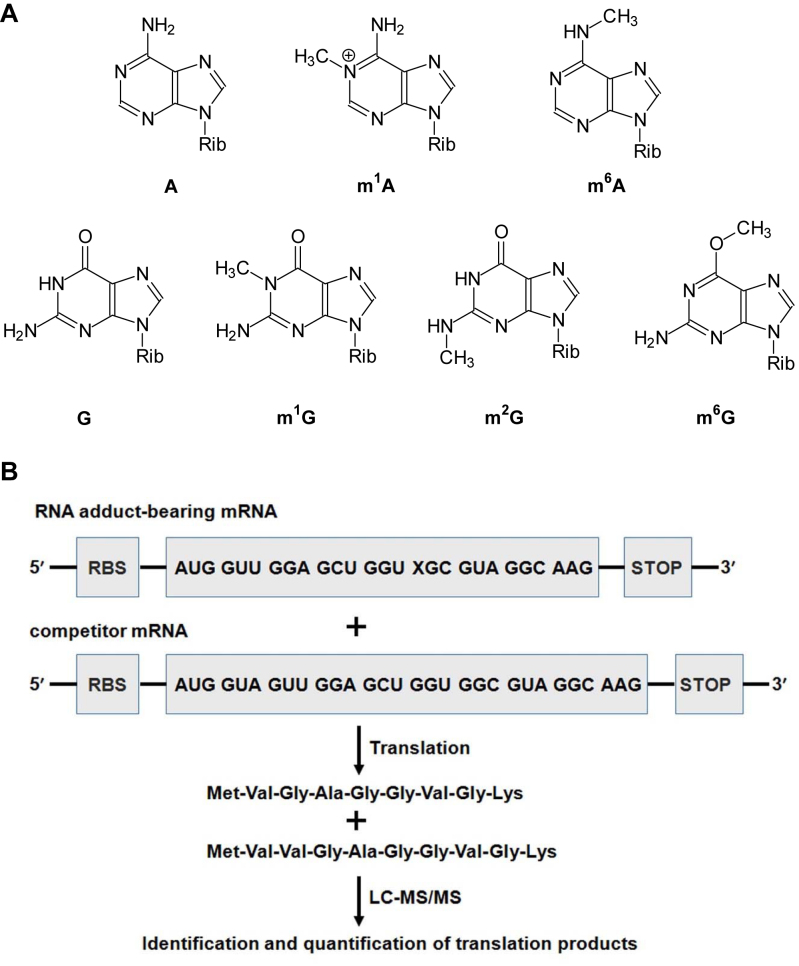
Abstract
Reversible methylation of the N6 or N1 position of adenine in RNA has recently been shown to play significant roles in regulating the functions of RNA. RNA can also be alkylated upon exposure to endogenous and exogenous alkylating agents. Here we examined how regio-specific methylation at the hydrogen bonding edge of adenine and guanine in mRNA affects translation. When situated at the third codon position, the methylated nucleosides did not compromise the speed or accuracy of translation under most circumstances. When located at the first or second codon position, N1-methyladenosine (m1A) and m1G constituted robust blocks to both Escherichia coli and wheat germ extract translation systems, whereas N2-methylguanosine (m2G) moderately impeded translation. While m1A, m2G and N6-methyladenosine (m6A) did not perturb translational fidelity, O6-methylguanosine (m6G) at the first and second codon positions was strongly and moderately miscoding, respectively, and it was decoded as an adenosine in both systems. The effects of methylated ribonucleosides on translation could be attributed to the methylation-elicited alterations in base pairing properties of the nucleobases, and the mechanisms of ribosomal decoding contributed to the position-dependent effects. Together, our study afforded important new knowledge about the modulation of translation by methylation of purine nucleobases in mRNA.
INTRODUCTION
Recent transcriptome-wide mapping studies have revealed the widespread occurrence of 5-methylcytidine, N1-methyladenosine (m1A) and N6-methyladenosine (m6A) in messenger RNA (mRNA) (1–3). In addition, proteins involved in the deposition, removal and recognition of m6A have been discovered and found to play important roles in modulating the stability, localization and translational efficiencies of mRNA (4–9). Thus, reversible methylation at the N1 and N6 positions of adenosine is thought to assume significant roles in gene regulation.
Aside from those natural methylations that are important in gene regulation, RNA can also be inadvertently alkylated upon exposure to alkylating agents that are ubiquitously present in the environment and in living cells (10). Such exposure gives rise to the conjugation of alkyl groups to the ring nitrogen as well as the exocyclic oxygen and nitrogen atoms of all nucleobases (11). In this context, owing to the lack of secondary structure and nucleobase protection from hydrogen bonding, nucleobases in mRNA are thought to be particularly susceptible to alkylation (12).
It has been well documented that alkylation in DNA may perturb the flow of genetic information by altering the efficiency and fidelity of DNA replication and transcription (13–15), and recent studies also revealed that modified nucleosides in mRNA could modulate translation. For instance, pseudouridylation could promote read-through translation via increased miscoding of a pseudouridylated stop codon (16), and depurination or nucleobase oxidation in mRNA could stall ribosome and produce truncated peptide/protein products (17–21). A few studies have also been conducted for investigating how the efficiency and accuracy of ribosomal decoding are influenced by alkylation in mRNA (12,22,23). No systematic study, however, has yet been conducted for assessing the effect of alkylated ribonucleosides on translational perturbation. Herein, we developed a quantitative mass spectrometry-based assay to assess the extents to which a single m1G, m2G, m6G, m1A or m6A (Figure 1) at defined codon positions in mRNA templates affects the speed and accuracy of translation mediated by prokaryotic and eukaryotic translation systems.
