Abstract
Chromatin reorganization is necessary for pluripotent stem cells, including embryonic stem cells (ESCs), to acquire lineage potential. However, it remains unclear how ESCs maintain their characteristic chromatin state for appropriate gene expression upon differentiation. Here, we demonstrate that chromodomain helicase DNA-binding domain 2 (Chd2) is required to maintain the differentiation potential of mouse ESCs. Chd2-depleted ESCs showed suppressed expression of developmentally regulated genes upon differentiation and subsequent differentiation defects without affecting gene expression in the undifferentiated state. Furthermore, chromatin immunoprecipitation followed by sequencing revealed alterations in the nucleosome occupancy of the histone variant H3.3 for developmentally regulated genes in Chd2-depleted ESCs, which in turn led to elevated trimethylation of the histone H3 lysine 27. These results suggest that Chd2 is essential in preventing suppressive chromatin formation for developmentally regulated genes and determines subsequent effects on developmental processes in the undifferentiated state.
INTRODUCTION
In multicellular organisms, proper expression of lineage-specific genes is essential for various types of cell differentiation. These genes are regulated through global chromatin reorganization in undifferentiated cells. Embryonic stem cells (ESCs), which are derived from the inner cell mass of a blastocyst, possess the potential to differentiate into multiple lineages and maintain their preparatory state for the expression of differentiation-associated genes that respond to differentiation signals. Such responses are enabled by loosening of the chromatin structure in ESCs and less repressive heterochromatin compared with that in differentiated cells (1). Regulation of the chromatin structure has been shown to be essential to maintain the differentiation potential in ESCs.
A known modulator of structural changes in chromatin is post-translational modification of the histone tail in histones H3 and H4. High levels of histone post-translational modifications associated with active transcription, such as hyperacetylation of H3 and H4, have been observed in undifferentiated ESCs (2). Moreover, markings with active trimethylation of histone H3 lysine 4 (H3K4me3) and repressive trimethylation of histone H3 lysine 27 (H3K27me3) are two characteristics of developmentally regulated genes in ESCs. This coexistence of active and repressive epigenetic regulators is called a bivalent state (3). Studies on histone-modifying enzymes involved in the bivalent state, such as Polycomb repressive complex 2 (PRC2), have revealed that these histone modifications and modifying enzymes are essential for the differentiation of ESCs (4,5). In addition to histone modifications, characteristic histone variants have been found to mark distinctive transcriptional states and play an integral role in proper cell differentiation (6). In particular, while the histone H3 variant H3.3 was first indicated to incorporate into a transcriptionally activated region (7), recent studies have revealed that it also provides a foothold for transcription-suppressing modifications such as H3K27me3 and trimethylation of histone H3 lysine 9 (H3K9me3) in the heterochromatin of ESCs (8–10). H3.3 knockdown was also shown to reduce the levels of H3K27me3 enrichment in the bivalent state and to alter the developmental potential of ESCs (9). Therefore, the histone variant H3.3 itself plays a fundamental role in regulation of the differentiation potential in ESCs.
Histone modifications are recognized by chromatin regulators such as adenosine triphosphate-dependent chromatin-remodeling enzymes for sliding, evicting, assembling, spacing and replacing nucleosomes for proper gene expression (11–13). These chromatin-remodeling enzymes are expressed abundantly in ESCs, and some have been indicated to be necessary for the functions of ESCs (14–16). For example, chromodomain helicase DNA-binding domain (Chd)1 incorporates H3.3 into nucleosomes of Drosophila embryos and is essential for maintaining open chromatin and pluripotency in mouse ESCs (mESCs) (17,18).
Shen et al. and others have also reported the requirement for Chd2, a member of the SNF2 family of chromatin-remodeling enzymes, for the normal differentiation of mouse myogenic and neural progenitor cells (19,20). Chd2-deficient mice have been demonstrated to exhibit a general growth delay and perinatal lethality (21). These studies suggest that Chd2 plays an intrinsic role in normal mammalian development. However, its role in regulating developmental gene expression is poorly understood.
In this study, we examined the function of Chd2 in the regulation of differentiation potential using mESCs. We found that Chd2 is essential for the appropriate expression of developmentally regulated genes during differentiation, but it does not affect gene expression in the undifferentiated state. Furthermore, chromatin dynamics regulated by the interplay among Chd2, Oct3/4 and H3.3 in the bivalent state determines subsequent effects on differentiation processes.
MATERIALS AND METHODS
Cells
The mouse ESC lines EB5 and ZHBTc4 were kindly provided by Dr Hitoshi Niwa (22,23). The cells were cultured in Glasgow minimum essential medium (Sigma-Aldrich) containing 10% fetal bovine serum, 1000 U/ml recombinant leukemia inhibitory factor (LIF) (Nacalai Tesque Inc.), 1 mM sodium pyruvate (Nacalai Tesque Inc.), 0.1 mM non-essential amino acids (Nacalai Tesque Inc.), 0.1 mM β-mercaptoethanol (Nacalai Tesque Inc.) and penicillin/streptomycin (Nacalai Tesque Inc.). They were cultured on gelatin-coated plates at 37°C in a 5% CO2 incubator.
Differentiation of ESCs was performed by the induction of embryoid body (EB) formation in vitro. ESCs were suspended in medium without LIF and drops of 400 cells/20 μl were transferred to the lid of a bacterial grade Petri dish (hanging drop method) (24). These hanging drops were incubated for 2 days and subsequently 50 EBs were transferred to differentiation medium with 2% methylcellulose (Nacalai Tesque Inc.) in each well of 24-well culture plates. The EBs were cultured further and collected at 4 days after the withdrawal of LIF.
Generation of Chd2mut/mut ESC lines
Chd2 genomic loci were disrupted using the CRISPR/Cas9 system (25). Sequences of the form N20-NGG in the assigned region were first extracted using Bowtie software (version 0.12.8) (26). Then, unique sequences, such that their variations with less than three mismatches do not exist outside the considered region, were selected as candidates for gRNAs. We predicted potential off-target sites using the 14 bp on the 3′ end of each guide sequence with Bowtie, as in previous studies (26–28). A gRNA sequence targeting exon 3 of Chd2 (chr. 7: 80, 664, 568–80, 664, 590) was cloned into pSpCas9 (BB)-2A-GFP (PX458, Addgene). The plasmid was transfected into ESCs using Xfect mESC Transfection Reagent (ClonTech), in accordance with the manufacturer's instructions. After 24 h, green fluorescent protein (GFP)-positive cells were sorted into 96-well plates to establish single cell clones that were subjected to genotyping by polymerase chain reaction (PCR). Chd2WT/mut (heterozygote mutant of Chd2) clones were obtained by detection of a frameshift mutation in one Chd2 allele using Sanger sequencing. Primers are listed in Supplementary Table S1. Next, to disrupt another allele of Chd2, pSp-gRNA-Cas9 (BB)-2A-GFP was transfected into Chd2WT/mut ESCs with plasmid-based donor repair templates, which contained the homology arms of both upstream and downstream of Chd2 exon 3 and the insertion reporter sequence of puro/2A/Halo. Transfected cells were cultured for 7 days in the presence of 2 μg/ml puromycin. The cells were isolated into single cells and then subjected to PCR genotyping to select Chd2mut/mut clones. The potential off-target sequences were predicted in five genomic lesions by a previously reported method, and off-target sequences were confirmed by Sanger sequencing. Primers are listed in Supplementary Table S1. For rescue experiments, the pTETmKO2-Chd2 cDNA plasmid, which we had constructed previously (19), was stably transfected into Chd2mut/mut ESC lines and monomeric Kusabira-Orange 2 (mKO2)-positive cells were selected as rescue clones by fluorescence-activated cell sorting.
Immunoblotting and immunoprecipitation
Western blot analysis and immunoprecipitation (IP) were performed as described previously (29). Each experiment was performed three times and each signal intensity was measured by CS Analyzer (Ato). Each value was normalized to Hsp90 protein levels. Simple western analysis was performed using Wes system (ProteinSimple), in accordance with the manufacturer's instructions. For immunoblotting, the primary antibodies included rabbit anti-Oct3/4 (H134, Santa Cruz Biotechnology, 1:1000), anti-Nanog (RCAB001P, Repro Cell, 1:1000), anti-Sox2 (EPR3131, Abcam, 1:1000), anti-Hsp90 (H114, Santa Cruz Biotechnology, 1:5000), rat anti-Chd2 (8H3, hybridoma supernatant, 1:100) (30), anti-H3.3 (4H2D7, hybridoma supernatant, 1:1000) (19), anti-H3.1 (1D4F2, hybridoma supernatant, 1:1000) (19), anti-H3 (1G1, hybridoma supernatant, 1:1000) (31) and anti-Brg1 (4E5, hybridoma supernatant, 1:1000) (32). Secondary antibodies were horseradish peroxidase-conjugated anti-rabbit or anti-rat IgGs (GE Healthcare, 1:5000). For IP, cleared lysates were incubated with 4 μg of antibody against Chd2 for 12 h. In these experiments, to eliminate the involvement of DNA in the interactions, cell extracts were treated with DNase (1 μl; Promega), micrococcal nuclease (1 μl; New England Biolabs) and ribonuclease A (1 μl; Nacalai Tesque) at 37°C for 40 min before IP, and then chromatin was confirmed to be digested into mono-nucleosome size.
