Abstract
DNA methylation in repetitive elements (RE) suppresses their mobility and maintains genomic stability, and decreases in it are frequently observed in tumor and/or surrogate tissues. Averaging methylation across RE in genome is widely used to quantify global methylation. However, methylation may vary in specific RE and play diverse roles in disease development, thus averaging methylation across RE may lose significant biological information. The ambiguous mapping of short reads by and high cost of current bisulfite sequencing platforms make them impractical for quantifying locus-specific RE methylation. Although microarray-based approaches (particularly Illumina's Infinium methylation arrays) provide cost-effective and robust genome-wide methylation quantification, the number of interrogated CpGs in RE remains limited. We report a random forest-based algorithm (and corresponding R package, REMP) that can accurately predict genome-wide locus-specific RE methylation based on Infinium array profiling data. We validated its prediction performance using alternative sequencing and microarray data. Testing its clinical utility with The Cancer Genome Atlas data demonstrated that our algorithm offers more comprehensively extended locus-specific RE methylation information that can be readily applied to large human studies in a cost-effective manner. Our work has the potential to improve our understanding of the role of global methylation in human diseases, especially cancer.
INTRODUCTION
DNA repetitive elements (RE), which account for about 50% of the human genome, are relics of transposons and can proliferate and mobilize throughout the genome (1). Alu element (Alu) and long interspersed element-1 (LINE-1) represent the two most abundant human RE sequences (2,3), with new insertions occurring in approximately one out of 20 births for Alu and out of 200 for LINE-1 (3,4). Alu and LINE-1 often target protein-coding genes for insertion (5), which may cause genomic instability and contribute to the development of human diseases, particularly cancer (6–8). DNA methylation in RE is a key mechanism defending against these transposition activities, and thus maintaining genomic integrity in humans (5,9–11).
DNA methylation refers to the addition of a methyl group to DNA, usually the fifth carbon atom of a cytosine ring at the ‘CG’ dinucleotide sequence. Decreased DNA methylation in RE, also widely referred as global hypomethylation, plays an important role in tumorigenesis (12–14). Over 90% of methylated CpG sites in the human genome occur in RE, particularly Alu and LINE-1 (15). Therefore given their genome-wide ubiquity and rich CpG content, bulk estimates of methylation in Alu and/or LINE-1 methylation throughout the genome (16) have been widely used as surrogate measures of global DNA methylation content in most human studies (17,18). Global hypomethylation is predominantly observed in human tumor and surrogate tissues, particularly blood from cancer patients (19–21). Accumulating evidence shows that Alu/LINE-1 methylation at specific genomic loci vary and exert distinct biological and/or pathological effects in cancer (22–28), suggesting that using mean values of methylation in RE as surrogates of global methylation may lead to biological information loss and hindering scientists from elucidating the distinct biological roles of DNA methylation in locus-specific RE. Indeed, previous investigations into the roles of RE methylation in cancer have been substantially inconsistent for both tissue (29) and blood (18), highlighting the inadequacy of studying mean Alu/LINE-1 methylation. We therefore suspect that the inconsistent results from previous studies were at least partially due to the inability to assess RE methylation levels at specific loci.
Whole-genome sequencing may plausibly allow us to study locus-specific RE methylation. However, single-base resolution sequencing of locus-specific RE is not optimal as the repeats create ambiguities in alignment and assembly, which produce biases and errors when interpreting results (30). Furthermore, profiling methylation in RE using sequencing is even more challenging as it produces higher mapping errors due to the reduced complexity reads from bisulfite conversion (31,32). Finally, sequencing genome-wide methylation remains prohibitively expensive.
In recent years, microarrays with optimized probes, such as the Infinium HumanMethylation450 BeadChip (HM450) and the upgraded Infinium MethylationEPIC BeadChip (EPIC) (33), have been widely used for robust genome-wide DNA methylation investigations in human studies. These array-based DNA methylation data may provide a cost-effective opportunity to study the role of locus-specific RE methylation in relation to cancers and other chronic diseases. However, RE coverage of Infinium methylation arrays are still limited and the profiled CpGs in RE are generally sparse. We therefore developed a predictive algorithm to computationally extend RE methylation based on the Infinium methylation array data. We further evaluated the prediction performance of our algorithm and demonstrated the algorithm's clinical utilities by exploring the biological implications of locus-specific Alu/LINE-1 methylation in cancer. To facilitate calculations, we developed an R package, REMP (Repetitive Element Methylation Prediction), available in Bioconductor repository.
MATERIALS AND METHODS
Data sources
First, for RE identification and annotation, we used the RepeatMasker (34) and NCBI RefSeqGene databases (35) to identify and annotate candidate RE loci for methylation prediction. We obtained the RepeatMasker Library (build hg19) and RefSeqGene annotation database (build hg19) through the R package AnnotationHub (36) (record number AH5122 and AH5040, respectively).
Second, for algorithm development and validation, we used data on HapMap (The International HapMap Project) lymphoblastoid cell line (LCL) GM12878, a Tier-1 sample from a female Utah resident with ancestry from Northern and Western Europe (37,38). There are extensive publicly-accessible methylation data on GM12878, making it an ideal sample for model development and validation. The HM450 data, Reduced Representation Bisulfite Sequencing (RRBS) (39), and Whole Genome Bisulfite Sequencing (WGBS) (40) data on GM12878 were downloaded from the ENCODE (The Encyclopedia of DNA Elements) (41); the EPIC data were the means of three technical replicates of GM12878 obtained from R package minfiDataEPIC (42). The NimbleGen SeqCap Epi 4M CpGiant (NimbleGen) (43) profiling data are courtesy of Roche Sequencing. Raw NimbleGen sequencing data processing followed the manufacturer's recommended workflow (44). For NimleGen, RRBS, and WGBS the processed BAM files of two replicates were united into a single dataset using R package methylKit (45). The ratio of methylated read counts (i.e. count of cytosine) to sequencing depth (i.e. count of cytosine + thymine) was calculated to represent methylation level. CpG sites with greater than 30 × sequencing depth were retained.
Finally, for algorithm application to clinical samples, we used The Cancer Genome Atlas (TCGA) database. We focused on four common types of cancer in the US (46): breast invasive carcinoma (BRCA, 90 tumor samples), Prostate adenocarcinoma (PRAD, 50 tumor samples), Lung squamous cell carcinoma (LUSC, 40 tumor samples), and Colon and rectal adenocarcinoma (COAD, 38 tumor samples). We selected primary tumor tissue with available paired normal solid tissue collected from the same individual. Processed and normalized (level 3) HM450 methylation data and RNA-Seq gene expression data were downloaded from the TCGA open-access database using the R package TCGAbiolinks (47).
Development of prediction algorithm
Structure of algorithm
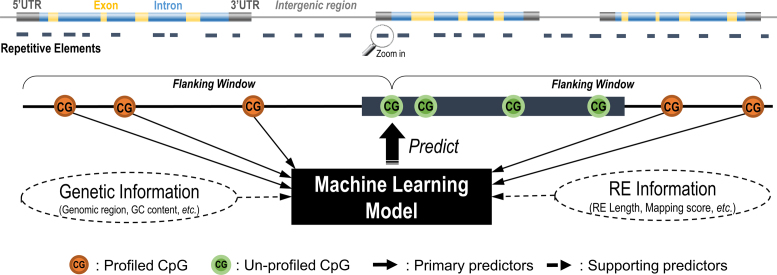
Previous studies have shown that the methylation levels of two nearby CpG sites are more likely to be co-methylated (48–51). Therefore, we proposed to predict the methylation levels of the target CpGs in RE using neighboring profiled CpGs within a flanking window (Figure 1). Within the flanking window of target RE CpGs with at least two neighboring profiled CpGs were considered to improve prediction reliability. Based on previous work in predictor prioritization (50) and our extensive experiments in selecting contributive predictors, we constructed the following primary predictors for each target CpG:
Methylation level of the closest and second-closest profiled CpGs in the flanking region of the target CpG.
Genomic distance in base pair (bp) from the closest and second-closest profiled CpGs to the target CpG.
Mean and variance of methylation levels at all neighboring profiled CpGs.
Mean and variance of genomic distance between all neighboring profiled CpGs and the target CpG.
Figure 1.
Diagram of the RE methylation prediction algorithm. For each un-profiled CpGs identified within a RE sequence, the neighboring profiled CpGs are identified within a given flanking window, where the primary and supporting predictors are collected. Those profiled CpGs in RE with sufficient neighboring information are included as a set for model training whereas CpGs not profiled in RE will be predicted using the trained model.