Figure 1.
Experimental outline. (A) Chemical structures of A, m1A, m6A, G, m1G, m2G and m6G. ‘Rib’ indicates ribose. (B) A schematic diagram illustrating the procedures employed for assessing the impact of the methylated ribonucleosides on translation. ‘X’ indicates a methylated adenosine or guanosine. ‘RBS’ and ‘STOP’ designate the ribosome binding site and the stop codon, respectively. Only the mRNA containing an m1G, m2G, m6G or G at the first position of sixth codon of the mRNA, as well as the wild-type peptide product MVGAGGVGK and the competitor peptide product MVVGAGGVGK are shown.
MATERIALS AND METHODS
Materials
Unmodified oligoribonucleotides (ORNs) and oligodeoxyribonucleotides (ODNs) were purchased from Integrated DNA Technologies, and the modified ORNs used in this study were obtained from Thermo Scientific Dharmacon. All enzymes and chemicals, unless otherwise specified, were purchased from New England Biolabs and Sigma-Aldrich, respectively. [γ-32P]ATP was obtained from Perkin Elmer.
Translation template preparation
The translation templates were prepared using a two-piece splint ligation procedure as previously described (Supplementary Figure S1) (24). We first constructed mRNA templates containing a single methylated ribonucleoside or its unmodified counterpart at the first position of the codon for the reconstituted Escherichia coli translation system. To this end, a 28-mer ORN (5′-GGUUGGAGCUGGUXGCGUAGGCAAGUAA-3′, X = m1G, m2G, m6G or G) was phosphorylated on the 5′ terminus using T4 polynucleotide kinase (T4 PNK) and ligated to a 24-mer ORN (5′-GGGAAUUCUAAGGAGGAUAUACAU-3′) using a 36-mer template ODN (5′-CGCCACCAGCTCCAACCATGTATATCCTCCTTAGAA-3′) and T4 DNA ligase (24). The ligation products were purified using RNA Clean & Concentrator™-5 Kit (Zymo Research) according to the manufacturer's instructions. Using the same method, we prepared translation templates containing a single m1G, m2G, m6G or G at the second and third positions of the codon for the reconstituted E. coli translation system, where the 28-mer ORNs were 5′-GGUUGGAGCUGGUGXCGUAGGCAAGUAA-3′ and 5′-GGUUGGAGCUGGUGGXGUAGGCAAGUAA-3′, respectively. We also prepared translation templates containing a single m1A, m6A or A at the first, second and third positions of the codon for the reconstituted E. coli translation system, where the 28-mer ORNs were 5′-GGUUGGAGCUGGUGGGXCUCGCAAGUAA-3′, 5′-GGUUGGAGCUGGUGGG GXCUCCAAGUAA-3′ and 5′-GGUUGGAGCUGGUGGGGGXCUCAAGUAA-3′, respectively. In this context, because m6A is known to be situated in a consensus sequence of GG(m6A)CU (1), we placed the two methylated adenosine derivatives in this specific sequence context. A competitor mRNA template was constructed by using a 31-mer ORN (5′-GGUAGUUGGAGCUGGUGGCGUAGGCAAGUAA-3′) in lieu of the 28-mer ORN. The translation templates for the cell-free wheat germ extract system were prepared in a similar way, except that the sequences of the ORN and the template ODN were 5′-GGGAGAGCCACCAU-3′ and 5′-CGCCACCAGCTCCAACCATGGTGGCTCTCCC-3′, respectively.
Normalization of translation templates
To determine the relative concentrations of translation templates for reactions with the reconstituted E. coli translation system, the aforementioned 52-mer methylated nucleoside-bearing RNA ligation products, their unmodified counterparts or the 55-mer competitor RNA ligation products were individually premixed with an equal amount of 45-mer RNA product, which was used as a reference and the mixtures were radiolabeled with the use of [γ-32P]ATP and T4 PNK. The radiolabeled products were separated using 12% denaturing polyacrylamide gel (acrylamide:bis-acrylamide = 29:1) containing 7 M urea, and the bands of interest were analyzed using Typhoon 9410 phosphorimager and ImageQuant software (GE Healthcare), as described elsewhere (Supplementary Figures S2 and 3). The normalization of translation templates for experiments using the cell-free wheat germ extract system was conducted in a similar fashion.
In vitro bacterial translation assay
The 52-mer methylated ribonucleoside-bearing or the corresponding unmodified control mRNA was premixed with the 55-mer competitor mRNA at a molar ratio of 6:1 (modified RNA or control/competitor). The mRNA mixtures were then employed as templates for in vitro translation reactions with PURExpress®In Vitro Protein Synthesis Kit (New England Biolabs), following the manufacturer's instructions. A typical reaction contained ∼5 μg of translation template, 20 U of RNase inhibitor, 10 μl of manufacturer's solution A and 7.5 μl of solution B in a 25 μl mixture and incubated at 37°C for 1 h.
