Abstract
The FANCJ DNA helicase is linked to hereditary breast and ovarian cancers as well as bone marrow failure disorder Fanconi anemia (FA). Although FANCJ has been implicated in the repair of DNA double-strand breaks (DSBs) by homologous recombination (HR), the molecular mechanism underlying the tumor suppressor functions of FANCJ remains obscure. Here, we demonstrate that FANCJ deficient human and hamster cells exhibit reduction in the overall gene conversions in response to a site-specific chromosomal DSB induced by I-SceI endonuclease. Strikingly, the gene conversion events were biased in favour of long-tract gene conversions in FANCJ depleted cells. The fine regulation of short- (STGC) and long-tract gene conversions (LTGC) by FANCJ was dependent on its interaction with BRCA1 tumor suppressor. Notably, helicase activity of FANCJ was essential for controlling the overall HR and in terminating the extended repair synthesis during sister chromatid recombination (SCR). Moreover, cells expressing FANCJ pathological mutants exhibited defective SCR with an increased frequency of LTGC. These data unravel the novel function of FANCJ helicase in regulating SCR and SCR associated gene amplification/duplications and imply that these functions of FANCJ are crucial for the genome maintenance and tumor suppression.
INTRODUCTION
Genomic instability in the form of chromosomal aberrations, gene amplification and loss of heterozygosity (LOH) are commonly found in various types of cancer (1,2). DNA double-strand breaks (DSBs) that are generated endogenously or by exogenous sources are considered as the primary cause of chromosome instability (3,4). DSBs that arise during replication are preferentially repaired by sister chromatid recombination (SCR) (5), a recombinational repair pathway that utilizes neighbouring intact sister chromatid as a template. Since the copied information is accurate, SCR is potentially an accurate pathway in repairing the DSBs and in maintaining the genome integrity (6–8). Tumor suppressor genes such as BRCA1, BRCA2, RAD51C and XRCC2 have been shown to control DSB repair by SCR (9–12), implying that dysregulation in SCR could lead to tumorigenesis.
Fanconi anemia (FA) pathway plays an important role in the repair of DNA inter-strand crosslinks (ICLs). To date, 21 genes including BRCA1, BRCA2 and FANCJ have been identified to participate in the FA pathway of ICL repair (13–15). FANCJ helicase was originally identified as a BRCA1 interacting C-terminal helicase (BACH1) (16), and was subsequently identified as a gene mutated in the J complementation group of FA (17–19), a rare chromosome instability genetic disorder characterized by bone marrow failure, skeletal abnormalities and predisposition to various types of cancer (20–22). In addition, monoallelic mutations in FANCJ confer familial breast and ovarian cancer similar to BRCA1 and BRCA2 germline mutations but with lower penetrance (23–25). FANCJ localizes to the sites of DSBs (16,26), and cells deficient in FANCJ exhibit significant sensitivity to DSB inducing agents (19,27–29). A biochemical study shows that FANCJ helicase unwinds D-loop (30), an intermediate in the homologous recombination (HR) pathway of DSB repair. Direct measurement of HR by a reporter also indicates that FANCJ participates in the repair of DSBs (19). Recent studies imply that FANCJ participates in the generation of ssDNA for the initiation of HR (29). Given that FANCJ unwinds D-loop structures (30), it is likely that FANCJ may also participate in the termination of HR by promoting synthesis dependent strand annealing (SDSA) mechanism of DSB repair. However, the precise role and the mechanism by which FANCJ participates in the repair of DSBs via HR remains elusive.
In mammalian cells, gene conversions typically extend <100 bp termed short-tract gene conversions (STGC) (31–33). However, minority of HR events entail several kilobases of nascent strand synthesis that are defined as long-tract gene conversions (LTGC) (11,34,35). LTGC is an error-prone form of HR as it could lead to tandem gene duplication and gene amplifications that are frequently found in many types of cancer (35,36). Mammalian cells lacking RAD51 paralogs (RAD51C, XRCC2 and XRCC3) exhibit reduced HR and an increased frequency of LTGC events in response to I-SceI endonuclease induced DSBs (10,11). Notably, the BRCA1 tumor suppressor has been shown to suppress LTGC at a site specific replication fork barrier as well as at I-SceI induced DSBs (5,9). However, the molecular mechanism by which RAD51 paralogs or BRCA1 control mammalian gene conversions (GC) is unclear. Here, using a previously characterized SCR reporter, we demonstrate that FANCJ deficient human and hamster cells exhibit a reduction in overall GC. Notably, GC tracts were biased in favour of LTGC in FANCJ depleted cells compared to control cells. Qualitative analysis shows that GFP-triplication, an outcome of LTGC was abundantly present in FANCJ depleted cells. Our analyses of FANCJ mutant that is deficient in its interaction with BRCA1 and helicase dead mutants reveal mechanistic insights into the fine balance of STGC and LTGC at the damaged chromatin. Moreover, cells expressing FANCJ pathological mutants exhibited defective SCR with an increased frequency of LTGC. Collectively, our data reveal that FANCJ dysfunction leads to aberrant amplification during SCR which might contribute to FA and breast/ovarian cancer predisposition.
MATERIALS AND METHODS
Cell lines, cell culture and transfections
U2OS SCR18, U2OS SCR 35S and V79B SCR55 were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in humidified air containing 5% CO2. All plasmid transfections for transient expression in U2OS SCR18 and U2OS SCR35S were performed at 300 V and 950 μF and in V79B SCR55 at 250V and 950 μF using a Bio-Rad Gene Pulsar X cell.
DNA constructs
The pcDNA3-myc-His FANCJ cDNA was a kind gift from Dr. Sharon Cantor. FANCJ shRNA#1 and #2 constructs (pRS shFANCJ) were generated using the previously reported sequences (16) (Supplementary Table S1) and cloned into the pRS shRNA vector. FANCM and BRCA1 shRNA plasmids were generated using the previously reported sequences (37,38) (Supplementary Table S1) and cloned into the pRS shRNA vector. Hemagglutinin (HA)-6xHis-tagged human wild-type (wt) FANCJ and its mutant constructs were generated by overlap extension PCR method using primers sequences indicated in Supplementary Table S2 and cloned into the modified pcDNA3β vector using EcoRV and XhoI restriction sites. The FANCJ shRNA resistant wt and mutant constructs were designed by introducing silence mutations in the FANCJ shRNA#1 sequence (Supplementary Table S2). These primers were used for restriction free cloning with PfuTurbo polymerase. The design and construction of SCR reporter and I-SceI expression vector has been described previously (35).
Cell survival assays
Cell survival assays were carried out as described previously (39). Briefly, cells were mock-treated or treated with MMC for 6–8 h, followed by recovery in fresh medium. 4–5 days post recovery, cell survival was monitored by MTT assay using microplate reader (VersaMax ROM version 3.13). Percent cell survival was calculated as treated cells/untreated cell × 100.
G2/M accumulation assay
G2/M accumulation assays were carried out as described previously (12). Exponentially growing U2OS cells transfected with either control shRNA or FANCJ shRNA plasmids were incubated in 100 ng/ml MMC for 6–8 h and later recovered in fresh media. For rescue experiments, the wt FANCJ construct was transfected 24 h post FANCJ shRNA plasmid transfection followed by incubation in MMC. 48–72 h post recovery, cells were collected and single-cell suspensions were fixed overnight with 70% ethanol in PBS at –20°C. After centrifugation, the cells were incubated with 0.10 mg/ml RNaseA (Fermentas) in PBS at 42°C for 4 h and then incubated for 10 min with propidium iodide (PI) (1 mg/ml) in the dark. A total of 1 × 104 cells were analysed by Verse flow cytometer (BD Biosciences). Aggregates were gated out, and percentages of cells with 2N and 4N DNA content were calculated using FACSDiva version 6.1.1 software (BD Biosciences).
Recombination assays
HR assays were performed as described previously (11). In brief, U2OS SCR18 cells were transfected with either FANCJ shRNA plasmid or various FANCJ cDNAs (WT or mutant) or control pcDNA3β vector. After 24 h, 2 × 106 cells were further transfected with 24 μg of I-SceI expression plasmid. Forty eight hours later, GFP+ cells were scored by FACS analysis using BD biosciences Verse flowcytometer. In each experiment, the percentage of GFP+ cells (either I-SceI induced or uninduced) was measured in triplicate samples, and I-SceI-transfected values were corrected for transfection efficiency (∼60–70%). The spontaneous GFP+ frequency (<0.01%) was subtracted from this value to obtain the I-SceI-induced GFP+ frequency. To quantitatively analyze the SCR events, U2OS SCR18 cultures transiently transfected with either I-SceI or control pcDNA3β vector, were trypsinized, counted, and replated at 2 × 105 to 4 × 105 cells per 10 cm plate. 24 h post plating, 10 μg/ml blasticidin (BSD) was added and colonies were selected for 2 weeks. Later, the cells were fixed with fixative (ethanol:acetone), stained with crystal violet and the colonies were counted for analysis. Plating efficiency was determined in parallel for each experiment by plating defined numbers of cells, allowing colonies to form without antibiotic selection. The absolute I-SceI induced BsdR+ frequency was corrected for transfection efficiency and plating efficiency, followed by negating the spontaneous BsdR+ frequency (was always <0.001%). To qualitatively analyse the SCR events, Southern blotting was performed as described previously (11). In U2OS SCR35S cells, GFP+ and GFP+ RFP+ frequencies were measured 72 h post-transfection by FACS using BD FACS ARIA FUSION in triplicates and corrected for transfection efficiency and background events. Data represent the mean of at least three independent experiments with SD values indicated by error bars. Statistical analysis was by two-tailed paired t-test (unknown variance).