Cell cycle analysis
Cell cycle analysis was performed by flow cytometry. Cells were collected by trypsinization and then centrifuged for 5 min at 190 × g. The cells were resuspended in 1 ml of phosphate-buffered saline (PBS) with Hoechst 33342 (Nacalai Tesque Inc.) and incubated for 15 min at room temperature. Stained cells were analyzed by a cell sorter (SH800; Sony) and divided into each cell cycle phase (G1, S and G2/M phases) based on the intensity of Hoechst 33342. The proportion of cells in each cell cycle phase was then calculated.
RNA-seq library preparation
Total RNAs from ESCs were obtained as described previously with biological duplicates (19). Library preparation and sequence analysis were performed in accordance with a protocol described previously (33).
RNA isolation, reverse transcription and RT-qPCR
Total RNA was isolated and reverse-transcribed using PrimeScript Reverse Transcriptase (Takara Bio Inc.) as described previously (19). qPCR was performed using Thunderbird qPCR Mix (Toyobo Co., Ltd.). Primers are listed in Supplementary Table S1. The data were normalized to Gapdh expression levels and are presented as the mean ± standard deviation of three independent experiments.
Teratoma formation
All animal experiments were approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University, Japan. Nonobese diabetic (NOD)/severe combined immune deficient (SCID) mice were purchased from Charles River Laboratories International, Inc. Teratomas were produced by the injection of 1 × 106 cells subcutaneously into the flanks of NOD/SCID mice. Tumor tissue samples that had developed by 3 weeks were surgically dissected and fixed in 4% formaldehyde before embedding in paraffin. Sections were stained with hematoxylin and eosin with a standard protocol.
Chromatin immunoprecipitation
ChIP assays were performed as described previously with some modifications (29). Briefly, 1 × 107 ESCs were crosslinked in 0.5% formaldehyde and suspended in ChIP buffer (5 mM PIPES, 200 mM KCl, 1 mM CaCl2, 1.5 mM MgCl2, 5% sucrose, 0.5% NP-40 and protease inhibitor cocktail; Nacalai Tesque Inc.). The samples were sonicated for 5 s three times at 70% maximum amplitude with a Sonicator (VCX130; Sonics & Materials, Inc.) and digested with micrococcal nuclease (1000 U/ml; New England Biolabs) at 37°C for 40 min. Then, 10 mM ethylenediaminetetraacetic acid (EDTA) was added to stop the reaction, and the samples were centrifuged at 15 000 × g for 10 min at 4°C. The supernatant was incubated with a rat monoclonal antibody against H3.3 (1E4A3, hybridoma supernatant, 2 μg) and mouse monoclonal antibodies against H3K4me3 (16H10, provided by Dr H. Kimura, 2 μg) (34), H3K27me3 (1E7, provided by Dr H. Kimura, 2 μg) (35) and H3K27ac (9E2H10, provided by Dr H. Kimura, 2 μg) (34) prebound to magnetic beads at 4°C overnight with rotation. The immunocomplexes were eluted from beads using elution buffer (50 mM Tris–HCl, pH 8.0, 10 mM EDTA and 1% sodium dodecyl sulphate), followed by washing with ChIP buffer and TE buffer (both twice). Crosslinks were reversed by treatment with proteinase K (Nacalai Tesque Inc.) at 50°C for 3 h and then at 65°C overnight. DNA was purified using a QIAquick PCR purification kit (Qiagen). qPCR analysis of immunoprecipitated DNA was performed using Thunderbird qPCR Mix (Toyobo Co.). Relative enrichment was defined as the ratio of the quantity of amplified PCR product relative to 10% of the input genomic DNA. Quantifications represent the mean of three independent experiments ± standard deviation. The primers are listed in Supplementary Table S1.
ChIP-seq and data analysis
ChIP libraries of H3K27me3, H3K4me3, H3K27ac and H3.3 were prepared using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs) with biological duplicates. The libraries were sequenced on Illumina HiSeq 1500 sequencers. The ChIP-seq data of Chd2 and Oct3/4 in mESCs were obtained from a public database (ENCSR531HWD and SRS000521) (36,37). Sequence reads were aligned to the reference mouse genome (mm9) using Bowtie 2 software (version 2.2.3) with default options (38). To analyze the ChIP-seq data, we counted the number of ChIP-seq unique tags in each 1 kbp width and 100 bp sliding window on the genome and normalized the data as reads per kbp of transcript per million mapped reads (RPKM) (39). To visualize ChIP-seq data with Integrative Genomics Viewer (IGV; version 2.3), signal intensities (SIs) were calculated as the RPKM of the ChIP sample with the RPKM of the input subtracted. The heatmap was created by hierarchical clustering (Ward's linkage with Euclid distance) using SIs of three histone modifications in a ±5 kbp window, consisting of 200 bp × 50 signal bins, around the transcription start site (TSS). The SIs were calculated using agplus (40). Three gene subsets were, as defined by Mikkelsen et al. (41), bivalent genes with H3K4me3 and H3K27me3, active genes with only H3K4me3 and high expression over the 80th percentile, and silent control genes with no modifications or expression (Supplementary Table S2). The binding sites of Chd2 and Oct3/4 were identified by MACS (version 2.0.10) (42) using the options --summit -p 7e-3 and --broad -p 1e-3. Peaks were annotated according to their proximity to TSSs using HOMER (43). The motif enrichment analysis was also carried out using HOMER with default options (43). The targeting genes were identified within ±50 kbp from each binding site. We clustered Chd2 binding genes into active, bivalent and other states using the intensities of histone modifications around TSS using ChIP-seq data of WT mESCs. Then, Gene Ontology analysis in each cluster was performed with clusterProfiler (44).
To compare the intensities of ChIP-seq signals between different ChIP samples prepared from different inputs, we performed between-sample normalization. First, means of counts on genomic windows (TSSs ± 5 kbp) were calculated for all genes in all inputs and ChIP samples. Next, between-sample normalization using the median of ratios of normalized values to geometric means of the values (geometric normalization) (45) was applied to correct the different library sizes for all input and ChIP counts. Finally, the (log-transformed) normalized input counts were subtracted from the corresponding (log-transformed) normalized ChIP counts.
Scatter plot of ChIP-seq
The reads were counted for H3K27me3, H3.3 and the input ChIP-seq within 2 kbp of the TSSs of all genes using the featureCounts software (44) for biological duplicate samples. The counts were normalized using the R library DESeq2 (46). The genes were categorized into two groups: those with higher H3K27me3 levels upon Chd2 depletion (red: H3K27me3high:Chd2−) and those with lower ones (blue: H3K27me3low:Chd2−) (similarly for H3.3). The log2 fold changes of the normalized counts from input for WT and Oct3/4 depleted ESCs were plotted for each group.
Proximity ligation assay (PLA)
Proximity ligation was performed using a Duolink In Situ PLA (Sigma-Aldrich) in accordance with the manufacturer's instructions, except that the oligonucleotide-conjugated anti-rabbit probe was used at 1:40 and the anti-rat probe conjugated by a Duolink In Situ Probemaker was used at 1:160. Three images were taken from each sample with a fluorescence microscope (BZ-X700; Keyence). The primary antibodies included rat monoclonal antibody against Chd2 (1:200) and H3.3 (1:1000) described above, and rabbit polyclonal antibodies against H3K27me3 (07-449, Millipore, 1:1000) and H3K4me3 (07-473, Millipore, 1:1000).
Statistical analysis
Statistical significance in RT-qPCR, ChIP-qPCR and PLA data was evaluated using Student's t-test.