In vitro eukaryotic translation assay
The 42-mer methylated ribonucleoside-bearing or the corresponding unmodified control mRNA was premixed with the 45-mer competitor mRNA at a molar ratio of 6:1 (modified RNA or control/competitor) and used as templates for in vitro eukaryotic translation assay. Translation reactions were performed using the wheat germ extract cell-free system (Promega) according to the manufacturer's instructions, with minor modifications. A typical reaction contained ∼5 μg of translation template, 40 U of RNase inhibitor, 1% (v/v) of protease inhibitor cocktail (Sigma-Aldrich), 4 μl of 1 mM amino acid mixture (Promega) and 4 μl of wheat germ extract in a 50 μl mixture and incubated at 25°C for 30 min.
Peptide extraction
The peptide products were isolated from the in vitro translation reactions as described previously (25,26). Briefly, the reaction mixture was extracted with an equal volume of methanol by votexing for 15 s, followed by the addition of an equal volume of n-butanol and votexing for 30 s. The resulting mixture was incubated at −80°C for 1 h and then centrifuged at 13 000 rpm at room temperature for 5 min to separate the peptides from proteins. The resulting supernatant was dried with Speed-vac and redissolved in water for mass spectrometric analysis.
Mass spectrometry analysis
The peptide products were analyzed by LC-MS using an EASY-nLC 1200 system coupled with a Q Exactive Plus Hydrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Peptides were separated by using a homemade trapping column (150 μm × 50 mm) and a separation column (75 μm × 120 mm), packed with ReproSil-Pur C18-AQ resin (3 μm in particle size, Dr Maisch HPLC GmbH, Germany). The peptide mixture was initially loaded onto the trapping column with a solvent mixture of 0.1% formic acid in CH3CN/H2O (2:98, v/v) at a flow rate of 3.0 μl/min. The peptides were then separated using a 90-min linear gradient of 4–40% acetonitrile in 0.1% formic acid and at a flow rate of 230 nl/min. To identify the translation products of mRNA carrying a single m1G, m2G or m6G at any position of the codon used in this study, the mass spectrometer was set up for monitoring the fragmentation of the [M+2H]2+ ions of 10-amino acid peptide (MVVGAGGVGK), 9-amino acid peptides (MVGAGXVGK, where ‘X’ designates any of the 20 canonical amino acids) as well as the possible truncated peptides including MVGAG, MVGAGG, MVGAGGV and MVGAGGVG. To identify the translation products of mRNA carrying a single m1A or m6A at the first codon position used in this study, the mass spectrometer was set up for monitoring the fragmentation of the [M+2H]2+ ions of 10-amino acid peptide (MVVGAGGVGK), 9-amino acid peptides (MVGAGGXRK, where ‘X’ indicates any of the 20 natural amino acids) as well as the possible truncated peptides including MVGAGG, MVGAGGT and MVGAGGTR. To identify the translation products of mRNA carrying a single m1A or m6A at the second codon position used in this study, the mass spectrometer was set up for monitoring the fragmentations of the [M+2H]2+ ions of 10-amino acid peptide (MVVGAGGVGK), 9-amino acid peptides (MVGAGGXSK, where ‘X’ designates any of the 20 natural amino acids) as well as the possible truncated peptides including MVGAGG, MVGAGGD and MVGAGGDS. To identify the translation products of mRNA carrying a single m1A or m6A at the third codon position used in this study, the mass spectrometer was set up for monitoring the fragmentation of the [M+2H]2+ ions of 10-amino acid peptide (MVVGAGGVLK), 9-amino acid peptides (MVGAGGXLK, where ‘X’ designates any of the 20 canonical amino acids) as well as the possible truncated peptides including MVGAGG, MVGAGGG and MVGAGGGL.
RESULTS
We developed a novel mass spectrometry-based assay to investigate the effects of methylated purine nucleosides in mRNA on translation (Figure 1B). To this end, we prepared mRNA templates harboring a single, site-specifically inserted methylated ribonucleoside (m1G, m2G, m6G, m1A or m6A, Figure 1A) as well as the corresponding control and competitor mRNA templates. The five methylated nucleosides were chosen for assessing the regioisomeric effects of nucleobase methylation on translation, and m1A and m6A were selected also because of their recently discovered functions in gene regulation. In addition, ribosomal decoding of mRNA necessitates strict Watson–Crick base pairing between the codon and anticodon, particularly at the first and second nucleotide positions of the codon (27–29). Thus, the use of these methylated nucleosides will allow us to assess systematically how installation of a methyl group to the Watson-Crick hydrogen bonding edge of the two purine nucleosides affects translational fidelity and efficiency.
The control mRNA templates comprised of a ribosome-binding site (RBS), the AUG start codon, sequences encoding eight amino acids and a stop codon. The RBS sequences were AAGGAG and GCCACC for prokaryotic and eukaryotic translation reactions, respectively (Figure 1B and Supplementary Figure S4). The modified mRNA templates are the same as the control except that the adenosine or guanosine at the first, second or third position of the sixth codon was replaced with the corresponding methylated nucleoside, thereby facilitating the determination of the position-dependent effects of the regioisomeric methylated adenosine and guanosine derivatives on translation. Relative to the unmodified control mRNA, the competitor mRNA harbors an additional codon immediately downstream of the start codon (Figure 1B).