Crossover/non crossover recombination analysis
Crossover/non crossover recombination events were analysed as described previously (11). Briefly, I-SceI transfected U2OS SCR18 cells were plated at a density of 5 × 105 cells per 15 cm dishes to yield single-cell colonies. 48 h later, cells were pulse treated (24 h) with 10 μg/ml BSD and subsequently grown in normal media lacking BSD. After two weeks, sectored colonies containing GFP+ and GFP− cells were identified by microscopic screening and these colonies were isolated and expanded for Southern blotting analysis.
Immunoprecipitation, western blotting and antibodies
Immunoprecipitation and immunoblotting was carried out as described previously (40). Briefly, cells were harvested and lysed in NTEN buffer (0.5% NP40, 50 mM Tris–HCl pH 8.0, 2 mM EDTA, 150 mM NaCl and 10% glycerol) supplemented with a complete protease inhibitor cocktail (Roche). For the immunoprecipitation assays, 1mg protein from the cell lysates was incubated with anti-HA (Roche) antibody and separated using protein G beads. The proteins were resolved on a 6% SDS-PAGE gel and transferred onto PVDF membrane (Millipore). The membranes were blocked using 5% skimmed milk in TBST buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, and 0.1% Tween-20) and incubated with primary antibody for 12 h at 4°C. The primary antibodies against FANCM (1:50, sc-101389), BRCA-1 (1:100, sc-6954), MCM3 (1: 500, sc-365616) that were used for western blot analysis were purchased from Santa Cruz. Anti-MRE11 (1:2000, 611366) antibodies were obtained from BD Biosciences; and the rabbit polyclonal antibody for FANCJ (1:1000, ab49657) was obtained from Abcam. Anti-HA antibody (1:2000, 12CA5) was obtained from Roche. The membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies, developed by chemiluminescence, and imaged using Chemidoc (GE healthcare LAS 4000).
RESULTS
FANCJ controls the fine balance between STGC and LTGC
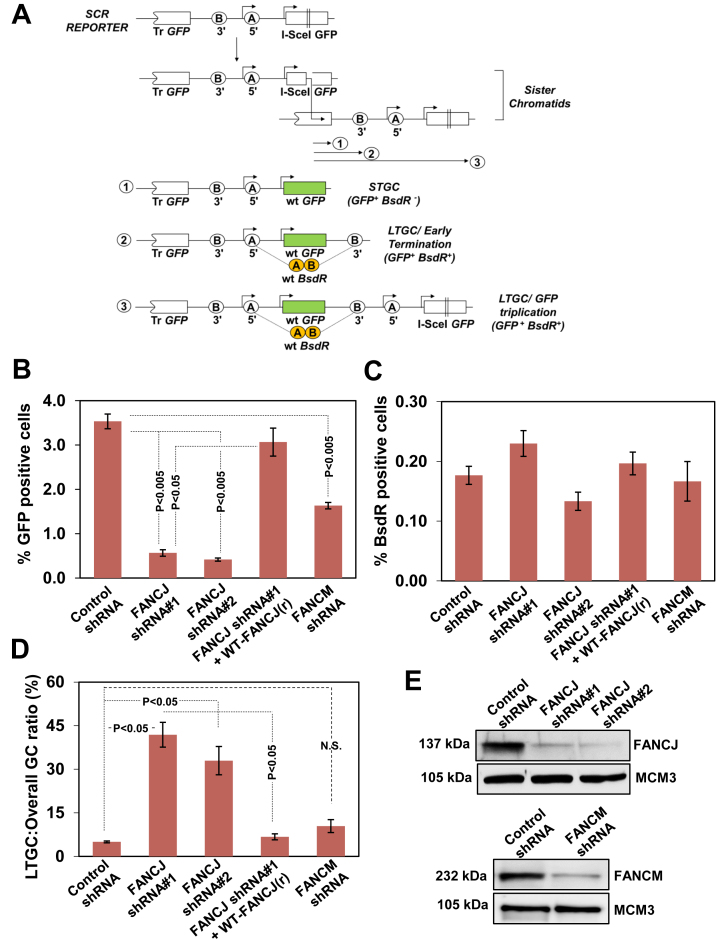
To study SCR in mammalian cells, a reporter was designed and constructed as described previously (35) (Figure 1A). The SCR reporter contains two mutated copies of the gene encoding GFP; the first one is 5′ truncated and the second one is full length but is interrupted by the insertion of 18 bp I-SceI recognition site. I-SceI induced gene conversion generates wt GFP, and the GFP+ cells can be quantified by flow cytometry. HR mediated STGC could arise by intrachromatid or interchromatid recombination. As earlier proposed (34,41), GFP-triplication could arise by LTGC or crossing over during SCR. However, crossover associated recombination events are highly suppressed in somatic cells (9,11,34). Thus, GFP-triplication in our assay reflects SCR/LTGC events. To distinguish between STGC and LTGC (GFP-triplication), two artificial exons (A and B) coding for blasticidin (BSD) resistance gene BsdR were introduced in a non-productive orientation between two GFP copies. In the context of STGC, SCR reporter remains un-rearranged and the cell remains sensitive to BSD (BsdR–) (Figure 1A, outcome 1). However, the GC tract of ≥ 1,031 bp results in the duplication of the BsdR exon B, allowing splicing between exon A with neighboring duplicated exon B to generate wtBsdR mRNA, rendering the cell resistant to BSD (BsdR+) (Figure 1A, outcome 2). If the GC extends ≥ 3, 250 bp, it results in GFP-triplication, duplicating the entire BsdR cassette (Figure 1A, outcome 3). The distinction of STGC and LTGC in our assay entails a GC of <1,031 bp for STGC and >1,031 bp for LTGC.
Figure 1.
FANCJ dysfunction skews gene conversion in favour of LTGC. (A) Structure of the SCR reporter. Filled arrow heads represent promoters corresponding to GFP and BsdR cassette. The circles depict 5′ and 3′ exon of BsdR cassette. The diagram shows two sister chromatids engaged in unequal SCR. The three outcomes of I-SceI induced SCR are shown: gene conversion that terminates before copying BsdR exon B (1, STGC); gene conversion that duplicates BsdR exon B but terminates prior to generating a third GFP copy (2, LTGC/early termination); and gene conversion that duplicates entire BsdR cassette, generating a third GFP copy (3, LTGC/GFP triplication). Wild-type GFP is shaded in green and wtBsdR mRNA is shaded in orange. Tr GFP, truncated GFP. (B) I-SceI induced GFP+ frequencies (total GFP; overall GC) in control shRNA, FANCJ shRNA#1, FANCJ shRNA#2, FANCJ shRNA#1 and its resistant wtFANCJ (r) and FANCM depleted U2OS SCR18 cells. (C) Frequencies of I-SceI induced BsdR+ colonies for the same experiment shown in panel B. (D) Ratio of I-SceI-induced BsdR+/GFP+ frequencies (LTGC/overall GC, expressed as a percentage) from the experiment whose results are shown in panels B and C. (E) Immunoblotting for FANCJ and FANCM protein levels after depletion of FANCJ and FANCM.
To investigate whether FANCJ controls the fine balance between STGC and LTGC, we used previously established U2OS (U2OS SCR18) cells containing a single chromosomally integrated SCR reporter. Transient transfection of U2OS SCR18 cells with hFANCJ shRNA plasmids (FANCJ shRNA#1 and FANCJ shRNA#2) yielded maximum depletion of endogenous FANCJ which resulted in heightened sensitivity and G2/M accumulation upon mitomycin C (MMC) induced ICL damage (Supplementary Figure S1A and S1D). To authenticate the specificity of shRNAs in depleting FANCJ, we generated shRNA#1 resistant construct (wt-FANCJ(r)). Western blotting analysis revealed that expression of wt-FANCJ(r) (HA-6xHis-FANCJ) was very similar to endogenous levels of FANCJ (Supplementary Figure S1C). Transfection with FANCJ shRNA #1(r) construct rescued U2OS cells from MMC induced DNA damage sensitivity and G2/M accumulation (Supplementary Figure S1B and S1D). I-SceI induced overall GC is measured by GFP+ cells. The I-SceI induced BsdR+ (LTGC) events are a subset of I-SceI induced GFP+ cells. Hence, the ratio of BsdR+ GFP+ cells to the overall GFP+ cells (LTGC/overall GC) represents the I-SceI induced GC resolving as LTGC. This ratio was found to be between 5% and 7% in mammalian cells (10,11,42). To study the HR efficiency, FANCJ depleted with shRNA#1 and control vector cells were transfected with I-SceI encoding plasmid, and 48 h after transfection, GFP+ cells were measured by flow cytometry. Depletion of FANCJ caused >3 fold reduction in GFP+ cells (<1%) compared to control cells (∼4%) (Figure 1B). The decrease in HR was rescued by transfecting the cells with FANCJ shRNA #1(r) plasmid (Figure 1B). Interestingly, although there was >3-fold reduction in HR frequency in FANCJ depleted cells, the absolute BsdR+ events were very similar to control cells (Figure 1C). The ratio of BsdR+ GFP+ events/total GFP+ events, a direct measure of LTGC was ∼5% in control cells. Strikingly, this ratio was increased to 6–8-fold in FANCJ depleted U2OS cells (Figure 1D). Consistently, transfection of cells with FANCJ shRNA #1(r) plasmid was able to rescue elevated LTGC events in FANCJ depleted cells (Figure 1D). We tested whether depletion of FANCM, another helicase/translocase in the FA pathway affects the balance of STGC and LTGC. Consistent with earlier report (43) depletion of FANCM caused significant reduction in overall HR (Figure 1E, lower panel and 1B). Interestingly, LTGC ratio was very similar to control cells in FANCM depleted cells, indicating that FANCJ but not FANCM controls the LTGC bias (Figure 1B–D). To validate our observation, we used U2OS cells that contain SCR reporter (U2OS SCR35S) where STGC and LTGC can be directly measured by FACS analysis (9). In this reporter, BsdR cassette was replaced by RFP encoding cassette and both overall GC and LTGC can be measured by scoring for GFP+ and GFP+ RFP+ cells, respectively. Using this modified SCR reporter, we made a similar observation as that of U2OS SCR18 cells (Supplementary Figure S2). These data suggest that FANCJ controls the overall GC and suppress LTGC during SCR.