We also constructed the following supporting predictors to better model local genomic characteristics of the target CpGs and their relationships with RE methylation:
RE CpG density: CpG density is correlated with DNA methylation across various tissues (49,52). For CpGs in RE, methylation level showed a reverse U-shaped relationship with increasing CpG density (53). We defined RE CpG density as the number of CpGs within RE divided by the length of RE.
RE length: Full-length RE sequences tend to be more active, usually representing more recently-evolved elements (particularly for LINE-1) (54). Increasing DNA methylation has been shown to correlate with younger evolutionary age of RE (55).
Smith-Waterman (SW) score: The RepeatMasker database employed a SW alignment algorithm (56) to computationally identify Alu and LINE-1 sequences in the reference genome. A higher score indicates fewer insertions and deletions in query RE sequences compared to consensus RE sequences. We included this factor to account for potential bias induced by SW alignment.
Number of neighboring profiled CpGs: More neighboring CpG profiles results in more reliable and informative primary predictors. We included this predictor to account for potential bias due to profiling platform design.
Genomic region of the target CpG: It is well-known that methylation levels differ by genomic regions. Our algorithm included a set of seven indicator variables for genomic region (as annotated by RefSeqGene) including: 2000 bp upstream of transcript start site (TSS2000), 5′UTR (untranslated region), coding DNA sequence, exon, 3′UTR, protein-coding gene, and noncoding RNA gene. Note that intron and intergenic regions can be inferred by the combinations of these indicator variables.
For a given flanking window size, we generated these predictors and trained a model to predict methylation levels of the target CpGs. We considered the following approaches:
Naïve method: This approach takes the methylation level of the closest neighboring CpG profiled by HM450 or EPIC as that of the target CpG. We treated this method as our ‘control’.
Support Vector Machine (SVM) (57): SVM has been extensively used for predicting methylation status (methylated vs. unmethylated) (58–63). We considered two different kernel functions to determine the underlying SVM architecture: the linear kernel and the radial basis function (RBF) kernel (64).
Random Forest (RF) (65): A competitor of SVM, RF recently demonstrated superior performance over other machine learning models in predicting methylation levels (50).
A 3-time repeated 5-fold cross validation was performed to determine the best model parameters for SVM and RF using the R package caret (66). The search grid was Cost = (2−15, 2−13, 2−11, …, 23) for the parameter in linear SVM, Cost = (2−7, 2−5, 2−3, …, 27) and γ = (2−9, 2−7, 2−5, …, 21) for the parameters in RBF SVM, and the number of predictors sampled for splitting at each node (3,6,12) for the parameter in RF.
We also evaluated and controlled the prediction reliability when performing model extrapolation out of training data. Quantifying prediction reliability in SVM is challenging and computationally intensive (67). In contrast, prediction reliability can be readily inferred by Quantile Regression Forests (QRF) (68) (available in the R package quantregForest (69)). Briefly, by taking advantage of the established random trees, QRF estimates the full conditional distribution for each of the predicted values. We therefore defined prediction error using the standard deviation (SD) of this conditional distribution to reflect variation in the predicted values. Less reliable RF predictions (results with greater prediction error) can be trimmed off (RF-Trim).
Performance evaluation
To evaluate and compare the predictive performance of different models, we conducted an external validation study. We prioritized Alu and LINE-1 for demonstration due to their high abundance throughout the genome as well as their biological relevance. We chose the HM450 as the primary platform for evaluation. We traced model performance using incremental window sizes from 200 to 2000 bp for Alu and LINE-1 and employed two evaluation metrics: Pearson's correlation coefficient (r) and root mean square error (RMSE) between predicted and profiled CpG methylation levels. Predicted RE methylation using the HM450 and EPIC were validated by NimbleGen. To account for evaluation bias (caused by the inherent variation between the HM450/EPIC and the sequencing platforms), we calculated ‘benchmark’ evaluation metrics (r and RMSE) between both types of platforms using the common CpGs profiled in Alu/LINE-1 as the best theoretically possible performance the algorithm could achieve. Since the EPIC covers twice as many CpGs in Alu/LINE-1 as the HM450 (Table 1), we also used EPIC to validate the HM450 prediction results.
Table 1. Alu/LINE-1 coverage with single-base profiling platforms and predictions.
| # of RE | # of RE CpGs | # of genes covereda | # of RE subfamilies covered | |
|---|---|---|---|---|
| Alu | ||||
| Profiling Platforms | ||||
| HM450 | 12255 | 13155 | 14276 | 37 |
| EPIC | 21300 | 23784 | 19856 | 40 |
| NimbleGen | 1289 | 2463 | 2178 | 31 |
| RRBS | 2985 | 5902 | 3266 | 34 |
| WGBS | 929874 | 3652457 | 40870 | 41 |
| Prediction | ||||
| GM12878 (HM450) | 33407 | 202731 | 22561 | 41 |
| GM12878 (EPIC) | 80131 | 481780 | 31163 | 41 |
| Breast cancer (BRCA) | 38848 | 235533b | 22924 | 41 |
| Lung cancer (LUSC) | 37647 | 225367b | 22786 | 41 |
| Colon cancer (COAD) | 34605 | 209313b | 21046 | 41 |
| Prostate cancer (PRAD) | 36224 | 219007b | 21533 | 41 |
| LINE-1 | ||||
| Profiling Platforms | ||||
| HM450 | 8309 | 9797 | 7399 | 115 |
| EPIC | 24713 | 29404 | 15558 | 116 |
| NimbleGen | 4667 | 13376 | 4617 | 115 |
| RRBS | 753 | 2023 | 663 | 94 |
| WGBS | 586345 | 2141737 | 31928 | 117 |
| Prediction | ||||
| GM12878 (HM450) | 4597 | 22968 | 4140 | 109 |
| GM12878 (EPIC) | 10133 | 31374 | 9544 | 115 |
| Breast cancer (BRCA) | 9174 | 44185b | 6897 | 116 |
| Lung cancer (LUSC) | 8928 | 43308b | 6824 | 115 |
| Colon cancer (COAD) | 6768 | 34595b | 5388 | 113 |
| Prostate cancer (PRAD) | 7715 | 37096b | 6021 | 115 |
aRefSeq genes, including gene proximal promoter region (i.e. 2000 bp upstream of the transcription start site).
bCpG sites with reliable prediction across >80% of the samples were retained.
Proof of concept
We designed a proof-of-concept study to test whether predicted Alu/LINE-1 methylation can correlate with the evolutionary ages of Alu/LINE-1 from the HapMap LCL GM12878 sample. The evolutionary age of Alu/LINE-1 is inferred from the divergence of copies from the consensus sequence as new base substitutions, insertions, or deletions accumulate in Alu/LINE-1 through ‘copy and paste’ retrotransposition activity. Older Alu/LINE-1 copies are in general inactive since more mutations were induced (partially by CpG methylation). Younger Alu/LINE-1, especially currently active RE, have fewer mutations and thus CpG methylation is a more important defense mechanism for suppressing retrotransposition activity. Therefore, we would expect DNA methylation level to be lower in older Alu/LINE-1 than in younger Alu/LINE-1. We calculated and compared the average methylation level across three evolutionary subfamilies in Alu (ranked from young to old): AluY, AluS and AluJ, and five evolutionary subfamilies in LINE-1 (ranked from young to old): L1Hs, L1P1, L1P2, L1P3 and L1P4. We tested trends in average methylation level across evolutionary age groups using linear regression models.
Applications in clinical samples
Next, to demonstrate our algorithm's utility, we set out to investigate (a) differentially methylated RE in tumor versus normal tissue and their biological implications and (b) tumor discrimination ability using global methylation surrogates (i.e. mean Alu and LINE-1) versus the predicted locus-specific RE methylation. To best utilize data, we conducted these analyses using the union set of the HM450 profiled and predicted CpGs in Alu/LINE-1, defined here as the extended CpGs.
For (a), differentially methylated CpGs in Alu and LINE-1 between tumor and paired normal tissues were identified via paired t-tests (R package limma (70)). Tested CpGs were grouped and identified as differentially methylated regions (DMR) using R package Bumphunter (71) and family wise error rates (FWER) estimated from bootstraps to account for multiple comparisons. Regulatory element enrichment analyses were conducted to test for functional enrichment of significant DMR. We used DNase I hypersensitivity sites (DNase), transcription factor binding sites (TFBS), and annotations of histone modification ChIP peaks pooled across cell lines (data available in the ENCODE Analysis Hub at the European Bioinformatics Institute). For each regulatory element, we then calculated the number of overlapping regions amongst the significant DMR (observed) and 10 000 permuted sets of DMR markers (expected). We calculated the ratio of observed to mean expected as the enrichment fold and obtained an empirical p-value from the distribution of expected. We then focused on gene regions and conducted KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis using hypergeometric tests via the R package clusterProfiler (72). To minimize bias in our enrichment test, we extracted genes targeted by the significant Alu/LINE-1 DMR and used genes targeted by all bumps tested as background. False discovery rate (FDR) <0.05 was considered significant in both enrichment analyses.
