Abstract
Imprinted genes are regulated by allele-specific differentially DNA-methylated regions (DMRs). Epigenetic methylation of the CpGs constituting these DMRs is established in the germline, resulting in a 5-methylcytosine-specific pattern that is tightly maintained in somatic tissues. Here, we show a novel epigenetic mark, characterized by strand-specific hemimethylation of contiguous CpG sites affecting the germline DMR of the murine Peg3, but not Snrpn, imprinted domain. This modification is enriched in tetraploid cortical neurons, a cell type where evidence for a small proportion of formylmethylated CpG sites within the Peg3-controlling DMR is also provided. Single nucleotide polymorphism (SNP)-based transcriptional analysis indicated that these epigenetic modifications participate in the maintainance of the monoallelic expression pattern of the Peg3 imprinted gene. Our results unexpectedly demonstrate that the methylation pattern observed in DMRs controlling defined imprinting regions can be modified in somatic cells, resulting in a novel epigenetic modification that gives rise to strand-specific hemimethylated domains functional for genomic imprinting. We anticipate the existence of a novel molecular mechanism regulating the transition from fully methylated CpGs to strand-specific hemimethylated CpGs.
INTRODUCTION
In the mouse, one hundred fifty one genes are known to be imprinted (see http://www.mousebook.org/imprinting-gene-list). As in other mammals, most of these imprinted genes are found in clusters distributed throughout the whole murine genome (1), and they are co-regulated by cis-acting imprinting control elements that contain allele-specific, differentially DNA-methylated regions, hereafter referred to as DMRs. Epigenetic cytosine methylation in the CpGs constituting these DMRs specifically occurs in the germline, and these 5-methylcytosine (5mC)-modified regions are referred to as germline DMRs as opposed to somatic DMRs generated after fertilization (2). The methylation pattern acquired in the germline is tightly maintained in all somatic tissues due to the activity of DNMT1, a DNA methyltransferase that methylates the newly synthesized strand after DNA replication (3,4).
Besides 5mC (5), 5-formylcytosine (5fC) can also be found as a stable DNA modification in mammalian genomes (6). This molecule is generated by sequential oxidation of 5mC which can be converted to 5-hydroxymethylcytosine (5hmC) and then to 5fC by the TET (ten eleven translocation) proteins (7). To study whether the methylation pattern of DMRs controlling genomic imprinting can be altered and whether 5fC can be present in these DMRs we focused on the Peg3 and Snrpn imprinted domains. This analysis was primarily focused on neurons from the mouse cerebral cortex as these imprinted domains contain genes that are expressed in neurons (8–10). We were particularly interested in determining whether cytosine modification in cortical neurons could be altered in response to changes in ploidy level, as a small proportion of cortical neurons contain double the normal amount of DNA in their nuclei (11).
In the vertebrate nervous system, a subset of differentiating projection neurons replicate their genome as they differentiate and then remain as tetraploid (4C) neurons in the adult brain (11–13). The mechanism leading to neuronal tetraploidy depends on the activation of the neurotrophin receptor p75 (11,12), which induces the activation of p38MAPK and the subsequent phosphorylation of E2F4 (14), a key regulator of the cell cycle. After DNA replication, Cdk1inactivation prevents differentiating 4C neurons from entering mitosis (15), while p27Kip1 expression in these cells avoids further rounds of endoreplication (16). Somatic 4C neurons show extensive dendritic arbors and enlarged cell bodies (12), express the immediate early genes Erg-1 and c-Fos (11), known to respond to neuronal activity (17,18), and constitute specific neuronal populations that innervate defined target areas (12), thus indicating that 4C neurons are fully functional.
In this study, we show that the germline DMR of Peg3, but not Snrpn, imprinting domain show a specific hemimethylated pattern in subpopulations of both diploid (2C) and 4C cortical neurons, being enriched in the latter group. We conclude that, in contrast to previous belief, germline DMRs can undergo covalent modifications that give rise to a novel epigenetic mark, characterized by strand-specific CpG hemimethylation. This epigenetic modification participates in the maintenance of the monoallelic expression pattern of the Peg3 imprinted gene.
MATERIALS AND METHODS
Mice
Male C57BL/6J mice from embryonic day 11 (E11) and postnatal day 15 (P15) were used in this study. Embryos were staged as described by Kaufman (19). In embryos, sex was determined by genomic PCR, using cromosome Y-specific primers (see Table 1). DBA/2J mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and bred to C57BL/6J mice to produce P15 mice of hybrid genetic background. Experimental procedures were approved by the CSIC Bioethics Committee, and they were performed in accordance with the European Union guidelines.
Table 1. Oligonucleotides used in this study.
| Primer | Sequence (5’ → 3’) | Modification | Purpose | Annealing temp. (°C) |
|---|---|---|---|---|
| Peg3 up-A | TTTTGTAGAGGATTTTGATAAGGAG | Cloning | 55 | |
| Peg3 down-A | CAATCTAATACACCCACACTAAACC | Cloning/Pyrosequencing | 55 | |
| Peg3 down-A-biot | CAATCTAATACACCCACACTAAACC | 5’-biotinilated | Pyrosequencing | 55 |
| Peg3 seq-A | ATGTTTATTTTGGGTTGGTGG | Pyrosequencing | ||
| Peg3 up-B | GAGGAGAAGCGGAGAGATGT | Enzyme-qPCR | 60 | |
| Peg3 down-B | CACAGCACTCTACGCACACA | Enzyme-qPCR | 60 | |
| Peg3 probe | AGACTGCCGAGGTCGG | FAM/TAMRA | Enzyme-qPCR | |
| Peg3 up-C | AGAYGTTGGGGAGTTAGGAG | Hairpin-bisulfite PCR | 56 | |
| Peg3 down-C | YAAAAAATATCCACCCTAAACTAATAAC | Hairpin-bisulfite PCR | 56 | |
| Peg3 hairpin | GCCGAGTCTGACTTTTTTGTCAGACT | Hairpin | ||
| Peg3 up-D | AATGGCACATGCCTGGAACT | 5’-biotinilated | SNP pyrosequencing | 58 |
| Peg3 down-D | CGATGAGTGGCCTTGTGTCA | SNP pyrosequencing | 58 | |
| Peg3 seq-B | CTCCTGTTCACTTCTTTGAGAGAC | SNP pyrosequencing | N/A | |
| Peg3 seq-C | TGAGGGTCTCACTATGTAGGTGT | SNP pyrosequencing | N/A | |
| Snrpn up-A | TTAGAGGGATAGAGATTTTTGTATTG | Cloning/ Pyrosequencing | 56 | |
| Snrpn down-A | CTAAAATCCACAAACCCAACTAAC | Cloning | 56 | |
| Snrpn down-A-biot | CTAAAATCCACAAACCCAACTAAC | 5’-biotinilated | Pyrosequencing | 56 |
| Snrpn seq | GTATGTGTAGTTATTGTTTGGGA | Pyrosequencing | 56 | |
| Chromosome Y-1 | GCATTTGCCTGTCAGAGAGAG | Sex determination | 58 | |
| Chromosome Y-2 | ACTGCTGCTGCTTTCCAACTA | Sex determination | 58 | |
| 5fC forward | MTMAMCCAMAACMAMAAAMA | 5fC-containing DNA | 57 | |
| 5fC reverse | CCATACCACCCATCACATCA | 5fC-containing DNA | 57 | |
| 5fC forward post-bisulfite | ACACTGACGACATGGTTCTACACTCACTTACAATCACAAACA | NGS | 57 | |
| 5fC reverse post-bisulfite | TACGGTAGCAGAGACTTGGTCTCCATACCACCCATCACATCA | NGS | 57 | |
| HhaI forward | CAGAGGACCCTGACAAGGAG | HhaI restriction assay | 59 | |
| HhaI reverse | AGTTCAGATGGTGTTTGGGG | HhaI restriction assay | 59 | |
| N/A: not applicable. |
Primary and secondary antibodies
The mouse anti-NeuN monoclonal antibody (Millipore) was used at 1/1000 and the Alexa Fluor 488 goat anti-mouse antibody was diluted 1/500 for nuclei sorting.