FANCJ is a highly conserved protein in metazoans with a 74% and 75% identity of mouse and hamster FANCJ with human FANCJ, respectively (Supplementary Figure S3). To test whether hamster FANCJ controls SCR, we depleted FANCJ in previously characterized V79B SCR55 cells (11) and measured the SCR outcome. FANCJ depletion was maximal similar to U2OS cells (Supplementary Figure S4A), and upon depletion, these cells were sensitive to MMC (Supplementary Figure S4A). In response to I-SceI induced DSBs, control cells displayed >0.3% GFP+ cells. In contrast, a 3-fold reduction in overall GC was observed in FANCJ deficient cells (Supplementary Figure S4B). Interestingly, although the frequency of BsdR+ cells in FANCJ depleted cells was similar (Supplementary Figure S4C), the probability of GC resolving as LTGC was elevated ∼5-fold in FANCJ depleted cells when compared with control shRNA and FANCJ shRNA(r) transfected cells (Supplementary Figure S4D). These results clearly suggest that FANCJ helicase controls the fine balance of STGC and LTGC during I-SceI induced SCR.
FANCJ deficient cells exhibit qualitative alterations in LTGC products
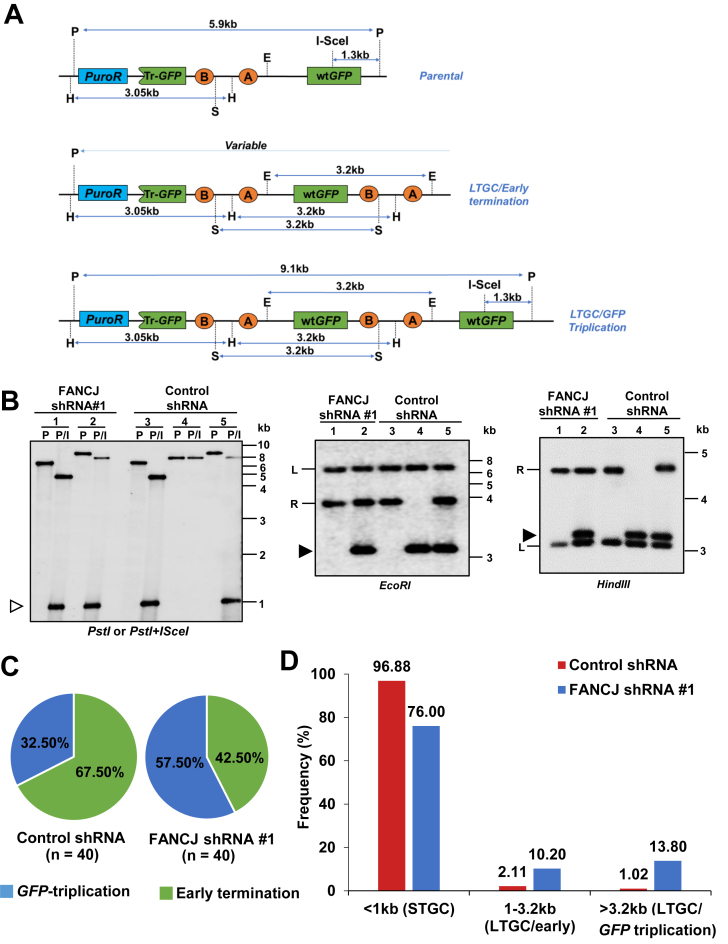
The GC tract that involves copying BsdR exon B (1031 bp) but is terminated before duplicating the third GFP copy represent early terminating LTGC events (LTGC/early termination). However, tract lengths extending ≥3.2 kb generate GFP-triplication (LTGC/GFP-triplication). We can distinguish ‘early terminating LTGC’ versus ‘GFP-triplication’ events within SCR reporter by Southern blot analysis of BsdR+ colonies. Figure 2A shows the expected restriction fragments of SCR reporter in Southern blots hybridized with a GFP cDNA probe for the parental reporter (Figure 2A, upper panel), for an early terminating LTGC (Figure 2A, middle panel) and for the GFP-triplication outcome (Figure 2A, lower panel).
Figure 2.
FANCJ depletion induces qualitative dysregulation in the SCR events. (A) Structure of the SCR reporter and the predicted DNA fragment sizes in Southern blotting using GFP cDNA probe from the parental (upper panel), LTGC/early termination (middle panel) and LTGC/GFP-triplication (lower panel) outcomes. P, PstI; H, HindIII; S, SacI; E, EcoRI; puro, puromycin; Tr-GFP, truncated GFP. (B) Southern blot analysis of parental U2OS SCR18 clone, lanes 1 and 3; a GFP-triplication outcome in FANCJ depleted and control cells (lanes 2 and 5) and LTGC/early termination event that was predominantly seen in control cells (lane 4). The lane numbers in each panel represents the same clone. The restriction enzymes used are shown under each panel. (Left panel) P, PstI digest; P/I, PstI + I-SceI digest; open arrowhead, 1.3-kb fragment released by PstI + I-SceI double digestion. In digests with EcoRI and HindIII the filled arrowhead indicates a 3.2-kb amplification product corresponding to duplication of part or all of the BsdR cassette. ‘L’ and ‘R’ indicate left and right arms of the reporter, respectively. (C) Gene conversion tract length frequency distribution in FANCJ depleted and control cells. Relative frequencies of I-SceI induced early terminating LTGC (green) and GFP-triplication (blue) events in FANCJ deficient and control cells. The number in parentheses shows the number of colonies analyzed. (D) Mean frequencies of gene conversion tract lengths in FANCJ depleted (blue bars) and control cells (red bars). These frequencies are obtained by combining the data shown in Supplementary Figure S5A, S5C and Figure 2C to indicate relative frequencies of STGC, LTGC/early termination and LTGC/GFP-triplication.
To test whether FANCJ depleted cells show qualitative dysregulation in LTGC, we examined I-SceI induced BsdR+ colonies from FANCJ depleted and control cells. Digestion of parental SCR reporter with PstI generates 5.9 kb fragment, whereas PstI and I-SceI yield 4.6 and 1.3 kb fragments (Figure 2A and B, lane 1 and 3). Either EcoRI or SacI digests generate two bands of varying length corresponding to ‘left’ and ‘right’ arm of the SCR reporter (Figure 2A and B, lane 1 and 3). HindIII digestion generates a 3.05 kb of left arm and a varying fragment of right arm (Figure 2A and B, lane 1 and 3). In the context of GFP-triplication, the size of PstI increases to 9.1 kb and the PstI + I-SceI digestion yields 7.8 and 1.3 kb fragments (Figure 2A, lower panel and B, lanes 2 and 5). The presence of intact I-SceI site in the third GFP indicates that GFP-triplication event occurred by SCR, since I-SceI site will be lost after the generation of DSB. Digestion with EcoRI, SacI or HindIII generates unperturbed left and right arms and a 3.2 kb fragment corresponding to the duplication of BsdR cassette. A proportion of LTGC events can get terminated prior to copying the third GFP (<2.9 kb) which are termed as ‘early terminating’ LTGC events that are characterized by shorter PstI fragment, absence of I-SceI site and loss of right arm of the reporter (Figure 2A, middle panel and B, lane 4).
Data shown in Supplementary Figure S6 and Figure 2C shows that in the control cells, 67.5% of LTGC events were early terminating and 32.5% entailed GFP-triplication. Strikingly, in the FANCJ depleted cells, LTGC events predominantly resolved as GFP-triplications (57.5%). Taking data from Supplementary Figure S5 and Figure 2C, we derived frequencies of SCR associated STGC (tract length of <1 kb), early terminating LTGC (tract length between 1 and 3.2 kb) and GFP-triplication events (tract length of >3.2 kb) in control and FANCJ depleted cells (Figure 2D). As shown in Figure 2D, 76% of GC events represented STGC in FANCJ depleted cells compared to ∼97% in control cells. The 24% of LTGC events that were observed in FANCJ depleted cells resulted predominantly in GFP-triplication outcome. In contrast, two-thirds of LTGC events entailed early terminating LTGC events, and one-third resolved as GFP-triplications in control cells (Figure 2D). These data clearly suggests that FANCJ plays a crucial role in the regulation of GC and limits the extent of copying during SCR.