For b), we employed conditional logistic regression with elastic net penalties (R package clogitL1) (73) to select locus-specific Alu and LINE-1 methylation for discriminating tumor and normal tissue. Missing methylation data due to insufficient data quality were imputed using KNN imputation (74). We set the tuning parameter α = 0.5 and tuned λ via 10-fold cross validation. To account for overfitting, 50% of the data were randomly selected to serve as the training dataset with the remaining 50% as the testing dataset. We constructed one classifier by using the selected Alu and LINE-1 to refit the conditional logistic regression model, and another using the mean of all Alu and LINE-1 methylation as a surrogate of global methylation. Finally, using R package pROC (75), we performed receiver operating characteristic (ROC) analysis and computed the area under the ROC curves (AUC) to compare the performance of each discrimination method in the testing dataset via DeLong tests (76).
RESULTS
Single-base methylation profiling approaches
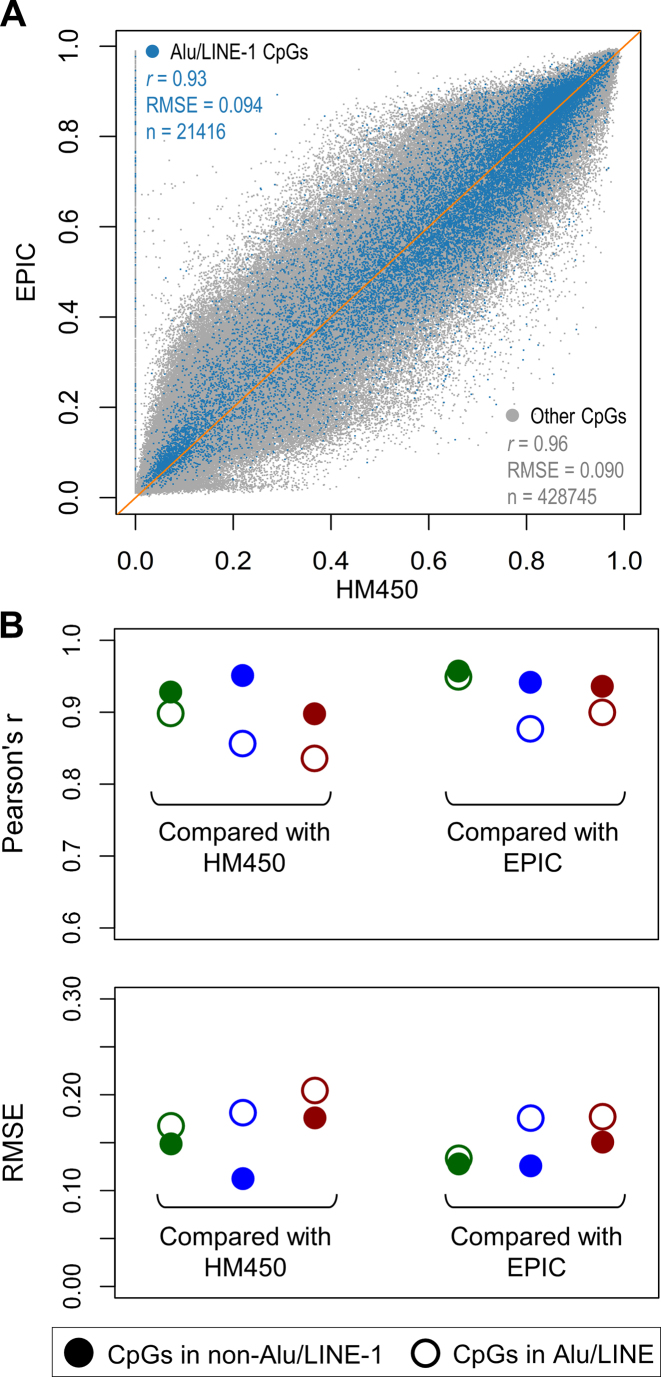
Based on the reference genome and the RepeatMasker library, about 35% of all 28 million CpG sites are in Alu (∼25%) and LINE-1 (∼10%). The RepeatMasker repeat library mapped 1 175 329 Alu and 923 315 LINE-1 loci in the UCSC hg19 reference genome assembly, corresponding to 9.9% and 16.4% of the human genome respectively. Most Alu and LINE-1 reside in intergenic (48.3% and 60.5%, respectively) or gene intronic regions (40.0% and 32.0%, respectively) (Supplementary Figure S1). Using the HapMap LCL GM12878 sample, we investigated the CpG coverage in Alu and LINE-1 among the four single-base methylation profiling approaches, i.e. HM450/EPIC, NimbleGen, RRBS, and WGBS. While all approaches save WGBS suffered from depleted coverage in Alu and LINE-1, all platforms cover a variety of Alu/LINE-1 subfamilies (Table 1). HM450/EPIC achieved the second highest coverage, significantly higher than NimbleGen and RRBS. To evaluate the reliability of profiled CpGs in Alu/LINE-1, we calculated inter-platform correlation and error and compared concordance between Alu/LINE-1 CpGs vs non-Alu/LINE-1 CpGs (with high concordance indicating robust methylation profiling). We observed that the HM450/EPIC achieved high concordance with correlations of 0.93 vs 0.96 and errors of 0.094 vs 0.090 for Alu/LINE-1 versus non-Alu/LINE-1 CpGs (Figure 2A), respectively. Hence with HM450/EPIC as the benchmark, concordance of NimbleGen was the highest, whereas in RRBS and WGBS correlations decreased and errors increased among Alu/LINE-1 CpGs (Figure 2B), suggesting potential measurement bias due to the ambiguous mapping of reads. Therefore, we opted to use the HM450/EPIC as the input data source for prediction and NimbleGen as the validation data source.
Figure 2.
Reliability of the profiling platforms interrogating CpG sites in Alu and LINE-1. If probes or reads targeting RE regions such as Alu and LINE-1 are affected by ambiguous mapping, methylation readings on these CpGs are more likely to yield different values for the same sample across different platforms. (A) Plot showing high correlation between CpGs profiled using both HM450 and EPIC, with CpGs in Alu/LINE-1 showing slightly smaller r and larger RMSE (root mean square error). (B) Evaluation of the reliability of the three sequencing-based platforms (using Infinium methylation arrays as the benchmark): NimbleGen (green), RRBS (blue), and WGBS (red). NimbleGen shows the highest concordance between both Alu/LINE-1 and non-Alu/LINE-1 CpGs.
Predicting locus-specific methylation of Alu and LINE-1 in GM12878
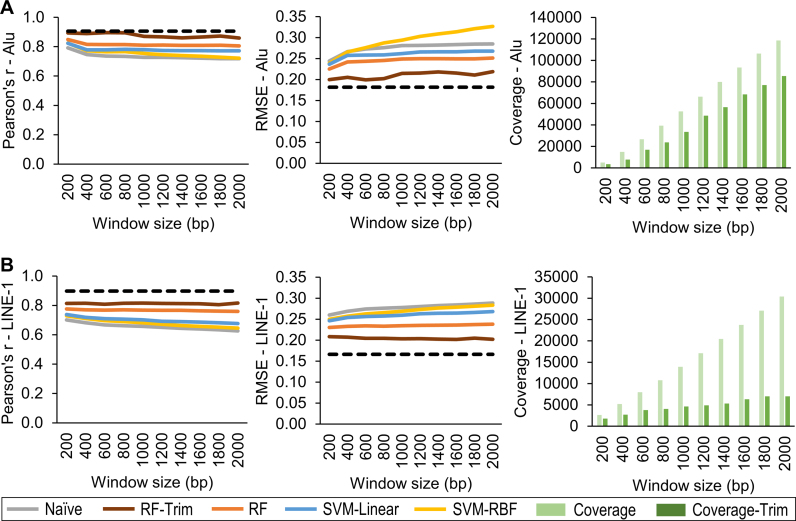
Validation results showed that RF had the best prediction performances. After trimming off less reliable predictions (RF-Trim, error ≤ 1.7), it achieved higher correlations and lower errors that approached the best theoretically possible performance. As window size increased above 1000 bp, prediction performances for Alu declined (Figure 3A) and the number of reliable predictions for LINE-1 leveled off (Figure 3B). These observations were consistent with the previous findings that two nearby CpG sites within 1000 bp are more likely to be co-methylated (48–51,77). We observed similar prediction performance using the EPIC (Supplementary Figure S2). We further validated the HM450 predicted results using the EPIC. RF-Trim (error ≤ 1.7) achieved the highest accuracy with Person's correlation coefficient (r) = 0.86 and 0.89 and root mean square error (RMSE) = 0.12 and 0.12 for Alu and LINE-1, respectively (Supplementary Figure S3). The cutoff of 1.7 for prediction error in RF-Trim is empirical, to balance the tradeoff between coverage and accuracy (i.e. more stringent prediction error threshold led to higher accuracy but lower Alu/LINE-1 coverage, Supplementary Figure S3).