The methylated ribonucleoside-bearing mRNA or the respective unmodified mRNA was premixed individually with the competitor mRNA at defined molar ratios and employed as templates for in vitro translation. The resulting peptide products were isolated from the translation reaction mixture and subjected to mass spectrometric analyses (Figure 1B). The miscoding potential of a modified ribonucleoside during translation is determined by the signal intensity of mutant peptide product(s) over the total signal intensity of all peptide products arising from the translation of methylated nucleoside-bearing mRNA. The translation bypass efficiency (TBE), which characterizes the degree to which a methylated ribonucleoside affects the speed of mRNA translation, is quantified by normalizing the ratio of the total signal observed for the methylated mRNA template to that for the competitor mRNA template against the ratio obtained from the corresponding control experiment.
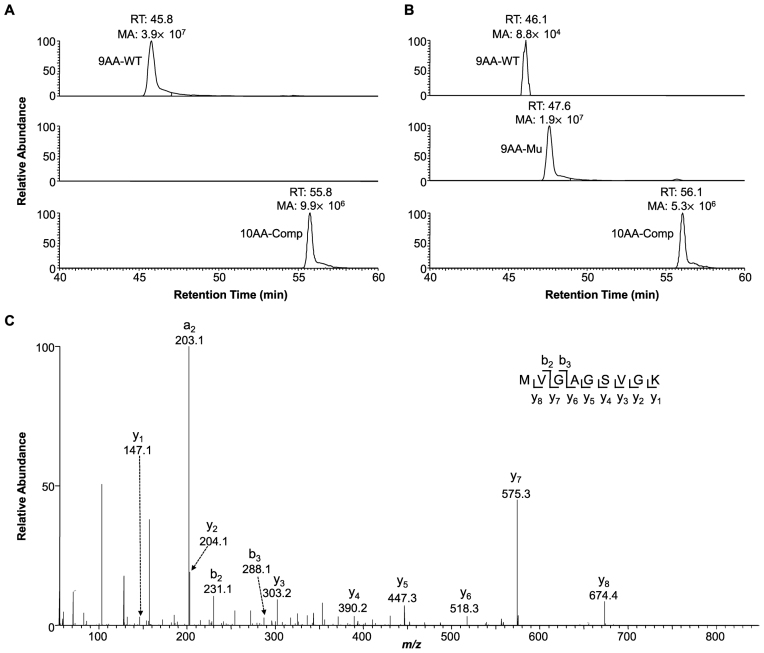
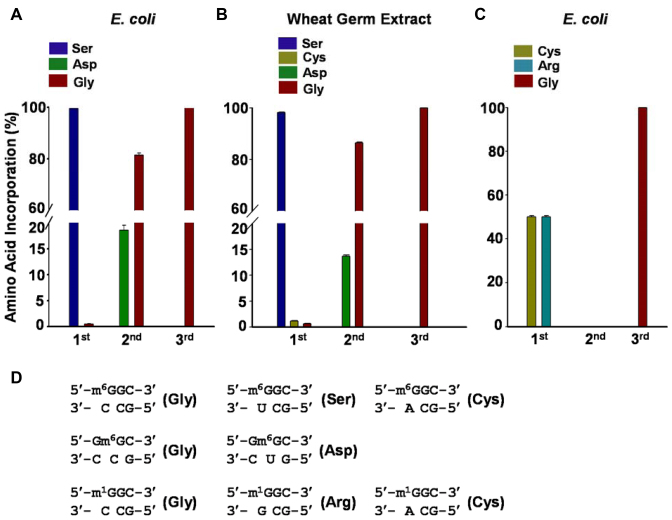
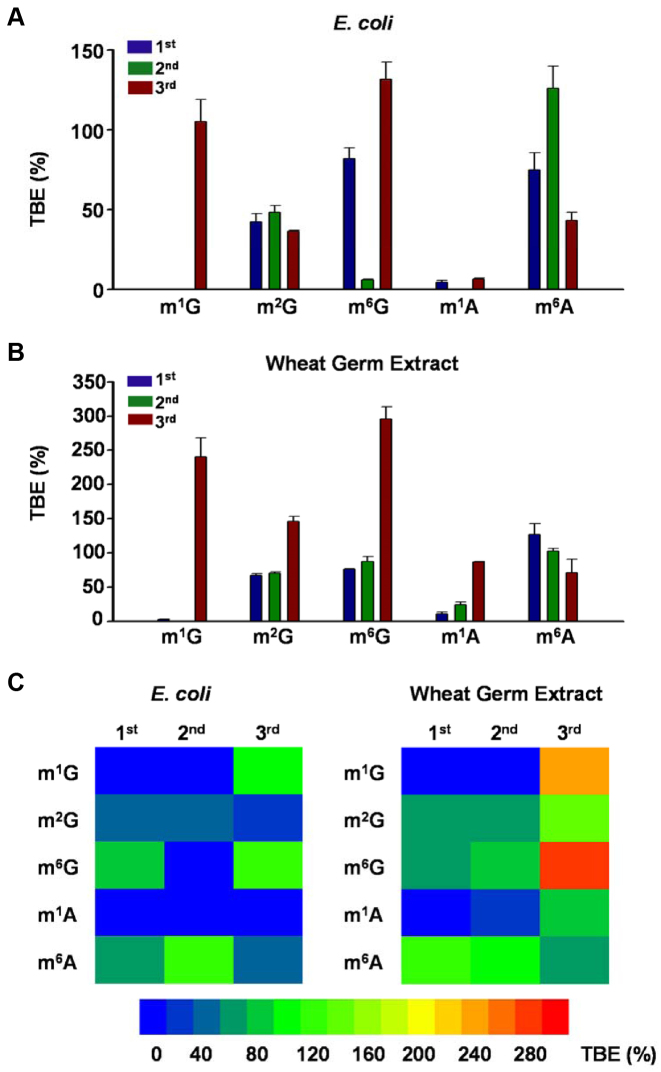
By using this method, we first examined how m1G, m2G and m6G affect the efficiency and fidelity of translation using a reconstituted translation system that supplies E. coli ribosome and its associated general translation factors (30). Analyses of the peptide products by electrospray ionization-mass spectrometry (ESI-MS) and tandem MS (MS/MS) revealed that, as expected, only the wild-type peptide product MVGAGGVGK and the competitor peptide product MVVGAGGVGK were detectable in the control experiments where the unmodified control mRNA was co-translated with the competitor mRNA (Figure 2A and Supplementary Figure S5). In addition, our results showed that m6G at the first position of the sixth codon (m6GGC) was decoded by the ribosome predominantly as an adenosine, which is reflected by the misincorporation of serine at a frequency of ∼99%. In addition, significant decoding of m6G as an adenosine was also observed when the methylated nucleoside is situated at the second codon position (i.e. Gm6GC), as manifested by the detection of ∼18% misincorporation of aspartic acid at this codon position (Supplementary Figure S6). In contrast, the placement of m6G at the third position of the codon (i.e. GGm6G) did not lead to any detectable mutant peptide products (Figures 2B, C and 3A). While m6G did not alter appreciably the translation efficiency when placed at the first or third codon position, its presence at the second codon position markedly reduced the translation efficiency, as indicated by a ∼6% TBE value (Figure 4A).
Figure 2.
Representative LC-MS results for monitoring the influence of m6G on translation. The relative abundances of the wild-type peptide product MVGAGGVGK (i.e. 9AA-WT), the mutant product MVGAGSVGK (i.e. 9AA-Mu) and the competitor peptide product MVVGAGGVGK (i.e. 10AA-Comp) from the reconstituted Escherichia coli translation reactions, where GGC and m6GGC were used as the sixth codon of the unmodified (A) and the m6G-bearing (B) mRNA templates, respectively. ‘RT’, retention time; ‘MA’, peak area found in the selected-ion chromatogram for monitoring the formation of the [M+2H]2+ ions of the peptide products. (C) The MS/MS of the [M+2H]2+ ion of the mutant peptide product MVGAGSVGK from the reconstituted E. coli translation reaction, where m6GGC was the sixth codon of the m6G-bearing mRNA template.