RESULTS
Chd2 depletion is independent of the capacity for self-renewal in undifferentiated mESCs
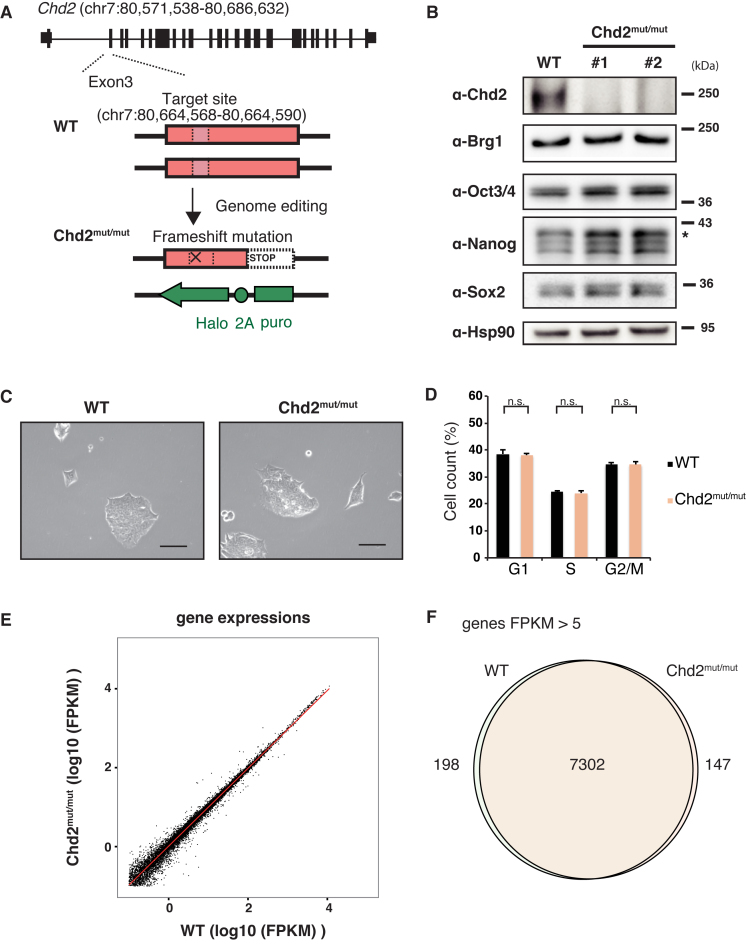
Because chromatin-remodeling factors, such as Chd1 and INO80, are required for maintenance of the self-renewal and pluripotency of ESCs (17,47), we explored the role of Chd2 in undifferentiated ESCs. First, to generate Chd2-depleted ESC lines, we disrupted both Chd2 alleles by the genome editing method (25). Using the CRISPR/Cas9 system, we introduced a frameshift mutation into exon 3 of one allele and inserted inactive sequences into exon 3 of the other allele by homologous recombination (Figure 1A and Supplementary Figure S1A–C). We obtained two independent clones. Western blot analysis demonstrated depletion of Chd2 protein in these cells (Figure 1B and Supplementary Figure S2A). Off-target sequences predicted in advance were confirmed to be intact by Sanger sequencing (Supplementary Figure S1D). Next, we examined the effects of Chd2 depletion in the undifferentiated state by comparing wild-type (WT) and Chd2-disrupted (Chd2mut/mut) mESCs. Chd2 depletion led to minimal morphological changes in ESCs under bright field observation (Figure 1C). Moreover, flow cytometric analysis revealed that the proportion of Chd2mut/mut ESCs in each stage of the cell cycle did not significantly differ from that of WT ESCs (Figure 1D). To determine the effect of Chd2 depletion in detail, we performed whole transcriptome profiling of WT and Chd2mut/mut ESCs by RNA sequencing (RNA-seq) (Figure 1E). Analysis of the data indicated that the expression level of each gene did not change drastically, with only 52 genes downregulated and 29 genes upregulated in Chd2mut/mut mESCs compared with their levels in WT mESCs (Supplementary Table S3). Expressed gene profiles of fragments per kilobasepair of transcript per million mapped reads (FPKM) greater than five were almost the same between WT and Chd2mut/mut mESCs (Figure 1F). In addition, Western blot analysis with quantification revealed that protein expression levels of master transcription factors in ESCs, such as Oct3/4 and Nanog, remained unchanged upon Chd2 depletion (Figure 1B and Supplementary Figure S2A). These results suggest that Chd2 might not affect the capacity for self-renewal and expansion of undifferentiated ESCs.
Figure 1.
Chd2 depletion is independent of the capacity for self-renewal in undifferentiated mESCs. (A) Scheme of generating Chd2mut/mut ESCs using the CRISPR/Cas9 system (25). The gRNA target site (chr. 7: 80, 664, 568–80, 664, 590) is indicated by a pink box. Details are shown in Supplementary Figure S1A. (B) Depletion of endogenous Chd2 protein in ESCs using the CRISPR/Cas9 system. Western blot analyses of WT and Chd2mut/mut ESCs using antibodies against Chd2, Brg1, Oct3/4, Nanog, Sox2 and Hsp90 are shown. (C) Morphologies of WT and Chd2mut/mut ESCs were not significantly changed under a bright field. Scale bars = 50 μm. (D) Cell cycle analysis of WT and Chd2mut/mut ESCs by flow cytometry. The population of cells in each cell cycle phase (G1, S or G2/M) was calculated based on the intensity of Hoechst 33342. Data represent the mean of three independent experiments ± standard deviation. n.s., P > 0.05. (E) Scatter plot showing the expression levels of all genes in undifferentiated WT and Chd2mut/mut ESCs. RNA-seq datasets were obtained and FPKM values were calculated. The red line indicates a regression line. (F) Venn diagram showing the overlap between genes with FPKM > 5 in WT and Chd2mut/mut ESCs.
Chd2 maintains the expression potential of developmentally regulated genes upon differentiation
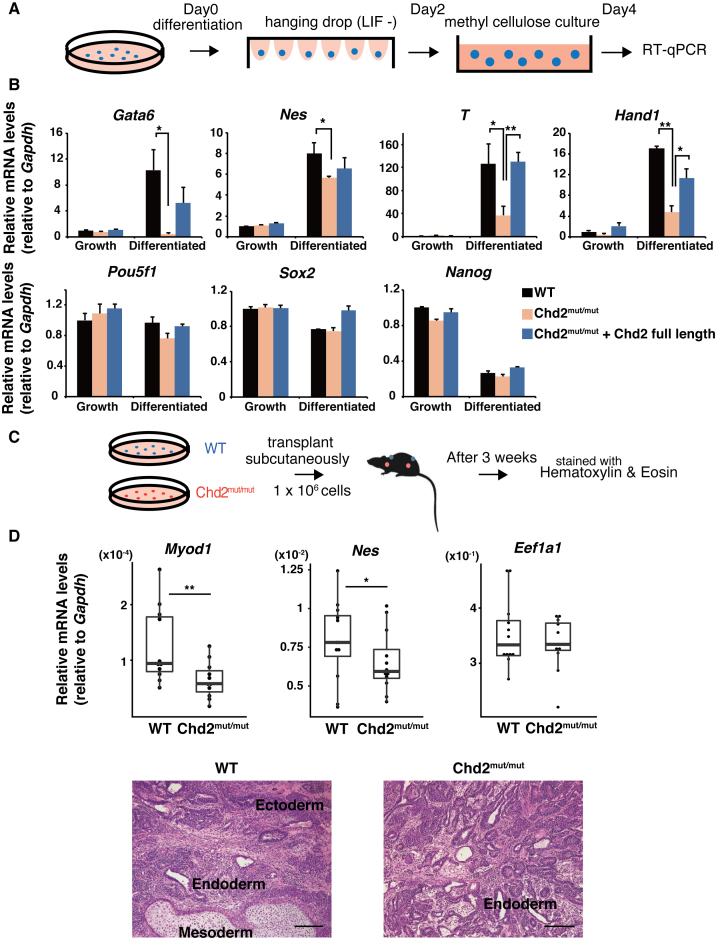
Previous studies have shown that some chromatin-remodeling factors and histone modifiers are required for ESC differentiation, despite their small effect on pluripotency factor gene expression (12,13,48,49). To evaluate the developmental potential of Chd2mut/mut ESCs, we analyzed gene expression during differentiation of EBs in vitro (50) (Figure 2A). RT-qPCR (n = 3) showed limited upregulation for early differentiation markers, such as endoderm markers Gata6 (P = 0.018) and Foxa2 (P = 0.046), mesoderm markers T (P = 0.039) and Hand1 (P = 0.003), and ectoderm marker Nes (P = 0.047), in EBs derived from Chd2mut/mut ESCs at day 4 after the withdrawal of LIF (Figure 2B and Supplementary Figure S3A). We observed downregulation of pluripotency markers in Chd2mut/mut EBs similar to that in WT EBs, such as Pou5f1 (P = 0.06), Nanog (P = 0.13) and Sox2 (P = 0.35) (Figure 2B). To confirm a Chd2-specific function upon ESC differentiation, we rescued the depletion of Chd2 expression via exogenous introduction of full-length Chd2 cDNA in Chd2mut/mut ESCs (Supplementary Figure S3B–D). As expected, RT-qPCR showed that the introduction of full-length Chd2 restored the expression of differentiation markers to levels comparable to those in WT EBs upon differentiation (Figure 2B). Next, we examined the effect of Chd2 depletion at the later stage of differentiation. We injected WT and Chd2mut/mut ESCs subcutaneously into immunodeficient mice and observed teratoma formation after 3 weeks (Figure 2C and Supplementary Figure S4A). Histological analysis showed that both Chd2mut/mut and WT ESCs formed teratomas containing cells representative of all three embryonic germ layers, but Chd2mut/mut teratomas contained predominantly glandular tissue and less skeletal muscles and neural tubes compared with WT teratomas (Figure 2D and Supplementary Figure S4B). To confirm these changes, we isolated RNA from WT and Chd2mut/mut teratomas. RT-qPCR (n = 10) analysis confirmed the downregulation of marker genes for myogenic and neural differentiation, such as Myod1 (P = 0.004) and Nes (P = 0.01), in Chd2mut/mut teratomas. The expression levels of a housekeeping gene, Eef1a1, were similar in WT and Chd2mut/mut ESCs (Figure 2D and Supplementary Figure S4C). Collectively, these results demonstrate that Chd2 is required for the proper upregulation of developmentally regulated genes upon differentiation.
