Abstract
Super-enhancers (SEs) are cis-regulatory elements enriching lineage specific key transcription factors (TFs) to form hotspots. A paucity of identification and functional dissection promoted us to investigate SEs during myoblast differentiation. ChIP-seq analysis of histone marks leads to the uncovering of SEs which remodel progressively during the course of differentiation. Further analyses of TF ChIP-seq enable the definition of SE hotspots co-bound by the master TF, MyoD and other TFs, among which we perform in-depth dissection for MyoD/FoxO3 interaction in driving the hotspots formation and SE activation. Furthermore, using Myogenin as a model locus, we elucidate the hierarchical and complex interactions among hotspots during the differentiation, demonstrating SE function is propelled by the physical and functional cooperation among hotspots. Finally, we show MyoD and FoxO3 are key in orchestrating the Myogenin hotspots interaction and activation. Altogether our results identify muscle-specific SEs and provide mechanistic insights into the functionality of SE.
INTRODUCTION
Transcription factors (TFs) shape the precise gene expression pattern required for a cell to perform its unique functions by binding cis-acting regulatory elements known as enhancers and by recruiting coactivators and RNA polymerase II (RNA Pol II) to target genes. DNA high-throughput sequencing technologies have enabled the identification of putative mammalian enhancers on a broad scale based on their enrichment of the histone modifications of mono-methylation of histone H3 at lysine 4 (H3K4me1) and acetylation of histone H3 at lysine 27 (H3K27ac), and the binding of coactivators such as p300 (1). More recently, the term super-enhancer (SE) was used to define densely spaced clusters of active enhancers that encompass large open chromatin domains richly decorated with H3K27ac modifications and abundant in cell type- and identity-specific TF binding motifs that enable TFs to bind cooperatively (2,3). Regions with intensive TF co-localization are defined as hotspots or epicenters and constitute a key feature of SEs; these hotspots form regulatory hubs where cell-intrinsic and extrinsic signaling cues are interpreted in a highly lineage- and context-dependent manner (4,5). So far SEs have been reported in a plethora of cell types (6–8), but their unique biological relevance is still a matter of debate. The functionality of SEs, particularly the existence of functional features uniquely associated with SEs, such as cooperativity between individual enhancers, remains to be tested by robust experimentation (6). Initial steps toward achieving this goal have been taken; for example, two very recent reports (9,10), demonstrated opposite views of whether SE is associated with any unique biological relevance. Shin H et al. showed that a mammary-specific SE driving Wap expression during pregnancy can act as a unique regulatory ensemble with a temporal and functional hierarchy among individual enhancer constituents (9). Conversely, Hay D et al. believed that each of the five constituent enhancers within α-globin SE acts independently without clear evidence of synergistic or higher-order effects (10). There is thus an imperative need to examine SE function on more loci and in more cell types to gain a better understanding of how the individual constituent enhancers contribute to maximal SE activity through functional cooperativity or hierarchy. To achieve the goal, genome editing through the CRISPR-Cas9 system offers the opportunity to manipulate specific genomic loci in mammalian cell lines with relative ease but the large size of some constituent enhancers could preclude the testing of their cooperativity in SEs. Considering the role of hotspots as assembly hub for key TFs and their relative small size (4,5), we reason that the cooperation of constituent enhancers may occur at the level of hotspots thus they serve as an ideal paradigm for genetic dissecting of enhancer interactions. In this study we tested the notion using skeletal myoblast (MB) differentiation as a model system.
MB differentiation is a key step in the skeletal muscle formation during the embryonic muscle development or postnatal muscle regeneration (11,12). For example, upon adult muscle injury, the quiescent adult muscle stem cells are rapidly activated to become MBs, undergo proliferative expansion and differentiate into myotubes (MTs) which eventually fuse to form new myofibers. The differentiation of MBs into MTs is a well-studied process owing to an excellent mouse MB cell line C2C12. Upon withdrawal of the serum in culture medium, the proliferating C2C12 MBs would exit the cell cycle and activate the differentiation program to induce the fusion; during the process many TFs act in a sequential manner to activate the transcriptional reprogramming (11,12). The DNA-binding basic helix-loop-helix (bHLH) protein, MyoD, the master TF governing the myogenic differentiation activates the early target gene, Myogenin, which work together and cooperate with other TFs, then induce a myriad of muscle specific genes such as those encoding structural proteins, Myosin heavy chain (MyHC) and alpha-Actin (α-Actin) (11–13). It is thus not surprising to find that MyoD is enriched in enhancers of differentiating MTs and speculated to play a critical role in the assembly of the enhancers (14,15). However, our knowledge of how MyoD drives the genome-wide assembly events together with other known and unknown co-factors remains incomplete. Besides, it remains to be tested whether MyoD also plays a central role in SEs as well, the illumination of which can only become possible after a systematic identification of SEs in myogenic differentiation is completed.
Here, we utilized C2C12 differentiation as a paradigm for functional dissection of SEs, aiming to provide critical insights into the spatiotemporal complexity of enhancers during myogenic differentiation. We demonstrated distinct SE landscapes and their progressive decommissioning/assembly during the course of differentiation. Taking advantage of the available ChIP-seq datasets, we identified a myriad of TFs that co-bind with MyoD thus are critical for driving the hotspot formation, among which we showed that FoxO3 represents a previously unknown ancillary TF that is recruited to the MyoD binding sites through their physical association and subsequently stabilizes MyoD binding. We further demonstrated the importance of MyoD and FoxO3 interaction and cooperativity in driving global hotspot formation and SE activation. Furthermore, we dissected hotspot interactions on an SE associated with Myogenin locus using both classical reporter assay and CRISPR-Cas9 genomic editing, which unveiled distinct and dynamic roles of individual SE hotspots in driving Myogenin gene activation as well as the key function of MyoD and FoxO3 in orchestrating the hotspot interaction and hierarchy.
MATERIALS AND METHODS
Cell culture
Mouse C2C12 (CRL-1772) MBs were obtained from American Type Culture Collection (ATCC) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/ml penicillin and 100 μg of streptomycin (1% Pen/Strep) at 37°C in 5% CO2. For harvesting −24 h MBs, cells were grown at a low confluency (∼50%). For myogenic differentiation, 2 × 105 cells were seeded in 6-well plates and shifted to DMEM containing 2% horse serum (differentiation medium, DM) when reaching 90% confluence (considered as DM 0 h). 10T1/2 (CCL-226) and HEK293T cells were cultured in DMEM supplemented with 10% FBS.
Plasmid
To construct the hotspot luciferase reporters, a Myogenin minimal promoter (16) was amplified from C2C12 genomic DNA and inserted into pGL3 basic vector (Promega) using Xho I and Hind III sites. Individual hotspot was then cloned into the downstream of the firefly luciferase gene using BamH I and Sal I sites. For generating the reporters with hotspot combination and deleting MyoD binding motif on H3 or H4 reporter, fusion polymerase chain reaction (PCR) was used by employing overlapping annealing sequences as described before (17). HA-tagged MyoD expression plasmids were kind gift from Dr Slimane Ait-Si-Ali. Flag-tagged FoxO3 expression plasmid was a kind gift from Dr Ping Hu.
Luciferase reporter assay
All the cells were transfected using Lipofectamine 2000 (Life Technologies). The Renilla plasmid (Promega) was co-transfected as a normalization control. At 24 h post-transfection, C2C12 cells were differentiated for 48 h and the luciferase activity was measured using the Dual-Glo Luciferase Assay system (Promega) according to the manufacturer's guidelines.
ChIP and sequential ChIP
ChIP assays were performed as previously described (18,19). For ChIP, C2C12 cells were crosslinked with 1% formaldehyde at room temperature for 10 min. For sequential ChIP, cells were double crosslinked with 1 mM DSG for 45 min and then for 10 min by 1% formaldehyde. In both cases, crosslinking reaction was quenched by addition of 0.125M glycine for 10 min. Chromatin was fragmented using sonicator, followed by incubation with 5 μg of antibodies at 4°C for overnight. Antibodies for MyoD (Santa Cruz Biotechnology, sc-304X, rabbit polyclonal), FoxO3 (Santa Cruz Biotechnology, sc-48348X, rabbit polyclonal) and histone H3-K27 acetylation (Abcam, ab4729, rabbit polyclonal) were used for ChIP, or normal rabbit IgG (Santa Cruz Biotechnology, sc-2027) was used as negative control. For sequential ChIP, the first ChIP was performed as described above and the immunocomplexes were eluted with 50 μl of 10 mM Dithiothreitol (DTT) at 37°C for 30 min. The eluted supernatant was diluted for 20 times, followed by incubation with antibodies for the second ChIP or IgG. Immunoprecipitated genomic DNA was resuspended in 50 μl of water. PCRs were performed with 1 μl of immunoprecipitated DNA as template with SYBR Green Master Mix (Life Technologies) and products were analyzed by qRT-PCR on a 7900HT system (Life Technologies). Primers used are listed in Supplementary Data S1.
Chromatin conformation capture (3C) PCR
Chromosome conformation capture (3C) was performed as previously described (20). Briefly, 2 × 107 C2C12 cells were fixed by adding 2% formaldehyde at room temperature for 10 min, followed by quenching the crosslinking reaction by adding 0.125 M glycine for 10 min. Cells were lysed with lysis buffer (10 mM Tris–HCl pH 8.0, 10 mM NaCl, 0.2% Igepal CA630, with protease inhibitor cocktail freshly added) rocking on ice for 10 min. Next, nuclei were centrifuged and re-suspended in CutSmart Buffer (NEB). The extracted chromatin was then digested with 400 units of EcoRI (NEB). On the next day, digested chromatin was ligated by 100 units of T4 DNA Ligase in 10 times of the initial volume at 16°C for 4 h. The chromatin was subsequently de-crosslinked at 65°C for overnight. On the next day, protein was removed by incubating with 300 μg of protease K (Life Technologies) at 55°C for 2 h, and RNA was removed by 300 μg of RNaseA (Life Technologies) at 37°C for 2 h. DNA was extracted by phenol/chloroform isolation, finally resuspended in 250 μl of 1× TE buffer. The digestion and ligation efficiencies were assessed before performing PCR. Probes were designed near enhancers and promoter. PCRs were performed with BAC clone RP24–343C13 as control, with SYBR Green Master Mix (Life Technologies) and products were analyzed by qRT-PCR on a 7900HT system (Life Technologies). The sequence and layout of probes were listed in Supplementary Data S1.
Genomic editing by CRISPR-Cas9
The CRISPR-Cas9 system was used to delete hotspots and knockout MyoD expression in C2C12 cell line following published protocols (21,22). Briefly, target-specific guide RNAs (gRNA) were designed using an online tool (http://crispr.mit.edu/). gRNAs were then cloned into Cas9 expressing pX330 plasmid (Addgene, 42230) using Bbs I site. C2C12 cells were transfected with two constructed plasmids using Lipofectamine 2000 (Life Technologies). A pSIREN plasmid was co-transfected for later screening. Forty-eight hours after transfection, the cells were treated with 2.5 μg/ml puromycin for 3 days and cultured in medium without puromycin for another 3 days. Cells were then suspended and diluted into 96-well plates to obtain single cell clones. Individual colonies were picked and validated by PCR and subsequent Sanger DNA sequencing. Double deletion of H2 and H3 was generated by deleting H3 in H2 KO cell line. Similarly, double deletion of H3 and H4 was generated by deleting H3 in H4 KO cell line. All the gRNAs and genotyping PCR primers are listed in Supplementary Data S1.
DNA pulldown
DNA pulldown assays were performed as previously described (23). In short, 2 × 107 cells were harvested and lysed, followed by precipitation of 400 μg lysate with 4 μg of biotinylated probe in streptavidin-agarose magnetic beads (Life technologies) for 2 h. DNA–protein complex was eluted from the beads and analyzed by western blotting. Primers used in generating biotinylated probe are listed in Supplementary Data S1. For immunoprecipitation with recombinant MyoD (rMyoD) protein, purified MyoD proteins was pre-incubated with DNA containing tandem repeats of E-box sequence or with scramble sequence together with the beads. Nuclear extracts from C2C12 MTs were then incubated with the beads pre-bound by MyoD and the DNA. The IP products were next subjected for western blot analysis.
qRT-PCR
Total RNAs from cells were extracted using TRIzol reagent (Life Technologies) according to the manufacturer's instructions and cDNAs were prepared using PrimeScript™ RT Master Mix kit (Takara, RR036A). Analysis of mRNA expression was performed with SYBR Green Master Mix (Life Technologies) as described on an 7900HT System (Life Technologies) (24). All primers are listed in Supplementary Data S1.