Figure 3.
Performance of RE methylation prediction algorithm in different prediction models. Comparison of correlation and RMSE between measured (NimbleGen) and predicted (based on HM450) values for five prediction models (Naïve, RF, RF-Trim, SVM-Linear, and SVM-RBF) relative to the best theoretically possible performance (dashed line). RF-Trim achieved the best performance for both Alu (A) and LINE-1 (B) and approach to the best theoretical level. Compared with RF, RF-Trim removed more unreliable predictions, leading to less coverage but superior performance. RF: random forest; SVM-Linear: support vector machine with linear kernel; SVM-RBF: support vector machine with radial basis function kernel.
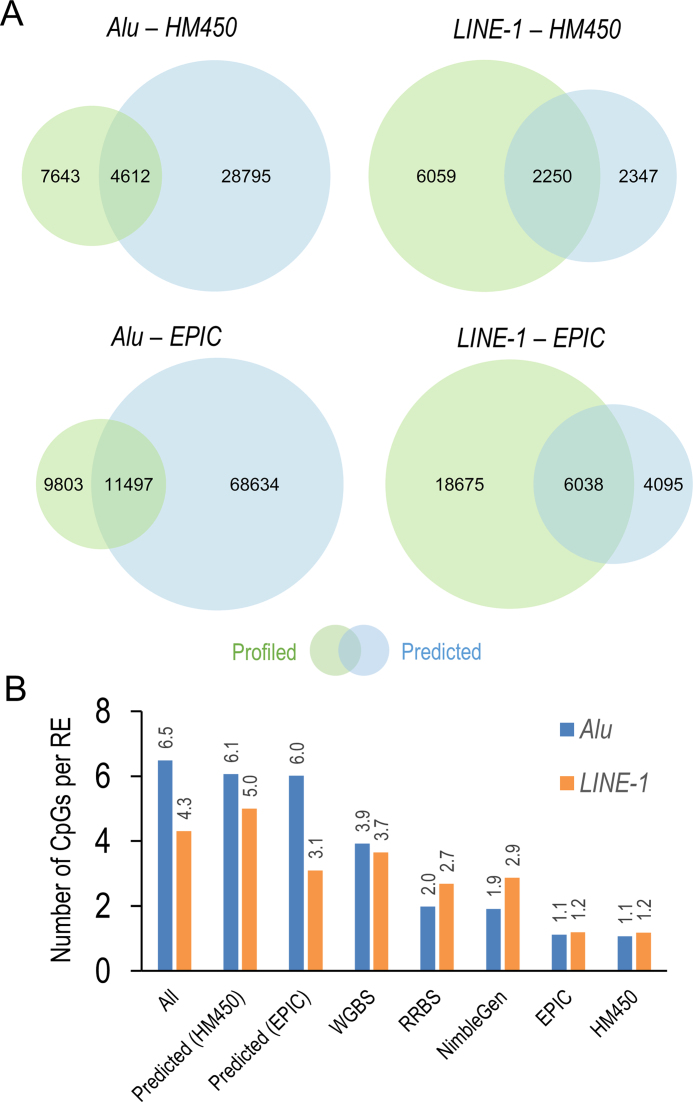
Taken altogether, RF-Trim with a 1000 bp window is our preferred method as it offers more accurate prediction and enables prediction quality control. Compared with the profiled Alu/LINE-1 methylation using the HM450/EPIC, our algorithm predicted 2.7–3.7 times as many Alu and about 20% more LINE-1; predictions based on the EPIC yielded nearly 2–3 times as many Alu/LINE-1 coverage than those based on the HM450 (Figure 4A). Moreover, our algorithm improved the CpG density in Alu/LINE-1. For example, using the HM450, each Alu contained 6.1 reliable predicted CpGs and each LINE-1 contained 5.0 reliable CpGs predicted, both 5–6 times higher than the HM450 and comparable with the average CpG density calculated in the full RE database (Figure 4B).
Figure 4.
Comparisons of Alu and LINE-1 coverage and CpG density using the prediction algorithm versus profiling platforms. (A) Alu and LINE-1 actual versus predicted coverage based on HM450 and EPIC. (B) Density of CpGs interrogated per Alu and LINE-1 locus of predicted vs. profiled values. The prediction algorithm enhanced CpG density by 5–6-fold, more comparable with the natural level of Alu/LINE-1 methylation.
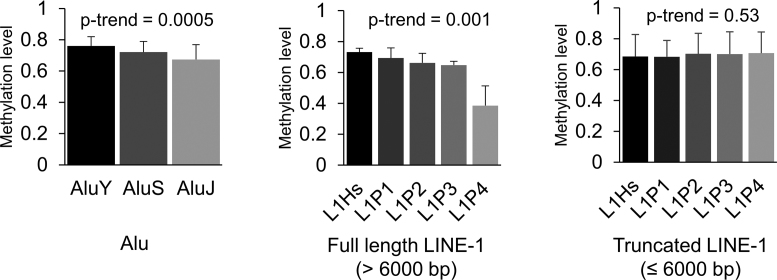
Proof-of-concept: methylation and evolutionary age of Alu and LINE-1
Using GM12878 data, we observed that HM450 predicted methylation level as associated with an inverse dose-response relationship with evolutionary age, indicating the defensive role of DNA methylation in RE (Figure 5). A similar relationship was evident among Alu and full-length (>6000 bp) LINE-1 but not truncated LINE-1; we found similar relationships using EPIC predicted values as well (Supplementary Figure S4).
Figure 5.
Inverse relationship between evolutionary ages of Alu and LINE-1 and mean methylation level based on predicted values. We considered three evolutionary subfamilies in Alu, from young to old: AluY, AluS, and AluJ, and five evolutionary subfamilies in LINE-1, from young to old: L1Hs, L1P1, L1P2, L1P3 and L1P4. The histograms and error bars represent the average and standard deviation of methylation level, respectively.
Application 1: predicting Alu and LINE-1 methylation enables more comprehensive differential methylation analyses
Using RF-Trim, we predicted about 37 000 Alu and 8000 LINE-1 across the genome in TCGA samples (Table 1). Most Alu and LINE-1 loci showed a unimodal distribution centered at a high methylation level (β ∼ 0.9) in both tumor and paired normal tissues, but was relatively lower and more widely dispersed in tumors (Supplementary Figure S5).