Figure 2.
Chd2 maintains the expression potential of developmentally regulated genes upon differentiation. (A) Scheme of EB formation analysis. EBs were induced by the hanging drop method and subsequently cultured in methylcellulose-containing medium. (B) RT-qPCR was performed on cDNA of EBs derived from WT ESCs, Chd2mut/mut ESCs and rescued ESCs generated by transfection of full-length Chd2 cDNA into Chd2mut/mut ESCs. The expression levels of pluripotency marker genes (Pou5f1, Nanog and Klf4) and differentiation marker genes (Gata6, Foxa2, T and Hand1) were evaluated at days 0 and 4. Transcript levels were normalized against Gapdh expression and values relative to those in WT ESCs at day 0 are shown (mean ± standard deviation of three independent experiments). *P < 0.05, **P < 0.01. See also Supplementary Figure S2A. (C) Scheme of teratoma analysis. WT and Chd2mut/mut ESCs were injected subcutaneously into each flank of NOD/SCID mice. Teratomas were collected after 3 weeks. (D) RT-qPCR of each lineage marker gene in WT and Chd2mut/mut teratomas (upper panel). Box plots show expression relative to Gapdh mRNA levels (n = 10). *P < 0.05, **P < 0.01. Histology of teratomas derived from WT and Chd2mut/mut ESCs (lower panel). Specimens were stained with hematoxylin and eosin (H&E). Observed tissues representing three embryonic germ layers are indicated. Scale bars = 200 μm.
Chd2-dependent developmental genes are in the bivalent state in undifferentiated mESCs
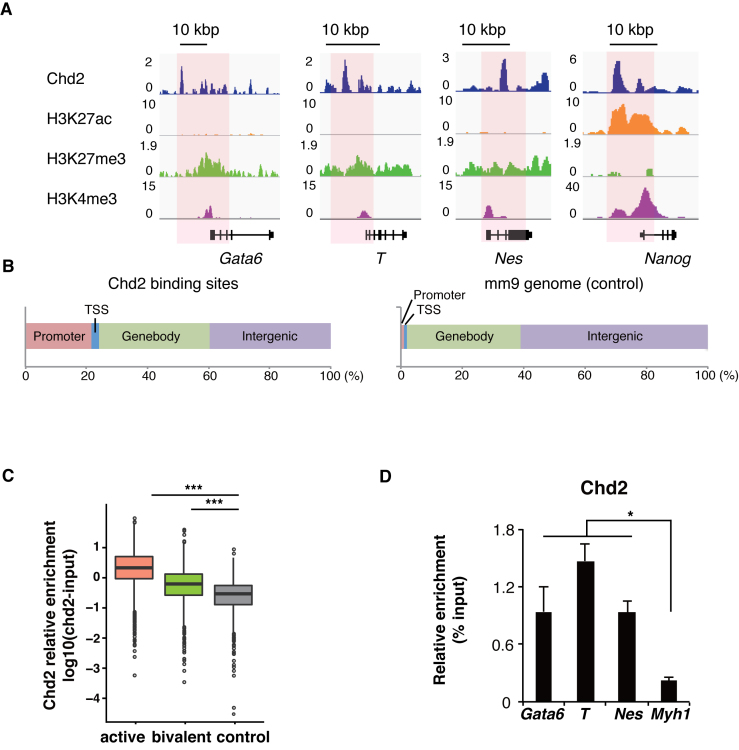
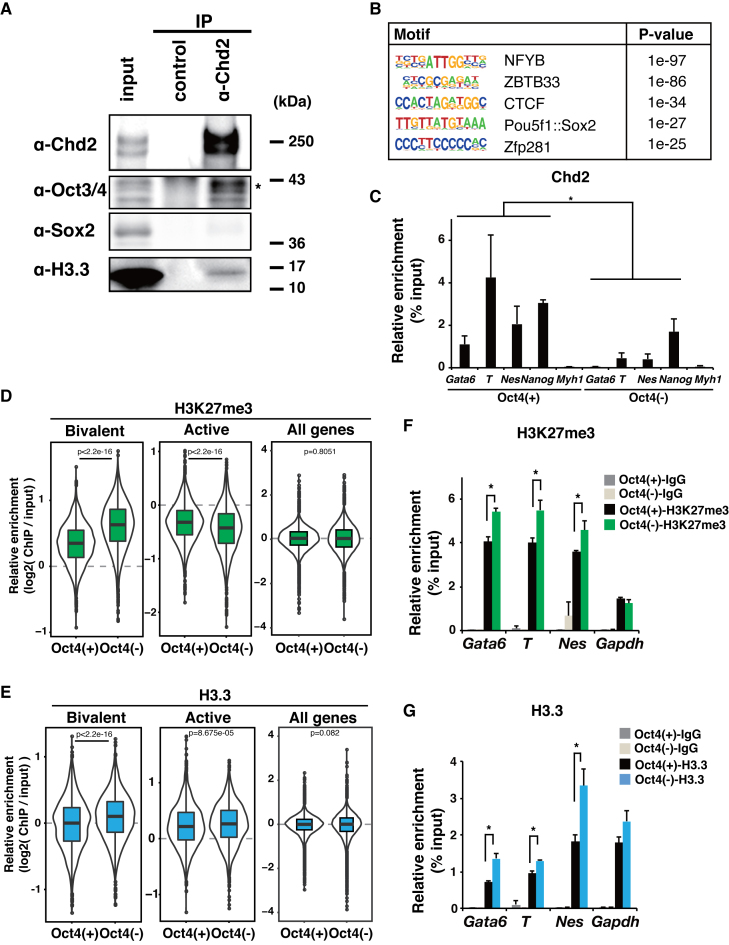
Next, we explored the chromatin state in Chd2mut/mut ESCs because Chd2 has been shown to be involved in chromatin regulation. Our data indicated that Chd2 did not affect the whole transcriptome profile of ESCs (Figure 1E and F). Previous studies also reported that no drastic change in the transcriptome was observed upon depletion of the histone modifiers Eed and Suz12, and also that of histone H3.3 itself (9,48). We first detected Chd2 binding around pluripotency-related genes such as Nanog using chromatin IP followed by sequencing (ChIP-seq). In addition, Chd2 binding was also observed around developmentally regulated genes such as Gata6, T and Nes at lower levels (Figure 3A). Next, Chd2-enriched regions were peak-called by model-based analysis for ChIP-seq (MACS) (42), and 60% of them were enriched in proximity to gene loci (Figure 3B). Moreover, Gene Ontology analysis on Chd2 binding genes showed that development-related terms such as cell fate commitment and differentiation were associated with these genes in the bivalent state (Supplementary Figure S5A and B). Moreover, developmentally regulated genes have been reported to be expressed at lower levels in ESCs, and the chromatin states of the reported genes were found to be bivalent, that is, marked with both active H3K4me3 and repressive H3K27me3 (3,41). Therefore, to examine genome-wide Chd2 co-enrichment with histone modifications, we performed ChIP-seq of histone modifications (H3K27ac, H3K27me3 and H3K4me3) in WT mESCs. As reported previously (3,41), the developmentally regulated genes, Gata6, T and Nes (Figure 2), were found to be located in bivalent loci, and pluripotency-related genes such as Nanog were marked with H3K4me3 (Figure 3A). We analyzed the Chd2 signal intensities by focusing on TSSs (±1 kbp) in the three gene subsets (active, bivalent and silent control genes) defined by Mikkelsen et al. (41). The data showed that the Chd2 signal intensity was higher at active and bivalent genes than at silent control genes (Figure 3C). Furthermore, we evaluated Chd2 enrichment at the developmentally regulated genes after differentiation by ChIP-qPCR with anti-Chd2 antibody using EBs. The results showed that the Chd2 enrichment level remained higher at the developmentally regulated genes such as Gata6, T and Nes, while it was low in the silent gene Myh1 (P = 0.026, 0.0012 and 0.0030) (Figure 3D). These findings indicated that Chd2 remained enriched after differentiation. A previous study suggested that Chd2 binding occurred at TSSs and was enriched at active genes (51). Here, we found that Chd2 was also enriched at bivalent genes, albeit at lower signal intensities and that some of the developmentally regulated genes suppressed by Chd2 depletion were in the bivalent state. These results showed that Chd2 is also bound to developmentally regulated gene loci in the bivalent state, in addition to active genes, in undifferentiated ESCs.
Figure 3.
Chd2-dependent developmental genes are in the bivalent state in undifferentiated mESCs. ChIP-seq data of histone modifications were obtained from WT ESCs and analyzed with the ENCODE dataset of Chd2 in mESCs. (A) Genome browser tracks representing Chd2, H3K27ac, H3K27me3 and H3K4me3 occupancy around bivalent genes (Gata6, T and Nes) and an active gene (Nanog). Read counts were normalized to the total number of reads for each dataset. (B) Left bar chart shows the distribution of Chd2-binding regions in the mm9 genome annotated by HOMER (43). Right bar chart shows the reference proportion of the mm9 genome. (C) ChIP-seq enrichment of Chd2 mapped around TSSs (±1 kbp) in each gene subset. (D) ChIP-qPCR assay using anti-Chd2 antibodies in EBs derived from WT ESCs. Both bivalent genes (Gata6, T and Nes) and a silent gene (Myh1) were analyzed. Recovery efficiency (mean ± standard deviation of three independent experiments) is expressed as enrichment relative to the input. *P < 0.05.