Oligonucleotides
Oligonucleotides used in this study are summarized in Table 1. Their specificity was verified by BLAST searches as well as by the actual PCR reaction.
Synthesis of non-methylated, hemimethylated, fully methylated, and methylformylated double-stranded DNA (dsDNA) for HhaI cleavage assays
Non-methylated and fully methylated dsDNA fragments were generated by PCR using genomic DNA isolated from cortical neurons. To this aim, dATP/dTTP/dGTP (0.2 mM each, Biotools) plus either CTP (0.2 mM, Biotools) or 5mCTP (0.2 mM, New England Biolabs) were used together with DreamTaq DNA Polymerase (1.25 units, Thermo) as well as the HhaI forward and HhaI reverse primers (0.4 μm each, Table 1) flanking the HhaI restriction sites from the Peg3- controlling DMR. Amplifications were performed using the following thermal cycle: 94°C for 3 min, 50× (94°C for 30 s, 59°C for 1 min, 72°C for 1 min), 72°C for 20 min. PCR products were cleaned using DCC Clean & Concentrator-5 (ZymoReasearch) following manufacturer's instructions. Hemimethylated and methylformylated dsDNA fragments were generated by PCR using fully methylated dsDNA. To this aim, dATP/dTTP/dGTP (0.2 mM each, Biotools) plus either CTP (0.2 mM, Biotools) or 5fCTP (0.2 mM, TriLink) were used together with DreamTaq DNA Polymerase (1.25 units, Thermo) as well as the HhaI forward and HhaI reverse primers (0.4 μm each) described above. Amplifications were performed using the following thermal cycle: 94°C for 3 min, 1× (94°C for 30 s, 59°C for 1 min, 72°C for 1 min), 72°C for 20 min. PCR products were cleaned using DCC Clean & Concentrator-5 (ZymoReasearch) following manufacturer's instructions. Non-methylated, hemimethylated, fully methylated, or methylformylated PCR products (10 μl each) were incubated for 1 h at 37°C in either the presence or absence of 20 U HhaI (Thermo Fisher Scientific). After incubation, reactions were inactivated for 20 min at 80°C and run in an agarose gel.
Synthesis of 5fC-containing DNA for sodium borohidride (BH)- and O-hydroxyethylamine (EA)-based bisulfite protection assays
DsDNA containing 5fC for BH/EA-based bisulfite protection assays was synthesized as previously described by (20). To this aim, template DNA (CTCACCCACAACCACAAACAAATTTAATACGATTAAATAATATTAATATATTATCGATTAGTAGGTTAAGTAAGGGTATTTGATGTGATGGGTGGTATGG) was PCR amplified with DreamTaq DNA Polymerase (Thermo) using the 5fC forward and 5fC reverse primers (Table 1), and commercially available 5fCTP (TriLink).
Cell nuclei isolation
Cell nuclei isolation was performed as described by López-Sánchez and Frade (11). Briefly, one hemicortex from mice or mouse embryos of the specified age, previously snap-frozen on dry ice and stored at –80°C, was placed in 2 ml of ice-cold, DNase-free PBS containing 0.1% Triton X-100 (Sigma) and protease inhibitor mixture (Roche) (nuclear isolation buffer). Cell nuclei were then isolated by mechanical disaggregation using a Dounce homogenizer. Undissociated tissue was removed by centrifugation at 200 × g for 1.5 min at 4°C. The supernatant was 3-fold diluted with nuclear isolation buffer and centrifuged at 400 × g for 4 min at 4°C. Supernatant with cellular debris was discarded, and the pellet incubated at 4°C in 50 μl of cold nuclear isolation buffer for at least 1 h, before mechanical disaggregation by gently swirl of the vial. The quality and purity of the isolated nuclei was analyzed microscopically after staining with 100 ng/ml DAPI. A similar procedure was used for the isolation of hepatocyte nuclei.
Cell nuclei sorting
Neuronal nuclear immunostaining was performed by adding primary and secondary antibodies to isolated unfixed nuclei containing 5% FCS and 1.25 mg/ml BSA. In control samples, the primary antibodies were excluded. Finally, the reaction was incubated O/N at 4°C in the dark. Immunostained nuclei (or hepatocyte nuclei) were filtered through a 40 μm nylon filter, and the volume adjusted with PBS containing propidium iodide (PI; Sigma) and DNase-free RNase I (Sigma) at a final concentration of 40 and 25 μg/ml, respectively, and incubated for 30 min at RT. The quality of the nuclei and specificity of immunostaining signal was checked by fluorescence microscopy. Nuclei sorting was performed with a FACSAria cytometer (BD Biosciences) equipped with a 488-nm Coherent Sapphire solid state and 633-nm JDS Uniphase HeNe air-cooled laser. Data were collected by using a linear digital signal process. The emission filters used were BP 530/30 for Alexa 488 and BP 616/23 for PI. Data were analyzed with FACSDiva (BD Biosciences). Gating was performed by removing doublets and selecting the NeuN-positive nuclei, as previously described (11). Cellular debris, which was clearly differentiated from nuclei due to its inability to incorporate PI, was gated and excluded from the analysis. For isolation of the neuronal population highly enriched in 4C DNA amount (∼90% purity), two rounds of sorting were performed. Cell nuclei from the first round of sorting were collected in Protein LoBind tubes (Eppendorf), while those from the second round were collected in DNA LoBind tubes (Eppendorf).
Genomic DNA isolation
Genomic DNA was isolated using DNeasy Blood & Tissue Kit (Qiagen) following the protocol described by the manufacturer. Two mice were used to generate each genomic DNA and at least three different genomic DNAs were employed in each experiment.
Whole genome amplification and CGH array
DNA was amplified using the GenomePlex WGA2 kit (Sigma Aldrich) according manufacturer's instructions. The amplified DNA was clean with phenol-chloroform using a standard protocol. CGH arrays were performed using the SurePrint G3 Mouse CGH Microarray Kit 180K (Agilent Technologies, Palo Alto, CA, USA) covering the whole genome with a 10.9 Kb overall median probe spacing following the manufacturer's protocol.
Methylation-sensitive restriction enzyme-based quantitative PCR (qPCR)
Quantification of CpG island methylation in the Peg3 DMR was performed in DNA isolated from 2C and 4C sorted neuronal nuclei as described above, using a modification of the procedure described by Hashimoto et al. (21). For each DNA sample, two parallel reactions were prepared, containing 15 ng of DNA (quantified by band intensity in agarose gels) in either the presence or the absence of 20 U HhaI (Thermo Fisher Scientific). All reactions were incubated for 5 h at 37°C in 20 μl total volume. After digestion, the reactions were inactivated by incubating the mixture at 80°C for 20 min. 1.14 μl of each reaction was added to 20 μl total volume PCR reaction containing 10 μl 2X TaqMan Universal PCR Master Mix, both primers at a concentration of 900 nM and a fluorescent probe at a concentration of 250 nM (for details, see list of primers in Table 1). Samples were amplified using an ABI Prism 7000 thermal cycler (Applied Biosystems) using the following conditions: 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of PCR consisting of 15 s at 95°C and 1 min at 60°C. Under these conditions, qPCR efficiency was 99.96% (amplification factor: 2). Quantification was performed within linear range (R2 of standard curve = 0.9918). In all cases, technical triplicates were performed (2C intra-assay mean CV = 18.53%; 4C intra-assay mean CV = 22,74%). Outliers were identified with the Dixon's Q test. Negative controls (DNA replaced by water) did not show amplification.