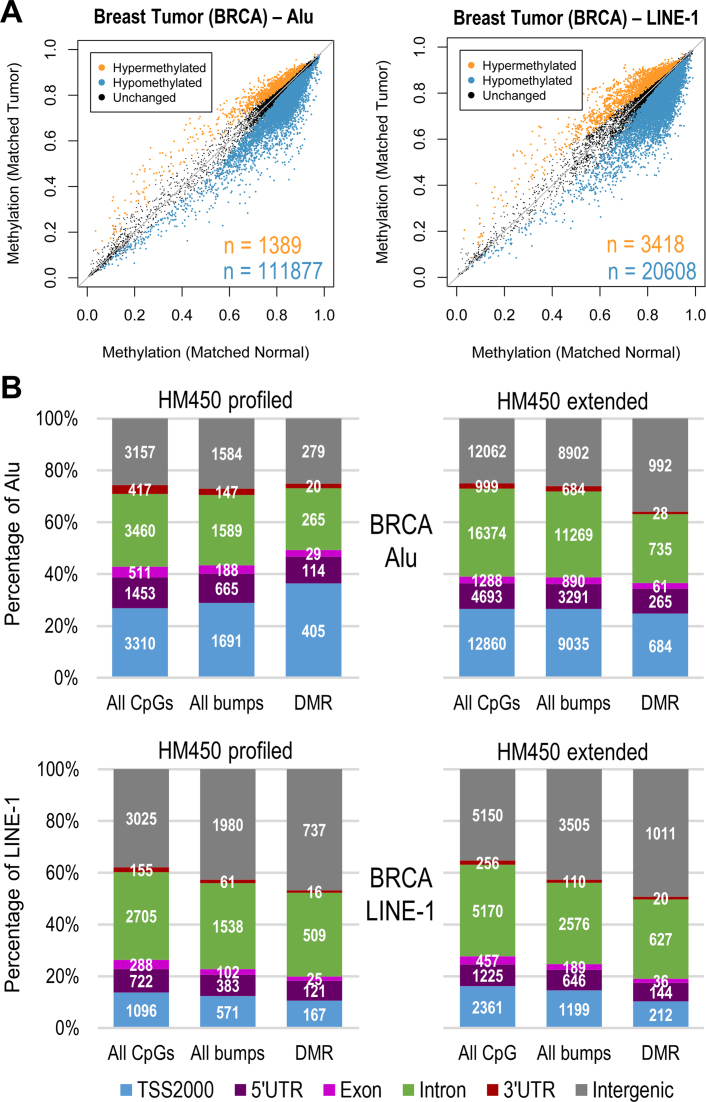
On average, around 77 000 extended (i.e. union set of profiled and predicted) CpGs (98%) in Alu and 15 000 (90%) in LINE-1 were hypomethylated across all four types of tumor tissues, with a general overall trend towards global hypomethylation (exemplified by breast cancer, Figure 6A, Supplementary Figure S6 for other cancers). In contrast, using only the profiled CpGs we found that ∼2500 (∼88% of profiled CpGs) in Alu or LINE-1 were hypomethylated. We conducted regional analysis to summarize significant DMR (FWER < 0.05) in Alu/LINE-1 using extended CpGs (see complete results in Supplementary Spreadsheet) and compared the results using profiled CpGs. The genomic distribution of all Alu/LINE-1 CpGs, all identified bumps, and significant DMR had similar proportions observed using both profiled and extended CpGs (exemplified by breast cancer, Figure 6B, Supplementary Figure S7 for other cancers). Therefore, it is unlikely that the prediction introduces any artificial bias towards specific genomic regions. Furthermore, due to the higher density of the predicted CpGs in Alu/LINE-1 there were more bumps detected using the extended CpGs compared to the profiled CpGs, particularly in Alu. Similarly compared to the profiled CpGs, the extended CpGs yielded nearly twice as many Alu/LINE-1 with significant DMR, especially in the intron and intergenic regions.
Figure 6.
Differentially methylated CpGs/regions in Alu and LINE-1. (A) Scatter plot comparing extended CpGs in Alu and LINE-1 between breast tumor and matched normal tissue; significant differences at Bonferroni P < 0.05 are colored and n is the number of CpGs (orange: hypermethylated; blue: hypomethylated). (B) Genome-wide break down of all CpGs tested, bumps formed using bumphunter, and significant DMR (FWER < 0.05) identified in breast cancer. Genomic distribution of extended CpGs was similar to profiled and identified more DMR of interest, especially in the intron and intergenic regions.
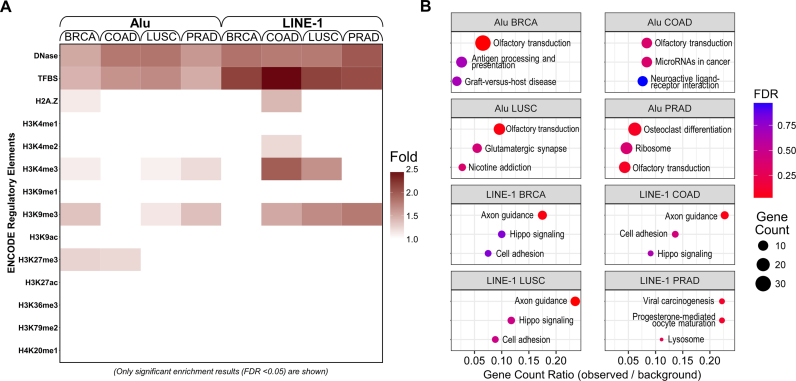
To explore the functional insights of locus-specific RE methylation in tumor tissue, we conducted the regulatory elements and KEGG enrichment analyses based on the significant hypo- and hyper-methylated Alu/LINE-1 DMR from the extended CpGs. Due to the limited number of hypermethylated DMR, only hypomethylated DMR yielded significant results. The enrichment can be found in regulatory elements including TFBS; active chromatin markers including DNase, H2A.Z and H3K4me3; and repressive chromatin markers such as H3K9me3 and H3K27me3. We found no enrichment found in the remaining active chromatin marks (H3K4me1, H3K9ac, H3K27ac, H3K36me3, H3K79me2 and H4K20me1) (Figure 7A). Common pathways across the four cancers were identified including olfactory transduction and axon guidance (Figure 7B). Higher enrichment fold in regulatory elements analysis and gene count ratio in KEGG analysis were observed in LINE-1 than in Alu, indicating a more active functional role for LINE-1 hypomethylation. Two full-length LINE-1 loci in the introns of SEMA3A (Semaphorin 3A) (a gene in the axon guidance pathway) were hypomethylated in breast, colon, and lung tumor tissues (Supplementary Figure S8). SEMA3A can inhibit angiogenesis and endothelial cell migration and its downregulation has been identified in breast cancer development (78). Using TCGA gene expression data, we confirmed that SEMA3A gene expression was significantly downregulated in breast tumor tissues in relative to the matched normal tissues. This could be attributed to the hypomethylated LINE-1 loci as we observed significant positive correlation between methylation at each of the LINE-1 loci and SEMA3A gene expression in the normal tissues, but substantially attenuated and non-significant correlation in the tumor tissues (Supplementary Figure S9).
Figure 7.
Regulatory element enrichment analysis and KEGG pathway enrichment analysis using significant hypomethylated Alu/LINE-1 DMR. Significant enrichment indicates Alu and LINE-1 DMR that are more likely to appear in regulatory elements (A) or KEGG pathways (B). Hypomethylation of Alu and LINE-1 may involve in cis-regulatory changes and potential transcription activation events in cancer-related pathways in tumor tissues. DNase: DNase I hypersensitivity sites; TFBS: transcription factor binding sites; H2A.Z: histone H2A variant; H3: histone H3; H4: histone H4; K: lysine; me1: monomethylation; me2: demethylation; me3: trimethylation; ac: acetylation.