Immunoprecipitation, immunoblotting and immunofluorescence staining
Immunoprecipitation was performed as described before (25). A total of 10 μg of antibodies against MyoD (Santa Cruz Biotechnology, sc-304X), or FoxO3 (Santa Cruz Biotechnology, sc-48348X), or HA (Santa Cruz Biotechnology, sc-805) were used. For immunoprecipitation with recombinant MyoD protein, purified proteins were pre-incubated with DNA containing tandem repeats of E-box sequence or with scramble sequence together with the beads. Nuclear extracts from C2C12 MTs were then incubated with the beads pre-bound by MyoD and DNA. The IP products were next subjected for Western blotting. For Western blot analysis, the following dilutions were used for each antibody: MyHC (Sigma; 1:2000), α-Tubulin (Sigma; 1:5000), Myf5 (Santa Cruz Biotechnology; 1:1000). Immunofluorescence staining on cultured cells was performed using antibodies against MyHC (Sigma; 1:350) and Myogenin (Santa Cruz Biotechnology, 1:600) as described before (19,26,27).
Sucrose gradient centrifugation
Whole cell extracts of C2C12 were obtained with cell lysis buffer (50 mM Tris–HCl pH7.6, 1% TritonX-100, 1 mM ethylenediaminetetraacetic acid, 10% glycerol, 1 mM DTT, 1 mM PMSF and protease inhibitor cocktail). For sucrose gradient analysis, 500–600 μl whole cell extracts was added on the top of a 13.5 ml 10–30% sucrose gradient. The sample was then centrifuged in a SW40Ti rotor (Beckman) at 38 000 Revolutions Per Minute (RPM) at 4°C for 16 h. The resulting gradient was then fractionated every 500 μl. Only odd-numbered fractions were extracted to avoid cross contamination between the two consecutive fractions. Samples were resolved on SDS-PAGE, followed by western blotting by MyoD or FoxO3 antibody.
ChIP-seq and RNA-seq datasets
A table summary of the publicly available datasets downloaded for this study can be found in Supplementary Table S1. Briefly, ChIP-seq data for H3K27ac, H3K4me1, H3K4me2, H3K18ac, H3K9ac, H4K12ac, H3K36me3, H3K27me3, PolII and p300 in MB and MT were collected from two previously published studies (14,28). Aligned sequence reads and identified read enriched regions (peaks) were downloaded. The aligned reads of ChIP-seq data for 12 TFs were collected from the ENCODE public repository (29), including Myogenin, MyoD, Fosl1, Cebpb, Ctcf, Usf1 and Max1 in MB, and MyoD, Tcf3, Tcf12, Cebpb, Usf1, Myogenin, Rest, E2f4, Max and Srf in MT. ChIP-seq data of Mef2d in MT (30), c-Jun in MB (14), Smad3 in MT (31), Yy1 in MB and MT (19), Myf5 and Snai1 in MB (32), Pbx in MB and MT (33) and Foxk1 in MB (34) were obtained from previous studies. Mus Musculus genome (UCSC mm9 assembly) was used for alignment of ChIP-seq reads when needed. All the coordinates mentioned in this study are mm9-based. RNA-seq datasets from MBs or the differentiating MTs at various time points (−24, 60, 120 and 168 hr) were obtained from a previous study (35). All the home generated ChIP-seq datasets can be found in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE93916 and GSE99207).
Analyses of TF motifs and peaks
SE or TE constituents were extended by 100 bp on both sides and used as the input regions to predict enriched motifs by HOMER (36). To scan for specific TF motifs, position-specific probability matrix of motifs for MyoD and FoxO3 was obtained from the TRANSFAC database (37). Utilizing FIMO (38), MyoD peaks and FoxO3 peaks in hotspots were scanned for MyoD and FoxO3 motif respectively. The whole hotspot region was used to scan for MyoD or FoxO3 motif, when MyoD or FoxO3 binding was not present at the hotspot. If a motif was detected within a region with P-value < 10−4, this motif is considered to be present at the region.
In order to unify the criteria for peak calling, aligned ChIP-seq reads for each TF were fed to MACS (version 2.0.9) (39), using threshold of q-value < 0.01 (−q 0.01) with the input ChIP-seq sample as the background. Other options were set as default. Since the number of Myogenin peaks in MB is <50, it was excluded from further analysis. All called and downloaded histone marks and TF ChIP-seq peaks were subjected to a filter to exclude the ENCODE blacklisted regions (40). When TF binding sites (TFBSs) within SEs or TEs were being studied, peaks falling in promoters and exons of coding genes were carefully excluded from the analysis. Promoters were defined as regions spanning ±2 Kb from transcription start sites (TSSs) in RefSeq annotation (June 2015).
To explore TF co-localization, all ChIP-seq peaks from all TFs were pooled in MB or MT respectively, with the ones overlapping each other merged as the same peak. Among all the pooled peaks, hotspots were defined as among the top 10% ranked by the number of harboring TFs. Regions bound by no less than 4 or 5 TFs in MB or MT, respectively, comprise <10% (7.7% and 10.0% for MB and MT) of the TF-bound genomic loci, and were thus defined as hotspots.
Analyses of ChIP-seq read density for TFs and histone marks
The ChIP-seq density (intensity or signal) was calculated by first aggregating all the ChIP-seq reads with their start coordinates overlapping every query region to a read count. Query regions were extended by 50 bp on both sides of enhancer constituents in calculation of histone mark ChIP-seq density, extended by 100 bp on both sides of enhancer constituents in calculation of TF ChIP-seq density or extended by 500 bp on both sides of pooled peaks in calculation of H3K27ac and Pol II ChIP-seq density. Then the aggregated read count were normalized to the length of the regions and the total mappable read count, resulting in RPKM (reads per kilo base per million mapped reads) as the measurement unit. For boxplot (Figures 1E–L, 2C, D, H, I, K, L, 3C, Supplementary Figures S1C, S2C and D and S3G and I), each RPKM value was then normalized by subtracting the RPKM of the input, with negative values adjusted to zeros if they comprised no more than 5% of all the data points. All RPKM in the figures refer to the background normalized RPKM unless otherwise mentioned. In signal heat maps (Figure 3B, Supplementary Figure S2F and G), read density in each 20 bp bin (x-axis) surrounding aligned centers (y-axis) was calculated. And in metagene plots (Figures 2G, 3A and Supplementary Figure S3J), the ChIP-seq reads were extended to 200 bp to intersect each 20 bp bin surrounding the region centers to smoothen the signal, after which the signal at each aligned bin was averaged and denoted as the height on the y-axis.
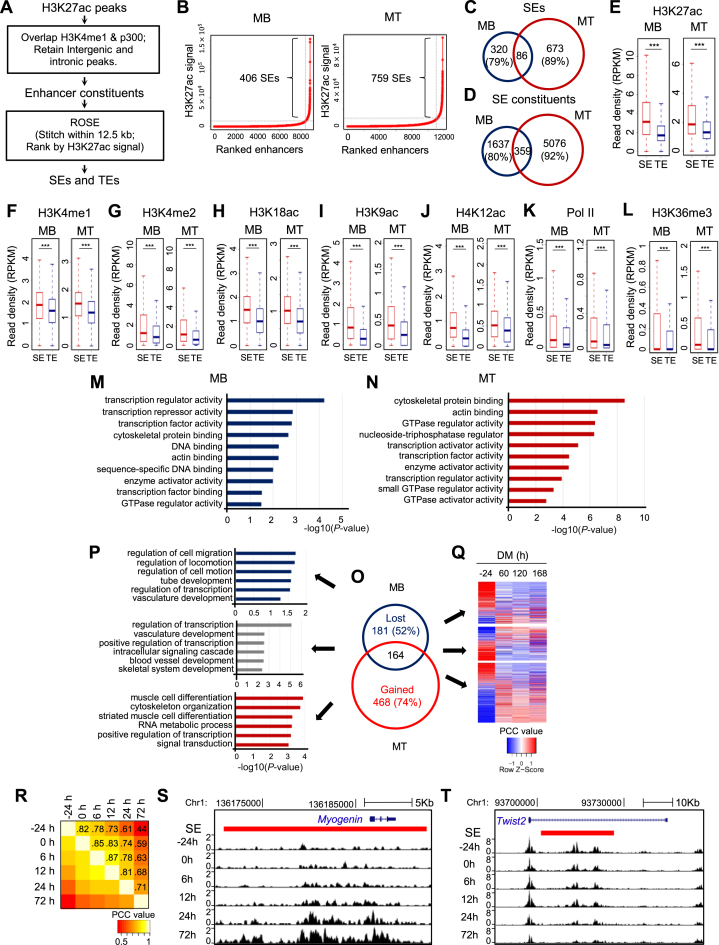
Figure 1.
Identification and characterization of SEs in skeletal muscle differentiation. (A) Illustration of the computational pipeline to identify TEs and SEs in MB and MT. (B) SEs were identified in a stage-specific manner in MB or MT utilizing ROSE, a program that stitches and ranks enhancers. When the background subtracted H3K27ac signal for enhancers was plotted in a ranked order, a clear geometric inflection point was revealed on the curve, with the ones above this point being super-enhancers (SEs), and the rest typical enhancers (TEs). (C and D) Shifted landscapes of SEs and SE constituents from MB to MT. A high portion was lost or gained during the differentiation. (E–L) Increased ChIP-seq intensity of enhancer marks (e.g. H3K27ac, H3K4me1, H3K4me2, H3K18ac, H3K9ac and H4K12ac), and indicators of active transcription (e.g. Pol II and H3K36me3) was observed on SE than TE constituents. ***P < 0.001. (M and N) Gene Ontology (GO) analysis of molecular functions for SE associated genes revealed enrichment of transcription regulator/factor activity in both MB and MT. The y-axis shows the top 10 enriched GO terms and the x-axis shows the enrichment significance P-values. (O) The number of SEs lost, maintained or gained as cells progressed from MB to MT. (P) GO analysis of the genes (top five) associated with the above categories of SEs. (Q) The differential expression patterns of the above genes were examined by RNA-seq collected from MB or MT at various time points in differentiation medium (DM). (R) H3K27ac ChIP-seq signals revealed SE landscape shifts with the progression of differentiation program from −24, 0, 6, 12, 24 and 72 hr in DM. The Pearson correlation coefficient (PCC) values are labeled on each grid to indicate the degree of correlation between the two consecutive stages. (S) Increasing H3K27ac density was observed for the SE (red bar) associated with Myogenin gene during the differentiation. (T) Decreasing H3K27ac density was observed for the SE associated with Twist2 gene during the differentiation.
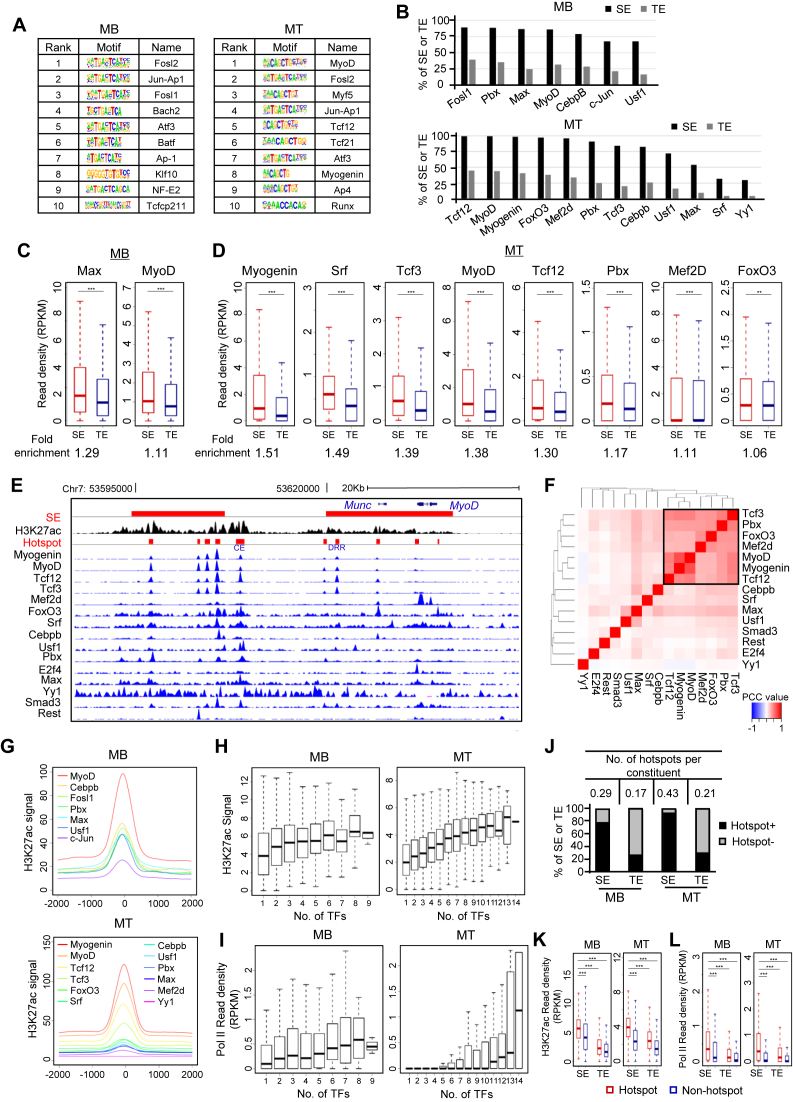
Figure 2.