DNA reduction with BH/EA
Genomic DNA and 5fC-containing synthetic DNA was reduced using BH as previously described by Booth et al. (20). Briefly, 5 μl of an aqueous sodium borohydride (Merk) solution (1 M), freshly prepared before every reduction, was added to 100–200 ng DNA (dissolved in 15 μl water). The reaction was vortexed and centrifuged, and held at room temperature in the dark for 1 h. The reactions were vortexed and centrifuged every 15 min to remove the bubbles that were generated, and the pressure was released by opening the lids. The reaction was finally quenched by slowly adding 10 μl of a solution containing 750 mM sodium acetate (Sigma), pH 5.0. Since violent release of hydrogen occurs during this quenching step, the reaction was held at room temperature for 10 min or until no further gas was released. EA protection of 5fC in genomic DNA and 5fC-containing synthetic DNA was performed with 10 mM O-ethylhydroxylamine (Sigma-Aldrich) in 100 mM MES buffer, pH 5.0, at 37°C for 2 h, as described by Song et al. (22). The DNA substrates were purified with Genomic DNA Clean & Concentrator™-10 (ZymoReasearch) and subjected to sodium bisulfite treatment as described below.
Bisulfite conversion
A volume of 20 μl DNA was converted using Epitect Bisulfite Kit (Qiagen). Bisulfite conversion was performed in 85 μl of Bisulfite Mix solution, and 35 μl of DNA Protect Buffer. The total reaction volume was 140 μl. The procedure was performed according to the manufacturer's instructions. A modified protocol was used for bisulfite conversion of borohydride-treated DNA. Briefly, 100–200 ng DNA (either treated with borohydride or untreated) was made up to 30 μl in water, and then combined with bisulfite mix (80 μl) and DNA protect buffer (30 μl). The reactions were then subjected to two cycles of the formalin-fixed, paraffin-embedded (FFPE) thermal cycle, and worked up as per the Qiagen Epitect Bisulfite Kit protocol for FFPE samples.
Bisulfite-converted DNA amplification, cloning, and Sanger sequencing
Primers were designed to match the DMRs of the imprinted Snrpn and Peg3 regions previously described by Lucifero et al. (8) (for sequences and annealing temperatures, see list of primers in Table 1). These primers are specific for the bisulfite-converted sequence, and they encompass 13 CpG sites within the Snrpn imprinted domain [corresponding to the CpG sites number 4 to 16 from Lucifero et al. (8)], and 12 CpG sites along the Peg3 imprinted domain [corresponding to the first 12 CpG sites from Lucifero et al. (8)]. PCR amplification was performed using standard procedures. PCR products were subcloned into the pGEM-Teasy vector (Promega), and Sanger sequenced.
Pyrosequencing analysis of bisultite converted DNA
Bisulfite-treated DNA was PCR amplified using Snrpn- or Peg3-specific primers, one of which was biotinylated at its 5’ end (for sequences and annealing temperatures, see list of primers in Table 1). The PCR product was then bound to streptavidin-coated Sepharose beads (GE-Healthcare) through the biotin tag and denatured to generate single-stranded DNA to allow annealing of an internal sequencing primer (16 pmol of each per reaction) (see list of primers in Table 1). Pyrosequencing was performed in a PSQ™96MA instrument (Biotage) using 25 μl of amplified DNA product and PyroMark Gold Q96 reagents (Qiagen). Pyrosequencing data analysis was done using the Pyro Q-CpG software (Biotage).
Next-generation sequencing (NGS) of bisulfite-converted DNA
DsDNA containing 5fC for BH/EA-based bisulfite protection assays was reduced with either BH or EA and subjected to bisulfite conversion as described above. PCR library amplification was performed with DNA Polymerase (Biotools) using the 5fC forward post-bisulfite and 5fC reverse post-bisulfite primers (Table 1). Illumina adapter sequences included at the 5’ end of the primers were used to prepare libraries, which were then sequenced on the Illumina Miseq platform (2 × 300 bp paired‐end reads). Sequences were aligned using the MiSeq Reporter software (Illumina) and BAM alignments were visualized using the Integrative Genomics Viewer platform. For methylated CpG level analysis within the Peg3 genomic region, including the Peg3 imprinted domain-controlling DMR (PCD), genomic DNA isolated from ∼2000 4C neuronal nuclei was subjected to bisulfite treatment and amplification with the Pico Methyl-Seq Library Prep kit (Zymo Research). The bisulfite-Seq library generated was sequenced on the Illumina HiSeq 2500 platform. Read mapping, processing, and analysis were performed as described previously (23).
Hairpin-bisulfite PCR
The presence of hemimethylated CpGs in the PCD was analyzed using a modification of the protocol described by Laird et al. (24). Briefly, 50 ng of DNA isolated from 4C sorted neuronal nuclei was digested with 2.5 U BbvI (Thermo Fisher Scientific) for 1 h at 37°C. Then, the hairpin linker (2 μg) was ligated to the cleaved DNA using 10 U T4 DNA ligase (Thermo Fisher Scientific). Hairpin-ligated genomic DNA was then subjected to bisulfite conversion using Epitect Bisulfite Kit (Qiagen). To avoid rapid fold-back renaturation of hairpin DNA, two extra thermal-denaturation steps were introduced without modifying the total incubation time indicated by the manufacturer (incubation conditions: 5 min at 95°C, 25 min at 60°C, 5 min at 95°C, 60 min at 60°C, 5 min at 95°C, 60 min at 60°C, 5 min at 95°C, 60 min at 60°C, 5 min at 95°C, 80 min at 60°C). Then, bisulfite-converted DNA was PCR amplified with the appropriate primers (see list of primers in Table 1) using standard conditions, with the exception that the concentration of MgCl2 was raised to 5 mM. PCR products were subcloned in the pGEM-Teasy Vector (Promega), and clones containing the appropriate inserts were Sanger sequenced.
SNP imprinting analysis
A polymorphism for SNP imprinting analysis was identified in intron 7 of mouse Peg3 gene using the Mouse Genome Informatics tool (The Jackson Laboratory) (http://www.informatics.jax.org/strains_SNPs.shtml). This SNP (NCBI accession number rs31442449), characterized by a nucleotide change from T (in C57BL/6J background) to C (in DBA/2J background), is located at bp 18 192 of the mouse Peg3 gene (NCBI accession number NC_000073, complement to bp 6 705 960–6 730 419). Total RNA was isolated using the RNAqueous-4PCR Kit (Ambion) from 2C and 4C neuronal nuclei derived from the cerebral cortex of P15 male littermates obtained from the mating of DBA/2J and C57BL/6J mice. Cerebral cortices were treated with Allprotect (Quiagen) following the protocol described by the manufacturer before nuclear isolation. RNA samples were treated with DNase I (Thermo Fisher Scientific) to remove traces of genomic DNA contamination. Synthesis of cDNA was performed using SuperScript III Reverse Transcriptase (Invitrogen) following manufacturer's instructions. PCRs were performed using Peg3-specific primers, one of which was biotinylated at its 5΄ end (for sequence and amplification conditions, see list of primers in Table 1). PCR products were pyrosequenced with an internal sequencing primer (see list of primers in Table 1) as indicated above.
Data analysis and statistics
Sanger sequencing results were analyzed for change in methylation pattern with the Quantification tool for Methylation Analysis (Quma) (http://quma.cdb.riken.jp/) (25). The software generates a panel of solid and open circles indicating methylated and unmethylated CpGs, respectively. Quantitative data were obtained from DNA analysis in triplicate from at least three different DNAs (obtained from two mice) for each experimental point, except in BH experiments with P20 liver, in which one DNA sample for each experimental point was used. Statistical analyses and graphics were performed using the Graphpad Prism 5 software package. Global methylation analysis was performed by pooling data from all CpG sites in each particular DMR. qPCR data were analyzed using 7000 System Sequence Detection Software version 1.2.3 (Applied Biosystems). The percentage of methylation in methylation-sensitive restriction qPCR assays was estimated by dividing the values obtained with the digested DNA aliquots by those of the non-digested DNA aliquots. The average values were obtained from thirteen (2C) and six (4C) independent experiments in triplicate. Statistical significance was determined using two-tailed Student's t test unless otherwise specified.