Chd2 prevents suppressive chromatin formation at Chd2-dependent developmental genes
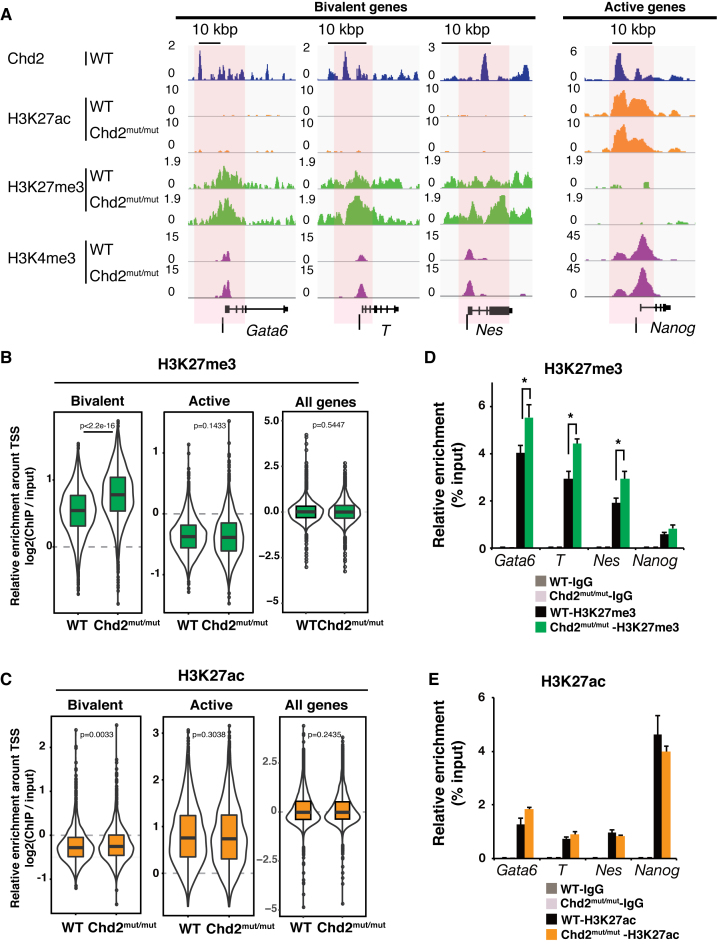
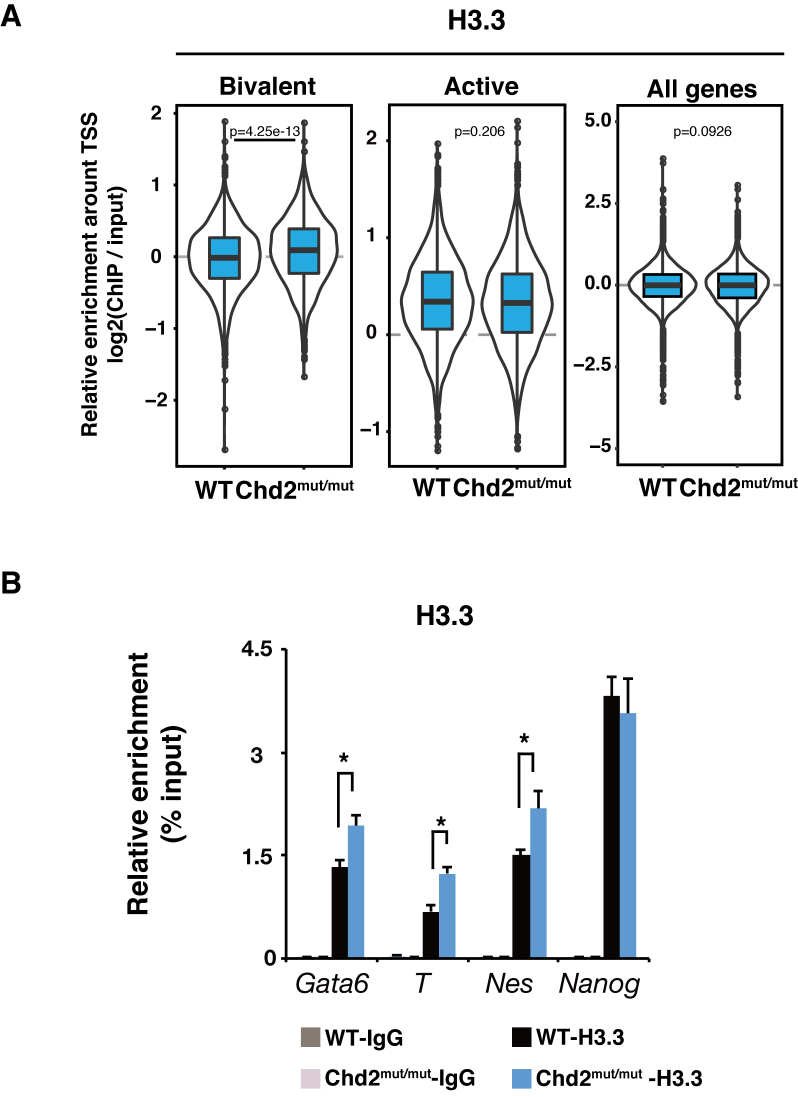
To understand how Chd2 depletion changes the chromatin landscape, we performed ChIP-seq of histone modifications in Chd2mut/mut ESCs and compared the results with those in WT ESCs. We found that repressive H3K27me3 mark enrichment was significantly increased at bivalent genes upon Chd2 depletion (P < 2.2e−16), while the enrichment levels showed no significant change globally (Figure 4A and B). Although Chd2 binding was also observed in the active state, we observed little change in H3K27ac and H3K4me3 enrichment at active genes (Figure 4C and Supplementary Figure S6A). The major effect of Chd2 depletion on histone modification was on H3K27me3 at the bivalent genes (Supplementary Table S4). ChIP-qPCR (n = 3) of histone modifications confirmed the increases of H3K27me3 levels around developmentally regulated genes [e.g. Gata6 (P = 0.036), T (P = 0.0076) and Nes (P = 0.0073)] and little change in H3K4me3 and H3K27ac levels around pluripotency-related genes [e.g. Nanog (P = 0.47 and 0.32, respectively)] (Figure 4D and E; Supplementary Figure S6B). These results are compatible with our data indicating that Chd2 depletion did not affect transcription in undifferentiated ESCs and suppressed the upregulation of developmentally regulated genes upon differentiation (Figures 1E, F and 2B, D). ChIP-qPCR showed that the enrichment of H3.3 and H3K4me3 was significantly suppressed at bivalent genes such as Gata6, T and Nes by Chd2 depletion after differentiation. This is consistent with the suppressed expression of these genes in Chd2mut/mut cells (Supplementary Figure S6C–E). Taken together, our data suggest that Chd2 functions to tune H3K27me3 levels and prevent suppressive chromatin formation at the bivalent-state chromatin in undifferentiated ESCs to maintain proper differentiation potential.
Figure 4.
Chd2 prevents suppressive chromatin formation at Chd2-dependent developmental genes. ChIP-seq data of histone modifications were compared between WT and Chd2mut/mut ESCs. (A) Genome browser representing H3K27ac, H3K27me3 and H3K4me3 occupancy around bivalent state genes (Gata6, T and Nes) and an active state gene (Nanog) in WT and Chd2mut/mut ESCs. Black bars indicate the regions selected for ChIP-qPCR. (B) Box plots show H3K27me3 and (C), H3K27ac ChIP-seq signal intensities for the bivalent, active, and all genes at the TSS ± 5 kbp. The Wilcoxon test was used for statistical analysis. (D) ChIP-qPCR assay using anti-H3K27me3 and (E) anti-H3K27ac antibodies. Both bivalent genes (Gata6, T and Nes) and an active gene (Nanog) were analyzed in WT and Chd2mut/mut ESCs. Recovery efficiency (mean ± standard deviation of three independent experiments) is expressed as enrichment relative to the input. *P < 0.05.