Identification of TF assembly and TF hotspots. (A) In silico prediction of TF motifs within SE constituents in MB and MT identified key regulators in the respective cell stage. (B) Compared with TEs, a larger percentage of SEs are occupied by the indicated TFs by analyzing available ChIP-seq datasets. (C and D) TFs showed higher read density at SEs than TEs in both MB and MT. ChIP-seq read density at each SE or TE constituents extended by 100 bp on both sides was calculated. Fold-enrichment below each plot was calculated as the ratio of the mean value between the read density at SE and TE constituents. *P < 0.05, **P < 0.01 and ***P < 0.001. (E) Genome browser snapshot to show co-localization of TFs on the two SEs (red bars) associated with MyoD locus. (F) Analysis for TF co-association in MT. A combinational module was identified in the square. The color code indicates the PCC between two TFs at their binding sites. (G) Metagene plots were generated to show the TFs cobound to proximal genomic loci. (H and I) Enhancer activity elevates with the increasing number of binding TFs as measured by H3K27ac signal or Pol II read density. (J) SEs are more enriched with TF hotspots compared with TEs. Y-axis represents the percentage of SEs or TEs harboring at least one hotspot (Hotspot+) or no hotspot (Hotspot−). The number of hotspots per enhancer constituent was calculated (Table on top). (K and L) Hotspot regions showed significantly higher level of H3K27ac or Pol II density compared to non-hotspot genomic loci. ***P < 0.001.
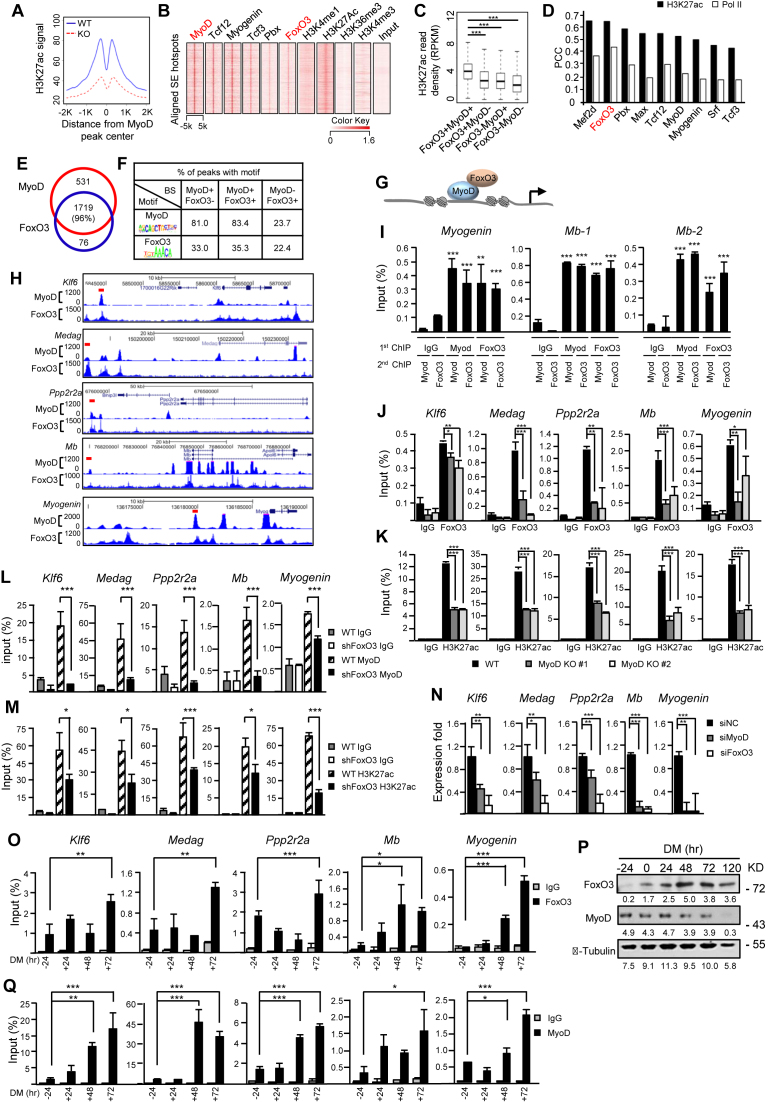
Figure 3.
FoxO3 and MyoD coordinate the genome-wide activation of SE hotspots during myoblast (MB) differentiation. (A) MyoD deletion decreased the activity at MyoD-bound hotspots within SEs compared to the wild-type (WT) control. H3K27ac ChIP-seq signal in WT or KO surrounding the aligned MyoD peak center in WT SE hotspots was calculated to measure the enhancer activity. (B) FoxO3 and some known co-factors of MyoD bind to MyoD peaks in SE hotspots in MT. (C) Genomic loci within SEs bound by both FoxO3 and MyoD displayed the highest H3K27ac density compared with FoxO3 or MyoD only sites in MT. ***P < 0.001. (D) PCC between H3K27ac or Pol II and TF ChIP-seq density revealed that the FoxO3 binding displayed the highest correlation with SE activity among the TFs. (E) A total of 96% of FoxO3 peaks within SE hotspots overlap with MyoD binding. (F) On the FoxO3-MyoD co-bound enhancer hotspots, a much higher percentage of MyoD binding E-box motif (5’-CAGGTG-3’) than FoxO3 binding Fork head motif (5’-GTAAACA-3’) was detected. (G) Schematic illustration of MyoD recruitment of FoxO3 to the hotspot region. (H) Snapshots of examples of MyoD/FoxO3 co-bound hotspots (red bar) and their associated genes. (I) Sequential ChIP showed both MyoD and FoxO3 bound on the same region in one hotspot associated with Myogenin, or two hotspots associated with Mb (Myoglobin) gene. (J and K) Decreased FoxO3 or H3K27ac binding on six selected examples of hotspots associated with Klf6, Medag, Ppp2r2a, Mb and Myogenin was detected in MyoD knockout (KO) cell lines #1 and #2 as compared to WT control. (L and M) Knockdown of FoxO3 by shRNA decreased MyoD or H3K27ac binding on the above SEs. (N) Knockdown of either MyoD or FoxO3 by siRNA reduced the expression of the above genes. (O) ChIP-PCR was performed to detect the enrichment of FoxO3 on the above hotspots in the differentiating myotubes (MTs) at −24, 24, 48 and 72 hr. (P) The expression of MyoD and FoxO3 in differentiating C2C12 was detected by western blotting using α-Tubulin as loading control. (Q) ChIP-PCR was performed to detect the enrichment of MyoD on the above hotspots at DM −24, 24, 48 and 72 hr. All qRT-PCR and ChIP-PCR data were normalized to GAPDH mRNA and ChIP input respectively and represent the average of three independent experiments ±s.d. *P < 0.05, **P < 0.01 and ***P < 0.001.
For TF ChIP-seq data, input DNA datasets from GSM915172 and GSM915181 were used as normalization control; for histone mark, p300 and Pol II ChIP-seq data, input DNA datasets from GSE25308 were used; and for H3K27ac ChIP-seq datasets generated by us, two previously published IgG controls (GSM1117984 and GSM1117985) (19) were used. According to the ChIP-seq guidelines and practices published by ENCODE (40), both ‘Input DNA’ and ‘IgG control’ are acceptable control samples in analyzing ChIP-seq datasets.
Analyses of Pearson correlation coefficient (PCC)
To investigate the SE landscape shifting along time (Figure 1R), all the SE constituent regions in six time points were pooled and the correlation of H3K27ac signal between each pair of time points at pooled peaks was calculated. The color of each grid represents the Pearson correlation coefficient (PCC) value, which is labeled on the grid. To identify the co-association module among TFs (Figure 2F and Supplementary Figure S2E), all the pooled ChIP-seq peaks overlapping extended SE constituents (500 bp on both sides) were examined as bound or not bound by each one of the TFs. The PCC was calculated and showed by the color between each pair of TFs to measure the co-occurrence correlation. To explore the correlation between TF and enhancer activity (Figure 3D), each SE constituent was extended by 500 bp on both sides, and the TF and histone mark ChIP-seq density were calculated as described above.
Statistical information
Welch's t-test was used in comparing histone mark or transcription factor density in figures of boxplot. Statistical comparison of two groups was done by Student's t-test or one-way analysis of variance with repeated measurements, followed by Tukey's post hoc test for multiple groups. (*P < 0.05, **P < 0.01 and ***P < 0.001).
RESULTS
Identification and characterization of SEs in myoblast differentiation
Various surrogate marks of enhancers alone or in combinations have been used for defining enhancer regions in previous studies (6). Taking advantage of the published ChIP-seq datasets (14,28) (Supplementary Table S1), we performed enhancer identification in both C2C12 proliferating MBs and fully differentiated MTs based on H3K27ac, H3K4me1 and p300 signals (Figure 1A). Briefly, intergenic and intronic H3K27ac peak regions, overlapped by H3K4me1 and p300 peaks were considered as the boundaries of enhancer constituents; using ROSE algorithm (3,41), the enhancer entities exhibiting high H3K27ac intensity above the cut-off point, i.e. the geometrical inflection point on the ranking curve (Figure 1B) were designated as SEs and the rest as typical enhancers (TEs). As a result, a total of 406 and 759 SEs together with 8389 and 11 052 TEs were identified in MB and MT, respectively (Supplementary Data S2). A high portion of SEs or SE constituents are only present in one stage, demonstrating the stage specificity and the shifting landscapes during cell differentiation (Figure 1C and D). Similarly, in line with previous findings (3), genomic regions spanned by SEs are significantly larger than TEs by one order of magnitude in both MB and MT (Supplementary Figure S1A) and are comprised of more enhancer constituents (Supplementary Figure S1B).
SEs have been shown to be associated with magnified signals of enhancer features compared to TEs (3). Expectedly, several histone marks known to be associated with enhancer activity (28) displayed higher occupancy on SEs than TEs, including H3K27ac (Figure 1E), H3K4me1 (Figure 1F), H3K4me2 (Figure 1G), H3K18ac (Figure 1H), H3K9ac (Figure 1I) and H4K12ac (Figure 1J). Increased intensity was also observed for Pol II (Figure 1K) and H3K36me3 (Figure 1L). On the contrary, as a mark associated with the poised state of enhancers (42), H3K27me3 signal was found to be lower in SEs than TEs (Supplementary Figure S1C). These results are in keeping with the notion that SEs are more active than TEs (2,3). Using proximity rule (3) to assign the neighboring expressed gene as the target gene under regulation by an SE, we also found SE-associated genes display higher expression than TE-associated ones (Supplementary Figure S1D).
SEs are known to drive high expression of cell-fate determining genes (2,6). In both MBs and MTs, SE-associated genes are enriched for molecular function GO terms (Supplementary Data S3) relevant to transcription (Figure 1M and N); this is in line with previous findings that TF encoding genes are enriched among SE-associated genes (3,43). To further look into SE orchestrated gene expression in skeletal muscle differentiation, we found SEs are decommissioned from 181 genes but gained on 468 genes during the transitioning from proliferating MB to terminally differentiated MT, and remain constitutively associated with 164 genes (Figure 1O). SE-lost genes are enriched for GO terms including ‘regulation of cell migration’ and ‘regulation of cell motion’ etc., while SE-gained are enriched for ‘muscle cell differentiation’ and ‘muscle cell development’ etc. (Figure 1P), suggesting cell and stage specific regulation of gene expression by SEs. To further investigate whether SEs could drive the expression of specific genes in the respective cell stage, analyzing available RNA-seq datasets (35) (Supplementary Table S1), we found that the majority of SE-lost genes (74%) are downregulated during the differentiation course from −24 to 60 hr, while SE-gained genes (69%) are upregulated (Figure 1Q). To further monitor the progressive remodeling of SE during the MB differentiation into MTs, we generated a time series of H3K27ac ChIP-seq data covering the differentiating course of −24, 0, 6, 12, 24 and 72 hr (Supplementary Data S2). As shown in Figure 1R, progressive temporal changes in enhancer usage were readily observed during the progression of differentiation. By examining the correlation of H3K27ac signals, a high correlation was observed between the two consecutive points but gradually decreases as the time interval increases. When examining the SE-associated genes, key TFs pertinent to the cellular state were found to be enriched (Supplementary Data S4). A gradual gain of H3K27ac signal was observed on the SEs associated with the key TFs driving the differentiation, for example, on the Myogenin SE (Figure 1S). The gain begins as early as 6 hr concomitant to Myogenin gene induction and continues to augment into 72 hr. Gradual loss of H3K27ac signal, on the other hand, was observed on SEs associated with genes determining the cell proliferation stage, for example (Figure 1T), on the Twist2 gene that inhibits C2C12 differentiation (44). Altogether the above findings support the notion that SE remodeling (decommissioning on some sites while assembly on others) occurs during the progression of MB cellular differentiation.