Application 2: predicting Alu and LINE-1 methylation improves power to discriminate tumor from normal tissue
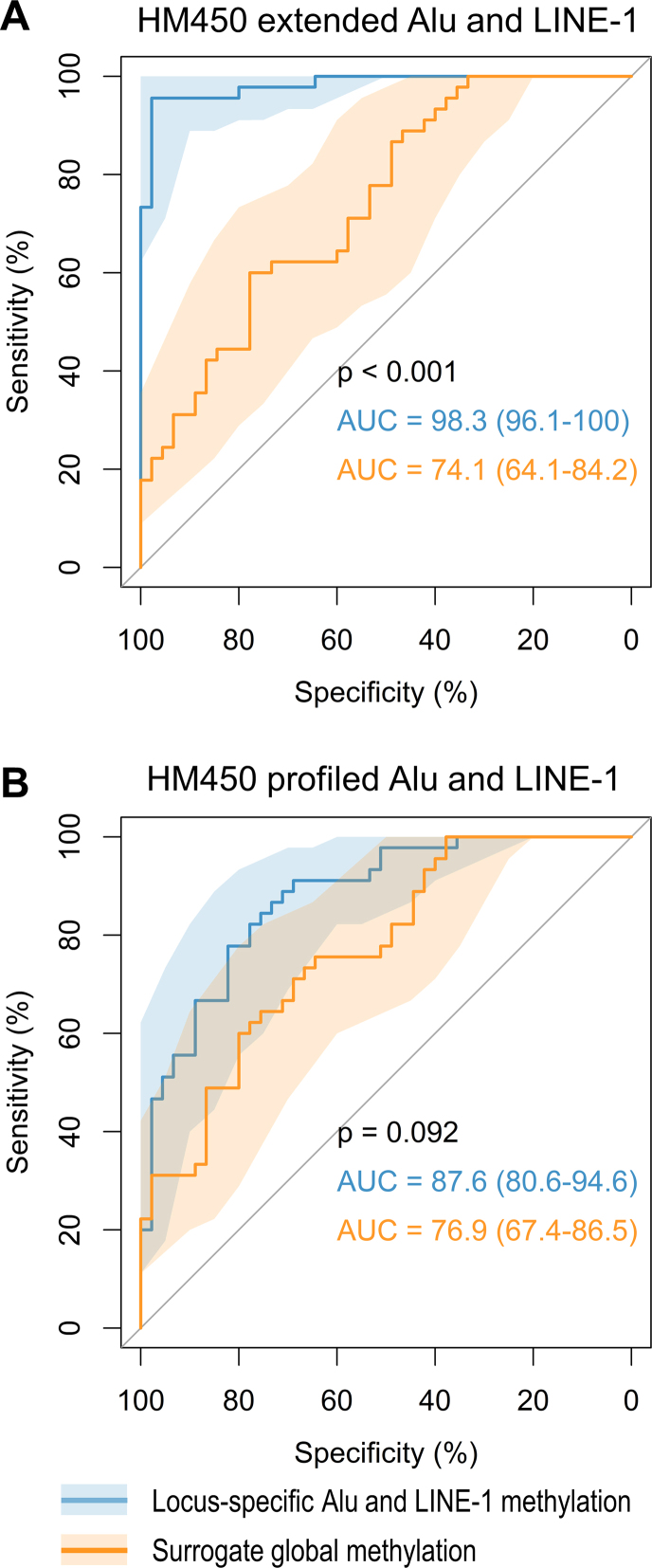
Finally, we implemented an ROC plot to compare the power of locus-specific Alu and LINE-1 methylation versus mean global methylation to discriminate between tumor and the paired normal samples. Mean methylation of CpGs in each Alu and LINE-1 locus were calculated to represent locus-specific methylation level. We demonstrated the discrimination power using extended or profiled Alu and LINE-1 in breast tumors, as other three tumors failed to yield convergent results due to limited sample sizes. The surrogate global methylation was computed by averaging all extended or profiled CpG methylation in Alu and LINE-1. We observed that locus-specific methylation achieved AUC of 98.3 (95% CI: 96.1–100.0), which was higher than that using the surrogate global methylation (74.1; 95% CI: 64.1–84.2; P < 0.001) in the extended Alu and LINE-1 (Figure 8A). For the profiled Alu and LINE-1 methylation, we observed lower AUC of 87.6 (95% CI: 80.6–94.6) for locus-specific methylation, which was again higher than the AUC using surrogate global methylation (76.9; 95% CI: 67.4–86.5), but not significantly so (Figure 8B).
Figure 8.
Discrimination power of locus-specific Alu/LINE-1 methylation vs surrogate global methylation. (A) extended Alu and LINE-1 methylation. (B) Profiled only. Shaded regions represent 95% confidence intervals of ROC curves. Locus-specific Alu and LINE-1 methylation achieved higher AUC than that using surrogate global methylation. Our predicted methylation achieved higher AUC than that using HM450-profiled methylation.
DISCUSSION
We developed a prediction algorithm and corresponding R package REMP to predict locus-specific RE methylation by mining methylation information from neighboring CpG sites profiled in Infinium methylation arrays. We validated the reliability of our algorithm using both sequencing (i.e. NimbleGen) and EPIC array (covering over 850 000 CpGs) data, further verifying the algorithm's prediction performance by demonstrating the inverse relationships between Alu/LINE-1 methylation and evolutionary age previously observed. More importantly, we tested the clinical use of our algorithm in TCGA data to examine epigenome-wide associations and distinguish tumor from normal tissues. Our algorithm may help address current challenges in studying the role of RE methylation in human diseases. It also directly addresses the assumption of a uniform methylation profile in RE with similar biological or pathological effects, which may have caused information loss in extant studies and hindered our understanding of the exact role that RE methylation plays in human diseases. Furthermore as technologies for epigenomic profiling continue to improve, our algorithm can serve as an important framework for later expanding RE coverage. This will enhance our ability to investigate relationships between RE epigenetic features and complex traits/diseases in a highly cost-effective manner in large clinical and population studies.
Our algorithm was mainly developed based on the HM450 and EPIC arrays, since compared to other sequencing-based approaches the array-based data were the most robust for Alu/LINE-1 measurement (higher coverage in some sequencing platforms, e.g. WGBS, notwithstanding). In addition, the Infinium methylation array is the ideal source to provide reliable neighboring information for methylation prediction. Previous attempts at predicting methylation suggested that incorporating extensive neighboring information such as profiled CpG sites, genomic positions, DNA sequence properties, and cis-regulatory elements could yield highly accurate predictions (50,58,79). However, in practice obtaining the requisite information is often impractical and infeasible. By leveraging the co-methylation features of neighboring CpGs and the structure of RE sequences, we devised a simpler predictive strategy and achieved high predictive performance for our algorithm. Our algorithm only relies on predictors that are easily extractable from DNA methylation profiling data, minimizing dependence on a reference genome and preserving individual variability in the human epigenome.
The predictive power of our algorithm was further confirmed by testing Alu/LINE-1 methylation in relation to evolutionary age. Alu and LINE-1 propagated in primate genomes over the past 65 and 80 million years, respectively, which resulted in phylogenetic trees of Alu/LINE-1 subfamilies with different evolutionary ages (80,81). One of our previous studies confirmed this inverse relationship by bisulfite-PCR-pyrosequencing 10 differentially-evolved RE subfamilies (82). In accordance with these findings the current study also confirmed this hypothesis from a more comprehensive genome-wide perspective, which further supports the reliability of our prediction results. This demonstrates the potential utility of our algorithm in studying more specific characteristics of RE methylation throughout the genome in connection with human diseases and other phenotypes, which may presently be impossible or impractical due to data limitations.
Our algorithm offers a more comprehensive perspective on the RE methylation landscape and biological implications of RE methylation on an epigenome-wide scale. The consistent enrichment of hypomethylated Alu and LINE-1 in regulatory regions (i.e. DNase and TFBS) across all four types of tested tumors highlights the potential cis-regulatory roles of Alu/LINE-1 methylation. This is supported by previous findings that RE derive a wide variety of gene regulatory regions, including DNase and TFBS in the human genome, demonstrating the effects of RE on regulating genes (83–85). The enrichment of hypomethylated Alu/LINE-1 in the histone modifications that we observed were largely consistent with a recent sequencing study of hypomethylated Alu in cancer cells (86). Specifically, the enrichment of hypomethylated Alu and LINE-1 in H3K4me3 (a marker for transcriptional activation) and H3K9me3 (for transcriptional repression) suggests possible involvement of RE methylation in transcription activation events in tumor (87). Ward et al. demonstrated that Alu and LINE-1 are responsible for transcriptional activation and enriched in regions marked by H3K4me3 (88). DNA hypomethylation in Alu and LINE-1 in H3K9me3 could be an indicator of decreased H3K9me3, suggesting a less transcriptionally repressive function as H3K9me3 is shown to promote persistent DNA methylation in RE (89). Furthermore, hypomethylated Alu/LINE-1 in breast and colon cancer were overrepresented in H2A.Z, a histone variant that can potentially alter nucleosome stability (90). It has been hypothesized that adequate genic methylation (mostly in RE) may stabilize translational control functions such as translation, ribosome biogenesis, RNA splicing, and protein localization by antagonizing H2A.Z deposition (91). Thus, hypomethylation of Alu/LINE-1 in H2A.Z may indicate dysfunction of translational control functions which are important to cancer etiology (92).
Our pathway-based analysis further supports the biological relevance of our predicted RE methylation. Olfactory transduction was one of the top-ranked pathways enriched by our predicted hypomethylated Alu in all four tumor tissues of interest. This pathway contains a large gene family of olfactory receptors (ORs), which have been found to be ectopically expressed in non-olfactory tissues (93) and for some ORs overexpressed in breast (94), colon (95), lung (96) and prostate tissues (97). In addition, we observed axon guidance pathway was significantly enriched with hypomethylated LINE-1 in breast, colon, and lung cancers. Axon guidance has been shown to play an important role in cancerogenesis (98). Further data analysis of the intronic locus-specific LINE-1 methylation and the host gene expression of SEMA3A supported the hypothesis that DNA methylation in intronic regions may potentially silence RE to maintain a gene's efficient transcription, and thus usually has a positive correlation with gene expression (99).
In the tumor-normal discrimination test, the improved AUC when using locus-specific Alu and LINE-1 methylation demonstrated the potential for information loss when using mean Alu and LINE-1 methylation (both widely used surrogate global methylation measures). In addition, the AUC using our extended Alu and LINE-1 methylation outperformed HM450 profiled methylation, further underscoring the valuable information added by our algorithm.