Chd2 regulates H3.3 occupancy in developmental gene loci
We next attempted to understand how Chd2 depletion leads to increased H3K27me3 levels at developmentally regulated genes. Banaszynski et al. reported that H3.3 depletion leads to reductions in the H3K27me3 levels at bivalent gene loci in mESCs (9). In addition, we and others previously demonstrated the involvement of Chd2 in the regulation of chromatin via incorporation of the histone variant H3.3 (19,52). Therefore, we hypothesized that Chd2 regulates H3.3 occupancy at developmentally regulated genes in ESCs. We analyzed the global association between H3.3 and H3K27me3 using a PLA (53,54) in WT and Chd2mut/mut mESCs. Substantial association of H3.3 with both H3K27me3 and H3K4me3 was observed in WT mESCs, which is consistent with previous studies (7–9). We found that the interaction between H3.3 and H3K27me3 was significantly increased in Chd2mut/mut mESCs compared with that in WT mESCs (P = 6.7 × 10−10) (Supplementary Figure S7A). Next, to determine whether Chd2 regulates local H3.3 occupancy at developmentally regulated genes in the bivalent state, we first evaluated genome-wide H3.3 incorporation by ChIP-seq of H3.3 in WT and Chd2mut/mut ESCs. The data revealed that H3.3 had accumulated at both bivalent and active genes in WT mESCs (Supplementary Figure S8A). These results showed that Chd2 depletion caused a significant increase in H3.3 enrichment at bivalent genes, which was in contrast to the small changes observed at active genes, while no significant change in enrichment levels was observed globally (Figure 5A). We further evaluated H3.3 density normalized with total nucleosome occupancy using ChIP-seq of total H3. The data showed that H3.3 density was still increased at the bivalent genes upon Chd2 depletion (P = 0.0006), whereas a drastic change of H3.3 density was not detected at the active genes and genome-wide control (P = 0.96). Although not statistically significant, H3 enrichment was also found to be increased (Supplementary Figure S7B and C). Overall, these findings indicate that Chd2 regulates the proportion of H3.3 in chromatin at developmentally regulated genes in the bivalent state. The increase of H3.3 enrichment at developmentally regulated genes was confirmed by ChIP-qPCR (n = 3) [e.g. Gata6 (P = 0.016),T (P = 0.0045) and Nes (P = 0.017)] (Figure 5B). Overall, these findings indicate that Chd2 regulates the proportion of H3.3 in the nucleosomes at developmentally regulated genes in the bivalent state.
Figure 5.
H3.3 occupancy in the bivalent state is regulated by Chd2. (A) Box plots show H3.3 ChIP-seq signal intensities for bivalent, active and all genes at the TSS ± 5 kbp in WT and Chd2mut/mut ESCs. The Wilcoxon test was used for statistical analysis. (B) ChIP-qPCR assay using an anti-H3.3-specific antibody. Both bivalent state genes (Gata6, T and Nes) and an active state gene (Nanog) were analyzed in WT and Chd2mut/mut ESCs. Recovery efficiency (mean ± standard deviation of three independent experiments) is expressed as enrichment relative to the input. *P < 0.05.
Chd2 is associated with Oct3/4 for H3.3 deposition in developmental gene loci
Because Chd2 has not been shown to bind to specific DNA sequences, we next investigated a carrier for recruitment of Chd2 into the genome. We have recently reported that Chd2 could be recruited by the myogenic transcription factor MyoD in myogenic progenitor cells (19). Therefore, we hypothesized that Chd2 might interact with a master transcription factor in ESCs. We screened for binding factors of Chd2 in mESCs using the candidate approach by co-IP of Chd2. Oct3/4, but not Sox2, was identified as a Chd2 co-IP product in mESCs (Figure 6A and Supplementary Figure S8B). We further performed reciprocal co-IP of Oct3/4 and identified Chd2 as an Oct3/4 co-IP product (Supplementary Figure S8C). Next, we further examined whether the Oct3/4 binding sequence was present in the Chd2-binding regions around bivalent genes using HOMER (43). As we expected, the motif annotated to Pou5f1 binding sequences was significantly enriched (P = 1 × 10−27) in Chd2-binding regions around bivalent genes (Figure 6B). To confirm the co-recruitment of Chd2 and Oct3/4, we analyzed the genome-wide distribution of Oct3/4 using the ChIP-seq data (37). We identified Oct3/4 ChIP-seq peaks using MACS and found that a significant proportion (43%) of Chd2-binding genes overlapped with Oct3/4-binding genes. The overlapping genes included Chd2-dependent developmentally regulated genes (e.g. Gata6, T and Nes) (Supplementary Figure S8D). Moreover, the data showed Oct3/4 binding around TSSs of both bivalent and active genes (Supplementary Figure S8A). These findings suggest that Oct3/4 might be involved in Chd2 functions. Next, to evaluate Oct3/4 involvement in H3.3 deposition which was found to be correlated with Chd2 as described above, we performed PLA using ZHBTc4 cells in which Oct3/4 expression can be suppressed by doxycycline treatment (22). Western blot analysis showed a reduction in the Oct3/4 protein level after 48 h of doxycycline treatment, with little change in Chd2 and H3.3 expression (Supplementary Figure S9A). These results showed that the interaction between H3.3 and H3K27me3 was significantly increased upon Oct3/4 depletion (P = 0.015) in contrast to the decrease of the interaction between Chd2 and H3K27me3 (P = 0.0096) (Supplementary Figure S9B). Moreover, to evaluate the requirement of Oct3/4 for the recruitment of Chd2, ChIP-qPCR of Chd2 was performed at the bivalent genes with and without Oct3/4 depletion. The results showed that the recruitment of Chd2 was diminished by Oct3/4 depletion (P = 0.013, 0.037 and 0.037), suggesting that Oct3/4 was required for the recruitment of Chd2 to the bivalent genes (Figure 6C). Finally, we performed ChIP-seq of H3K27me3 and H3.3. The results revealed an increase in H3K27me3 and H3.3 enrichment at bivalent genes upon Oct3/4 depletion, which was almost identical to the effect of Chd2 depletion (Figure 6D and E). Although slight changes were detected at the active genes, the major effects of Oct3/4 depletion on H3K27me3 and H3.3 were observed at the bivalent genes according to effect sizes (Supplementary Table S4). ChIP-qPCR (n = 3) confirmed the increases of H3K27me3 and H3.3 levels around developmentally regulated genes such as Gata6, T and Nes (Figure 6F and G). Moreover, significant overlaps were observed between the genes with higher enrichment levels by Chd2 depletion and by Oct3/4 depletion in both H3K27me3 and H3.3 ChIP-seq (Supplementary Figure S10A–D). These findings suggest that Oct3/4 is involved in Chd2 recruitment to regulate H3.3 occupancy at bivalent genes.
Figure 6.
Chd2 is associated with Oct3/4 for H3.3 deposition in developmental gene loci. (A) Immunoprecipitations were performed from WT ESC extracts using a Chd2-specific antibody or IgG as a control. Western blot analyses of the products using antibodies against Chd2, Oct3/4, Sox2 and H3.3 are shown. (B) Motif analysis of the Chd2-binding region around the bivalent genes (±50 kbp) using HOMER (43). (C) ChIP-qPCR assay using anti-Chd2 antibodies at bivalent genes (Gata6, T, Nes, Nanog and Myh1) in WT and Oct3/4-depleted mESCs. Recovery efficiency (mean ± standard deviation of three independent experiments) is expressed as enrichment relative to the input. *P < 0.05. (D) Box plots show H3K27me3 and (E) H3.3 ChIP-seq signal intensities for bivalent and active state genes at the TSS ± 5 kbp in ZHBTc4 cells with or without doxycycline treatment. The Wilcoxon test was used for statistical analysis. (F). ChIP-qPCR assay using anti-H3K27me3 and (G) anti-H3K27ac antibodies. Both bivalent genes (Gata6, T and Nes) and an active gene (Nanog) were analyzed in WT and Oct3/4-depleted ESCs. Recovery efficiency (mean ± standard deviation of three independent experiments) is expressed as enrichment relative to the input. *P < 0.05.
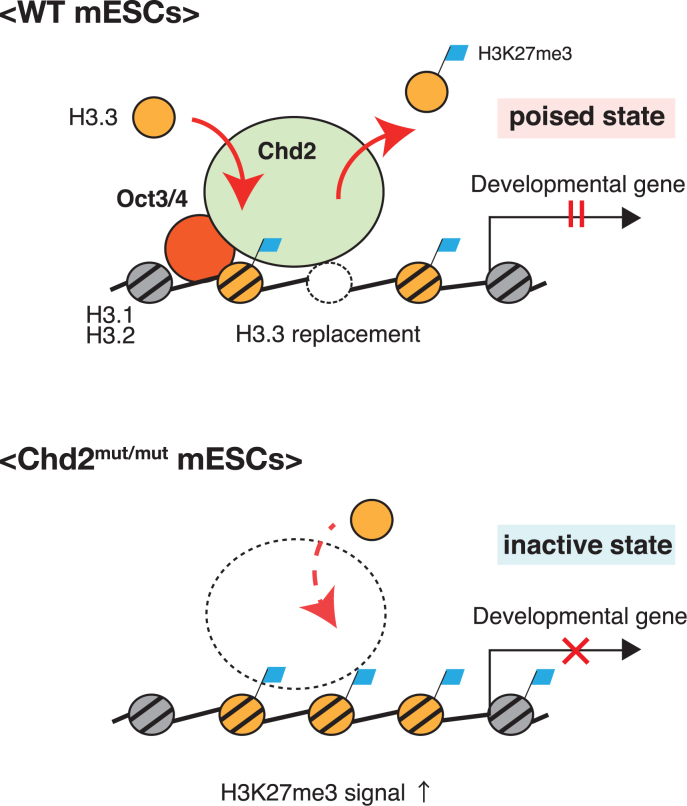
Taken together, these results indicate that Chd2 associates and may collaborate with Oct3/4 in the regulation of H3.3 deposition at developmentally regulated genes in ESCs. Because a previous study showed that H3K27me3 enrichment requires H3.3 at bivalent genes (9), the increase of H3.3 enrichment appears to lead to the subsequent increase of H3K27me3 enrichment at developmentally regulated genes in Chd2mut/mut ESCs. In our model, developmentally regulated genes are localized in the bivalent state that is regulated by Chd2. Bivalent-state chromatin may be maintained in a flexible form by Chd2 regulating the incorporation or eviction of H3.3 with appropriate histone modification levels in undifferentiated ESCs. This bivalent state of chromatin may be changed to the suppressive state with H3.3 accumulation and elevated H3K27me3 levels induced by Chd2 depletion. Thus, Chd2-mediated maintenance of the bivalent state enables proper gene expression in response to differentiation stimuli and subsequent normal differentiation (Figure 7).