Figure 3.
The effects of m6G and m1G in mRNA on the translational fidelity. (A and B) Quantification of the peptides carrying an a glycine (Gly), serine (Ser) or aspartic acid (Asp) at the sixth codon of the m6G-bearing mRNA templates during the reactions mediated by Escherichia coli (A) or the wheat germ extract (B) translation systems. (C) Quantification of the peptides carrying a glycine (Gly), cysteine (Cys) or arginine (Arg) at the sixth codon of the m1G-bearing mRNA templates produced from the reactions mediated by the wheat germ extract translation systems. ‘1st’, ‘2nd’ and ‘3rd’ indicate the first, second and third positions of the codon, respectively. The data represent the mean and standard error of results from three independent experiments. (D) The codon-anticodon pair involved in the generation of translation products for the m6G- and m1G-bearing mRNAs.
Figure 4.
Effects of methylated adenosine and guanosine derivatives in mRNA on translation efficiency. Shown are translation bypass efficiencies (TBEs) of m1G, m2G, m6G, m1A and m6A in the translation reactions mediated by the reconstituted Escherichia coli (A) and wheat germ extract (B) translation systems. A summary of TBE values are also displayed in heatmaps (C). ‘1st’, ‘2nd’ and ‘3rd’ indicate the first, second and third positions of the codon, respectively. The data represent the mean and standard error of results from three independent experiments.
Different from the findings made for m6G, an m1G present at the first or second codon position completely inhibited translation mediated by the E. coli translation machinery, though its presence at the third position of the codon did not result in any significant alteration in the speed or accuracy of E. coli translation (Figure 4A). On the other hand, m2G, regardless of being situated at the first, second or third position of the codon, did not induce any mutations during translation mediated by the E. coli translation apparatus; however, they constituted moderate impediments to translation, with the TBE values being ∼42, ∼48 and ∼36%, respectively (Figure 4A).
We next investigated the effects of m1G, m2G or m6G on eukaryotic translation using a cell-free wheat germ extract system. Reminiscent of what we observed for the E. coli system, m6G at the first and second positions of the trinucleotide codon was again decoded as an adenosine at frequencies of ∼99 and ∼13%, respectively, though no perturbation in translation fidelity was observed when it is located at the third position of the codon (Figure 3B; Supplementary Figures S7 and 8). Interestingly, we also observed a very low frequency of Gly→Cys substitution (∼1.5%) at the m6GGC codon, for which the m6G was decoded as a uridine (Figure 3B and D). In addition, we did not detect any alteration in translation efficiency when this modified nucleoside is situated at the first or second position of the codon (Figure 4B and C). However, instead of impeding translation, the placement of m6G at the third position of the codon enhanced wheat germ extract-mediated translation, with a TBE value of ∼ 300% (Figure 4B and C).