Several features and caveats of our algorithm are worth noting. First, our algorithm was not designed to cover whole-genome RE methylation, but rather to provide a reliable extension of RE methylation profiled using Infinium methylation arrays, which prioritize CpG interrogations in genes and functional regions. The predicted CpG sites maintained a similar genomic distribution as those profiled in the arrays, thus offering extended information on the biological roles of RE methylation in transcriptional regulation, identifying biomarkers of diseases, and devising useful clinical tools with minimal artificial bias. Second, our algorithm's performance can be influenced by methylation data quality and patterns of RE methylation in different tissues. However our algorithm allows for convenient evaluation and control of prediction reliability using the forest-based model to ensure prediction quality. Incorporating prediction reliability control may lead to missing data, posing potential challenges to downstream data analysis, however imputation techniques such as K Nearest Neighbor (KNN) imputation (74) can be applied to obtain more complete data if needed. Third, the test of our algorithm's clinical utility was conducted only on TCGA HM450 data due to the lack of more advanced data, such as that from the EPIC array. Further investigations in larger human studies using such data to validate the clinical utility of our algorithm are warranted. Fourth, our algorithm was designed to predict all types of RE methylation. However, our validation and clinical application tests only focused on the two most common human RE, Alu and LINE-1, due to their predominance throughout the human genome. The algorithm can be used on other human RE such as long terminal repeats and tandem repeats (100).
In conclusion, the proposed algorithm can be applied to the widely used methylation profiling platforms and extend RE CpG coverage in a highly cost-effective manner. More importantly it promotes genome-wide, locus-specific RE methylation association analyses in large human population and clinical studies by providing extended coverage of locus-specific RE methylation. This allows for more precise investigations into the tumorigenic (and potentially other etiological) roles of RE methylation, improving the accuracy of epigenetic studies. Our work may drive further investigations on how DNA methylation in RE may differ in their cis- and/or trans-effects on genomic stability, such as increasing mutation rates or aberrant gene expression, and identify novel RE loci that may exert important biological and pathological effects for cancer early detection and diagnosis.
AVAILABILITY
REMP is available for download at Bioconductor: http://bioconductor.org/packages/REMP.
RepeatMasker Library (build hg19) and RefSeqGene annotation database (build hg19) are available through the R package AnnotationHub, record number = AH5122 and AH5040, respectively.
GM12878 HM450 data are available at ENCODE: http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeHaibMethyl450/
GM12878 EPIC data are available in R package minfiDataEPIC.
GM12878 RRBS data are available at ENCODE: http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeHaibMethylRrbs/
GM12878 WGBS data are available at ENCODE: https://www.encodeproject.org/experiments/ENCSR000AJI/
GM12878 NimbleGen data are available upon reasonable request and with permission of Roche Sequencing.
HM450 data of TCGA tumor tissue and paired normal tissue are available at GDC Data Portal: https://portal.gdc.cancer.gov/
Regulatory element data available in the ENCODE Analysis Hub at the European Bioinformatics Institute: http://ftp.ebi.ac.uk/pub/databases/ensembl/encode/integration_data_jan2011/
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Xiaogang Su at the University of Texas at El Paso for his thoughtful comments and constructive suggestions on the choice and implementation of machine learning algorithms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01CA202936A to L.H., R21CA187869 to W.Z. and L.H., P30CA060553 Career Development Award to W.Z.] (in part). Funding for open access charge: Northwestern University Robert H. Lurie Comprehensive Cancer Center Rosenberg Research Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. et al. . Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 2. Rodic N., Burns K.H.. Long interspersed element-1 (LINE-1): passenger or driver in human neoplasms. PLoS Genet. 2013; 9:e1003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cordaux R., Batzer M.A.. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009; 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xing J., Zhang Y., Han K., Salem A.H., Sen S.K., Huff C.D., Zhou Q., Kirkness E.F., Levy S., Batzer M.A. et al. . Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009; 19:1516–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Slotkin R.K., Martienssen R.. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007; 8:272–285. [DOI] [PubMed] [Google Scholar]
- 6. Hancks D.C., Kazazian H.H. Jr. Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 2012; 22:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Batzer M.A., Deininger P.L.. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002; 3:370–379. [DOI] [PubMed] [Google Scholar]
- 8. Beck C.R., Garcia-Perez J.L., Badge R.M., Moran J.V.. LINE-1 elements in structural variation and disease. Annu. Rev. Genomics Hum. Genet. 2011; 12:187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Morgan H.D., Sutherland H.G., Martin D.I., Whitelaw E.. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999; 23:314–318. [DOI] [PubMed] [Google Scholar]
- 10. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002; 16:6–21. [DOI] [PubMed] [Google Scholar]
- 11. Qu G., Dubeau L., Narayan A., Yu M.C., Ehrlich M.. Satellite DNA hypomethylation vs. overall genomic hypomethylation in ovarian epithelial tumors of different malignant potential. Mut. Res. 1999; 423:91–101. [DOI] [PubMed] [Google Scholar]
- 12. Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009; 1:239–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robertson K.D. DNA methylation, methyltransferases, and cancer. Oncogene. 2001; 20:3139–3155. [DOI] [PubMed] [Google Scholar]
- 14. Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002; 21:5400–5413. [DOI] [PubMed] [Google Scholar]
- 15. Beisel C., Paro R.. Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 2011; 12:123–135. [DOI] [PubMed] [Google Scholar]
- 16. Yang A.S., Estecio M.R., Doshi K., Kondo Y., Tajara E.H., Issa J.P.. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004; 32:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lisanti S., Omar W.A., Tomaszewski B., De Prins S., Jacobs G., Koppen G., Mathers J.C., Langie S.A.. Comparison of methods for quantification of global DNA methylation in human cells and tissues. PLoS One. 2013; 8:e79044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brennan K., Flanagan J.M.. Is there a link between genome-wide hypomethylation in blood and cancer risk. Cancer Prev. Res. 2012; 5:1345–1357. [DOI] [PubMed] [Google Scholar]
- 19. Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007; 8:286–298. [DOI] [PubMed] [Google Scholar]
- 20. Lu X.-J., Xue H.-Y., Qi X., Xu J., Ma S.-j. LINE-1 in cancer: multifaceted functions and potential clinical implications. Genet Med. 2015; 18:431–439. [DOI] [PubMed] [Google Scholar]
- 21. Barchitta M., Quattrocchi A., Maugeri A., Vinciguerra M., Agodi A.. LINE-1 hypomethylation in blood and tissue samples as an epigenetic marker for cancer risk: a systematic review and meta-analysis. PLoS One. 2014; 9:e109478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pobsook T., Subbalekha K., Sannikorn P., Mutirangura A.. Improved measurement of LINE-1 sequence methylation for cancer detection. Clin. Chim. Acta. 2011; 412:314–321. [DOI] [PubMed] [Google Scholar]
- 23. Phokaew C., Kowudtitham S., Subbalekha K., Shuangshoti S., Mutirangura A.. LINE-1 methylation patterns of different loci in normal and cancerous cells. Nucleic Acids Res. 2008; 36:5704–5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xie H., Wang M., Bonaldo Mde F., Smith C., Rajaram V., Goldman S., Tomita T., Soares M.B.. High-throughput sequence-based epigenomic analysis of Alu repeats in human cerebellum. Nucleic Acids Res. 2009; 37:4331–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xie H., Wang M., de Andrade A., Bonaldo Mde F., Galat V., Arndt K., Rajaram V., Goldman S., Tomita T., Soares M.B.. Genome-wide quantitative assessment of variation in DNA methylation patterns. Nucleic Acids Res. 2011; 39:4099–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szpakowski S., Sun X., Lage J.M., Dyer A., Rubinstein J., Kowalski D., Sasaki C., Costa J., Lizardi P.M.. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene. 2009; 448:151–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nusgen N., Goering W., Dauksa A., Biswas A., Jamil M.A., Dimitriou I., Sharma A., Singer H., Fimmers R., Frohlich H. et al. . Inter-locus as well as intra-locus heterogeneity in LINE-1 promoter methylation in common human cancers suggests selective demethylation pressure at specific CpGs. Clin Epigenet. 2015; 7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luo Y., Lu X., Xie H.. Dynamic Alu methylation during normal development, aging, and tumorigenesis. Biomed. Res. Int. 2014; 2014:784706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weisenberger D.J., Campan M., Long T.I., Kim M., Woods C., Fiala E., Ehrlich M., Laird P.W.. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005; 33:6823–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Treangen T.J., Salzberg S.L.. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 2012; 13:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stevens M., Cheng J.B., Li D., Xie M., Hong C., Maire C.L., Ligon K.L., Hirst M., Marra M.A., Costello J.F.. Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Res. 2013; 23:1541–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hansen K.D., Timp W., Bravo H.C., Sabunciyan S., Langmead B., McDonald O.G., Wen B., Wu H., Liu Y., Diep D. et al. . Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 2011; 43:768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L. et al. . High density DNA methylation array with single CpG site resolution. Genomics. 2011; 98:288–295. [DOI] [PubMed] [Google Scholar]
- 34. Smit A.F.A., Hubley R., Green P.. RepeatMasker Open-4.0. 2013. [Google Scholar]
- 35. Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M. et al. . RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014; 42:D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morgan M., Carlson M., Tenenbaum D., Arora S.. Annotationhub: Client to access annotationhub resources. 2016; R package version 2.6.4.