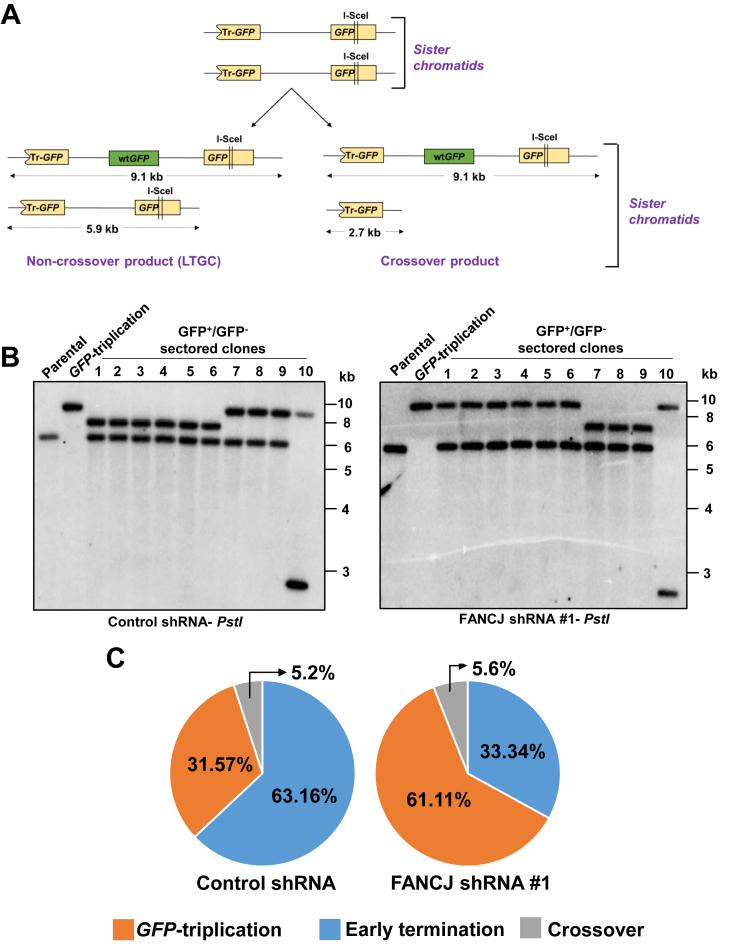
GFP-triplication in FANCJ depleted cells arises by LTGC
Elevated GFP-triplication events that were observed in FANCJ depleted cells could arise by crossover (CO) recombination between sister chromatids or by LTGC. CO between sister chromatids alters the structure of donor sister by truncation of GFP reporter (Figure 3A). However, LTGC mechanism of GFP-triplication arises by extended copying during repair synthesis leaving the donor sister chromatid intact (Figure 3A). We captured the donor sister cells after pulse exposure to blasticidin as described previously (11). Using this method we retrieved 19 sectored (GFP+/GFP–) colonies from control cells and 18 sectored colonies from FANCJ depleted cells. These colonies were subjected to Southern blotting analysis using PstI digested genomic DNA. In control cells, 6/19 (31.5%) colonies exhibited bands corresponding to 9 and 6 kb, indicating LTGC mediated GFP-triplications; 12/19 (63.1%) colonies revealed ∼8 and 6 kb bands corresponding to early terminating LTGC events and 1/19 (5.2%) showed a CO recombination event with 9 and 2.9 kb fragments (Figure 3B and C). In FANCJ depleted cells, 61% of colonies (11/18) exhibited 9 and 6 kb bands, suggesting that GFP-triplications arises by LTGC; 33.3% (6/18) colonies revealed early terminating events and 1/18 (5.6%) showed CO recombination product (Figure 3B and C). These data clearly indicate that GFP-triplications that were abundantly seen in FANCJ depleted cells arise by LTGC.
Figure 3.
Analysis of crossover and non-crossover SCR events in U2OS SCR18 cells. (A) Illustration of crossover and LTGC products during SCR. (B) Southern blotting analysis of PstI digested sectored GFP+/GFP– colonies from control (left panel) and FANCJ depleted cells (right panel). Images depict representative ten colonies each from control and FANCJ depleted cells. (C) Frequency distribution of GFP-triplication, early termination and crossover recombination events during SCR in control and FANCJ depleted cells.
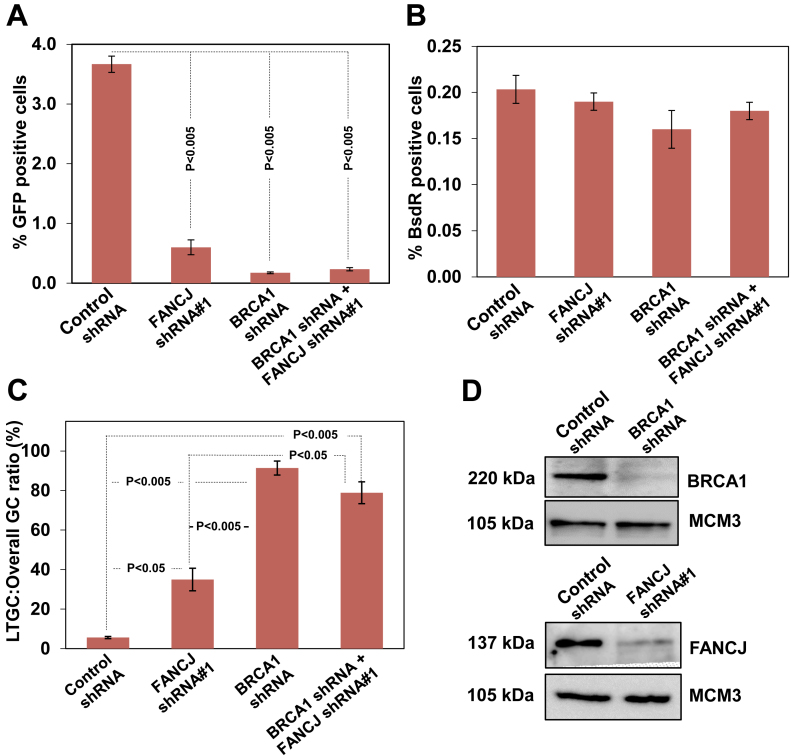
The SCR regulation by FANCJ is BRCA1 dependent
BRCA1 plays an upstream role in the repair of DSBs by HR (44). BRCA1 dysfunction skews GC in favor of LTGC-associated GFP-triplication events (9). To understand whether FANCJ functions in the BRCA1 pathway of LTGC suppression, we examined SCR events by depleting BRCA1 and FANCJ independently and by co-depletion. Compared to FANCJ depleted cells, BRCA1 depletion caused more severe HR defect (Figure 4A). However, co-depletion of BRCA1 and FANCJ had similar reduction in HR as that of BRCA1 alone depleted cells (Figure 4A and D), implying that FANCJ has a downstream role in BRCA1 mediated HR regulation. Consistent with earlier study (9), BRCA1 dysfunction resulted in LTGC bias and the LTGC frequency was 2–3 folds higher than FANCJ depleted cells (Figure 4C). However, this increase in LTGC was very similar to FANCJ and BRCA1 co-depleted cells (Figure 4C), suggesting that FANCJ participate in BRCA1 mediated HR and in controlling the balance between STGC and LTGC.
Figure 4.
FANCJ and BRCA1 function in a common pathway to suppress LTGC. (A) I-SceI induced GFP+ frequencies in control shRNA, FANCJ and BRCA1 single and co-depleted cells. (B) Frequencies of I-SceI induced BsdR+ colonies for the same experiment shown in panel A. (C) Ratio of I-SceI-induced LTGC/overall GC from the experiment whose results are shown in panels A and B. (D) BRCA1 and FANCJ protein levels after depletion of BRCA1 and FANCJ, respectively.