Our results showed that m1G and m2G were recognized by the wheat germ extract translation system in a very similar fashion as that by the E. coli translational machinery, albeit with some exceptions. First, the m1G in the m1GGC codon was decoded by the ribosome as a cytidine or uridine and directed the misincorporation of arginine and cysteine, respectively, at frequencies of 50% each (Figure 3C and D and Supplementary Figure S9). Second, the placement of m1G and m2G at the third position of the codon increased markedly the yield of full-length translation product, with the TBE values being ∼240 and 150%, respectively (Figure 4B and C). Third, the presence of m2G at the first and second positions of the codon only slightly diminished the translation efficiency in the wheat germ extract system, with the TBE values being ∼67% and ∼71%, respectively (Figure 4B and C).
Having examined the effects of m1G, m2G and m6G on translation, we next asked how m1A and m6A affect translation. We found that, in both translation systems, the introduction of a single m1A or m6A into any position of the codon was not mutagenic. However, m1A could strongly impede translation when placed at any of the three codon positions for the E. coli system (with the TBE values being <10%, Figure 4A and C) and when placed at the first and second positions of the codon for the wheat germ extract system (with the TBE values being 10–24%, Figure 4B and C). The presence of m1A at the third position, however, did not alter the translation efficiency of the wheat germ extract system. The results also revealed that m6A at the first and second codon positions exerted negligible impact on the alterations of the peptide products in the prokaryotic or eukaryotic translation system (Figure 4). In addition, m6A at the third position of the codon did not substantially perturb translation in the wheat germ extract system, though it resulted in a moderate repression (by ∼57%) of translation mediated by the reconstituted E. coli translation system (Figure 4A and B).
DISCUSSION
In this study, we assessed systematically how methylated adenosine and guanosine derivatives in mRNA templates affect the speed and accuracy of translation mediated by prokaryotic and eukaryotic systems. We were able to draw several important conclusions about how ribosomal decoding is affected by the locations of methyl group on the nucleobase and by the positions of the methylated nucleosides in the trinucleotide codon. Additionally, we were able to compare the prokaryotic and eukaryotic translational machineries with respect to their recognition of the methylated nucleosides.
When situated at the first and second positions of the trinucleotide codon, the regioisomeric methylated nucleosides displayed different effects on the fidelity and efficiency of translation. Methylation at the N1 position of adenosine or guanosine exerted pronounced blockage effects on translation mediated by both E. coli and wheat germ extract translation systems, regardless of whether the methylated nucleosides are placed at the first or second codon position (Figure 4). This observation is not surprising from the standpoint that the addition of a methyl group to the center of the Watson–Crick hydrogen bonding edge would disrupt base pairing between the methylated nucleobase in mRNA template and the anticodon base in aa-tRNA at the decoding center of the ribosome. Unlike m1A which does not give rise to decoding error, m1G situated at the first codon position were decoded as a C or U by the wheat germ extract translation machinery (Figure 3). Methylations at the major-groove O6 position of guanine or the N6 position of adenine elicited distinct effects on translation fidelity (Figure 3): while the accuracy of ribosomal decoding was not affected by the replacement of A with m6A, m6G was decoded incorrectly as an A, and the magnitude of the decoding error for m6G was dependent on whether the modified nucleoside is situated at the first (98–99% by both translation systems) or second (18.6 and 13.6% by the E. coli and wheat germ extract systems, respectively) position of the trinucleotide codon (Figure 3). Although no apparent inhibition on translation was observed for m6A, marked inhibition of translation was only detected for m6G for the E. coli translation system when the modified nucleoside was located at the second codon position (Figure 4). Methylation at the minor-groove N2 position of guanine did not compromise the fidelity of translation, and it only moderately repressed translation when situated at the first or second position of the trinucleotide codon (Figures 3 and 4). Our results also uncovered the absence of blockage or miscoding effects from any of the methylated nucleosides when placed at the third position of the trinucleotide codon, except that GGm1A is a strong block to the E. coli translation system (Figures 3 and 4). The above findings are in agreement with the X-ray crystal structure of the E. coli 30S ribosome complex, which revealed that the minor groove of the first and second base pairs of the codon-anticodon helix is closely monitored by the ribosome via interaction with the 16S ribosomal RNA through A1493 and both A1492 and G530, respectively (27,28). Unlike the findings made in the E. coli translation system, we found that, in wheat germ extract system, m1G and m6G at the third position of the codon substantially promoted translational efficiency (Figure 4). The exact reason underlying the elevated translation efficiency is unclear, though we speculate that the methylation at the third position of the trinucleotide codon may stimulate the efficiency of translocation by promoting the movement of tRNA from P site to E site after nascent peptide bond formation. In this vein, it is worth noting that elevated levels of m6A in mRNA transcripts were previously found to stimulate translation in a rabbit reticulocyte system, but not in a wheat germ extract system (31). These findings suggested that, under some circumstances, the impact of methylated ribonucleosides in mRNA on translation efficiency of eukaryotic systems may be species-specific (31). With a few exceptions as discussed above, the impact of the methylated nucleosides on translation mediated by the E. coli and wheat germ extract systems (Figures 3 and 4) are very similar, which is in accordance with the notion that ribosomal decoding is a highly conserved process (27,28,32).