RESULTS
Reduced methylation levels in the Peg3 imprinted domain-controlling DMR (PCD) of cortical neurons
To explore the impact of neuronal tetraploidization on the methylation status of specific germline DMRs controlling the expression of neuron-expressed genes (26) we focused on the Peg3 imprinted domain (Figure 1A) (8,9). For this analysis, we used cell nuclei from 2C and 4C neurons (identified by NeuN-specific immunoreactivity), isolated by FACS from the mouse cerebral cortex (Supplementary Figure S1), a structure that in adult mice contains around 3% of 4C neurons (11). Postnatal day 15 (P15) was chosen for this analysis, as Peg3 is crucial for lactation and postnatal growth (27).
Figure 1.
Scheme showing the genomic structure of murine Peg3 and Snrpn imprinted domains. (A) Genes constituting the Peg3 imprinted domain are shown. Apeg3, Peg3, Usp29 and Zfp264 show paternal expression whereas Zim1, Zim2 and Zim3 are expressed from the maternal-derived chromosome. The Peg3 imprinting center, which contains the Peg3-controlling DMR, is located within the Peg3 promoter region. Methylated CpG sites from the Peg3-controlling DMR are represented by solid circles. Open circles represent non-methylated CpGs from the Peg3-controlling DMR. (B) Genes constituting the Snrpn imprinted domain are shown. All transcriptional units show paternal expression. The Snrpn imprinting center, which contains the Snrpn-controlling DMR, is located within the Snrpn promoter region. Methylated CpG sites from the Snrpn-controlling DMR are represented by solid circles. Open circles represent non-methylated CpGs from the Snrpn-controlling DMR.
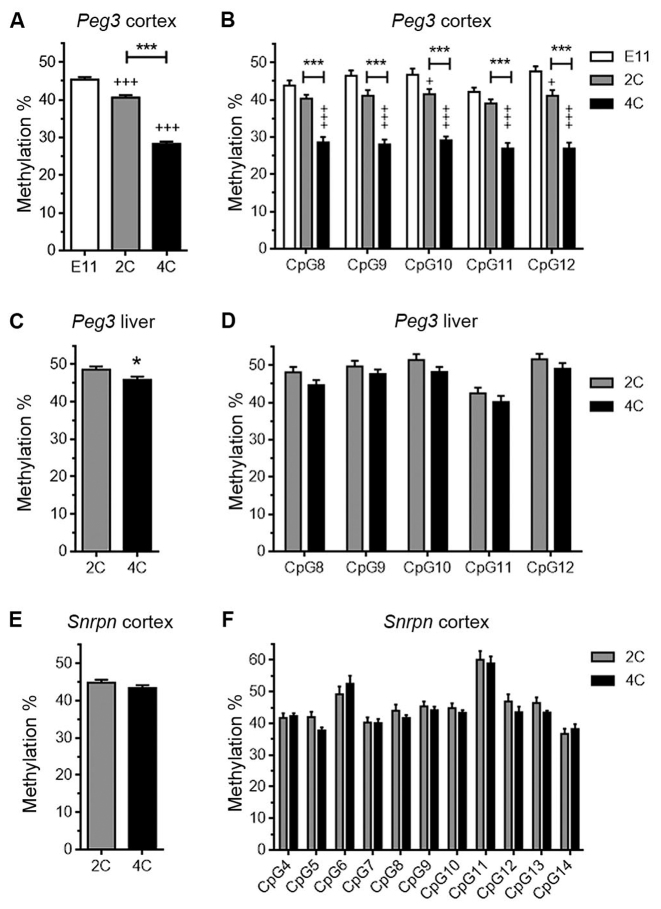
The proportion of methylated CpG sites within the PCD was analyzed in bisulfite-treated genomic DNA from either 2C or 4C neuronal nuclei. Bisulfite-treated genomic DNA from embryonic day 11 (E11) telencephalic neuroepithelium, a structure that contains mostly neural precursors, was used as a control. Bisulfite-modified genomic DNA was PCR amplified with primers specific for the Peg3 domain (see Table 1), and the amplified bands were pyrosequenced using a specific internal primer (Figure 2A). This analysis indicated that the global percentage of methylated CpGs in the PCD from 2C cortical neurons was significantly lower when compared with the E11 telencephalic neuroepithelium (Figure 3A). In addition, the methylation level observed in the PCD in the 4C cortical neurons was dramatically reduced with respect to 2C neurons (Figure 3A and B; see also Supplementary Figure S2A for Sanger sequencing). The decrease in CpG methylation observed in 4C neurons does not derive from their tetraploid status since the PCDs of 4C hepatocytes isolated from P20 livers show just a minimal reduction in the global levels of methylation compared with 2C (Figure 3C and D). The decrease in methylation observed in 4C neurons does not derive from partial genome duplication of the maternal chromosome in these neurons either. In support, whole genome amplification from 2C and 4C neuronal nuclei (22 000–27 000 on average) obtained from P15 cerebral cortex, followed by comparative genomic hybridization (CGH), demonstrated that the genome of 4C cortical neurons is fully duplicated (Supplementary Figure S3). Targeted NGS of the Peg3 genomic domain from 4C neuron-derived genomic DNA demonstrated that the subdomain of the PCD analyzed in this study specifically contains a lower proportion of 5mCs (Supplementary Figure S4).
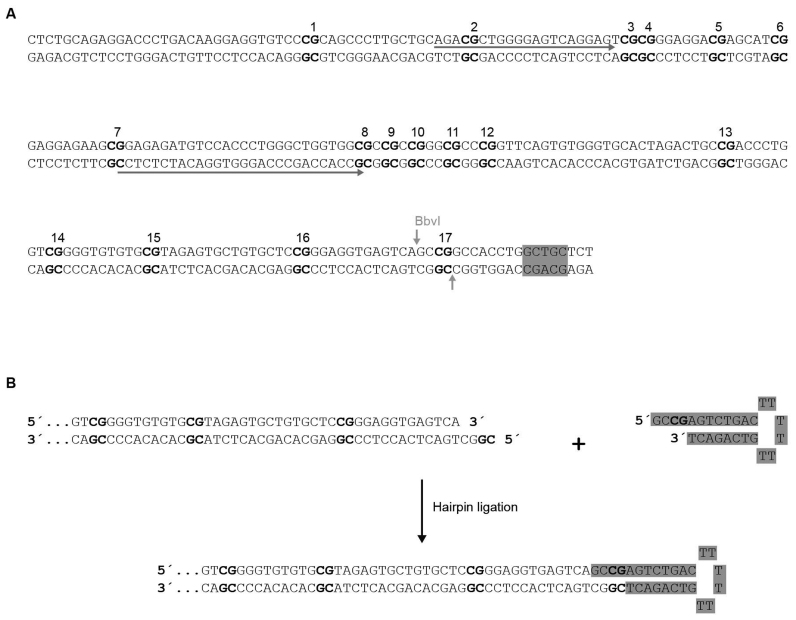
Figure 2.
Genomic DNA sequences corresponding to the Peg3- and Snrpn-imprinting control regions analyzed in this study. (A and B) CpG sites are numbered and shown in bold, and HhaI restriction sites are indicated with purple rectangles. For qPCR amplification in (A), genomic DNAs were either cut with HhaI or left untreated. Solid purple arrows represent the sequences where the primers for qPCR were designed. Dashed purple line corresponds to the sequence used for the Taqman probe. For methylation analysis in both (A) and (B), genomic DNAs were bisulfite-converted and PCR amplified using modified primers (Cs substituted by Ts) corresponding to the sequences indicated by blue and orange solid arrows, and either cloned for Sanger sequencing [see Supplementary Figure S2] or subjected to pyrosequencing. In this latter case the downstream primers were 5’-biotinylated (solid circles), and pyrosequencing was performed with the primers indicated by dashed blue arrows (modified by substituting Cs by Ts in its sequence).