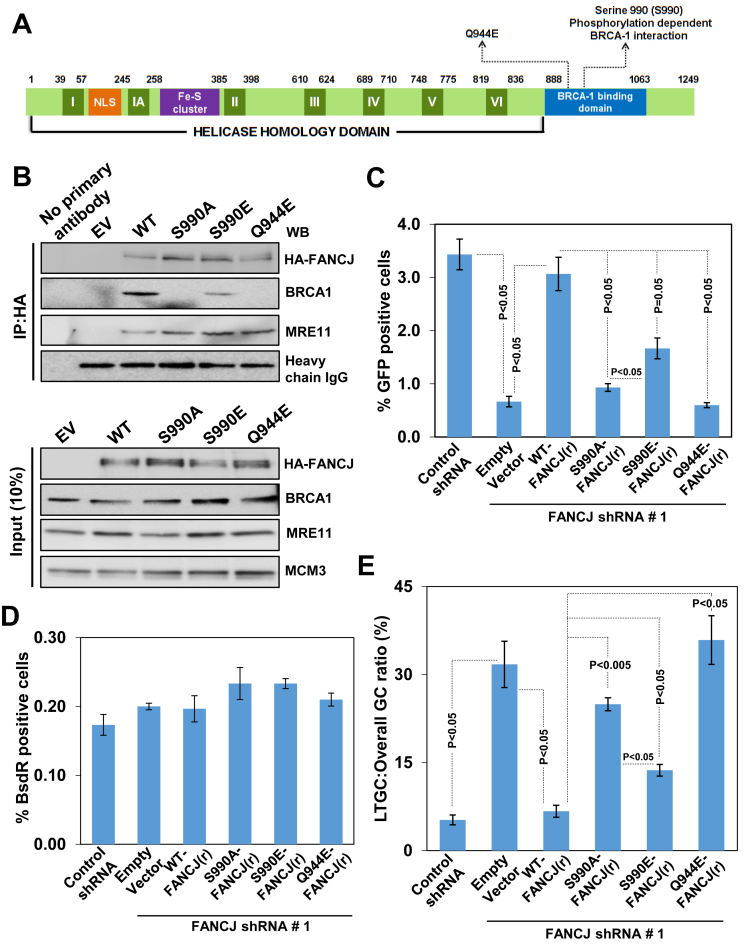
FANCJ S990 is known to undergo phosphorylation by CDK and this phosphorylation is required for FANCJ interaction with BRCA1 (45) (Figure 5A). However, the precise role of FANCJ interaction with BRCA1 during HR is unclear. To address this, we generated FANCJ shRNA#1 resistant FANCJ S990A and a phosphomimicking FANCJ S990E mutant as described in materials and methods. The steady state levels of both the mutants were comparable to wt FANCJ levels (Figure 5B). Our immunoprecipitation studies revealed that FANCJ S990A was defective with BRCA1 interaction (Figure 5B). In contrast, FANCJ S990E was partially competent to interact with BRCA1 (Figure 5B). FANCJ Q944E pathological mutant was identified in a breast cancer patient (46) and this mutation is located close to the site of S990 which undergoes phosphorylation and interacts with BRCT domain of BRCA1. Although FANCJ Q944E mutant protein levels were comparable to wt FANCJ, this mutant was defective with BRCA1 interaction. In agreement with a previous study (47), all the mutants were proficient in interacting with MRE11 (Figure 5B). Next, we examined the role of these FANCJ mutants in SCR regulation. After 24 h of FANCJ depletion, FANCJ mutant plasmids were co-transfected with I-SceI expression plasmids and SCR events were analysed. Interestingly, transient expression of FANCJ S990A caused a ∼4-fold reduction in overall GC and ∼5-fold increase in LTGC compared to control shRNA or wt FANCJ (r) cells (Figure 5C–E). The FANCJ S990E that retained partial interaction with BRCA1 exhibited significant reduction in HR and increase in LTGC (Figure 5C, D and E). In contrast, FANCJ Q944E that was incompetent to bind BRCA1 exhibited ∼5 fold reduction in I-SceI induced HR compared to control cells (Figure 5C). Interestingly, expression of this mutant skewed the HR toward >5-fold increase in LTGC compared to control cells (Figure 5E). Together, these results suggest that FANCJ interaction with BRCA1 is crucial for the FANCJ mediated overall HR and in suppressing LTGC.
Figure 5.
SCR regulation by FANCJ is dependent on its interaction with BRCA1. (A) Schematic diagram of FANCJ depicting the conserved helicase domains (I–VI), NLS motif, Fe-S cluster and the BRCA1 binding domain. The S990 phosphorylation site and breast cancer associated Q944E residue is indicated. (B) Interaction of BRCA1 with wt and FANCJ mutants in the U2OS SCR18 cells. HA-6xHis-FANCJ was measured using HA specific antibody. (C) I-SceI induced GFP+ frequencies in U2OS SCR18 cells transfected with FANCJ shRNA#1 along with empty vector (EV) and the plasmids that express FANCJ shRNA resistant wtFANCJ, FANCJ S990A, FANCJ S990E and FANCJ Q944E. (D) Absolute frequencies of BsdR+ colonies for the experiment shown in panel C. (E) Ratio of LTGC:Overall GC obtained from the data shown in panels C and D.
FANCJ ATPase/helicase activity is essential for controlling the SCR events
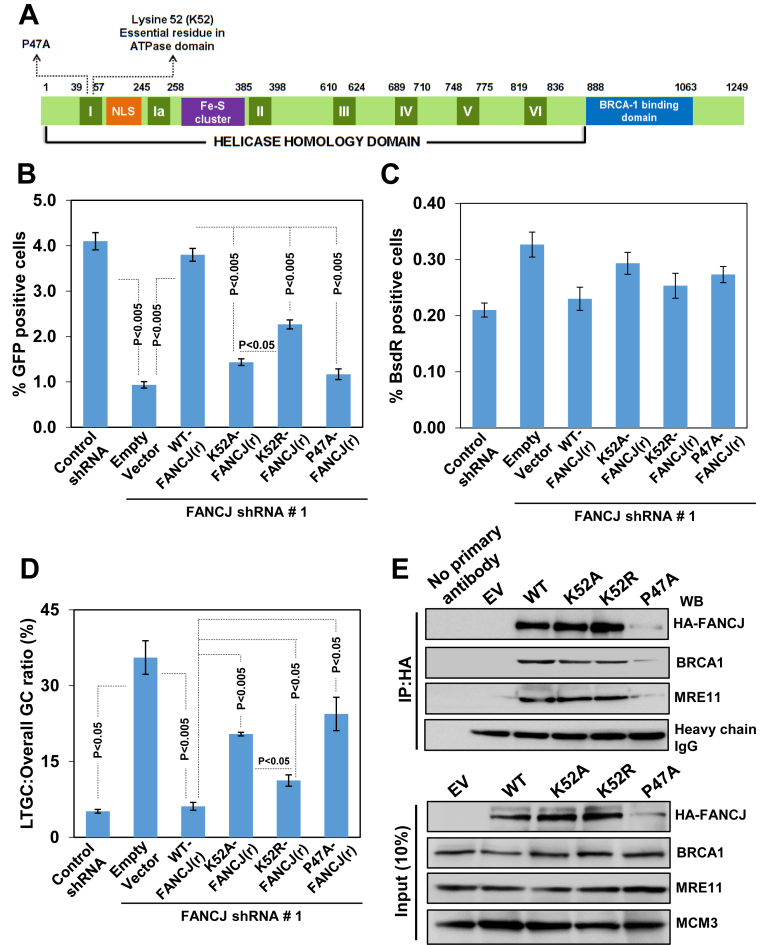
FANCJ helicase possesses conserved Walker A and B motifs that are essential for ATPase and helicase functions. To test whether the helicase activity of FANCJ is critical for the SCR regulation, we generated shRNA resistant Walker A motif K52A which is defective for ATP binding, and K52R which can bind but not hydrolyze ATP (Figure 6A). The steady state levels of these mutants were very similar to wtFANCJ levels (Figure 6E). Expression of FANCJ K52A caused ∼4-fold reduction in I-SceI induced HR (Figure 6B) and ∼4-fold increase in the ratio of LTGC to overall GC compared to control shRNA or shRNA resistant wt FANCJ (r) cells (Figure 6D). In a parallel experiment, we measured the HR in cells expressing FANCJ K52R and found a significant reduction in GFP+ cells compared to control cells (Figure 6B). The HR events were biased towards ∼2-fold increase in LTGC events in cells expressing FANCJ K52R when compared with control cells (Figure 6B–D). To test whether FANCJ Walker motif lysine mutants are competent to bind BRCA1, we performed interaction studies and find that both FANCJ K52A and K52R mutants were proficient in binding to BRCA1 (Figure 6E). These data suggest that ATP hydrolysis driven helicase activity of FANCJ is crucial for the SCR regulation.
Figure 6.
Helicase/ATPase activity of FANCJ is required for SCR regulation. (A) Schematic diagram of FANCJ depicting the conserved helicase domains (I–VI), NLS motif, Fe–S cluster and the BRCA1 binding domain. The Walker A motif K52 and breast cancer associated P47A mutant residues are indicated. (B) I-SceI induced GFP+ frequencies in U2OS SCR18 cells transfected with FANCJ shRNA#1 along with empty vector (EV) and the plasmids that express FANCJ shRNA resistant wtFANCJ, FANCJ K52A, FANCJ K52R and FANCJ P47A. (C) Absolute frequencies of BsdR+ colonies for the experiment shown in panel B. (D) Ratio of LTGC:Overall GC obtained from the data shown in panels B and C. (E) Ability of Walker motif mutants of FANCJ interaction with BRCA1. HA-6xHis-FANCJ was measured using HA specific antibody.
The FANCJ P47A mutation has been identified in a patient with early onset of breast cancer (16). This mutation resides within the Walker A motif and the purified protein exhibit reduced ATPase and helicase activity (48). We generated shRNA resistant FANCJ P47A mutant and examined its role in SCR. In contrast to wt protein, a reduced abundance in the FANCJ P47A mutant was observed (Figure 6E). Expression of FANCJ P47A pathological mutant caused a ∼4-fold reduction in HR (Figure 6B) and >4-fold increase in the LTGC frequency compared to control cells (Figure 6D). The observed HR/SCR defect with this mutant could be largely due to protein instability. However, this mutant was competent to bind BRCA1 (Figure 6E).