It is worth comparing the results obtained from the present work with previously published data from the replication and/or transcription studies for the same set of methylated nucleobases in DNA. In this vein, O6-methylguanine does not strongly block DNA replication or transcription, and it miscodes preferentially as an adenine during these processes (33,34). In addition, N1-methylguanine exhibits strong inhibitory and mutagenic effects on DNA replication, whereas N1-methyladenine only compromises strongly the efficiency, but not the fidelity of DNA replication (35). Moreover, N2-methylguanine does not affect the efficiency or fidelity of DNA replication in E. coli cells (36). Thus, the stalling and miscoding properties of the methylated adenine and guanine bases are very similar when they are encountered by polymerases during replication or transcription and by ribosomes during translation, particularly when the methylated nucleosides are situated at the first and second positions of the trinucleotide codon.
It is known that the repair and replicative bypass O6-methylguanine in DNA can be influenced by sequence contexts (33,37). Likewise, a recent study showed that the local sequences surrounding m6A-modified codon could affect the magnitude of translational effects (38). Thus, some differences in effects of m6A in mRNA on translation observed between the present and previous studies (23) might be attributed to the use of different sequence contexts for the mRNA templates.
RNA contains more than 100 distinct types of natural modifications, which can modulate the structure and functions of RNA (39). Among them, m6A is the most abundant internal modification in mRNA that can mediate post-transcriptional regulation of gene expression via its binding proteins (8,40). m1A is another prevalent mRNA modification in eukaryotic cells, though the biological functions of m1A in mRNA remain elusive (2,3). The lack of miscoding potential of m6A and m1A on translation is in keeping with their roles in regulating RNA function. In addition, blockage to translation mediated by natural methylation at the N1 position of adenine may constitute an alternative mechanism for post-transcriptional regulation of gene expression.
The results from our study showed that translational bypass of some methylated ribonucleosides (e.g. m1G and m6G) can lead to amino acid substitutions in proteins, whereas that of others can stall translational machinery. While amino acid changes in proteins may disrupt their functions, stalling of translation is known to activate ribosome-based mRNA surveillance mechanism known as no-go decay, which leads to the degradation of mRNA and immature polypeptides (19,21,41,42). Thus, our results suggest that inadvertent methylation arising from environmental exposure or endogenous metabolism may justify the need for an evolutionarily conserved mRNA quality control system (42) and possibly RNA repair mechanism(s). In the latter respect, it is of note that E. coli AlkB was documented to be capable of removing simple methyl groups from N-alkylated ribonucleosides (43,44) and some human orthologs of AlkB are known to be involved in the demethylation of m6A and m1A (5,6,45).
It is also worth discussing the novelty of the method developed in the present study. Through the incorporation of mass spectrometry into the workflow, the in vitro translation assay described here enables rapid and unambiguous identification and quantification of translation products. With the use of competitor mRNA template as an internal standard, the method facilitates the outcome from a single biological experiment to be employed for interrogating quantitatively the degrees to which the efficiency and fidelity of mRNA translation are modulated by methylated ribonucleosides. We envision that the method should be generally amenable for assessing how other modified ribonucleosides modulate translation.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R21 ES025392 to Y. W., T32 ES018827 for supporting X.D.]; Scientific Research Foundation of Hunan University [531107050937 to Y.C.]. Funding for open access charge: NIH [R21 ES025392].
Conflict of interest statement. None declared.
REFERENCES
- 1. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 2. Li X., Xiong X., Wang K., Wang L., Shu X., Ma S., Yi C.. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol. 2016; 12:311–316. [DOI] [PubMed] [Google Scholar]
- 3. Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M.S., Dai Q., Di Segni A., Salmon-Divon M., Clark W.C. et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016; 530:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G. et al. N 6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vagbo C.B., Shi Y., Wang W.L., Song S.H. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C.. N 6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.B.. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015; 526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drablos F., Feyzi E., Aas P.A., Vaagbo C.B., Kavli B., Bratlie M.S., Pena-Diaz J., Otterlei M., Slupphaug G., Krokan H.E.. Alkylation damage in DNA and RNA–repair mechanisms and medical significance. DNA Repair. 2004; 3:1389–1407. [DOI] [PubMed] [Google Scholar]
- 11. Liu S., Wang Y.. Mass spectrometry for the assessment of the occurrence and biological consequences of DNA adducts. Chem. Soc. Rev. 2015; 44:7829–7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simms C.L., Zaher H.S.. Quality control of chemically damaged RNA. Cell Mol. Life Sci. 2016; 73:3639–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shrivastav N., Li D., Essigmann J.M.. Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis. 2010; 31:59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bregeon D., Doetsch P.W.. Transcriptional mutagenesis: causes and involvement in tumour development. Nat. Rev. Cancer. 2011; 11:218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. You C., Wang Y.. Mass spectrometry-based quantitative strategies for assessing the biological consequences and repair of DNA adducts. Acc. Chem. Res. 2016; 49:205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Svidritskiy E., Madireddy R., Korostelev A.A.. Structural basis for translation termination on a pseudouridylated stop codon. J. Mol. Biol. 2016; 428:2228–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tanaka M., Chock P.B., Stadtman E.R.. Oxidized messenger RNA induces translation errors. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shan X., Chang Y., Lin C.L.. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASEB J. 2007; 21:2753–2764. [DOI] [PubMed] [Google Scholar]