Identification of TF assembly and TF hotspots in SEs
The fundamental question in understanding enhancer function is to dissect the TF/co-factor binding events spatiotemporally and to identify key TFs driving enhancer activation/deactivation. Applying HOMER (36), we predicted an array of TFs enriched within SE constituents in MBs and MTs (Figure 2A). A large proportion of the top-ranked motifs are previously known key regulators in the respective cell stage, for example, MyoD and Myogenin in MT; and Tcf12 (Transcription factor 12), the known heterodimer partner for MyoD (45) (Figure 2A). It also reveals factors with previously unexplored function such as Atf3 (Activating transcription factor 3) in both MBs and MTs. Luckily in C2C12 cells we could collect a number of ChIP-seq datasets for TFs (14 and 15 from MB and MT, respectively) from various sources (Supplementary Table S1) for further examination of TF binding. A large number of the TFs (7 in MB and 12 in MT) exhibit enriched binding in the enhancer regions (over 10% of the total binding peaks residing in the enhancers) (Supplementary Figure S2A and B). We found that a higher percentage of SEs than TEs are associated with these TFs (Figure 2B). For example, 99% of SEs in MT contains MyoD binding peaks compared to 44% of TEs. For enhancer constituents, which are comparable in length, 46.6% of them in SEs are bound by MyoD, compared to 38.9% in TEs; each SE constituent is associated with 0.64 MyoD peak on average, compared to 0.42 for TE constituent (Supplementary Table S2). Furthermore, some of these TFs also display higher local signals in SEs than in TEs (Figure 2C and D). In MT, eight TFs exhibit significantly higher signal within SE, with Myogenin and MyoD showing 1.51- and 1.38-fold enrichment (Figure 2D and Supplementary Figure S2D), suggesting that these TFs are positively correlated with SE activity. In MB, Max (Myc-associated factor X) and MyoD (Figure 2C) but not other examined TFs (Supplementary Figure S2C) are more enriched in SE constituents than TE. Convergence of multiple TFs is a key feature of enhancers especially notable in SEs (2). Indeed, intensive co-localization of TFs was uncovered in both MB and MT. Shown in Figure 2E is a snapshot of two SEs identified 31 249 and 3619 bp upstream of MyoD TSS in MT, in which multiple TFs bind in close proximity. To examine the degree of co-association between TFs, a heat map was generated based on the correlation of binding loci between each TF pair within all the SE repertoires. One distinct combinational module was identified in MT by unsupervised clustering (Figure 2F). In addition to the expected intimate associations among MyoD with Myogenin, Tcf3 and Tcf12, we observed its association with Pbx (Pre-B cell leukemia transcription factor), which was recently shown to co-occupy muscle regulatory regions with MyoD (33); these TFs were also retrieved in searches for MyoD protein interactome (46,47); In addition, we found an association with Mef2d (Myocyte enhancer factor 2d), which is a regulatory partner of MyoD in regulating myogenic gene expression (47). Interestingly, FoxO3 (Forkhead box 3) exhibits association with the above TFs in the same module. Similar TF associations and a combinational module were also observed in MB (Supplementary Figure S2E). Consistent with the previous report showing c-Jun participates in the assembly of enhancer in proliferating MBs (14), we also found c-Jun cobinds with MyoD in the combinational module in MB. The degree of TF co-localization is also shown in a metagene plot (Figure 2G), in which the tight shape of the peaks indicates that the binding sites of most TFs cluster within a very short distance. This co-localization tendency was again demonstrated by generating heat maps showing focal signal for TFs around the aligned center of the pooled peaks (Supplementary Figure S2F and G). Furthermore, we found that the activity of enhancers measured by H3K27ac (Figure 2H) and Pol II (Figure 2I) density is positively correlated with the number of bound TFs. Taken together, these results identify the TFs that cooperate extensively to open the chromatin and establish the SE profiles in muscle cells.
Next, we defined a short genomic region co-bound by no less than 4 or 5 of the above analyzed TFs in MB or MT, respectively as a TF hotspot, which has led to the genome-wide identification of a total of 7790 and 22 827 hotspots in MB and MT with the median size of 428 and 403 bp (Supplementary Data S5). Among these, 603 hotspots in MB are located in SEs, with around 75.4% of SEs (306/406) being occupied by at least one hotspot (25.4% for TEs (2134/8389)); 2342 SE hotspots in MT were detected to occupy 94.6% of SEs (718/759) compared to 29.7% for TEs (3281/11 052) (Figure 2J). Each SE constituent has 0.29 and 0.43 hotspot on average in MB and MT respectively, while each TE constituent only 0.17 and 0.21 (Figure 2J), further demonstrating that SEs are highly enriched for the hotspots defined by these available TF ChIP-seq. In addition, H3K27ac (Figure 2K) and Pol II (Figure 2L) densities within hotspots are significantly higher than within non-hotspots (occupied by <4 or 5 TFs in MB and MT) in SEs, indicating these hotspots are super active centers that may orchestrate the entire SE functionality.
FoxO3 and MyoD coordinate the global activation of hotspots
Consistent with its known function as a key factor in driving MB differentiation, MyoD was found in 88.6% of the genome-wide hotspots (20 233/22 827) in MT and for hotspots within SE regions, this number increases to 96.1% (2250/2342), suggesting that MyoD is pivotal in driving SE hotspot assembly events. Indeed, the deletion of MyoD by CRISPR-Cas9 in the cells (Supplementary Figure S3A–D) induced extraordinary changes at SE landscape; 584 SEs in WT were lost while 257 new ones were gained (Supplementary Figure S3E and F). The deletion not only sharply attenuated the enhancer activity at the WT SE hotspots (Figure 3A), but also resulted in the activation of genes that acquired de novo SEs in KO cells (Supplementary Figure S3G). Interestingly enough, Myf5, another myogenic bHLH protein, which may act redundant in the specification and differentiation of skeletal muscle cells (48,49), was only slightly downregulated in the MyoD KO cells (Supplementary Figure S3H), suggesting that Myf5 could not compensate for the loss of MyoD and they may have distinct roles in regulating SEs. Moreover, in agreement with the reported role of MyoD in priming the enhancers prior to differentiation (14,15,50), we found among the 2533 SE constituents bound by MyoD in MT, 1127 (44.5%) are already bound by MyoD in MB. It was suggested that MyoD initiates enhancer assembly by recruiting other ancillary factors. Indeed, several known co-factors of MyoD, such as Tcf12, Myogenin, Tcf3 (45), Pbx (33) are enriched and co-localized at the MyoD binding sites within the SE hotspots in MT (Figure 3B). Interestingly, all the examined TFs are to some extend associated with higher SE activity compared to MyoD or the TF alone binding (Figure 3C and Supplementary Figure S3I); among them we noticed FoxO3 whose co-binding with MyoD is evident and results in a significantly higher (1.55- or 1.50-fold) SE activity compared to MyoD or FoxO3 alone binding (Supplementary Table S3). The crucial roles of FoxO3 in global enhancer regulation are emerging in recent years (51,52), but its possible involvement of enhancer regulation in muscle cells remains unknown thus attracted our attention. In fact, FoxO3 binding intensity in MT correlates best with SE activity as measured by H3K27ac or Pol II ChIP-seq density, with the PCC reaching 0.65 with H3K27ac, which is even higher than that of MyoD (Figure 3D). Among 1719 SE hotspots co-bound by MyoD and FoxO3 in MT (Figure 3E), 972 (53.9%) are pre-bound by MyoD in MB, suggesting FoxO3 binds the MyoD-primed hotspots upon differentiation. In addition, the H3K27ac signal is higher on the primed hotspots upon FoxO3 binding compared with non-FoxO3 bound sites (Supplementary Figure S3J), hinting FoxO3 binding amplifies the activity of MyoD primed hotspots and this was not found to be a general phenomenon for the other examined TFs (Supplementary Figure S3J and Table S4). We found 95.9% (1719/1793) of the FoxO3-bound hotspots loci are also bound by MyoD (Figure 3E), indicating that MyoD binding may be necessary for the recruitment of FoxO3 protein. Indeed, when further examining the binding motifs in the peaks (Figure 3F), we found that among FoxO3 peaks that overlap with MyoD peaks, only 35.3% harbor FoxO3 motifs (5’-GTAAACA-3’), but a striking 83.4% has MyoD motifs. However, among non-MyoD overlapping peaks 22.4 and 23.7% harbored FoxO3 or MyoD motif, which is relatively comparable. Hence, a large portion of FoxO3 binding on the co-bound regions may be recruited through MyoD, instead of binding through its own motif (Figure 3G). Altogether the above findings raised the speculation that FoxO3 represents a previously unknown co-factor of MyoD in driving hotspot/SE assembly thus warranted further elucidation.
To experimentally test the above MyoD/FoxO3 cooperative model, we first confirmed that indeed both MyoD and FoxO3 can be detected on the selected hotspots (one associated with Myogenin and two with Mb (Myoglobin) loci) by performing sequential ChIPs in MT with either MyoD or FoxO3 antibody first. Consistent with the results from ChIP-seq analyses (Figure 3H), we detected high enrichment of both TFs on the same hotspot region (Figure 3I). The binding of FoxO3 was significantly reduced (Figure 3J) in MyoD KO cells (Supplementary Figure S3B) without affecting the FoxO3 level (Supplementary Figure S3C and D); H3K27ac signal was also reduced on the same sites (Figure 3K), suggesting that MyoD is necessary for FoxO3 binding on the hotspot and subsequent activation of the region. Interestingly, knocking down FoxO3 by shRNA also reduced MyoD binding (Figure 3L) and H3K27ac intensity (Figure 3M) on the co-bound hotspots, suggesting FoxO3 could help stabilize the MyoD binding. However, it is known that FoxO3 could also transcriptionally regulate MyoD (53,54) and indeed we found evident downregulation of MyoD in the shFoxO3 cells (Supplementary Figure S4A and B); to distinguish the two modes of regulation, we found ectopic expression of MyoD in the shFoxO3 cells could not rescue the H3K27ac reduction (Supplementary Figure S4C), suggesting the phenotype caused by FoxO3 loss may not be a result of the decreased MyoD level and FoxO3 has an active role in stabilizing MyoD binding. Altogether the above data support a model in which MyoD recruits FoxO3 on the hotspots and cooperatively they contribute to the activation of SEs. In line with this model, knockdown of either MyoD or FoxO3 reduced the expression of the neighboring genes associated with the above analyzed SEs (Figure 3N); and the ectopic expression of MyoD in the shFoxO3 cells again did not rescue the reduction in the gene expression (Supplementary Figure S4D) as well. Finally, when examining the temporal dynamics of MyoD and FoxO3 binding during the differentiation course, FoxO3 binding increases on most sites (Figure 3O) as its level elevates at 24 hr (FoxO3 protein was found to be low in the proliferating MBs as shown in Figure 3P) and continues to increase into late differentiation stage (e.g. 72 hr) (Figure 3O); MyoD binding that is pre-existent on some sites in MB is augmented showing a concomitant increase (Figure 3Q) despite its unchanged expression upon differentiation, suggesting an active recruitment by FoxO3. As expected, knock-down of FoxO3 blocked the C2C12 differentiation (Supplementary Figure S4E and F), which phenocopied the effect of MyoD loss (data not shown). These data again reinforce the dynamic interplay of MyoD and FoxO3 in driving the hotspot assembly/SE activation during the myogenic differentiation process.