Iron–sulfur (Fe–S) cluster domain of FANCJ helicase is critical for SCR regulation
Many DNA metabolizing proteins, including DNA glycosylases, primases and helicases contain iron–sulfur (Fe–S) cluster, and this domain appears to have crucial role in the regulation of these enzymes (49,50). FANCJ helicase also possesses a single Fe–S cluster and has been implicated in the functional activities of FANCJ (49). Fe–S cluster domain generally contains four conserved cysteine residues which participate in the coordination of Fe metal ion. C283, C298, C310 and C350 are the four conserved cysteines that are present in the Fe–S domain of FANCJ helicase (Supplementary Figure S3 and S7A). To investigate the functional role of FANCJ Fe–S cluster in SCR regulation, we generated shRNA resistant FANCJ C350A and C350S mutants. Analyses of the steady state levels of these mutants indicated that FANCJ C350A mutant is unstable (Supplementary Figure S7E), whereas FANCJ C350S mutant protein levels were very similar to that of the wt FANCJ (Supplementary Figure S7E). Expression of FANCJ C350A resulted in HR/SCR dysfunction (Supplementary Figure S7B, C and D), a defect that could be largely attributed to unstable protein. In a parallel experiment, we measured the SCR events with the cells that express FANCJ C350S mutant and found that cysteine to serine substitution largely rescues the SCR defect (Supplementary Figure S7B–D). Interaction studies revealed that both FANCJ C350A and C350S mutants are proficient in binding to BRCA1 (Supplementary Figure S7E).
Alanine 349 to proline mutation has been identified in a FA patient (18), and this residue is immediately adjacent to conserved cysteine 350 residue within the Fe–S cluster of FANCJ. To test the role of FANCJ A349P pathological mutant in the SCR regulation, we generated shRNA resistant mutant and analysed the steady state levels. In contrast to wt FANCJ, A349P substitution in FANCJ resulted in partially unstable protein (Supplementary Figure S7E). Expression of FANCJ A349P caused ∼4 fold HR/SCR defect, a phenotype that may be largely due to protein instability (Supplementary Figure S7B–D). Although this mutant was less stable, it was competent to interact with BRCA1 (Supplementary Figure S7E). Together, these data suggest that FANCJ Fe–S cluster is critical for the stability of the protein and may play a role in the DSB repair and in terminating extended repair synthesis during SCR.
DISCUSSION
Despite FANCJ helicase functions have been implicated in downstream steps of ICL repair and also in the HR mediated repair of DSBs (19,51), the precise role and the molecular mechanism by which FANCJ controls HR is unclear. Moreover, the factors and the mechanism(s) that regulate the outcome of HR are less understood. In the current study, we demonstrate that FANCJ helicase participates in the repair of DSBs by HR. Strikingly, our data shows that FANCJ but not FANCM determines the fine balance between STGC and LTGC during SCR. Qualitative analyses of the GC tracts reveal that FANCJ terminates extensive repair synthesis during SCR to suppress GFP-triplication events. The SCR regulation by FANCJ was dependent on its interaction with BRCA1 tumor suppressor. In addition, the helicase activity of FANCJ was required for efficient HR and suppression of LTGC. These data reveal a novel function of FANCJ helicase in controlling SCR and SCR associated gene amplification/duplications and suggest that these functions may be crucial for the tumor suppression.
The role of FANCJ in SCR regulation and in suppressing LTGC events between sister chromatids appears to be conserved across different species. Indeed, our studies extended to hamster cells reveal that FANCJ fine tunes the HR outcome by terminating the GC in favor of STGC. Consistent with our reporter analysis, Southern blotting data from the BsdR+ clones of U2OS cells show those LTGC events are skewed towards more of GFP-triplications in FANCJ deficient cells. These observations are made with depletion of FANCJ, and it is highly likely that the proportion of GFP-triplications would be further elevated with biallelic mutations in FANCJ. Infact, our earlier studies reveal a bimodal distribution of GC tracts with a frequency of 97% of LTGC events resolving as GFP-triplication events in cells lacking RAD51C (11).
How does FANCJ regulates overall HR? One possibility is that FANCJ could participate in BRCA1 mediated resection of DSBs. Indeed, a reduction in RPA loading on to chromatin has been observed in FANCJ depleted cells (52). Moreover, we find that BRCA1 depletion shows more severe reduction in HR than FANCJ depleted cells, indicating that BRCA1 is epistatic to FANCJ. These data suggest that FANCJ participate in the BRCA1 pathway of HR regulation likely by controlling BRCA1 mediated end resection and defective end resection could account for the reduction in overall HR in FANCJ deficient cells. How could we explain the GC bias in favour of LTGC in FANCJ depleted cells? SDSA pathway of DSB repair invokes a model in which displaced nascent strand pairs with the second end of broken chromosome to complete the DSB repair (53–55). Conceivably, if the ends are not sufficiently resected, the displaced nascent strand fails to anneal to the second end of the broken chromosome. In this scenario, disengaged nascent strand could reinvade the sister chromatid and initiate repair synthesis. Multiple cycles of this reinvasion could generate GC tracts of several kilobases. A similar mechanism has been proposed for the increase in LTGC events observed in BRCA1 deficient cells (9). FANCJ could also play an independent and downstream role in displacing the nascent strand from the D-loop and thereby facilitate SDSA mechanism of DSB repair. Consistent with this hypothesis, a biochemical study shows that purified FANCJ helicase unwinds D-loop structures (30). It is likely that failure in the FANCJ mediated nascent strand disengagement could lead to continued synthesis accounting for increased GFP-triplications observed in FANCJ depleted cells. In addition, displacement of RAD51 from nascent strand can be another mechanism by which repair synthesis is terminated during SDSA type of repair. Indeed, FANCJ has been shown to catalyse stripping of RAD51 from the ssDNA (56).
FANCJ is dispensable for the FANCD2 activation during FA pathway mediated ICL repair and plays a downstream role in repairing the DSBs via HR in the later steps of ICL repair (19). The role of FANCJ in SCR regulation appears to be independent of its participation in ICL repair. The involvement of FANCJ in ICL repair is dependent on its association with mismatch repair proteins but independent of its interaction with BRCA1 (28,51). A study demonstrates that BRCA1 plays a distinct role in ICL repair which is independent of its established role in HR (57). Our data clearly shows that FANCJ participates in the BRCA1 pathway of SCR regulation, suggesting that FANCJ must have evolved with distinct functions in ICL and DSB repair. Recent studies identify multiple roles of FANCJ in genome maintenance by participating in G4 DNA resolution (58–62), microsatellite stability (63,64) and rescuing cells from UV induced lesions (65) which are independent of its role in FA pathway of ICL repair. FANCM translocase/helicase is crucial for sensing the ICL lesion and loading of FA core complex proteins during ICL repair (22). Notably, depletion of FANCM did not exhibit LTGC bias during SCR but displayed moderate HR reduction. Recent studies indicate that FANCM interacts with end resection machinery and participate in end resection at ICLs (15,66,67). The moderate reduction in HR in our assay could be attributed to its role in end resection.
BRCA1 plays a crucial role in determining the choice between HR and NHEJ pathway of DSB repair by participating in the resection of DSBs (68–71). FANCJ exists in the BRCA1-B complex by its direct interaction with BRCA1 (16,26). FANCJ phosphorylation at S990 is essential for its interaction with BRCA1 through BRCT motifs (45,51). In our studies, expression of shRNA resistant FANCJ S990A mutant caused reduction in overall GC and an increase in LTGC events. This phenotype was partially corrected by the expression of phosphomimicking mutant of FANCJ (FANCJ S990E). This data suggest that FANCJ interaction with BRCA1 is critical for the FANCJ mediated SCR regulation and in the suppression of LTGC during SCR. The FANCJ Q944E mutation has been identified in a breast cancer patient (46) and we find that expression of this mutant cause defect in HR and an imbalance in the STGC/LTGC. The FANCJ Q944 is in the close proximity to S990 of FANCJ which is required for FANCJ interaction with BRCA1 upon phosphorylation. Perhaps, FANCJ Q944 residue also may be important for its interaction with BRCA1 to engage in end resection step prior to HR initiation. FANCJ lysine 1249 has been shown to be acetylated and this modification contributes to processing of DSB ends to facilitate HR (72). It will be interesting to test whether FANCJ acetylation is critical for the fine regulation of STGC and LTGC during SCR. Notably, BRCA1-C complex that contains CtIP and MRN complex proteins have been implicated in end resection (73–76). It is unclear whether an interplay between BRCA1-B and BRCA1-C complexes exist in vivo for the resection of DSB ends. However, further studies are required to delineate the mechanism underlying the BRCA1-FANCJ mediated end resection.
Although lack of BRCA1 and RAD51 paralogs cause HR defect and loss of STGC/LTGC balance (9–11), the precise mechanism underlying this phenotype remains to be elucidated. FANCJ being a helicase and effector protein could directly participate in the initial and late stages of HR to influence the HR outcome. Our analysis with Walker A motif lysine mutants revealed interesting phenotypes. FANCJ K52A mutant which lacks ATP binding exhibits a severe reduction in overall GC with a greater increase in LTGC. In contrast, ATP binding competent FANCJ K52R mutant was partially functional. Indeed, FANCJ K52R mutant was devoid of ATPase and helicase activity (30,48,77) and dominant negative expression of this mutant caused delay in the repair of IR induced DSBs (16). Notably, FANCJ K52A and K52R mutants were competent to bind BRCA1, suggesting that in addition to BRCA1 interaction, helicase activity of FANCJ is crucial for SCR regulation. It is likely that FANCJ ATPase and helicase activity may be required for BRCA1 mediated end resection or FANCJ could directly participate in nascent strand displacement via its helicase function. FANCJ P47A missense mutation has been identified in a breast cancer patient and this residue is in the highly conserved Walker A motif (16). The FANCJ P47A mutant protein was defective with ATPase and helicase functions in vitro (48). Consistent with earlier study (16), we find that FANCJ P47A mutant was unstable and the SCR defect associated with this mutant could be largely attributed to its instability.