- 19. Gandhi R., Manzoor M., Hudak K.A.. Depurination of Brome mosaic virus RNA3 in vivo results in translation-dependent accelerated degradation of the viral RNA. J. Biol. Chem. 2008; 283:32218–32228. [DOI] [PubMed] [Google Scholar]
- 20. Calabretta A., Kupfer P.A., Leumann C.J.. The effect of RNA base lesions on mRNA translation. Nucleic Acids Res. 2015; 43:4713–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simms C.L., Hudson B.H., Mosior J.W., Rangwala A.S., Zaher H.S.. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 2014; 9:1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hudson B.H., Zaher H.S.. O 6-Methylguanosine leads to position-dependent effects on ribosome speed and fidelity. RNA. 2015; 21:1648–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoernes T.P., Clementi N., Faserl K., Glasner H., Breuker K., Lindner H., Huttenhofer A., Erlacher M.D.. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 2016; 44:852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kershaw C.J., O’Keefe R.T.. Splint ligation of RNA with T4 DNA ligase. Methods Mol. Biol. 2012; 941:257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hackett P.B., Petersen R.B., Hensel C.H., Albericio F., Gunderson S.I., Palmenberg A.C., Barany G.. Synthesis in vitro of a seven amino acid peptide encoded in the leader RNA of Rous sarcoma virus. J. Mol. Biol. 1986; 190:45–57. [DOI] [PubMed] [Google Scholar]
- 26. Raney A., Baron A.C., Mize G.J., Law G.L., Morris D.R.. In vitro translation of the upstream open reading frame in the mammalian mRNA encoding S-adenosylmethionine decarboxylase. J. Biol. Chem. 2000; 275:24444–24450. [DOI] [PubMed] [Google Scholar]
- 27. Ogle J.M., Brodersen D.E., Clemons W.M. Jr, Tarry M.J., Carter A.P., Ramakrishnan V.. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001; 292:897–902. [DOI] [PubMed] [Google Scholar]
- 28. Ogle J.M., Murphy F.V., Tarry M.J., Ramakrishnan V.. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002; 111:721–732. [DOI] [PubMed] [Google Scholar]
- 29. Rozov A., Westhof E., Yusupov M., Yusupova G.. The ribosome prohibits the G*U wobble geometry at the first position of the codon-anticodon helix. Nucleic Acids Res. 2016; 44:6434–6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., Ueda T.. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001; 19:751–755. [DOI] [PubMed] [Google Scholar]
- 31. Heilman K.L., Leach R.A., Tuck M.T.. Internal 6-methyladenine residues increase the in vitro translation efficiency of dihydrofolate reductase messenger RNA. Int. J. Biochem. Cell Biol. 1996; 28:823–829. [DOI] [PubMed] [Google Scholar]
- 32. Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M.. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011; 334:1524–1529. [DOI] [PubMed] [Google Scholar]
- 33. Delaney J.C., Essigmann J.M.. Context-dependent mutagenesis by DNA lesions. Chem. Biol. 1999; 6:743–753. [DOI] [PubMed] [Google Scholar]
- 34. Burns J.A., Dreij K., Cartularo L., Scicchitano D.A.. O 6-methylguanine induces altered proteins at the level of transcription in human cells. Nucleic Acids Res. 2010; 38:8178–8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Delaney J.C., Essigmann J.M.. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:14051–14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shrivastav N., Fedeles B.I., Li D., Delaney J.C., Frick L.E., Foti J.J., Walker G.C., Essigmann J.M.. A chemical genetics analysis of the roles of bypass polymerase DinB and DNA repair protein AlkB in processing N2-alkylguanine lesions in vivo. PLoS One. 2014; 9:e94716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Delaney J.C., Essigmann J.M.. Effect of sequence context on O6-methylguanine repair and replication in vivo. Biochemistry. 2001; 40:14968–14975. [DOI] [PubMed] [Google Scholar]
- 38. Choi J., Ieong K.W., Demirci H., Chen J., Petrov A., Prabhakar A., O’Leary S.E., Dominissini D., Rechavi G., Soltis S.M. et al. N 6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 2016; 23:110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cantara W.A., Crain P.F., Rozenski J., McCloskey J.A., Harris K.A., Zhang X., Vendeix F.A., Fabris D., Agris P.F.. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011; 39:D195–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwartz S., Agarwala S.D., Mumbach M.R., Jovanovic M., Mertins P., Shishkin A., Tabach Y., Mikkelsen T.S., Satija R., Ruvkun G. et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013; 155:1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Doma M.K., Parker R.. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006; 440:561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shoemaker C.J., Green R.. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012; 19:594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aas P.A., Otterlei M., Falnes P.O., Vagbo C.B., Skorpen F., Akbari M., Sundheim O., Bjoras M., Slupphaug G., Seeberg E. et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003; 421:859–863. [DOI] [PubMed] [Google Scholar]
- 44. Ougland R., Zhang C.M., Liiv A., Johansen R.F., Seeberg E., Hou Y.M., Remme J., Falnes P.O.. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell. 2004; 16:107–116. [DOI] [PubMed] [Google Scholar]
- 45. Liu F., Clark W., Luo G., Wang X., Fu Y., Wei J., Wang X., Hao Z., Dai Q., Zheng G. et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell. 2016; 167:816–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.