Figure 3.
Reduced methylation levels in the PCD of cortical neurons. (A) Global pyrosequencing analysis of the CpG sites present in the PCD of E11 telencephalic neuroepithelial cells, and 2C and 4C cortical neurons. +++P < 0.005 (versus E11); ***P < 0.005 (versus 2C). (B) Pyrosequencing analysis of individual CpG sites (numbered as in Figure 2A) in the PCD of E11 telencephalic neuroepithelial cells, and 2C and 4C cortical neurons. +++P < 0.005; +P < 0.05 (versus E11); ***P < 0.005 (versus 2C). (C and E) Global pyrosequencing analysis of the CpG sites present in the PCD of 2C and 4C hepatocytes (C) and in the SCD of 2C and 4C cortical neurons (E). *P < 0.05 (versus 2C). (D and F) Pyrosequencing analysis of individual CpG sites (numbered as in Figure 2A) in the PCD of 2C and 4C hepatocytes (D) and in the SCD (numbered as in Figure 2B) of 2C and 4C cortical neurons (F).
The reduction in the global percentage of methylated CpGs in 4C neurons is specific for the Peg3 imprinted region as the Snrpn imprinting domain-controlling DMR (SCD) (8,28), also known to regulate a genomic domain (Figure 1B) comprising genes with neuronal expression (10), showed similar levels of global methylation in both 2C and 4C cortical neurons (Figure 3E). This conclusion was further supported by the analysis with specific primers (Figure 2) of individual CpGs within the SCD, either by pyrosequencing (Figure 3F) or Sanger sequencing (Supplementary Figure S2B).
Strand-specific reduction of methylation levels in the PCD of cortical neuron subpopulations
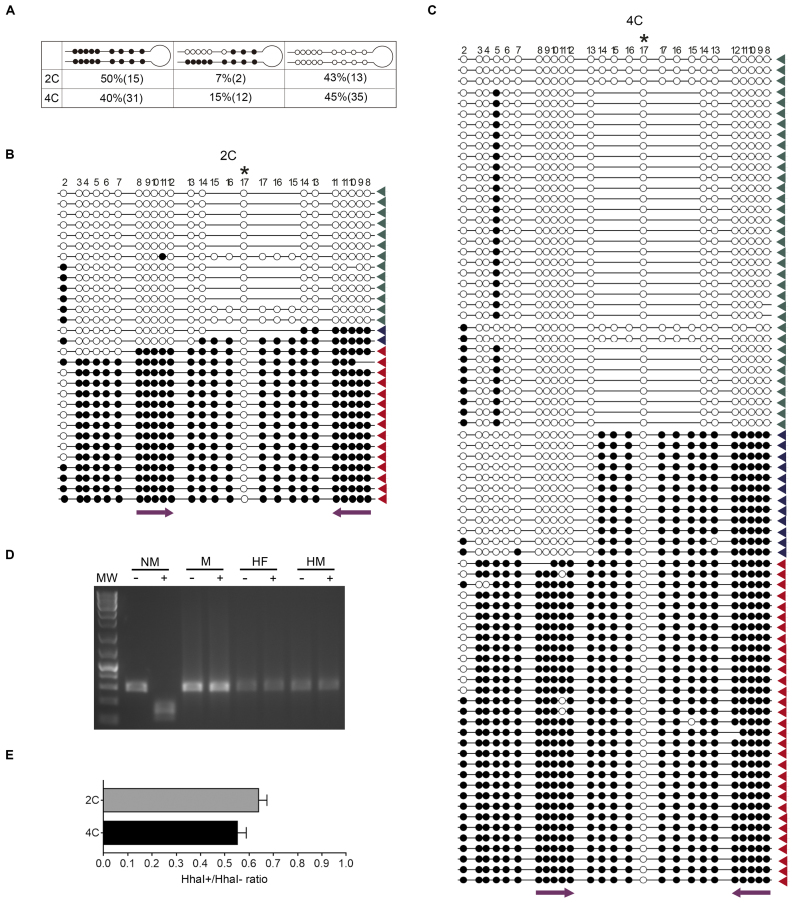
To identify the reason behind the reduction of CpG methylation levels within the PCD from cortical neurons, we used a variation of the hairpin-bisulfite PCR method (24). This technique is able to assess bisulfite protection patterns on complementary strands of individual DNA molecules. To this end, we used a hairpin linker that is ligated to BbvI-cleavaged genomic DNA obtained from 2C and 4C neuronal nuclei (Figure 4A and B). This approach results in the attachment of the complementary strands of individual DNA molecules, which are subsequently denatured and subjected to bisulfite conversion, followed by PCR amplification with Peg3-specific primers upstream of the cleavage sequence (Figure 4A). After Sanger sequencing of the PCR fragments, the presence of bisulfite protected or unprotected cytosines in both DNA strands is analyzed (Figure 5B and C). This analysis demonstrated that 15% of the Peg3-controlling DMRs display strand-specific demethylation in the 4C neurons (Figure 5A). In addition, a reduced proportion of strand-specific demethylated DMRs (7%) was detected in the PCD of 2C neurons, an observation consistent with the comparatively lower proportion of methylated DNA in these latter cells (Figure 3A). In both types of neurons, demethylation affected the CpGs located on the Peg3 sense strand. We therefore concluded that a portion of the CpGs from the PCD in the methylated chromosome, comprising at least CpGs #8–13 (Figure 4A), is asymmetrically methylated in the P15 cortical neurons. This analysis also indicated that CpG sites #14–17 are methylated on both strands (Figure 5A–C), as expected from the differential chromosome methylation associated with the imprinting process. Overall, these results indicate that the observed decrease of methylation in cortical neurons (Figure 3A), roughly mimicking the proportion of hemimethylated DMRs (Figure 5A), is likely due to the asymmetric removal of 5mCs in a subdomain of the PCD from the methylated chromosome. Interestingly, this hemimethylated subdomain contains recognition motifs for Pax-5, p53, RAR-β:RXR-α, RXR-α, Egr3, VDR, FOXP3 and PXR-1:RXR-α transcription factors, as evidenced by the PROMO tool available at http://alggen.lsi.upc.es.
Figure 4.
Scheme showing the strategy for hairpin-bisulfite PCR. (A) Genomic DNA sequence corresponding to the domain of the Peg3 imprinting control region analyzed in this study. CpG sites are numbered and shown in bold. The BbvI recognition/cleavage site is indicated in gray. Sequences indicated by solid arrows correspond to the modified primers (Cs substituted by Ts) used for PCR. (B) Diagram illustrating the ligation of the BbvI-cleavaged genomic DNA with the hairpin (labeled in gray). The resulting DNA is bisulfite-converted and subjected to PCR amplification with the oligonucleotides indicated in (A). The resulting amplification bands were cloned and Sanger sequenced (see Figure 5B and C).
Figure 5.
Reduced methylation levels in the PCD of cortical neurons is strand-specific. (A) Percentage and absolute number of hairpin-bisulfite PCR clones [see (B) and (C)] with the indicated bisulfite protection patterns. Unmethylated CpG sequences are indicated by open circles and methylated CpG sequences are indicated by solid circles. (B and C) Sanger sequences of the hairpin-bisulfite PCR clones from genomic DNA derived from 2C neurons (B) and 4C (C). Green arrowheads: clones with non-methylated CpG sites #8–12, blue arrowheads: clones with asymmetrically methylated CpG sites #8–12, red arrowheads: clones with bimethylated CpG sites #8–12. Non-methylated CpG sequences are indicated by open circles and methylated CpG sequences are indicated by solid circles. Asterisk indicates the CpG sequence from CpG site #17 that is included in the hairpin (see Figure 4A), and therefore is not methylated. CpG site #2 is shown as the oligonucleotide used for PCR amplification was degenerated. The localization of CpG sites #8–12 is indicated with purple arrows. (D) HhaI restriction assay using non-methylated DNA (NM), fully methylated DNA (M), DNA formylated in one strand and methylated in the other strand (HF), and DNA methylated in one strand (HM). –: Mock-treated DNA, +: HhaI-treated DNA, MW: DNA molecular weight marker. (E) Genomic DNA from 2C and 4C neuronal nuclei, undigested (HhaI–) or digested with HhaI (HhaI+), was subjected to real time PCR amplification using Peg3-specific primers flanking the restriction sites (see Figure 2A). Bars represent the HhaI+/HhaI– ratio average (±S.E.M.). Values were not significantly below 0.5 (one-tailed, Student's t-test).