Although FANCJ has been considered as a bonafide tumor suppressor, the molecular mechanism by which FANCJ prevents cancer predisposition remains unclear. Data presented in the current study clearly identify that FANCJ helicase restricts pathological outcome of HR by suppressing LTGC. The observed LTGC in FANCJ deficient cells could arise via break-induced replication (BIR) as documented from studies using yeast model organism (78,79). LTGC could also arise by defective displacement of nascent strand or by defective annealing of displaced nascent strand to the second end of broken chromosome during SDSA mechanism of two ended DSB repair. However, further studies are required to understand the mechanism underlying the LTGC associated with SCR in FANCJ defective cells. Analyses of the breast and ovarian cancer genomes reveal complex genome rearrangements including tandem gene segmental duplications/amplifications (80–82). Such duplications are associated with loss of tumor suppressor genes, overexpression of oncogenes, and dysregulated cell cycle and DNA damage repair (82). Breaks arising during replication appear to be the primary cause of tandem gene segmental duplications/ampifications (36,83). Indeed, studies from Escherichia coli and yeast identified microhomology mediated segmental duplications arising during replicative stress (84–86). In addition, a recent study shows that in mammalian cells aberrant termination of HR could induce microhomology mediated complex rearrangements and BIR (87). It is highly likely that FANCJ could suppress LTGC during repair of breaks induced during replication. Notably, inactivation of BRCA1/BRCA2 tumor suppressor leads to elevated LTGC at Tus/Ter induced breaks during replication (5). It will be interesting to test whether FANCJ regulates the balance of STGC/LTGC during repair of replication stress induced DSBs using a similar system.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Sharon Cantor for providing FANCJ cDNA and Kaustuv Sanyal and his lab members for their help with Southern blot analyses. We thank Indian Institute of Science (IISc) FACS facility for assisting the work. We also thank Dipankar Nandi for allowing us to use the Centre for Infectious Disease Research Flow cytometer. We are grateful to Drs Jim Haber, Wolf-Heyer and Sandeep Burma for their useful discussions. We thank Sneha Saxena for stimulating discussions and critical reading of the manuscript.
Footnotes
Present address: Kumar Somyajit, NNF Center for Protein Research, University of Copenhagen, Faculty of Health and Medical Sciences, Copenhagen, Denmark.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology (DBT) [BT/ PR5846/BRB/10/1099/2012]; IISc-DBT partnership program [DBT/BF/PR/INS/IISc/2011–12]; IISc (to S.N.); DBT (to A.M.); CSIR, Bristol Meyers-Squibb fellowship from UK and Ranbaxy Science Foundation, India (to K.S.). Funding for open access charge: Department of Biotechnology, India.
Conflict of interest statement. None declared.
REFERENCES
- 1. Aguilera A., Gomez-Gonzalez B.. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008; 9:204–217. [DOI] [PubMed] [Google Scholar]
- 2. Negrini S., Gorgoulis V.G., Halazonetis T.D.. Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell. Biol. 2010; 11:220–228. [DOI] [PubMed] [Google Scholar]
- 3. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hartlerode A.J., Scully R.. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem. J. 2009; 423:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Willis N.A., Chandramouly G., Huang B., Kwok A., Follonier C., Deng C., Scully R.. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature. 2014; 510:556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moynahan M.E., Jasin M.. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell. Biol. 2010; 11:196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nagaraju G., Scully R.. Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair (Amst). 2007; 6:1018–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scully R., Livingston D.M.. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000; 408:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chandramouly G., Kwok A., Huang B., Willis N.A., Xie A., Scully R.. BRCA1 and CtIP suppress long-tract gene conversion between sister chromatids. Nat. Commun. 2013; 4:2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nagaraju G., Hartlerode A., Kwok A., Chandramouly G., Scully R.. XRCC2 and XRCC3 regulate the balance between short- and long-tract gene conversions between sister chromatids. Mol. Cell. Biol. 2009; 29:4283–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagaraju G., Odate S., Xie A., Scully R.. Differential regulation of short- and long-tract gene conversion between sister chromatids by Rad51C. Mol. Cell. Biol. 2006; 26:8075–8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Somyajit K., Subramanya S., Nagaraju G.. Distinct roles of FANCO/RAD51C protein in DNA damage signaling and repair: implications for Fanconi anemia and breast cancer susceptibility. J. Biol. Chem. 2012; 287:3366–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Somyajit K., Subramanya S., Nagaraju G.. RAD51C: a novel cancer susceptibility gene is linked to Fanconi anemia and breast cancer. Carcinogenesis. 2010; 31:2031–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michl J., Zimmer J., Tarsounas M.. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J. 2016; 35:909–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ceccaldi R., Sarangi P., D’Andrea A.D.. The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol. 2016; 17:337–349. [DOI] [PubMed] [Google Scholar]
- 16. Cantor S.B., Bell D.W., Ganesan S., Kass E.M., Drapkin R., Grossman S., Wahrer D.C., Sgroi D.C., Lane W.S., Haber D.A. et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001; 105:149–160. [DOI] [PubMed] [Google Scholar]
- 17. Levitus M., Waisfisz Q., Godthelp B.C., de Vries Y., Hussain S., Wiegant W.W., Elghalbzouri-Maghrani E., Steltenpool J., Rooimans M.A., Pals G. et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet. 2005; 37:934–935. [DOI] [PubMed] [Google Scholar]
- 18. Levran O., Attwooll C., Henry R.T., Milton K.L., Neveling K., Rio P., Batish S.D., Kalb R., Velleuer E., Barral S. et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet. 2005; 37:931–933. [DOI] [PubMed] [Google Scholar]
- 19. Litman R., Peng M., Jin Z., Zhang F., Zhang J., Powell S., Andreassen P.R., Cantor S.B.. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005; 8:255–265. [DOI] [PubMed] [Google Scholar]
- 20. Venkitaraman A.R. Tracing the network connecting BRCA and Fanconi anaemia proteins. Nat. Rev. Cancer. 2004; 4:266–276. [DOI] [PubMed] [Google Scholar]
- 21. Joenje H., Patel K.J.. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2001; 2:446–457. [DOI] [PubMed] [Google Scholar]
- 22. Kee Y., D’Andrea A.D.. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010; 24:1680–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seal S., Thompson D., Renwick A., Elliott A., Kelly P., Barfoot R., Chagtai T., Jayatilake H., Ahmed M., Spanova K. et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat. Genet. 2006; 38:1239–1241. [DOI] [PubMed] [Google Scholar]
- 24. Rafnar T., Gudbjartsson D.F., Sulem P., Jonasdottir A., Sigurdsson A., Jonasdottir A., Besenbacher S., Lundin P., Stacey S.N., Gudmundsson J. et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat. Genet. 2011; 43:1104–1107. [DOI] [PubMed] [Google Scholar]
- 25. Cantor S.B., Guillemette S.. Hereditary breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future Oncol. 2011; 7:253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Greenberg R.A., Sobhian B., Pathania S., Cantor S.B., Nakatani Y., Livingston D.M.. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006; 20:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bridge W.L., Vandenberg C.J., Franklin R.J., Hiom K.. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 2005; 37:953–957. [DOI] [PubMed] [Google Scholar]
- 28. Peng M., Litman R., Xie J., Sharma S., Brosh R.M. Jr, Cantor S.B.. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 2007; 26:3238–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suhasini A.N., Rawtani N.A., Wu Y., Sommers J.A., Sharma S., Mosedale G., North P.S., Cantor S.B., Hickson I.D., Brosh R.M. Jr. Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom's syndrome. EMBO J. 2011; 30:692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gupta R., Sharma S., Sommers J.A., Jin Z., Cantor S.B., Brosh R.M. Jr. Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J. Biol. Chem. 2005; 280:25450–25460. [DOI] [PubMed] [Google Scholar]
- 31. Sweetser D.B., Hough H., Whelden J.F., Arbuckle M., Nickoloff J.A.. Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion tracts in Saccharomyces cerevisiae reveals reversible mitotic conversion polarity. Mol. Cell. Biol. 1994; 14:3863–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elliott B., Richardson C., Winderbaum J., Nickoloff J.A., Jasin M.. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 1998; 18:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taghian D.G., Nickoloff J.A.. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol. 1997; 17:6386–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson R.D., Jasin M.. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000; 19:3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Puget N., Knowlton M., Scully R.. Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair (Amst). 2005; 4:149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Willis N.A., Rass E., Scully R.. Deciphering the code of the cancer genome: mechanisms of chromosome rearrangement. Trends Cancer. 2015; 1:217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xue Y., Li Y., Guo R., Ling C., Wang W.. FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum. Mol. Genet. 2008; 17:1641–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gorski J.J., James C.R., Quinn J.E., Stewart G.E., Staunton K.C., Buckley N.E., McDyer F.A., Kennedy R.D., Wilson R.H., Mullan P.B. et al. BRCA1 transcriptionally regulates genes associated with the basal-like phenotype in breast cancer. Breast Cancer Res. Treat. 2010; 122:721–731. [DOI] [PubMed] [Google Scholar]