Figure 7.
Model of Chd2 in regulation of the chromatin structure in the bivalent state for expression of developmentally regulated genes upon differentiation. Chd2 maintains a proper proportion of nucleosomes in the bivalent state for the expression of developmentally regulated genes upon differentiation.
DISCUSSION
In this study, we found that the loss of Chd2 in mESCs results in the decrease of expression in differentiation-related genes after differentiation through a shift to a suppressive chromatin structure. To analyze the role of Chd2 in this process, we depleted Chd2 by the genome-editing method in mESCs. Chd2 depletion altered the proportion of nucleosomes to increase H3.3 enrichment, leading to elevation of the H3K27me3 level in the bivalent state. These changes in the chromatin landscape in turn led to suppression of the expression of developmentally regulated genes upon differentiation and subsequent differentiation defects, despite having little effect on gene expression before differentiation. Taken together, our findings suggest that Chd2 plays a key role in maintaining the differentiation potential via the formation of a proper chromatin structure for appropriate gene expression in response to differentiation stimuli in ESCs.
We found that Chd2-depleted mESCs demonstrated a defect in the expression of early developmentally regulated genes, whereas a previous report showed that Chd2-deficient mice displayed perinatal lethality (21). In this previous study, the Chd2-deficient mice were not knockout mice but had instead been established from mESCs with retrotransposon-induced mutations. As a result, the truncated Chd2 protein might still have functioned on chromatin, causing a milder phenotype than that observed in the current study.
The reduction of H3.3 in mESCs was previously reported to cause a decrease in nucleosome turnover and H3K27me3 levels at bivalent loci with subsequent upregulation of developmental markers upon differentiation (9). Interestingly, these changes upon H3.3 depletion are consistent with the phenotype of our Chd2-depleted ESCs with enhanced H3.3 enrichment at bivalent loci. Previous studies have shown that chromatin-remodeling enzymes of the Chd family cause spaced nucleosomes (52,55). Therefore, the nucleosomes at the bivalent state in H3.3-depleted ESCs probably remain spaced due to the presence of Chd2. In this case, the lack of newly incorporated H3.3 may result in a decrease of nucleosome density and hence upregulation of developmental markers. In the current study, the loss of Chd2 might have caused less flexibility in nucleosome turnover and resulted in H3.3 accumulation because of other H3.3-incorporating factors such as Chd1 or Hira. Chd2 might regulate the balance of assembly or disassembly of H3.3 at the bivalent loci, thereby maintaining proper differentiation potential.
A previous study reported a decrease in the incorporation of H3.3 in myogenic progenitor cells at myogenic genes upon Chd2 reduction (19). Although the defect in differentiation potential was common to Chd2 depletion in both C2C12 cells and mESCs, the changes in H3.3 occupancy and histone modification at the bivalent loci were unexpectedly different. One reason for the differences between the two cell types might be the abundance of chromatin-remodeling enzymes in ESCs. Although H3.3 incorporation has been reported to be modulated by other proteins, such as the closely related chromatin-remodeling enzyme Chd1 and histone chaperone Hira in ESCs (8,18), Chd2 might be a critical factor for H3.3 incorporation at the bivalent loci in C2C12 cells. Because the function of Chd2 in preventing suppressive chromatin formation at bivalent loci might be shared in common in the two situations, the subsequent defect in differentiation potential upon Chd2 depletion was similarly observed in both cell types. Interactions between Chd2 and H3.3 have also been indicated in other cell types. Chd2 knockdown has been reported to be associated with significantly elevated H3 occupancy and reduced H3.3 enrichment in DNase-hypersensitive sites of K562 cells (51). In addition, a recent study revealed that Chd2 triggers the deposition of H3.3 at sites of DNA damage, which promotes efficient DNA repair in U2OS cells (52). These findings suggest that the contribution to H3.3 deposition by Chd2-mediated chromatin remodeling and subsequent effects might vary according to the genomic location and cell type.
We observed changes in the nucleosome configuration only in the bivalent state upon Chd2 depletion, although Chd2 binding was observed around both bivalent and active state genes. The effect of Chd2 depletion on H3.3 incorporation and H3K4me3 at active gene loci was not observed, which is consistent with the fact that transcription was not affected. This suggests that chromatin regulation at the active genes could be independent of the function of Chd2. The reason for these results might be the functional redundancy of other chromatin-remodeling enzymes that are highly expressed in ESCs. For example, INO80, a member of another family of chromatin-remodeling enzymes, maintains an open chromatin architecture and is thus essential for the core pluripotency transcription circuit (47). The mild phenotype of Chd2mut/mut ESCs might be a result of the abundant redundancy in the active state and the major contribution of Chd2 to the maintenance of bivalent state chromatin in undifferentiated ESCs. Furthermore, owing to the unexpectedly mild phenotype, we propose a model in which regulation of the chromatin structure in the bivalent state by chromatin-remodeling enzymes determines the differentiation potential of ESCs without involvement of the core pluripotency transcription circuit.
We also found that Oct3/4, one of the master transcription factors in ESCs, associated with Chd2. In addition to maintaining pluripotency as a member of the core pluripotency transcription circuit, Oct3/4 has been reported to regulate developmental genes in ESCs (56,57). Furthermore, Oct3/4 was previously shown to interact with other chromatin-remodeling enzymes such as INO80 at pluripotency-related genes in ESCs (47). The current study revealed that Oct3/4 participated in the regulation of the bivalent genes by interacting with chromatin-remodeling enzymes in ESCs. Furthermore, the recent study mentioned above showed that Chd2 is associated with the myogenic transcription factor MyoD in myogenic progenitor cells (19). This might be a common model in which cell-type-specific transcription factors regulate nucleosomes by recruiting Chd2 in individual cell types. Because not all Chd2 binding was co-localized with Oct3/4 in our results, other transcription factors might regulate Chd2 deposition in ESCs. Collectively, the comprehensive combinations among transcription factors and chromatin-remodeling enzymes expressed in ESCs might facilitate the appropriate chromatin landscape necessary to maintain the unique functions of ESCs.
We report that Chd2 prevents the formation of suppressive chromatin in the bivalent state, and that the Chd2-mediated chromatin structure is associated with the proper expression of developmentally regulated genes. Recently, Chd2 mutations have been detected in some human diseases, including pediatric neurodevelopmental disorders and some hematological malignancies in adults (58,59). The defect in differentiation potential due to Chd2 dysfunction in chromatin organization could be one conceivable pathogenesis of these diseases. Thus, our findings demonstrate a novel role of Chd2 in normal development and may shed light on the molecular mechanisms underlying certain types of developmental disorders and cancers.
ACCESSION NUMBER
ChIP-seq and RNA-seq datasets have been deposited with the following accession code: GSE96662.