The hemimethylation status described above was confirmed through methylation-sensitive, restriction enzyme-based quantitative PCR (21,29,30) (see Experimental Procedures). HhaI was chosen for this analysis, as this restriction enzyme does not cut fully- and hemi-methylated DNA (31) (Figure 5D), as well as methylformylated DNA (Figure 5D). Genomic DNAs isolated from either 2C or 4C neuronal nuclei were either mock digested or digested with HhaI. DNAs were then amplified through real time PCR using oligonucleotides flanking the HhaI cleavage sites in the PCD [CpGs #8 and #11 (Figure 2A)]. This assay demonstrated that ∼50% of genomic DNA remains uncleaved in both 2C and 4C cortical neurons (Figure 5E), thus indicating that half of the chromosomes are resistant to HhaI cleavage in the PCD. Overall, these results demonstrate that the reduction of methylation observed in the PCD is not due to full demethylation of the studied CpGs, but rather to partial, strand-specific demethylation of specific CpGs.
Reduced methylation in the PCD of cortical neurons is largely independent on the presence of 5fC
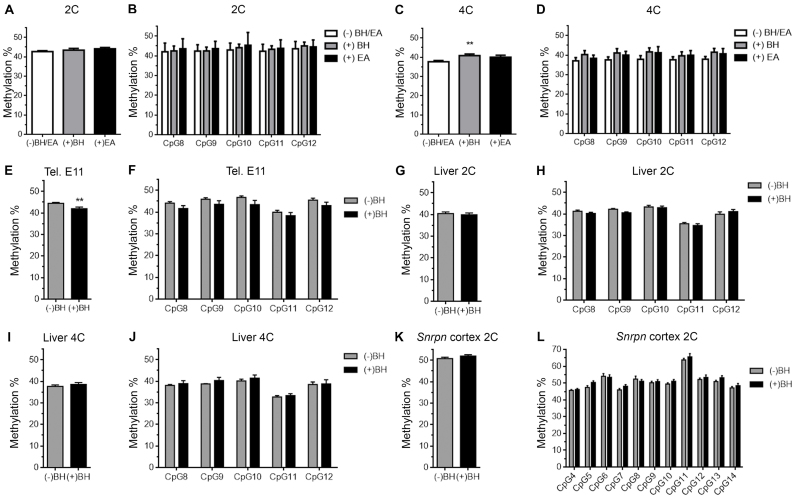
5fC can be found as a stable DNA modification in mammalian genomes (6), but it cannot be detected through bisulfite sequencing (32). To explore whether the hemimethylated CpG sites from the subdomain of the PCD could contain 5fC, genomic DNA from 2C and 4C neuronal nuclei was treated with BH or EA. These compounds are reducing agents that specifically transform 5fC into 5hmC (22,32), a molecule that is protected from bisulfite treatment. The reducing capacity of BH and EA was confirmed in our experiments, which included a control DNA containing 5fC in specific CpGs (20) (see Supplementary Figure S5). Neither BH nor EA treatment followed by bisulfite pyrosequencing was able to restore the proportion of global bisulfite-protected levels in the hypomethylated PCD subdomain from 2C neurons (Figure 6A,B). In contrast, BH was able to partially increase the global levels of bisulfite-protected CpGs in 4C neurons (Figure 6C), while EA treatment resulted in the tendency of a global increase of CpG methylation levels that was not statistically significant (Figure 6C). A BH- or EA-dependent increase in the proportion of bisulfite-protection in 4C neurons could not be confirmed when individual CpGs were analyzed either, although a statistically non-significant tendency could also be observed (Figure 6D). As a control, no significant increase was detected when similar analyses were performed with genomic DNA from E11 telencephalic neuroepithelium (Figure 6E and F), 2C hepatocytes (Figure 6G and H), and 4C hepatocytes (Figure 6I and J). Finally, no changes were observed when a similar analysis was performed in genomic DNA from 2C neurons using primers specific for the SCD both globally (Figure 6K) and in individual CpG sites (Figure 6L). Overall, these results suggest that a subdomain of the PCD contains differentially methylformylated CpGs in a subpopulation of 4C cortical neurons. This pattern, which represents a novel epigenetic label, is likely to derive from chemical modification of the cytosines lying on one of the two DNA strands.
Figure 6.
Reduced methylation levels in the PCD of 4C cortical neurons seems to be partially due to the presence of 5fC. (A) Global pyrosequencing analysis of the CpG sites present in the PCD, performed with untreated (white), BH- (gray), or EA-treated genomic DNA (black) from 2C cortical neurons. (B) Pyrosequencing analysis of individual CpG sites (numbered as in Figure 2A), performed with untreated (white), BH- (gray), or EA-treated genomic DNA (black) from 2C cortical neurons. (C) Global pyrosequencing analysis of the CpG sites present in the PCD, performed with untreated (white), BH- (gray), or EA-treated genomic DNA (black) from 4C cortical neurons. (D) Pyrosequencing analysis of individual CpG sites (numbered as in Figure 2A), performed with untreated (white), BH- (gray), or EA-treated genomic DNA (black) from 4C cortical neurons. (E, G and I) Global pyrosequencing analysis, performed with genomic DNA treated either with BH (black) or left untreated (gray), of the CpG sites present in the PCD of E11 telencephalic neuroepithelium (E), 2C hepatocytes (G), and 4C hepatocytes (I). (F, H and J) Pyrosequencing analysis, performed with genomic DNA treated either with BH (black) or left untreated (grey), of individual CpG sites (numbered as in Figure 2A) present in the PCD of E11 telencephalic neuroepithelium (F), 2C hepatocytes (H) and 4C hepatocytes (J). (K) Global pyrosequencing analysis, performed with genomic DNA treated either with BH (black) or left untreated (grey), of the CpG sites present in the SCD of 2C cortical neurons. (L) Pyrosequencing analysis, performed at P15 with genomic DNA treated either with BH (black) or left untreated (grey), of individual CpG sites (numbered as in Figure 2B) present in the SCD of 2C cortical neurons. **P < 0.01.