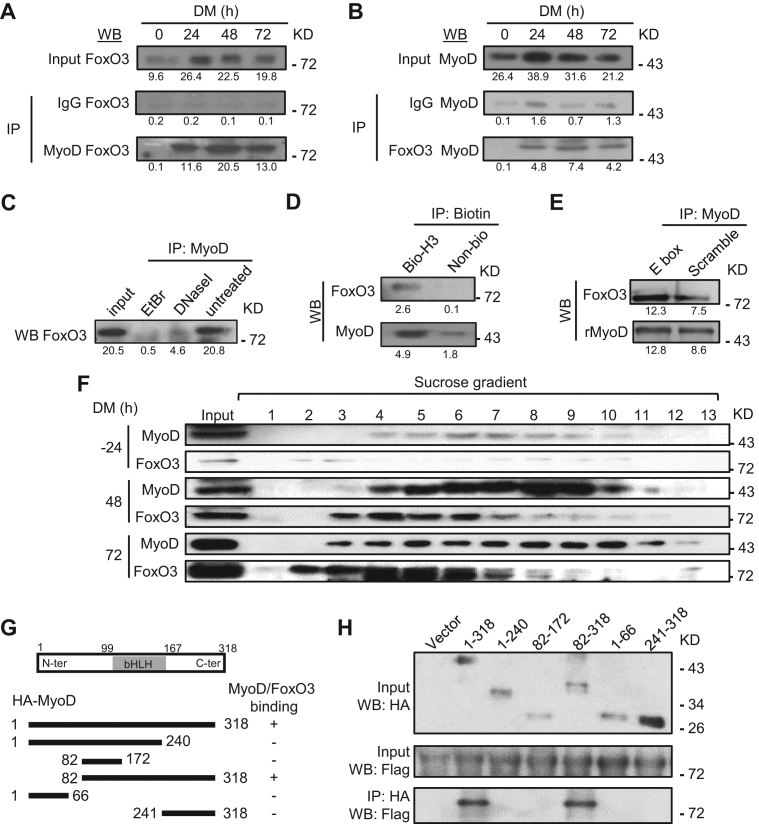
To further dissect whether the interaction of MyoD and FoxO3 is through physical binding between the two proteins, an evident association between the endogenous proteins was detected in the differentiating MTs by reciprocal co-immunoprecipitation assays using either FoxO3 (Figure 4A) or MyoD (Figure 4B) antibody. Interestingly, the binding was abolished upon Ethidium Bromide (EtBr) or DNase I treatment that disrupts the integrity of DNA structure (55) (Figure 4C), suggesting the binding is DNA dependent. To further examine whether the binding depends on the hotspot region, we performed DNA immunoprecipitation using biotin-labeled DNAs from the Myogenin hotspot region (445 bp long), a significantly higher amount of FoxO3 (26-fold) and MyoD (2.7-fold) were retrieved compared to the pull downs by non-biotin labeled control DNAs (Figure 4D). Furthermore, recombinant MyoD (rMyoD) protein was able to retrieve a significant amount of FoxO3 protein from MT lysates and the retrieval was enhanced (1.6-fold) when the protein was pre-incubated with DNAs containing MyoD binding E-box motif (Figure 4E), which is in line with the earlier finding that FoxO3 is recruited to MyoD primed sites thus its binding is dependent on the motif (Figure 3F). To strengthen the above results, FoxO3 and MyoD were eluted in the same fractions in the differentiating MTs at both 48 and 72 hr during the sucrose gradient elution (Figure 4F). Finally, to map the protein–protein interacting domain, we found that domain 82–318 of MyoD could interact with FoxO3 but domain 82–172 (bHLH DNA binding domain) or 241–318 failed, indicating both DNA binding and C-terminal domains of MyoD together are needed for the physical association with full length FoxO3 (Figure 4G and H). Altogether the above results have identified FoxO3 as a previously unknown co-regulator in assisting the hotspot formation/SE activation driven by MyoD.
Figure 4.
Physical association of MyoD and FoxO3 at the hotspots in the differentiating MTs. (A) C2C12 was harvested in DM at 0, 24, 48 and 72 h and the lysates were immunoprecipitated with IgG or MyoD antibody and then detected for FoxO3 by western blotting. Numbers below each blot indicate the intensity of each band as measured by Image-Pro Plus. (B) The reciprocal IP was performed with IgG or FoxO3 antibody followed by WB detection of MyoD. (C) The binding between MyoD and FoxO3 at 48 h was abolished by either DNase I or ethidium bromide (EtBr) treatment. (D) When immuniprecipitated by Biotin, biotin-labeled Myogenin hotspot 3 (H3), but not biotin-labeled control, was able to retrieve MyoD and FoxO3. (E) Recombinant MyoD (rMyoD) protein retrieved FoxO3 from MT lysates and the addition of E-box motif DNA increased the retrieval. (F) Sucrose gradient of C2C12 nuclear extracts at DM −24, 48 and 72 h followed by western blotting detected the elution profiles of MyoD and FoxO3, which were found in the same fractions (4–6 and 7) in DM 48 and 72 h. (G and H) The plasmid expressing various domains or the full-length (1-318) of HA-tagged MyoD was co-transfected with plasmid expressing Flag-tagged full length FoxO3. Lysates were pulled down by HA antibody and examined by western blotting using Flag antibody. For all the above panels, the representative images from two replicates are shown.
Dynamic cooperation among hotspots regulates Myogenin induction
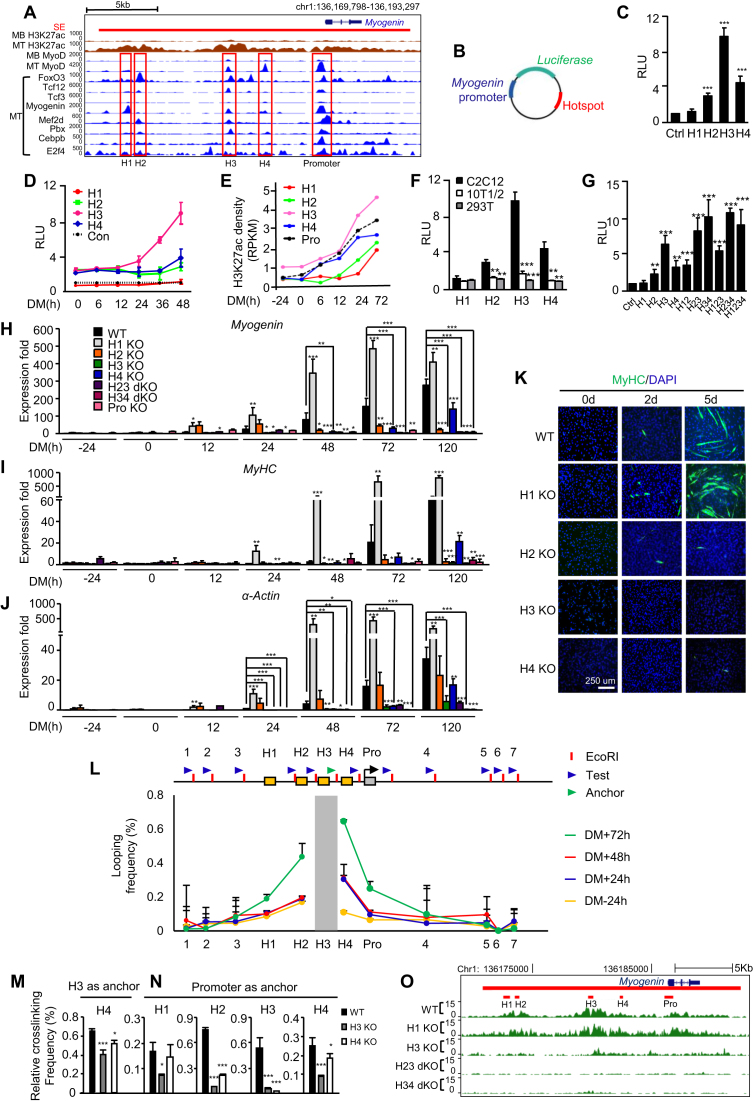
Cooperativity between enhancers at the level of hotspot was reported (5), but the functional significance needs further investigation. We thus looked into the interactions among hotspots within SE, reasoning that the interactions mediated through TFs/co-factors are key to SE activation. The SE associated with Myogenin gene was used as a paradigm for this investigation (Figure 5A). Myogenin is a key developmental regulator for skeletal muscle formation whose expression is restricted to skeletal muscle. Despite the accumulated work over the past 20 years, our knowledge of its strict temporal and spatial regulation in a tissue-specific manner remained incomplete (56). Early studies utilizing LacZ reporter mice identified the proximal promoter region (−130 to +18 bp relative to the TSS) is sufficient to ensure the proper temporal and spatial expression of the Myogenin during embryonic myogenesis, the level of the LacZ expression, however, was relatively weak (57,58); it was thus speculated that additional DNA elements beyond the proximal promoter are required for high-level of Myogenin expression (56). The presence of an immediate upstream regulatory region (−1092 to −130 bp) was also defined to drive the Myogenin expression (57,58). In addition, through comparative analysis of Myogenin genes from different genomes, three putative regulatory elements around −4.5, −5.5 and −6.5 kb upstream of the Myogenin TSS were identified but their functions in driving Myogenin expression remain to be experimentally tested. To this end, a very recent report confirmed that the −6.5 kb region indeed is bound by MyoD and drives Myogenin promoter activation (59). The SE spanning 21 751 bp region (chr1:136170803–136192554) encompassing Myogenin locus was defined using H3K27ac signal, within which two distal and two proximal non-promoter hotspots (H1, −13.8 kb; H2, −12.9kb; H3, −6.7 kb; H4, −4.1 kb relative to the TSS of Myogenin) were identified along with a promoter hotspot (−472 to +280 bp). Each of them is featured by various degrees of H3K27ac density as well as MyoD and FoxO3 binding. H3 displays the highest H3K27ac signal and sharp peaks of MyoD and FoxO3 binding as well as other hotspot enriched TFs; this is comparable to the binding profiles on the promoter hotspot. In fact, the H3K27ac and MyoD binding was already visible in MB stage albeit to a much lower level (Figure 5A). H4 has a sharp MyoD peak but lacking FoxO3 binding; H1 has relatively low MyoD binding but a visible Myogenin peak; H2 has the weakest MyoD binding but a sharp FoxO3 peak. These binding profiles triggered us to speculate that each hotspot is different with respect to their strength to activate the Myogenin gene promoter and the interaction may exist to achieve the optimal activation. To test this notion, we first employed the enhancer reporter assay by constructing a series of luciferase reporters in which each hotspot was fused to the minimal Myogenin promoter region (−133 to −1 bp to TSS) (Figure 5B). Consistent with the H3K27ac profile, H3 exhibited the highest activity in the differentiating C2C12 MTs. H4 with strong MyoD but very low FoxO3 binding showed lower activity than H3, hinting that FoxO3 binding is necessary for MyoD to achieve the maximal activation of the hotspot; Consistent with the key role of MyoD in hotspot activation, H1 exhibited almost no activity; H2, however, had 3-fold activation compared to the control reporter minus hotspot (Figure 5C). To further examine temporal dynamics of their activation during the course of differentiation, we found H3 was the first to be activated at 6 hr of differentiation and continued to increase sharply into 48 hr whereas H4 and H2 activities were not induced until 48 h; H1 strikingly exhibited lower activity than the control from the beginning, hinting an intriguing likelihood that it serves as a repressive element (Figure 5D). The above findings were solidified by quantifying signals of H3K27ac from Figure 1S to assess the temporal establishment of each hotspot. As shown in Figure 5E, indeed, establishment of H3 peak is already existent in MB stage and continued to increase along the differentiation course; H4 was quickly established as early as 6 h; H3K27ac was not evident on H2 until later differentiation phase; On H1, interestingly, accumulation of the signal was observed starting 24 hr and continued to increase into 72 hr, suggesting the permissive chromatin is needed even for a repressive element. Predictably, the above reporter activation was not observed when the reporters were transfected into murine fibroblast 10T1/2 cells or human embryonic kidney 293T cells (Figure 5F), indicating their activation is myogenic specific. To further dissect the possible combinational effect of the hotspots, various combinations were generated (Figure 5G). Interestingly, addition of H4 enhanced H3 activity almost additively (H34 versus H3); further addition of H2 (H234 versus H34) led to no further increase though. Analogously, addition of H2 also additively increased H3 activity (H23 versus H3). Strikingly, further addition of H1 (H123 versus H23) decreased the activation, again hinting that H1 is possibly a repressive element.
Figure 5.
Dynamic and hierarchical cooperation among hotspots governs Myogenin gene induction during differentiation. (A) ChIP-seq profiles for across the Myogenin locus in C2C12 MB or MT. The SE is shown as red bar. Four non-promoter hotspots and one promoter hotspot are highlighted in red rectangles. (B) Diagram of the plasmid constructed for luciferase reporter assays. The Myogenin minimal promoter (blue) was inserted upstream while individual hotspot (red) was inserted downstream of the luciferase gene (green). (C) The above reporter was transfected into C2C12 for 24 hr before the cells were switched to DM for 48 h and measured for the luciferase activity. The plasmid containing the minimal promoter was used as control (Ctrl). RLU: relative luciferase unit. (D) Temporal dynamics of the reporter activity during C2C12 differentiation was measured at various time points. (E) Quantification of H3K27ac ChIP-seq density at each hotspot and the promoter region. (F) The hotspot activity was measured in 10T1/2 or HEK293T cells to compare with C2C12. (G) Reporters with various combinations of hotspots were generated and the activities were measured. (H) Each hotspot alone or in combination was removed by CRISPR-Cas9 and the expression of Myogenin was measured at various time points. (I and J) The degree of differentiation was measured by the expression of MyHC and α-Actin markers and (K) the staining for MyHC by immunofluorescence. (L) A schematic of the Myogenin locus illustrating hotspots H1–4 (yellow boxes), the promoter (gray box), three upstream and four downstream control loci (1–7). Blue arrow heads indicate 3C primers designed at EcoRI sites (red bar) for each test region, while green arrow head indicates anchor primer for H3 region. Quantitative 3C-qPCRs were performed using the anchor and each primer to test the interaction between H3 (the anchor region, gray bar) and the test region. The looping frequency (%) was obtained by normalizing to the control BAC template. (M) The interaction between H3 and H4 in H3 or H4 KO was assessed by 3C-qPCR. (N) The interaction between each hotspot and the promoter was assessed by 3C-PCR in H3 or H4 KO cells. (O) H3K27ac ChIP-seq experiments were performed to measure the effect of hotspot deletion on the SE activity. All luciferase data represent the average of three independent experiments ± s.d. All qRT-PCR and ChIP-PCR data were normalized to GAPDH mRNA or ChIP input respectively and represent the average of three independent experiments ±s.d. *P < 0.05, **P < 0.01 and ***P < 0.001.
