. 2012; 40:2747–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Menafra R., Stunnenberg H.G.. MBD2 and MBD3: elusive functions and mechanisms. Front Genet. 2014; 5:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Irizarry R.A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P., Cui H., Gabo K., Rongione M., Webster M. et al. . The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009; 41:178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Doi A., Park I.-H., Wen B., Murakami P., Aryee M.J., Irizarry R., Herb B., Ladd-Acosta C., Rho J., Loewer S. et al. . Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009; 41:1350–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.