Maintenance of imprinting in the Peg3 domain of cortical neurons
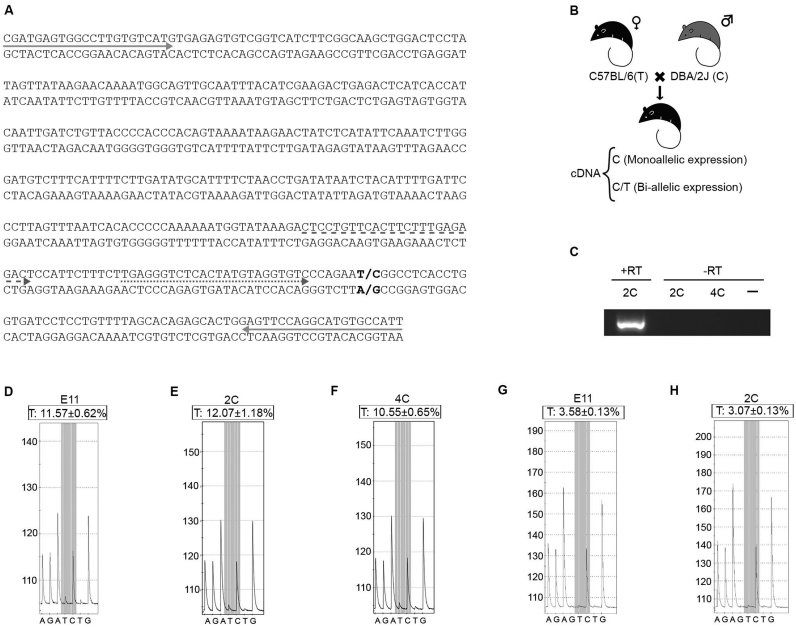
To study whether GpG hemimethylation participates in the maintenance of genomic imprinting in the Peg3 domain, we focused on the Peg3 gene as a read-out of the system. To this end, gene silencing in the maternal chromosome was analyzed using a single nucleotide polymorphism (SNP # rs31442449) located in intron 7 of the Peg3 gene, characterized by a nucleotide change from T in C57BL/6J mice to C in DBA/2J mice (Figure 7A). Female C57BL/6J mice and male DBA/2J mice were mated to generate hybrid mice heterozygous for the T/C polymorphism within the maternal/paternal chromosomes, respectively (Figure 7B). Nascent mRNA (enriched in unspliced transcripts) was then obtained from 2C and 4C neuronal nuclei, isolated by FACS from the cerebral cortex of the hybrid mice at P15. In addition, nascent RNA was also obtained from telencephalic neuroepithelial cell nuclei of E11 heterozygous embryos. These mRNAs were not contaminated by genomic DNA as no specific amplification was detected after 100 PCR cycles with the primers used in the subsequent analysis (Figure 7C). Furthermore, the results indicated absence of maternal expression (Figure 7 and Supplementary Figure S6), therefore ruling out the existence of genomic contamination. Pyrosequencing analysis of the cDNAs derived from these mRNAs indicated that the Peg3 gene remains silenced in the maternal chromosome, since only background signal was obtained for the T-containing Peg3 sequence in both 2C (Figure 7E) and 4C (Figure 7F) cortical neurons, which was similar to that observed in the E11 neuroepithelium (Figure 7D). The silencing of the maternal Peg3 gene in the E11 neuroepithelium and 2C neurons was confirmed by additional pyrosequencing using another primer that was closer to the SNP (Figure 7G,H). These results were confirmed in reciprocal crosses (female DBA/2J mice mated to male C57BL/6J mice) (Supplementary Figure S6).
Figure 7.
PCD hemimethylation can maintain the monoallelic expression pattern of the Peg3 imprinted gene. (A) Partial cDNA sequence from the Peg3 unspliced transcript (amplified using the primers indicated with solid arrows) is shown. Specific primers [dashed arrow: primer used for (D–F); dotted arrow: primer used for (G and H)] were used to pyrosequence the region containing SNP # rs31442449 (shown in bold). (B) Scheme of the breeding design using mice with a SNP in the Peg3 gene, and the expected cDNAs in case of either mono or biallelic expression. (C) PCR amplification with the primers indicated with solid arrows in (A) using mock reverse transcribed (-RT) or reverse transcribed (+RT) mRNAs from 2C or 4C neuronal nuclei or water (–). 50 PCR cycles were given to the +RT cDNA and 50 + 50 cycles were given to the –RT reaction. (D–F) Representative pyrograms from the cDNA sequence in the E11 telencephalic neuroepithelium (D), 2C (E), and 4C (F) cortical neurons, using the primer indicated with a dashed arrow in (A). The SNP (T/C) is shown in grey. T from position 6 is a negative control. Two different cDNAs were used for each analysis. Mean of the proportion of T4 (±S.E.M.) is shown. (G and H) Representative pyrograms from the cDNA sequence in the E11 telencephalic neuroepithelium (G) and 2C (H) cortical neurons, using the primer indicated with a dotted arrow in (A). The SNP (T/C) is shown in grey. G from position 4 (G4) and T7 are negative controls. Mean of the proportion of T7 (±S.E.M.) is shown.
DISCUSSION
In this study, we have shown that the methylation pattern of the DMR controlling the murine Peg3 imprinted domain can be altered in cortical neurons. In subpopulations of these cells, the normal pattern, characterized by symmetrical methylation of the constituting CpG sites that are present in the maternal allele, is changed to a strand-specific, hemimethylated pattern observed in a small segment of the Peg3-controlling DMR. This conclusion was reached after using five different approaches: bisulfite Sanger sequencing, bisulfite pyrosequencing, methylation-sensitive DNA cleavage followed by real time PCR, hairpin bisulfite PCR, and targeted NGS analysis. Since it takes place in a small region of the PCD inserted within a fully methylated domain (Figure 5A–C), we believe this strand-specific modification affects the maternal allele.
Genomic imprinting prevents parthenogenesis (33) and polyploidy (34,35) in mammalian embryos by regulating genes that are usually required for normal development (36). In addition, diploid/triploid mosaicism in somatic tissues may cause genetic syndromes with aberrant genomic imprinting (37). In this study, we have shown that the imprinting process in 4C cortical neurons is not altered. This indicates that 4C neurons generated during normal development (11,12) are unlikely to have mechanisms that disrupt imprinting.
Strand-specific CpG hemimethylation is not specific for tetraploid neurons since it can also be detected in diploid neurons. In addition, not all tetraploid neurons show this modification. The most likely explanation is that this epigenetic mark affects to a subset of cortical neurons, which are mainly tetraploid.
Our study challenges the current view that the methylation pattern of germline DMRs cannot be altered in somatic cells as they can contain hemimethylated and probably methylformylated subdomains. Further studies are needed to evaluate the biological impact of the modifications observed in the PCD of specific cortical neurons. Our results indicate that the CpG hemimethylation within the murine PCD specifically affects one of the two DNA strands. This suggests the existence of a novel molecular mechanism regulating the transition from fully methylated CpGs to strand-specific formylated CpGs. We note that the Peg3-controlling DMR is located within the Peg3 gene promoter, and that promoters are known to be enriched in 5fC (38). As also happens with full CpG formylation (39), CpG methylformylation might alter the DNA structure of the maternal Peg3 promoter. This may result in changes of Peg3 expression from the paternal chromosome due to the complex regulation that seems to affect to the PCD (40), while maintaining the imprinting capacity of the Peg3 domain. In addition, we cannot rule out that specific 5fC interactors with defined functions (41) or putative proteins interacting with hemimethylated CpGs can participate in the regulation of the PCD. The residual presence of 5fC within the murine PCD of 4C cortical neurons could also be interpreted as an intermediate state of the strand-specific demethylation process. Uncovering the molecular mechanism leading to strand-specific demethylation will facilitate our understanding of the function of this novel epigenetic label.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sergio Casas-Tintó for discussion about this work, Mark Strunk for his technical help, Simon Heath and Angelika Merkel for their help with the bioinformatic analysis, and Chaska Walton for scientific discussion and the final English version of the Manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fundación Ramón Areces [CIVP16A1815]; Spanish Ministry of Economy and Competitivity [SAF2015-68488-R]; PhD contract from the Spanish Ministry of Economy and Competitivity (to I.P.P.). Funding for open access charge: Spanish Ministry of Economy and Competitivity [SAF2015-68488-R].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wan L., Bartolomei M.S.. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv. Genet. 2008; 61:207–223. [DOI] [PubMed] [Google Scholar]
- 2. Nowak K., Stein G., Powell E., He L.M., Naik S., Morris J., Marlow S., Davis T.L.. Establishment of paternal allele-specific DNA methylation at the imprinted mouse Gtl2 locus. Epigenetics. 2011; 6:1012–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leonhardt H., Page A.W., Weier H.U., Bestor T.H.. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992; 71:865–873. [DOI] [PubMed] [Google Scholar]