The above data clearly suggested complex cooperative modes among the Myogenin hotspots loci and the prominent status of H3. To strengthen the above results from using the luciferase reporters which may not recapitulate the native chromatin state, genomic editing using CRISPR-Cas9 technology was employed to remove individual or combinational hotspots in C2C12 cells (Supplementary Figure S5A–C). Consistently, deletion of H3 completely abolished Myogenin induction from the beginning phase (Figure 5H and Supplementary Figure S5D); and blocked the entire differentiation process as assessed by the expression of MyHC and α-Actin markers (Figure 5I and J; Supplementary Figure S5E and F) as well as staining of Myogenin (Supplementary Figure S6A and B) and MyHC proteins (Figure 5K and Supplementary Figure S6C and D) and viewing of the cell morphology (Supplementary Figure S6E). In H4 KO cells, Myogenin induction did not appear until late stage around 48 hr and was much lower than WT levels; consequently, the myogenic differentiation was delayed but not blocked, suggesting H4 is important for Myogenin induction in the early phase. Conversely, deletion of H2 caused an elevation of the Myogenin induction in the early phase which gradually diminished after 12 hr, suggesting an intriguing possibility that H2 is a repressive element in the early differentiation but becomes essential for sustaining Myogenin expression in the later stage. Consequently, accelerated MT formation was observed in the beginning phase but could not proceed to terminal differentiation (Figure 5K and Supplementary Figure S6A–E). Interestingly but not unexpectedly, deletion of H1 stimulated an increased level of Myogenin induction from as early as 12 hr and the expression remained elevated than WT level into late differentiation (Figure 5H and Supplementary Figure S5D); accordingly, the entire myogenic differentiation was accelerated (Figure 5I–K and Supplementary Figures S5E-F and 6A–E). As expected, successive deletion of both H2 and H3 (H23 dKO) or H3 and H4 (H34 dKO) abolished Myogenin expression and the differentiation in an analogous fashion as H3 KO (Figure 5H–J; Supplementary Figures S5D–F and 6A–E).
To further illuminate whether the cooperation is mediated through physical interaction through chromosomal looping, 3C (chromosome confirmation capture) assay was performed followed by PCR to analyze the chromatin loop structure (Figure 5L). Detectable looping frequency between H3 and H4 or the promoter but not with the negative control regions upstream of H1 or downstream of Myogenin gene was observed upon early differentiation at 24 h and persisted throughout the differentiation whereas the interaction between H3 and H1 or H2 occurred in a later stage (Figure 5L–N and Supplementary Figure S6F), confirming H3 is the earliest to be activated then probably helped the establishment of H4 and others through their interactions. Consistent with the notion, H3 deletion significantly diminished its interaction with H4 and also drastically reduced the interaction of the other three hotspots with the promoter (Figure 5M and N), reinforcing its prominent status in orchestrating the assembly and function of the other three hotspots. H4 deletion, on the other hand, disrupted its interaction with H3; meanwhile, the interactions between the promoter and H2 or H3 was also reduced (Figure 5N and O), suggesting H4 activation proceeds and may be necessary for H2 activation and subsequent interaction with the promoter. In agreement with the above findings, in H3 KO MTs H3K27ac signal was largely reduced not only on the H3 region but also on all the other three hotspots (Figure 5O and Supplementary Figure S6G), reinforcing the initiating function of H3 for the entire SE. In H23 and H34 dKO cells, the H3K27ac signal on the other hand was almost ablated over the entire SE, suggesting the interaction of H3 with H2 and H4 is also needed for the full establishment of the SE activity. Consistent with its repressive role, in H1 KO cells the signal was evidently increased over the whole SE and promoter (Figure 5O and Supplementary Figure S6G). Altogether the above findings from the genomic deletion clearly indicate functional diversity and hierarchy of the four hotspots and their cooperation in driving the Myogenin gene induction during the course of MB differentiation.
MyoD and FoxO3 orchestrate the Myogenin hotspot interaction
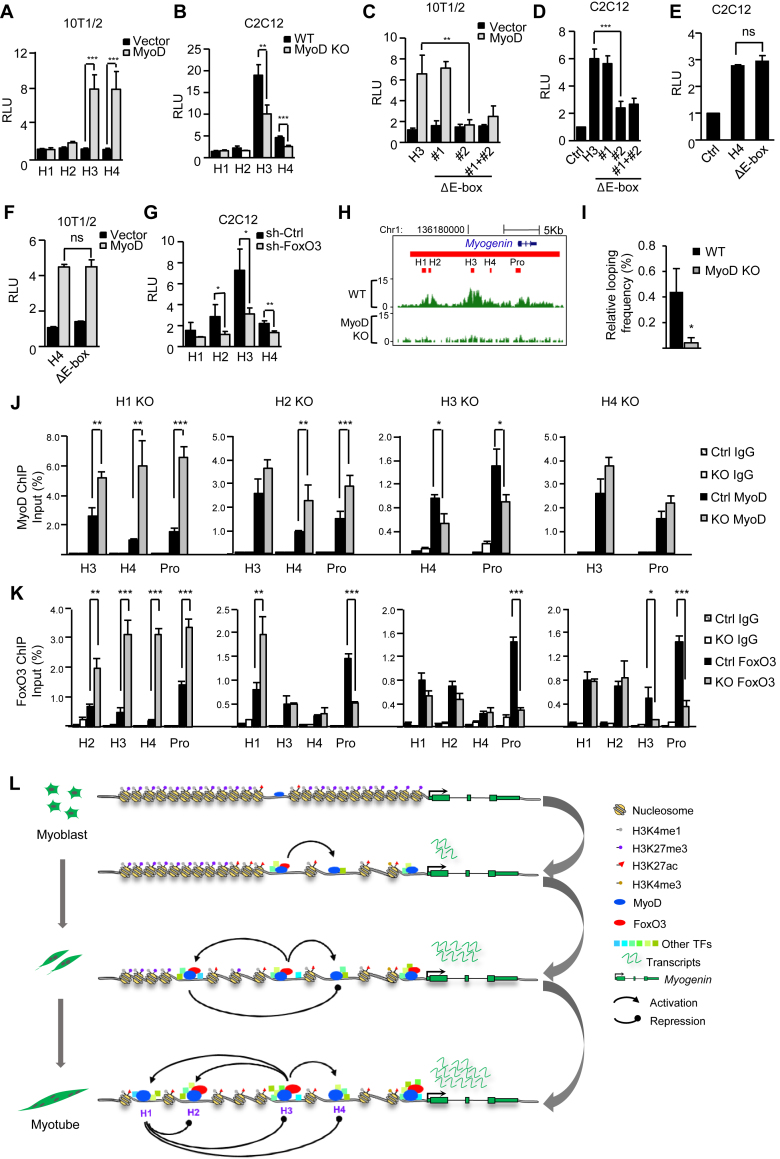
Given the uncovered roles of MyoD/FoxO3 in orchestrating the global hotspot assembly earlier in Figure 3, we next asked whether they are key to the hotspot interaction and activation on the Myogenin SE. To test the notion, we found H3 or H4 reporter was highly induced in 10T1/2 cells with MyoD overexpression (Figure 6A), suggesting MyoD is key to their activation. Consistent with what was seen earlier in C2C12 cells, the activation is not through changing FoxO3 levels (Supplementary Figure S7A). Interestingly, H1 and H2 reporters showed no such response to MyoD, which is consistent with their lacking of strong MyoD binding from ChIP-seq (Figure 5A). Expectedly, the activity of H3 or H4 in C2C12 MTs was markedly inhibited by MyoD deletion (Figure 6B), whereas H1 or H2 activity was unaffected. To further pinpoint the importance of MyoD binding in activating H3, two MyoD binding motifs identified through TRANSFAC database (60) in H3 (Supplementary Figure S7B) were removed individually or combined from the H3 reporter; deletion of # 2 or both but not # 1 abolished MyoD-dependent H3 reporter activation in 10T1/2 cells (Figure 6C); similarly, the # 2 deletion also inhibited the reporter activity in C2C12 MTs (Figure 6D), suggesting the essential role of the motif # 2 in mediating MyoD binding to H3 and its subsequent activation. When examining closely, we found indeed motif# 1 is slightly different from the known canonical E-box motif (CANNTG), which may explain its incompetence in mediating MyoD activation. Interestingly, such response to MyoD was not observed when the identified MyoD binding motif was removed from the H4 reporter (Figure 6E and F), which indicates the binding is not mediated through direct recruitment to this motif. When further examining the role of FoxO3 in controlling the hotspots activation, we found knockdown of FoxO3 in C2C12 cells markedly hampered the activity of H3 (2.3-fold), H2 (2.4-fold) and to a less extent on H4 (1.65-fold) (Figure 6G); FoxO3 knock-down also prevented the full degree of H3 and H4 activation by MyoD overexpression (Supplementary Figure S7C).
Figure 6.
MyoD and FoxO3 orchestrate the hotspot interactions. (A) Overexpression of MyoD in 10T1/2 cells induced the H3 or H4 hotspot reporter activities. (B) H3 or H4 reporter activity was decreased in MyoD KO cells. (C) Deletion of motif # 2 or both but not #1 diminished the induction of H3 reporter by MyoD overexpression in 10T1/2 cells. (D) The deletion of motif # 2 or both but not #1 diminished the H3 reporter activities in differentiating C2C12 MTs. (E) Deletion of the motif in H4 had no impact on H4 reporter activity in differentiating C2C12 MTs. ns, no significance. (F) The deletion of the motif had no impact on the induction of H4 activity by MyoD over-expression in 10T1/2 cells. (G) H2, H3 or H4 hotspot reporter activity was reduced in shFoxO3 MTs compared to the vector control. (H) H3K27ac ChIP-seq was performed in WT or MyoD KO cells to assess the impact of MyoD deletion on hotspot activity. (I) The interaction between H3 and H4 was decreased in MyoD KO cells as assessed by 3C-PCR. (J and K) The impact of each hotspot deletion on the enrichment of MyoD or FoxO3 on the hotspots and Myogenin promoter was examined by ChIP-PCR. All luciferase data were normalized to Renillia protein and represent the average of three independent experiments ± s.d. Relative Luciferase Unit (RLU) normalized to the vector or WT control is shown. All qRT-PCR and ChIP-PCR data were normalized to GAPDH mRNA and ChIP input respectively and represent the average of three independent experiments ±s.d. *P < 0.05, **P < 0.01 and ***P < 0.001. (L) Schematic illustration of MyoD/FoxO3 mediated hotspot interaction that orchestrates Myogenin SE activation during MB differentiation. The dominant H3 hotspot is primed in the MB by MyoD binding; upon early differentiation, binding of FoxO3 further amplifies its activity and subsequent interaction with H4; H3 next interacts with H2 and H1 to activate their formation; H1 appears to repress the other three hotspots thus acting as a brake. Altogether the hierarchical activation of the four hotspots orchestrates the Myogenin gene expression.
The above data suggested it is likely that H3 functions as a main platform for initial binding of MyoD which orchestrates the hierarchical interaction and assembly of all hotspots. In fact, MyoD was found on H3 prior to differentiation in the proliferating MB (Figure 5A), and MyoD deletion indeed abolished the establishment of H3K27ac on H3 as well as the entire SE region by ChIP-seq (Figure 6H). Consistent with the theory that MyoD guides the interaction between H3 and H4, the looping frequency between H3 and H4 was diminished in MyoD KO MTs (Figure 6I). TFs guide the physical interaction among hotspots, which in turn may promote further sharing and recruitment of TF to each hotspot. To further examine the importance of each Myogenin hotspot on MyoD/FoxO3 recruitment, as shown in Figure 6J, MyoD binding on H4 and the promoter was decreased in the H3 KO cells, suggesting MyoD binding on H4 is dependent on their interaction. On the contrary, binding of MyoD on H3 and the promoter was not affected in H4 KO cells. However, FoxO3 binding on H3 and promoter was markedly decreased by H4 deletion (Figure 6K), suggesting the essential function of H4 in FoxO3 recruitment. Strikingly, in H1 KO cells, MyoD and FoxO3 recruitment on H3, H4 and promoter were all significantly increased (Figure 6J and K), again reinforcing the idea that H1 functions as a brake to prevent over-loading of the key TFs and over-activation of the Myogenin gene. Similarly, H2 deletion increased MyoD binding on H4 but not on H3, suggesting H2 may act also as a brake for H4 antagonizing MyoD binding on H4; this possibly is crucial for the early phase of differentiation when H2 likely acts as a repressive element as implicated in Figure 5H. Altogether the above data support the central role of MyoD and FoxO3 in orchestrating the dynamic interactions among hotspots and their synergism in activating the Myogenin gene.