Supplementary Material
ACKNOWLEDGEMENTS
We thank T. Tachibana (Osaka City University) and H. Kimura (Tokyo Institute of Technology) for supplying antibodies, H. Niwa and D. Konno (RIKEN) for supplying the mESC lines (EB5 and ZHBtc4), H. Fujii, T. Ichinose, M. Kato, N. Urasaki and Y. Nakajo for technical support, and staff at the Advanced Computational Scientific Program of Research Institute for Information Technology, Kyushu University, and the National Institute of Genetics (NIG) for providing the high-performance computing resources.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JST CREST [JPMJCR16G1]; MEXT/JSPS KAKENHI [25116010, 25132709, 25118518, 26290064 to Y.O., 16H01219, 15K18457 to A.H., 16K18479, 16H01577, 16H01550 to K.M.]. Funding for open access charge: MEXT/JSPS KAKENHI [25116010, 25132709, 25118518, 26290064 to Y.O., 16H01219, 15K18457 to A.H., 16K18479, 16H01577, 16H01550 to K.M.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gaspar-Maia A., Alajem A., Meshorer E.. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 2011; 12:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T.. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006; 10:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. . A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 4. Pasini D., Bracken A.P., Hansen J.B., Capillo M., Helin K.. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007; 27:3769–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G.. Genome regulation by polycomb and trithorax proteins. Cell. 2007; 128:735–745. [DOI] [PubMed] [Google Scholar]
- 6. Hake S.B., Allis C.D.. Histone H3 variants and their potential role in indexing mammalian genomes: the ‘H3 barcode hypothesis’. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:6428–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wirbelauer C., Bell O., Schübeler D.. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 2005; 19:1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goldberg A.D., Banaszynski L.A., Noh K.-M., Lewis P.W., Elsaesser S.J., Stadler S., Dewell S., Law M., Guo X., Li X. et al. . Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010; 140:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Banaszynski L.A., Wen D., Dewell S., Whitcomb S.J., Lin M., Diaz N., Elsässer S.J., Chapgier A., Goldberg A.D., Canaani E. et al. . Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013; 155:107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Elsässer S.J., Noh K.-M., Diaz N., Allis C.D., Banaszynski L.A.. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015; 522:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Narlikar G.J., Sundaramoorthy R., Owen-Hughes T.. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013; 154:490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Petty E., Pillus L.. Balancing chromatin remodeling and histone modifications in transcription. Trends Genet. 2013; 29:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ho L., Crabtree G.R.. Chromatin remodelling during development. Nature. 2010; 463:474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D.J., Dash C., Bazett-Jones D.P., Le Grice S., McKay R.D.G., Buetow K.H. et al. . Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008; 2:437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Dieuleveult M., Yen K., Hmitou I., Depaux A., Boussouar F., Dargham D.B., Jounier S., Humbertclaude H., Ribierre F., Baulard C. et al. . Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature. 2016; 530:113–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hayashi M., Maehara K., Harada A., Semba Y., Kudo K., Takahashi H., Oki S., Meno C., Ichiyanagi K., Akashi K. et al. . Chd5 regulates MuERV-L/MERVL expression in mouse embryonic stem cells via H3K27me3 modification and histone H3.1/H3.2. J. Cell. Biochem. 2015; 117:780–792. [DOI] [PubMed] [Google Scholar]
- 17. Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M.J., Heidersbach A., Ramalho-Santos J., McManus M.T., Plath K., Meshorer E. et al. . Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009; 460:863–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Konev A.Y., Tribus M., Park S.Y., Podhraski V., Lim C.Y., Emelyanov A.V., Vershilova E., Pirrotta V., Kadonaga J.T., Lusser A. et al. . CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007; 317:1087–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harada A., Okada S., Konno D., Odawara J., Yoshimi T., Yoshimura S., Kumamaru H., Saiwai H., Tsubota T., Kurumizaka H. et al. . Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J. 2012; 31:2994–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen T., Ji F., Yuan Z., Jiao J.. CHD2 is required for embryonic neurogenesis in the developing cerebral cortex. Stem Cells. 2015; 33:1794–1806. [DOI] [PubMed] [Google Scholar]
- 21. Marfella C.G.A., Ohkawa Y., Coles A.H., Garlick D.S., Jones S.N., Imbalzano A.N.. Mutation of the SNF2 family member Chd2 affects mouse development and survival. J. Cell. Physiol. 2006; 209:162–171. [DOI] [PubMed] [Google Scholar]
- 22. Niwa H., Miyazaki J.-I., Smith A.G.. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000; 24:372–376. [DOI] [PubMed] [Google Scholar]
- 23. Ogawa K., Matsui H., Ohtsuka S., Niwa H.. A novel mechanism for regulating clonal propagation of mouse ES cells. Genes Cells. 2004; 9:471–477. [DOI] [PubMed] [Google Scholar]
- 24. Kurosawa H. Methods for inducing embryoid body formation: in vitro differentiation system of embryonic stem cells. J. Biosci. Bioeng. 2007; 103:389–398. [DOI] [PubMed] [Google Scholar]
- 25. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. . Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D.. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013; 31:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al. . DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harada A., Maehara K., Sato Y., Konno D., Tachibana T., Kimura H., Ohkawa Y.. Incorporation of histone H3.1 suppresses the lineage potential of skeletal muscle. Nucleic Acids Res. 2014; 43:775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harada A., Yoshimura S., Odawara J., Azuma M., Okada S., Nakamura M., Tachibana T., Ohkawa Y.. Generation of a rat monoclonal antibody specific for Chd2. Hybridoma. 2010; 29:173–177. [DOI] [PubMed] [Google Scholar]
- 31. Nozawa R.-S., Nagao K., Masuda H.-T., Iwasaki O., Hirota T., Nozaki N., Kimura H., Obuse C.. Human POGZ modulates dissociation of HP1α from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 2010; 12:719–727. [DOI] [PubMed] [Google Scholar]
- 32. Ohkawa Y., Harada A., Nakamura M., Yoshimura S., Tachibana T.. Production of a rat monoclonal antibody against Brg1. Hybridoma (Larchmt). 2009; 28:463–466. [DOI] [PubMed] [Google Scholar]
- 33. Odawara J., Harada A., Yoshimi T., Maehara K., Tachibana T., Okada S., Akashi K., Ohkawa Y.. The classification of mRNA expression levels bythe phosphorylation state of RNAPII CTD basedon a combined genome-wide approach. BMC Genomics. 2011; 12:516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kimura H., Hayashi-Takanaka Y., Goto Y., Takizawa N., Nozaki N.. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 2008; 33:61–73. [DOI] [PubMed] [Google Scholar]
- 35. Hayashi-Takanaka Y., Yamagata K., Wakayama T., Stasevich T.J., Kainuma T., Tsurimoto T., Tachibana M., Shinkai Y., Kurumizaka H., Nozaki N. et al. . Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 2011; 39:6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mouse ENCODE Consortium Stamatoyannopoulos J.A., Snyder M., Hardison R., Ren B., Gingeras T., Gilbert D.M., Groudine M., Bender M., Kaul R. et al. . An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 2012; 13:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. et al. . Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008; 133:1106–1117. [DOI] [PubMed] [Google Scholar]
- 38. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mortazavi A., Williams B.A., McCue K., Schaeffer L.. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Methods. 2008; 5:621–628. [DOI] [PubMed] [Google Scholar]
- 40. Maehara K., Ohkawa Y.. agplus: a rapid and flexible tool for aggregation plots. Bioinformatics. 2015; 31:3046–3047. [DOI] [PubMed] [Google Scholar]
- 41. Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R.P. et al. . Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007; 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 45. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:31–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang L., Du Y., Ward J.M., Shimbo T., Lackford B., Zheng X., Miao Y.-L., Zhou B., Han L., Fargo D.C. et al. . INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Stem Cell. 2014; 14:575–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chamberlain S.J., Yee D., Magnuson T.. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008; 26:1496–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lei I., West J., Yan Z., Gao X., Fang P., Dennis J.H., Gnatovskiy L., Wang W., Kingston R.E., Wang Z.. BAF250a protein regulates nucleosome occupancy and histone modifications in priming embryonic stem cell differentiation. J. Biol. Chem. 2015; 290:19343–19352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Itskovitz-Eldor J., Schuldiner M., Karsenti D., Eden A., Yanuka O., Amit M., Soreq H., Benvenisty N.. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol. Med. 2000; 6:88–95. [PMC free article] [PubMed] [Google Scholar]
- 51. Siggens L., Cordeddu L., Rönnerblad M., Lennartsson A., Ekwall K.. Transcription-coupled recruitment of human CHD1 and CHD2 influences chromatin accessibility and histone H3 and H3.3 occupancy at active chromatin regions. Epigenet. Chromatin. 2015; 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luijsterburg M.S., de Krijger I., Wiegant W.W., Shah R.G., Smeenk G., de Groot A.J.L., Pines A., Vertegaal A.C.O., Jacobs J.J.L., Shah G.M. et al. . PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non- homologous end-joining. Mol. Cell. 2016; 61:547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.-J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.-G. et al. . Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Meth. 2006; 3:995–1000. [DOI] [PubMed] [Google Scholar]
- 54. Hattori N., Niwa T., Kimura K., Helin K., Ushijima T.. Visualization of multivalent histone modification in a single cell reveals highly concerted epigenetic changes on differentiation of embryonic stem cells. Nucleic Acids Res. 2013; 41:7231–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lieleg C., Ketterer P., Nuebler J., Ludwigsen J., Gerland U., Dietz H., Mueller-Planitz F., Korber P.. Nucleosome spacing generated by ISWI and CHD1 remodelers is constant regardless of nucleosome density. Mol. Cell. Biol. 2015; 35:1588–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Young R.A. Control of the embryonic stem cell state. Cell. 2011; 144:940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ng H.-H., Surani M.A.. The transcriptional and signalling networks of pluripotency. Nat. Cell Biol. 2011; 13:490–496. [DOI] [PubMed] [Google Scholar]
- 58. Rodríguez D., Bretones G., Quesada V., Villamor N., Arango J.R., López-Guillermo A., Ramsay A.J., Baumann T., Quirós P.M., Navarro A. et al. . Mutations in CHD2 cause defective association with active chromatin in chronic lymphocytic leukemia. 2015; 126:195–202. [DOI] [PubMed] [Google Scholar]
- 59. Allen A.S., Consortium E., Berkovic S.F., Cossette P., Project E.P.G., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T. et al. . De novo mutations in epileptic encephalopathies. Nature. 2013; 501:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.