- 4. Constância M., Pickard B., Kelsey G., Reik W.. Imprinting mechanisms. Genome Res. 1998; 8:881–900. [DOI] [PubMed] [Google Scholar]
- 5. Suzuki M.M., Bird A.. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008; 9:465–476. [DOI] [PubMed] [Google Scholar]
- 6. Bachman M., Uribe-Lewis S., Yang X., Burgess H.E., Iurlaro M., Reik W., Murrell A., Balasubramanian S.. 5-Formylcytosine can be a stable DNA modification in mammals. Nat. Chem. Biol. 2015; 11:555–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y.. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011; 333:1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lucifero D., Mertineit C., Clarke H.J., Bestor T.H., Trasler J.M.. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002; 79:530–538. [DOI] [PubMed] [Google Scholar]
- 9. Huang J.M., Kim J.. DNA methylation analysis of the mammalian PEG3 imprinted domain. Gene. 2009; 442:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmauss C., Brines M.L., Lerner M.R.. The gene encoding the small nuclear ribonucleoprotein-associated protein N is expressed at high levels in neurons. J. Biol. Chem. 1992; 267:8521–8529. [PubMed] [Google Scholar]
- 11. López-Sánchez N., Frade J.M.. Genetic evidence for p75NTR-dependent tetraploidy in cortical projection neurons from adult mice. J. Neurosci. 2013; 33:7488–7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morillo S.M., Escoll P., de la Hera A., Frade J.M.. Somatic tetraploidy in specific chick retinal ganglion cells induced by nerve growth factor. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shirazi Fard S., Jarrin M., Boije H., Fillon V., All-Eriksson C., Hallböök F.. Heterogenic final cell cycle by chicken retinal Lim1 horizontal progenitor cells leads to heteroploid cells with a remaining replicated genome. PLoS One. 2013; 8:e59133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morillo S.M., Abanto E.P., Román M.J., Frade J.M.. Nerve growth factor-induced cell cycle reentry in newborn neurons is triggered by p38MAPK-dependent E2F4 phosphorylation. Mol. Cell. Biol. 2012; 32:2722–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ovejero-Benito M.C., Frade J.M.. Brain-derived neurotrophic factor-dependent cdk1 inhibition prevents G2/M progression in differentiating tetraploid neurons. PLoS One. 2013; 8:e64890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ovejero-Benito M.C., Frade J.M.. p27Kip1 participates in the regulation of endoreplication in differentiating chick retinal ganglion cells. Cell Cycle. 2015; 14:2311–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaczmarek L., Chaudhuri A.. Sensory regulation of immediate-early gene expression in mammalian visual cortex: implications for functional mapping and neural plasticity. Brain Res. Brain Res. Rev. 1997; 23:237–256. [DOI] [PubMed] [Google Scholar]
- 18. Knapska E., Kaczmarek L.. A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK?. Prog. Neurobiol. 2004; 74:183–211. [DOI] [PubMed] [Google Scholar]
- 19. Kaufman M.H. The Atlas of Mouse Development. 1992; San Diego: Academic Press Inc. [Google Scholar]
- 20. Booth M.J., Marsico G., Bachman M., Beraldi D., Balasubramanian S.. Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nat. Chem. 2014; 6:435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hashimoto K., Kokubun S., Itoi E., Roach H.I.. Improved quantification of DNA methylation using methylation-sensitive restriction enzymes and real-time PCR. Epigenetics. 2007; 2:86–91. [DOI] [PubMed] [Google Scholar]
- 22. Song C.X., Szulwach K.E., Dai Q., Fu Y., Mao S.Q., Lin L., Street C., Li Y., Poidevin M., Wu H. et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013; 153:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schuyler R.P., Merkel A., Raineri E., Altucci L., Vellenga E., Martens J.H., Pourfarzad F., Kuijpers T.W., Burden F., Farrow S. et al. Distinct trends of DNA methylation patterning in the innate and adaptive immune systems. Cell Rep. 2016; 17:2101–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laird C.D., Pleasant N.D., Clark A.D., Sneeden J.L., Hassan K.M., Manley N.C., Vary J.C. Jr, Morgan T., Hansen R.S., Stöger R.. Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumaki Y., Oda M., Okano M.. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008; 36:W170–W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Relaix F., Weng X., Marazzi G., Yang E., Copeland N., Jenkins N., Spence S.E., Sassoon D.. Pw1, a novel zinc finger gene implicated in the myogenic and neuronal lineages. Dev. Biol. 1996; 177:383–396. [DOI] [PubMed] [Google Scholar]
- 27. Curley J.P., Barton S., Surani A., Keverne E.B.. Coadaptation in mother and infant regulated by a paternally expressed imprinted gene. Proc. Biol. Sci. 2004; 271:1303–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miyazaki K., Mapendano C.K., Fuchigami T., Kondo S., Ohta T., Kinoshita A., Tsukamoto K., Yoshiura K., Niikawa N., Kishino T.. Developmentally dynamic changes of DNA methylation in the mouse Snurf/Snrpn gene. Gene. 2009; 432:97–101. [DOI] [PubMed] [Google Scholar]
- 29. Hua D., Hu Y., Wu Y.Y., Cheng Z.H., Yu J., Du X., Huang Z.H.. Quantitative methylation analysis of multiple genes using methylation-sensitive restriction enzyme-based quantitative PCR for the detection of hepatocellular carcinoma. Exp. Mol. Pathol. 2011; 91:455–460. [DOI] [PubMed] [Google Scholar]
- 30. Bai L., Yan P., Cao X., Jia L., Zhang C., Guo D.. Methylation-sensitive restriction enzyme nested real time PCR, a potential approach for sperm DNA identification. J. Forensic Leg. Med. 2015; 34:34–39. [DOI] [PubMed] [Google Scholar]
- 31. Bird A.P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J. Mol. Biol. 1978; 118:49–60. [DOI] [PubMed] [Google Scholar]
- 32. Plongthongkum N., Diep D.H., Zhang K.. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat. Rev. Genet. 2014; 15:647–661. [DOI] [PubMed] [Google Scholar]
- 33. Spindle A., Sturm K.S., Flannery M., Meneses J.J., Wu K., Pedersen R.A.. Defective chorioallantoic fusion in mid-gestation lethality of parthenogenone ⇔ tetraploid chimeras. Dev. Biol. 1996; 173:447–458. [DOI] [PubMed] [Google Scholar]
- 34. Devriendt K. Hydatidiform mole and triploidy: the role of genomic imprinting in placental development. Hum. Reprod. Update. 2005; 11:137–142. [DOI] [PubMed] [Google Scholar]
- 35. Bacquet C., Imamura T., Gonzalez C.A., Conejeros I., Kausel G., Neildez-Nguyen T.M., Paldi A., Gallardo M.H.. Epigenetic processes in a tetraploid mammal. Mamm. Genome. 2008; 19:439–447. [DOI] [PubMed] [Google Scholar]
- 36. Biniszkiewicz D., Gribnau J., Ramsahoye B., Gaudet F., Eggan K., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R.. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 2002; 22:2124–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rittinger O., Kronberger G., Pfeifenberger A., Kotzot D., Fauth C.. The changing phenotype in diploid/triploid mosaicism may mimic genetic syndromes with aberrant genomic imprinting: follow up in a 14-year-old girl. Eur. J. Med. Genet. 2008; 51:573–579. [DOI] [PubMed] [Google Scholar]
- 38. Neri F., Incarnato D., Krepelova A., Rapelli S., Anselmi F., Parlato C., Medana C., Dal Bello F., Oliviero S.. Single-base resolution analysis of 5-formyl and 5-carboxyl cytosine reveals promoter DNA methylation dynamics. Cell Rep. 2015; 10:674–683. [DOI] [PubMed] [Google Scholar]
- 39. Ngo T.T., Yoo J., Dai Q., Zhang Q., He C., Aksimentiev A., Ha T.. Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability. Nat. Commun. 2016; 7:10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perera B.P., Kim J.. Sex and tissue specificity of Peg3 promoters. PLoS One. 2016; 11:e0164158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Song J., Pfeifer G.P.. Are there specific readers of oxidized 5-methylcytosine bases?. Bioessays. 2016; 38:1038–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.