DISCUSSION
In this study, we dissected the super-enhancer functionality in the paradigm of MB differentiation using genome-wide profiling coupled with in-depth enhancer editing. We defined stage-specific SEs that drive the myogenic conversion and identified hotspots enriched for TFs. We uncovered the previously unknown function of FoxO3 in driving hotspot formation and SE activity as an accessory factor for MyoD. Finally, using Myogenin gene as a model, we dissected the interactions among hotspots within the SE region. We demonstrated that multiple modes of cooperation exist among the hotspots to achieve the optimal level of Myogenin gene activation during the course of differentiation (Figure 6L). Our findings thus favor the argument that many SEs defined on the basis of ChIP-seq experiments and specific algorithms are special biological entities.
A compendium of SEs in myogenic differentiation and novel regulators of SE activation
Although MB differentiation represents a relatively well-defined process during muscle stem cell lineage progression at the transcriptional level, study of SE regulation remained a gap to fill. Here our findings provide the compendium of SEs and hotspots, which will be useful for future discovery of SE-associated mechanisms that drive the MB differentiation into MT. For example, two SEs were found to be associated with MyoD gene (Figure 2E), whose regulatory elements have been well characterized before (61–65); the well-studied regulatory region DRR (distal regulatory region) happens to be one of the five hotspots in the proximal SE; the other known CE (core enhancer) regulatory region instead lies adjacent to the distal SE. Interestingly, if using H3K27ac alone without H3K4me1 and p300 marks for the definition of SEs, these two MyoD SEs would be merged into one encompassing both DRR and CE (data not shown), suggesting using different surrogate marks may cause slightly different results; therefore reinforcing that experimental validation is still necessary to confirm the functionality of SEs identified through the ChIP-seq marks.
Extending the previous study by Blum R. et al. (14), our findings not only demonstrated MyoD is enriched in SEs but also showed its unequivocal role in coordinating hotspot assembly/interaction and SE activation. We believe that MyoD binds to some dominant hotspots and primes them for further activation. Upon differentiation, these hotspots will be used as initiating platforms to interact with and assemble other hotspots through chromosomal looping that are guided by MyoD together with transcriptional regulators expressed later. The interactions on the other hand, further facilitate and amplify the enhancer activity of the initiating hotspot as well as others, resulting in the optimal induction of the genes. Also worthy to point out, Myf5, another key Myogenic regulatory factors (MRF) regulating skeletal muscle specification and differentiation (48,49) did not seem to compensate for the effect of MyoD loss in guiding the SE assembly. This is in line with a recent report (66) showing the two MRFs have distinct roles in the transcription of skeletal muscle program; Myf5 may functionally precede MyoD by recognizing chromatin prior to robust transcription of differentiation genes and induces histone acetylation without Pol II recruitment or robust gene activation, whereas MyoD subsequently binds to the same sites and induces histone acetylation, recruits Pol II and robustly activates gene transcription. It is thus very likely Myf5 will aid MyoD in priming the dominant hotspots in a SE prior to their activation. To define MyoD interactome on SEs, furthermore, our study is the first to collect all the available TF/co-factor ChIP-seq to identify the TFs that actually co-bind with MyoD genome-wide. Consistent with a recent report (33) and its known role in stabilizing MyoD binding (17), Pbx is found to be associated with MyoD in the same combinational module in both MB and MT; Max as the b-HLH-LZ Myc partner protein probably associates with MyoD to block its transactivation activity in MB (30); it interestingly became associated with Usf1 in MT, suggesting the dynamic usage of the same TF during the enhancer assembly. Usf1 was reported to bind the Myf5 enhancer region (31), but the implication of its genome-wide association with SE also remains unknown. Our findings thus unveiled unknown roles of many factors in the myogenic-specific hotspot and SE formation, which opens new avenues for future exploration.
Moreover, we for the first time revealed FoxO3 is an ancillary factor for MyoD. The connection between the two TFs in muscle cells was first reported by Hu et al. (53), who demonstrated that FoxO3 can regulate MyoD transcriptionally; its knock down thus impairs MyoD transcription and inhibits MB differentiation. However, ectopic expression of MyoD in the FoxO3 knockdown cells could only partially rescue the differentiation, suggesting the downregulation of MyoD may not fully explain the phenotype and other modes of interaction may exist between the two molecules. Here we demonstrate that the physical interaction of MyoD and FoxO3 proteins mediates their functions in the enhancer regulation. In our hands, ectopic expression of MyoD in shFoxO3 cells could hardly rescue the SE activation and gene expression, suggesting the critical role of FoxO3 in stabilizing MyoD on these sites. This was further substantiated by multiple lines of evidence from biochemistry assays (Figure 4), which demonstrated the presence of direct protein–protein binding between MyoD and FoxO3; the binding was enhanced by E-box motif, reinforcing the thinking that FoxO3 binding occurs after MyoD binds and primes the site. Therefore, we believe FoxO3 functions to stabilize and amplify the MyoD binding; still, further investigation is needed to find out how subsequently FoxO3 functions in the MyoD complex to activate enhancer and gene expression, which may very likely involve other co-factors as FoxO3 exhibits association with an array of TFs in the combinational module shown in Figure 2F. It is also likely FoxO3 involves the recruitment of p300 histone acetyltransferase since direct binding between p300 and FOXOs has been observed in many studies (67). Forkhead box O family of TFs such as FOXA1 can act as pioneer TF to open compacted chromatin. Our findings show that the level of FoxO3 is low in proliferating MBs and induced upon differentiation when it helps to refine or amplify the pre-established hotspot ‘code’. This is interestingly in agreement with two findings in colorectal adenocarcinoma cell line by Eijkelenboom et al. (51,52), where they demonstrated that instead of binding to compacted chromatin as a pioneer TF like FOXA1, FoxO3 binding regions are already in an open and potentially active state in colon carcinoma cells. Nevertheless, in addition to the MyoD dependent function, we also found many FoxO3 binding events independent of MyoD binding, suggesting FoxO3 may have other roles than acting as an ancillary factor for MyoD. In addition to MyoD, FoxO3 has been shown to interact with ASCL (68), also a bHLH transcription factor involved in both NPCs and myogenesis (69); this interaction also occurs in the enhancer regions, raising an interesting possibility that binding of FoxO3 and bHLH factor may be a general feature in directing cell state conversion. In addition, we must also point out that the function of FoxO3 in muscle cells is likely to be context dependent. While we found FoxO3 knockdown inhibited the C2C12 differentiation, a recent report (54) showed that FoxO3 knockout in muscle satellite cells (SCs) promoted their differentiation; this may be a result of SC’s inability to maintain quiescent stage while we have only dissected its direct role in the phase of MB differentiation. Altogether our findings suggest the dynamic existence of a mega complex in responsive to spatiotemporal cues that orchestrates hotspot assembly and activation; the identification of the participating components is only limited by the availability of the ChIP-seq datasets. Our study thus sets the stage for exciting future exploration aiming at dissecting the nature of muscle-specific enhancer regulation and advancing our understanding of mechanisms underpinning transcriptional reprogramming during myogenesis.
Myogenin gene expression is governed by multiple hotspots in a SE
Our results from using Myogenin gene as a paradigm for functional dissection provide a thorough understanding of how Myogenin gene is temporarily regulated at the enhancer level during the terminal differentiation. Myogenin is a key developmental regulator for skeletal muscle formation whose expression is restricted to skeletal muscle. We for the first time dissected the SE driving Myogenin gene expression and confirmed the earlier speculation that distal regulatory enhancers exist in addition to the proximal enhancer/promoter to confer the high expression of Myogenin during myogenesis (56). We showed this SE is a functional unit that is key to regulate Myogenin promoter activation throughout the course of differentiation and its functionality is driven by the cooperation among four hotspots. The simple beauty of the C2C12 system has allowed us a unique opportunity of dissecting the in-depth mechanism underpinning the SE regulation and laid a foundation for future explorations of the SE activity in vivo. It will be interesting to find out whether such regulatory mechanism also applies to the Myogenin regulation developmentally during embryogenesis and whether H3 still acts as a prominent hotspot driving the maximal Myogenin expression in vivo.
Key TFs drive the temporal dynamics of hotspot interaction
In terms of enhancing our understanding of the SE functionality, we have provided direct evidence that multiple constituent enhancers show cooperative behavior at the level of hotspots and such cooperativity is relevant for gene regulation, SEs can thus serve as regulatory hubs which dictate lineage-specific functions. As modeled in Figure 6L, although H3 is the main hotspot leading to productive induction of Myogenin expression, the existence of other hotspots may ensure the robustness and precision of its expression in the course of differentiation. The combinational action of multiple hotspots can be achieved spatially and/or temporally. H3 exerts the dominant impact on the Myogenin gene expression throughout the entire differentiation course, which may stem from the recruitment of MyoD on it prior to differentiation in the MB stage and as well as the subsequent recruitment of FoxO3; as a result, H3 deletion completely blocked Myogenin gene expression throughout the entire differentiation course. 3C-PCR (Figure 5L) also showed constitutive interaction between H3 and promoter, further confirming H3’s dominant role in activating the promoter. H4 appears to be especially important for achieving the optimal gene activation in the early stage as its deletion delayed but not blocked Myogenin expression. Interestingly, there is only MyoD but no FoxO3 binding on H4, indicating MyoD binding alone is sufficient to activate the hotspot but FoxO3 is needed to achieve the maximal activation. In the 3C assay, the interaction between H3 and H4 was seen throughout the early to late differentiation course, suggesting the intimate cooperation between them is necessary. H2 seems to be necessary for the continuous induction of Myogenin expression through the late differentiation. Interestingly, MyoD binding is very weak on this hotspot but FoxO3 is high, suggesting an interesting scenario that binding of FoxO3 in the later phase activates it and induces its interaction with H3 and promoter to sustain Myogenin promoter activation. In this case, FoxO3 recruitment to the region is probably through other factors. The most intriguing finding came from dissecting the role of H1, its deletion strikingly enhanced the Myogenin promoter expression throughout the differentiation, hinting that this hotspot acts as a brake for the entire SE. To our knowledge, this is the first report clearly demonstrating the negative effect of a hotspot. Currently we do not know how it acts as a repressive element; other factors that bind to this region may contribute to the phenomenon, for example, a very recent report from Shen et al. demonstrates that RACK7 recruits KDM5C to enhancer regions and negatively regulates enhancer activation (70). We are thus keen into finding out the underpinning mechanism to explain the repressive effect of H1.
The mode of the SE activation uncovered from the Myogenin gene resembles the very recent finding from studying SE regulation associated with Wap induction during pregnancy (9). In both cases, a dominant enhancer/hotspot was identified and primed long before the gene induction; and others are being assembled successively upon the inducing cues. We suspect this mode of SE regulation may apply to many genes especially those that need sharp induction upon external stimuli. Interestingly, in the case of Myogenin, the relatively proximal enhancers (H3 and H4) seem to be more potent than the distal ones, which was opposite to what was found on the Wap case.
Altogether, our data provides evidence for the notion that hotspots are central constituents in SEs. Hotspots serve to integrate external signals through TF colocalization on chromatin. TFs, on the other hand, through the formation of hotspots, cooperate in recruitment of coactivators, chromatin remodeling and establishment of an active epigenomic signature, as well as the activation of nearby genes during myogenesis. Within an SE, hotspots differ in their relative importance for the activity of SEs; not all of them are activating; some may be silent or even repressive at certain stage. The crosstalk between hotspots is thus important for reprogramming of the genome and such exquisite regulation of hotspots within an SE could allow a gene to be robustly responsive to differentiation cues. We imagine this could be a general mechanism that allows flexible response of tissue-specific gene expression with high precision and efficiency in the face of dynamic environmental changes and developmental cues. With the rapidly emerging epigenomics data, we expect more examples showing that many of these units defined on the basis of ChIP-seq experiments and specific algorithms are indeed special biological entities and more exciting modes of their function will be uncovered. Finally, unlike manipulating the entire enhancer or SE, the small size of the hotspot makes it feasible for genomic engineering for the purpose of dissecting enhancer function or using as therapeutic approaches for targeting enhancers whose deregulation is associated with human diseases (8,71).