- 39. Somyajit K., Basavaraju S., Scully R., Nagaraju G.. ATM- and ATR-mediated phosphorylation of XRCC3 regulates DNA double-strand break-induced checkpoint activation and repair. Mol. Cell. Biol. 2013; 33:1830–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Somyajit K., Saxena S., Babu S., Mishra A., Nagaraju G.. Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 2015; 43:9835–9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen W., Jinks-Robertson S.. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol. Cell. Biol. 1998; 18:6525–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xie A., Puget N., Shim I., Odate S., Jarzyna I., Bassing C.H., Alt F.W., Scully R.. Control of sister chromatid recombination by histone H2AX. Mol. Cell. 2004; 16:1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Howard S.M., Yanez D.A., Stark J.M.. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015; 11:e1004943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li M.L., Greenberg R.A.. Links between genome integrity and BRCA1 tumor suppression. Trends Biochem. Sci. 2012; 37:418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu X., Chini C.C., He M., Mer G., Chen J.. The BRCT domain is a phospho-protein binding domain. Science. 2003; 302:639–642. [DOI] [PubMed] [Google Scholar]
- 46. Cao A.Y., Huang J., Hu Z., Li W.F., Ma Z.L., Tang L.L., Zhang B., Su F.X., Zhou J., Di G.H. et al. Mutation analysis of BRIP1/BACH1 in BRCA1/BRCA2 negative Chinese women with early onset breast cancer or affected relatives. Breast Cancer Res. Treat. 2009; 115:51–55. [DOI] [PubMed] [Google Scholar]
- 47. Suhasini A.N., Sommers J.A., Muniandy P.A., Coulombe Y., Cantor S.B., Masson J.Y., Seidman M.M., Brosh R.M. Jr. Fanconi anemia group J helicase and MRE11 nuclease interact to facilitate the DNA damage response. Mol. Cell. Biol. 2013; 33:2212–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cantor S., Drapkin R., Zhang F., Lin Y., Han J., Pamidi S., Livingston D.M.. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:2357–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu Y., Brosh R.M. Jr. DNA helicase and helicase-nuclease enzymes with a conserved iron-sulfur cluster. Nucleic Acids Res. 2012; 40:4247–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thakur R.S., Desingu A., Basavaraju S., Subramanya S., Rao D.N., Nagaraju G.. Mycobacterium tuberculosis DinG is a structure-specific helicase that unwinds G4 DNA: implications for targeting G4 DNA as a novel therapeutic approach. J. Biol. Chem. 2014; 289:25112–25136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brosh R.M. Jr, Cantor S.B.. Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi anemia. Front. Genet. 2014; 5:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gupta R., Sharma S., Sommers J.A., Kenny M.K., Cantor S.B., Brosh R.M. Jr. FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007; 110:2390–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Paques F., Haber J.E.. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999; 63:349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heyer W.D., Ehmsen K.T., Liu J.. Regulation of Homologous Recombination in Eukaryotes. Annu. Rev. Genet. 2010; 44:113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. San Filippo J., Sung P., Klein H.. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008; 77:229–257. [DOI] [PubMed] [Google Scholar]
- 56. Sommers J.A., Rawtani N., Gupta R., Bugreev D.V., Mazin A.V., Cantor S.B., Brosh R.M. Jr. FANCJ uses its motor ATPase to destabilize protein-DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J. Biol. Chem. 2009; 284:7505–7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bunting S.F., Callen E., Kozak M.L., Kim J.M., Wong N., Lopez-Contreras A.J., Ludwig T., Baer R., Faryabi R.B., Malhowski A. et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell. 2012; 46:125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu Y., Shin-ya K., Brosh R.M. Jr. FANCJ helicase defective in Fanconi anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008; 28:4116–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. London T.B., Barber L.J., Mosedale G., Kelly G.P., Balasubramanian S., Hickson I.D., Boulton S.J., Hiom K.. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008; 283:36132–36139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sarkies P., Murat P., Phillips L.G., Patel K.J., Balasubramanian S., Sale J.E.. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012; 40:1485–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schwab R.A., Nieminuszczy J., Shin-ya K., Niedzwiedz W.. FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J. Cell Biol. 2013; 201:33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Castillo Bosch P., Segura-Bayona S., Koole W., van Heteren J.T., Dewar J.M., Tijsterman M., Knipscheer P.. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014; 33:2521–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Matsuzaki K., Borel V., Adelman C.A., Schindler D., Boulton S.J.. FANCJ suppresses microsatellite instability and lymphomagenesis independent of the Fanconi anemia pathway. Genes Dev. 2015; 29:2532–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barthelemy J., Hanenberg H., Leffak M.. FANCJ is essential to maintain microsatellite structure genome-wide during replication stress. Nucleic Acids Res. 2016; 44:6803–6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Guillemette S., Branagan A., Peng M., Dhruva A., Scharer O.D., Cantor S.B.. FANCJ localization by mismatch repair is vital to maintain genomic integrity after UV irradiation. Cancer Res. 2014; 74:932–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Murina O., von Aesch C., Karakus U., Ferretti L.P., Bolck H.A., Hanggi K., Sartori A.A.. FANCD2 and CtIP cooperate to repair DNA interstrand crosslinks. Cell Rep. 2014; 7:1030–1038. [DOI] [PubMed] [Google Scholar]
- 67. Unno J., Itaya A., Taoka M., Sato K., Tomida J., Sakai W., Sugasawa K., Ishiai M., Ikura T., Isobe T. et al. FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep. 2014; 7:1039–1047. [DOI] [PubMed] [Google Scholar]
- 68. Prakash R., Zhang Y., Feng W., Jasin M.. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 2015; 7:a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu C., Srihari S., Cao K.A., Chenevix-Trench G., Simpson P.T., Ragan M.A., Khanna K.K.. A fine-scale dissection of the DNA double-strand break repair machinery and its implications for breast cancer therapy. Nucleic Acids Res. 2014; 42:6106–6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ceccaldi R., Rondinelli B., D’Andrea A.D.. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016; 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Aparicio T., Baer R., Gautier J.. DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst). 2014; 19:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xie J., Peng M., Guillemette S., Quan S., Maniatis S., Wu Y., Venkatesh A., Shaffer S.A., Brosh R.M. Jr, Cantor S.B.. FANCJ/BACH1 acetylation at lysine 1249 regulates the DNA damage response. PLoS Genet. 2012; 8:e1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yu X., Wu L.C., Bowcock A.M., Aronheim A., Baer R.. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 1998; 273:25388–25392. [DOI] [PubMed] [Google Scholar]
- 74. Wong A.K., Ormonde P.A., Pero R., Chen Y., Lian L., Salada G., Berry S., Lawrence Q., Dayananth P., Ha P. et al. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene. 1998; 17:2279–2285. [DOI] [PubMed] [Google Scholar]
- 75. Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P.. Human CtIP promotes DNA end resection. Nature. 2007; 450:509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chen L., Nievera C.J., Lee A.Y., Wu X.. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008; 283:7713–7720. [DOI] [PubMed] [Google Scholar]
- 77. Wu Y., Sommers J.A., Suhasini A.N., Leonard T., Deakyne J.S., Mazin A.V., Shin-Ya K., Kitao H., Brosh R.M. Jr. Fanconi anemia group J mutation abolishes its DNA repair function by uncoupling DNA translocation from helicase activity or disruption of protein-DNA complexes. Blood. 2010; 116:3780–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Anand R.P., Lovett S.T., Haber J.E.. Break-induced DNA replication. Cold Spring Harb. Perspect. Biol. 2013; 5:a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Malkova A., Ira G.. Break-induced replication: functions and molecular mechanism. Curr. Opin. Genet. Dev. 2013; 23:271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Stephens P.J., McBride D.J., Lin M.L., Varela I., Pleasance E.D., Simpson J.T., Stebbings L.A., Leroy C., Edkins S., Mudie L.J. et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009; 462:1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A. et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012; 149:979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Menghi F., Inaki K., Woo X., Kumar P.A., Grzeda K.R., Malhotra A., Yadav V., Kim H., Marquez E.J., Ucar D. et al. The tandem duplicator phenotype as a distinct genomic configuration in cancer. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E2373–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Carr A.M., Lambert S.. Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J. Mol. Biol. 2013; 425:4733–4744. [DOI] [PubMed] [Google Scholar]
- 84. Slack A., Thornton P.C., Magner D.B., Rosenberg S.M., Hastings P.J.. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2006; 2:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Anand R.P., Tsaponina O., Greenwell P.W., Lee C.S., Du W., Petes T.D., Haber J.E.. Chromosome rearrangements via template switching between diverged repeated sequences. Genes Dev. 2014; 28:2394–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sakofsky C.J., Ayyar S., Deem A.K., Chung W.H., Ira G., Malkova A.. Translesion polymerases drive microhomology-mediated break-induced replication leading to complex chromosomal rearrangements. Mol. Cell. 2015; 60:860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hartlerode A.J., Willis N.A., Rajendran A., Manis J.P., Scully R.. Complex breakpoints and template switching associated with non-canonical termination of homologous recombination in mammalian cells. PLoS Genet. 2016; 12:e1006410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.