ACCESSION NUMBER
All datasets described in the paper have been deposited in NCBI Gene Expression Omnibus under access #GSE93916 and GSE99207.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: Conceived and designed the experiments: H.W., X.L.P., K.K.S., L.H. and H.S. Performed the experiments: K.K.S., L.H., Y.Z., S.Z., M.Y., and B. X. Analyzed the data: X.L.P., J.Z., P. H. and H. Y. Wrote the paper: H.W., X.L.P., K.K.S. and L.H. Reviewed and edited the manuscript: H.W., H.S., X.L.P., K.K.S. and L.H.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
General Research Funds (GRF) from the Research Grants Council (RGC) of the Hong Kong Special Administrative Region [14102315 and 14113514 to H.S.; 14133016, 14100415, 14116014, and 476113 to H.W.]; Focused Innovations Scheme: Scheme B to H.S. [Project Code: 1907307]; RGC Collaborative Research Fund (CRF) from RGC [Project Code: C6015- 14G to H.S. and H.W.]; 973 grant from the Ministry of Science and Technology of China [Project Code: 2014CB964700 to H.W. and P.H.]. Funding for open access charge: GRF grants from RGC of the Hong Kong Special Administrative Region.
Conflict of interest statement. None declared.
REFERENCES
- 1. Levine M., Cattoglio C., Tjian R.. Looping back to leap forward: transcription enters a new era. Cell. 2014; 157:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hnisz D., Schuijers J., Lin C.Y., Weintraub A.S., Abraham B.J., Lee T.I., Bradner J.E., Young R.A.. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell. 2015; 58:362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Siersbaek R., Rabiee A., Nielsen R., Sidoli S., Traynor S., Loft A., La Cour Poulsen L., Rogowska-Wrzesinska A., Jensen O.N., Mandrup S.. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014; 7:1443–1455. [DOI] [PubMed] [Google Scholar]
- 6. Pott S., Lieb J.D.. What are super-enhancers. Nat. Genet. 2014; 47:8–12. [DOI] [PubMed] [Google Scholar]
- 7. Long H.K., Prescott S.L., Wysocka J.. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016; 167:1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sur I., Taipale J.. The role of enhancers in cancer. Nat. Rev. Cancer. 2016; 16:483–493. [DOI] [PubMed] [Google Scholar]
- 9. Shin H.Y., Willi M., Yoo K.H., Zeng X., Wang C., Metser G., Hennighausen L.. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 2016; 48:904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hay D., Hughes J.R., Babbs C., Davies J.O., Graham B.J., Hanssen L.L., Kassouf M.T., Oudelaar A.M., Sharpe J.A., Suciu M.C. et al. Genetic dissection of the alpha-globin super-enhancer in vivo. Nat. Genet. 2016; 48:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bentzinger C.F., Wang Y.X., Rudnicki M.A.. Building muscle: molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 2012; 4:a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dumont N.A., Wang Y.X., Rudnicki M.A.. Intrinsic and extrinsic mechanisms regulating satellite cell function. Development (Cambridge, England). 2015; 142:1572–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G.J., Parker M.H., MacQuarrie K.L., Davison J., Morgan M.T., Ruzzo W.L. et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 2010; 18:662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blum R., Vethantham V., Bowman C., Rudnicki M., Dynlacht B.D.. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev. 2012; 26:2763–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blum R., Dynlacht B.D.. The role of MyoD1 and histone modifications in the activation of muscle enhancers. Epigenetics. 2013; 8:778–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yee S.-P., Rigby P.. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993; 7:1277–1289. [DOI] [PubMed] [Google Scholar]
- 17. Shevchuk N.A., Bryksin A.V., Nusinovich Y.A., Cabello F.C., Sutherland M., Ladisch S.. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 2004; 32:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen X., He L., Zhao Y., Li Y., Zhang S., Sun K., So K., Chen F., Zhou L., Lu L. et al. Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discov. 2017; 3:17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu L., Sun K., Chen X., Zhao Y., Wang L., Zhou L., Sun H., Wang H.. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J. 2013; 32:2575–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hagege H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forne T.. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2007; 2:1722–1733. [DOI] [PubMed] [Google Scholar]
- 21. Huang J., Liu X., Li D., Shao Z., Cao H., Zhang Y., Trompouki E., Bowman T.V., Zon L.I., Yuan G.C. et al. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev. Cell. 2016; 36:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu K.K. Analysis of protein-DNA binding by streptavidin-agarose pulldown. Methods Mol. Biol. 2006; 338:281–290. [DOI] [PubMed] [Google Scholar]
- 24. Yang Y., Zhou L., Lu L., Wang L., Li X., Jiang P., Chan L., Zhang T., Yu J., Kwong J.. A novel miR-193a-5p-YY1-APC regulatory axis in human endometrioid endometrial adenocarcinoma. Oncogene. 2013; 32:3432–3442. [DOI] [PubMed] [Google Scholar]
- 25. Zhou L., Wang L., Lu L., Jiang P., Sun H., Wang H.. A novel target of microRNA-29, Ring1 and YY1-binding protein (Rybp), negatively regulates skeletal myogenesis. J. Biol. Chem. 2012; 287:25255–25265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang L., Zhao Y., Bao X., Zhu X., Kwok Y.K., Sun K., Chen X., Huang Y., Jauch R., Esteban M.A. et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015; 25:335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou L., Sun K., Zhao Y., Zhang S., Wang X., Li Y., Lu L., Chen X., Chen F., Bao X. et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 2015; 6:10026. [DOI] [PubMed] [Google Scholar]
- 28. Asp P., Blum R., Vethantham V., Parisi F., Micsinai M., Cheng J., Bowman C., Kluger Y., Dynlacht B.D.. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:E149–E158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Consortium. E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sebastian S., Faralli H., Yao Z., Rakopoulos P., Palii C., Cao Y., Singh K., Liu Q.C., Chu A., Aziz A. et al. Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genes Dev. 2013; 27:1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mullen A.C., Orlando D.A., Newman J.J., Loven J., Kumar R.M., Bilodeau S., Reddy J., Guenther M.G., DeKoter R.P., Young R.A.. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011; 147:565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soleimani V.D., Yin H., Jahani-Asl A., Ming H., Kockx C.E., van Ijcken W.F., Grosveld F., Rudnicki M.A.. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol. Cell. 2012; 47:457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dell’Orso S., Wang A.H., Shih H.Y., Saso K., Berghella L., Gutierrez-Cruz G., Ladurner A.G., O'Shea J.J., Sartorelli V., Zare H.. The histone variant MacroH2A1.2 is necessary for the activation of muscle enhancers and recruitment of the transcription factor Pbx1. Cell Rep. 2016; 14:1156–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bowman C.J., Ayer D.E., Dynlacht B.D.. Foxk proteins repress the initiation of starvation-induced atrophy and autophagy programs. Nat. Cell Biol. 2014; 16:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K. et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006; 34:D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012; 22:1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loven J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A.. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013; 153:320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bulger M., Groudine M.. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011; 144:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saint-Andre V., Federation A.J., Lin C.Y., Abraham B.J., Reddy J., Lee T.I., Bradner J.E., Young R.A.. Models of human core transcriptional regulatory circuitries. Genome Res. 2016; 26:385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Franco H.L., Casasnovas J., Rodriguez-Medina J.R., Cadilla C.L.. Redundant or separate entities?–roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res. 2011; 39:1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tapscott S.J. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005; 132:2685–2695. [DOI] [PubMed] [Google Scholar]
- 46. Boyarchuk E., Robin P., Fritsch L., Joliot V., Ait-Si-Ali S.. Identification of MyoD interactome using tandem affinity purification coupled to mass spectrometry. J. Vis. Exp. 2016; 111:e53924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Singh K., Cassano M., Planet E., Sebastian S., Jang S.M., Sohi G., Faralli H., Choi J., Youn H.D., Dilworth F.J. et al. A KAP1 phosphorylation switch controls MyoD function during skeletal muscle differentiation. Genes Dev. 2015; 29:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Braun T., Rudnicki M.A., Arnold H.H., Jaenisch R.. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell. 1992; 71:369–382. [DOI] [PubMed] [Google Scholar]
- 49. Kablar B., Krastel K., Ying C., Asakura A., Tapscott S.J., Rudnicki M.A.. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 1997; 124:4729–4738. [DOI] [PubMed] [Google Scholar]
- 50. Blum R. Activation of muscle enhancers by MyoD and epigenetic modifiers. J. Cell Biochem. 2014; 115:1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Eijkelenboom A., Mokry M., de Wit E., Smits L.M., Polderman P.E., van Triest M.H., van Boxtel R., Schulze A., de Laat W., Cuppen E. et al. Genome-wide analysis of FOXO3 mediated transcription regulation through RNA polymerase II profiling. Mol. Syst. Biol. 2013; 9:638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eijkelenboom A., Mokry M., Smits L.M., Nieuwenhuis E.E., Burgering B.M.. FOXO3 selectively amplifies enhancer activity to establish target gene regulation. Cell Rep. 2013; 5:1664–1678. [DOI] [PubMed] [Google Scholar]
- 53. Hu P., Geles K.G., Paik J.H., DePinho R.A., Tjian R.. Codependent activators direct myoblast-specific MyoD transcription. Dev. Cell. 2008; 15:534–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dentice M., Ambrosio R., Damiano V., Sibilio A., Luongo C., Guardiola O., Yennek S., Zordan P., Minchiotti G., Colao A. et al. Intracellular inactivation of thyroid hormone is a survival mechanism for muscle stem cell proliferation and lineage progression. Cell Metab. 2014; 20:1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li W., Hu Y., Oh S., Ma Q., Merkurjev D., Song X., Zhou X., Liu Z., Tanasa B., He X. et al. Condensin I and II complexes license full estrogen receptor alpha-dependent enhancer activation. Mol. Cell. 2015; 59:188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Faralli H., Dilworth F.J.. Turning on myogenin in muscle: a paradigm for understanding mechanisms of tissue-specific gene expression. Comp. Funct. Genomics. 2012; 2012:836374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yee S.P., Rigby P.W.. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993; 7:1277–1289. [DOI] [PubMed] [Google Scholar]
- 58. Edmondson D.G., Cheng T.C., Cserjesi P., Chakraborty T., Olson E.N.. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol. Cell. Biol. 1992; 12:3665–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen K.W., Chang Y.J., Yeh C.M., Lian Y.L., Chan M.W., Kao C.F., Chen L.. SH2B1 modulates chromatin state and MyoD occupancy to enhance expressions of myogenic genes. Biochim. Biophys. Acta. 2017; 1860:270–281. [DOI] [PubMed] [Google Scholar]
- 60. Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K. et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006; 34:D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mousavi K., Zare H., Dell’orso S., Grontved L., Gutierrez-Cruz G., Derfoul A., Hager G.L., Sartorelli V.. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell. 2013; 51:606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tapscott S.J., Lassar A.B., Weintraub H.. A novel myoblast enhancer element mediates MyoD transcription. Mol. Cell. Biol. 1992; 12:4994–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kucharczuk K.L., Love C.M., Dougherty N.M., Goldhamer D.J.. Fine-scale transgenic mapping of the MyoD core enhancer: MyoD is regulated by distinct but overlapping mechanisms in myotomal and non-myotomal muscle lineages. Development. 1999; 126:1957–1965. [DOI] [PubMed] [Google Scholar]
- 64. Chen J.C., Love C.M., Goldhamer D.J.. Two upstream enhancers collaborate to regulate the spatial patterning and timing of MyoD transcription during mouse development. Dev. Dyn. 2001; 221:274–288. [DOI] [PubMed] [Google Scholar]
- 65. Asakura A., Lyons G.E., Tapscott S.J.. The regulation of MyoD gene expression: conserved elements mediate expression in embryonic axial muscle. Dev. Biol. 1995; 171:386–398. [DOI] [PubMed] [Google Scholar]
- 66. Conerly M.L., Yao Z., Zhong J.W., Groudine M., Tapscott S.J.. Distinct activities of Myf5 and MyoD indicate separate roles in skeletal muscle lineage specification and differentiation. Dev. Cell. 2016; 36:375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Calnan D.R., Brunet A.. The FoxO code. Oncogene. 2008; 27:2276–2288. [DOI] [PubMed] [Google Scholar]
- 68. Webb A.E., Pollina E.A., Vierbuchen T., Urban N., Ucar D., Leeman D.S., Martynoga B., Sewak M., Rando T.A., Guillemot F. et al. FOXO3 shares common targets with ASCL1 genome-wide and inhibits ASCL1-dependent neurogenesis. Cell Rep. 2013; 4:477–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang C., Wang M., Arrington J., Shan T., Yue F., Nie Y., Tao W.A., Kuang S.. Ascl2 inhibits myogenesis by antagonizing the transcriptional activity of myogenic regulatory factors. Development. 2017; 144:235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shen H., Xu W., Guo R., Rong B., Gu L., Wang Z., He C., Zheng L., Hu X., Hu Z. et al. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell. 2016; 165:331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Groschel S., Sanders M.A., Hoogenboezem R., de Wit E., Bouwman B.A., Erpelinck C., van der Velden V.H., Havermans M., Avellino R., van Lom K. et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014; 157:369–381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.