- 37. International HapMap Consortium The International HapMap Project. Nature. 2003; 426:789–796. [DOI] [PubMed] [Google Scholar]
- 38. International HapMap Consortium A haplotype map of the human genome. Nature. 2005; 437:1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meissner A., Gnirke A., Bell G.W., Ramsahoye B., Lander E.S., Jaenisch R.. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005; 33:5868–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M. et al. . Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fortin J., Hansen K.D.. minfiDataEPIC: Example data for the Illumina Methylation EPIC array. 2016; R package version 1.0.0.
- 43. Duhaime-Ross A. Revved-up epigenetic sequencing may foster new diagnostics. Nat. Med. 2014; 20:2. [DOI] [PubMed] [Google Scholar]
- 44. Roche Diagnostics Sequencing Solutions Technical Note: How To Evaluate NimbleGen SeqCap Epi Target Enrichment Data. 2014. [Google Scholar]
- 45. Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E.. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012; 13:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. American Cancer Society Cancer Facts and Figures 2017. Atlanta: American Cancer Society. 2017. [Google Scholar]
- 47. Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I. et al. . TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016; 44:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bell J.T., Pai A.A., Pickrell J.K., Gaffney D.J., Pique-Regi R., Degner J.F., Gilad Y., Pritchard J.K.. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011; 12:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A. et al. . DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006; 38:1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang W., Spector T.D., Deloukas P., Bell J.T., Engelhardt B.E.. Predicting genome-wide DNA methylation using methylation marks, genomic position, and DNA regulatory elements. Genome Biol. 2015; 16:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li Y., Zhu J., Tian G., Li N., Li Q., Ye M., Zheng H., Yu J., Wu H., Sun J. et al. . The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010; 8:e1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B. et al. . Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008; 454:766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Edwards J.R., O’Donnell A.H., Rollins R.A., Peckham H.E., Lee C., Milekic M.H., Chanrion B., Fu Y., Su T., Hibshoosh H. et al. . Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010; 20:972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rangwala S.H., Zhang L, Kazazian H.H. Jr. Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 2009; 10:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Price E.M., Cotton A.M., Penaherrera M.S., McFadden D.E., Kobor M.S., Robinson W.. Different measures of ‘genome-wide’ DNA methylation exhibit unique properties in placental and somatic tissues. Epigenetics. 2012; 7:652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smith T.F., Waterman M.S.. Identification of common molecular subsequences. J. Mol. Biol. 1981; 147:195–197. [DOI] [PubMed] [Google Scholar]
- 57. Cortes C., Vapnik V.. Support-Vector Networks. Mach. Learn. 1995; 20:273–297. [Google Scholar]
- 58. Zheng H., Wu H., Li J., Jiang S.W.. CpGIMethPred: computational model for predicting methylation status of CpG islands in human genome. BMC Med. Genomics. 2013; 6(Suppl. 1):S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. James P., Girijadevi R., Charles S., Pillai M.R.. MethFinder - A software package for prediction of human tissue-specific methylation status of CpG islands. Bioinformation. 2013; 9:61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fan S., Zhang M.Q., Zhang X.. Histone methylation marks play important roles in predicting the methylation status of CpG islands. Biochem. Biophys. Res. Commun. 2008; 374:559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bock C., Walter J., Paulsen M., Lengauer T.. CpG island mapping by epigenome prediction. PLoS Comput. Biol. 2007; 3:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fang F., Fan S., Zhang X., Zhang M.Q.. Predicting methylation status of CpG islands in the human brain. Bioinformatics. 2006; 22:2204–2209. [DOI] [PubMed] [Google Scholar]
- 63. Bock C., Paulsen M., Tierling S., Mikeska T., Lengauer T., Walter J.. CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure. PLoS genetics. 2006; 2:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vert J.P., Tsuda K., Scholkopf B.. Kernel Methods in Computational Biology. 2004; MIT Press. [Google Scholar]
- 65. Breiman L. Random Forests. Mach. Learn. 2001; 45:5–32. [Google Scholar]
- 66. Kuhn M. Caret package. J. Stat. Softw. 2008; 28:1–26.27774042 [Google Scholar]
- 67. Jiang B., Zhang X., Cai T.. Estimating the Confidence Interval for Prediction Errors of Support Vector Machine Classifiers. J. Mach. Learn. Res. 2008; 9:521–540. [Google Scholar]
- 68. Meinshausen N. Quantile regression forests. J. Mach. Learn. Res. 2006; 7:983–999. [Google Scholar]
- 69. Meinshausen N. quantregForest: Quantile Regression Forests. R package version. 2016; 1:3–5. [Google Scholar]
- 70. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jaffe A.E., Murakami P., Lee H., Leek J.T., Fallin M.D., Feinberg A.P., Irizarry R.A.. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 2012; 41:200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yu G., Wang L.G., Han Y., He Q.Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Reid S., Tibshirani R.. Regularization paths for conditional logistic regression: the clogitL1 package. J. Stat. Softw. 2014; 58: [PMC free article] [PubMed] [Google Scholar]
- 74. Troyanskaya O., Cantor M., Sherlock G., Brown P., Hastie T., Tibshirani R., Botstein D., Altman R.B.. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001; 17:520–525. [DOI] [PubMed] [Google Scholar]
- 75. Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Muller M.. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011; 12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. DeLong E.R., DeLong D.M., Clarke-Pearson D.L.. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988; 44:837–845. [PubMed] [Google Scholar]
- 77. Moen E.L., Zhang X., Mu W., Delaney S.M., Wing C., McQuade J., Myers J., Godley L.A., Dolan M.E., Zhang W.. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics. 2013; 194:987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mishra R., Thorat D., Soundararajan G., Pradhan S.J., Chakraborty G., Lohite K., Karnik S., Kundu G.C.. Semaphorin 3A upregulates FOXO 3a-dependent MelCAM expression leading to attenuation of breast tumor growth and angiogenesis. Oncogene. 2015; 34:1584–1595. [DOI] [PubMed] [Google Scholar]
- 79. Das R., Dimitrova N., Xuan Z., Rollins R.A., Haghighi F., Edwards J.R., Ju J., Bestor T.H., Zhang M.Q.. Computational prediction of methylation status in human genomic sequences. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:10713–10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kapitonov V., Jurka J.. The age of Alu subfamilies. J. Mol. Evol. 1996; 42:59–65. [DOI] [PubMed] [Google Scholar]
- 81. Smit A.F., Toth G., Riggs A.D., Jurka J.. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 1995; 246:401–417. [DOI] [PubMed] [Google Scholar]
- 82. Byun H.M., Motta V., Panni T., Bertazzi P.A., Apostoli P., Hou L., Baccarelli A.A.. Evolutionary age of repetitive element subfamilies and sensitivity of DNA methylation to airborne pollutants. Part Fibre Toxicol. 2013; 10:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jordan I.K., Rogozin I.B., Glazko G.V., Koonin E.V.. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003; 19:68–72. [DOI] [PubMed] [Google Scholar]
- 84. Thornburg B.G., Gotea V., Makalowski W.. Transposable elements as a significant source of transcription regulating signals. Gene. 2006; 365:104–110. [DOI] [PubMed] [Google Scholar]
- 85. Marino-Ramirez L., Jordan I.K.. Transposable element derived DNaseI-hypersensitive sites in the human genome. Biol Direct. 2006; 1:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jorda M., Diez-Villanueva A., Mallona I., Martin B., Lois S., Barrera V., Esteller M., Vavouri T., Peinado M.A.. The epigenetic landscape of Alu repeats delineates the structural and functional genomic architecture of colon cancer cells. Genome Res. 2017; 27:118–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K.. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837. [DOI] [PubMed] [Google Scholar]
- 88. Ward M.C., Wilson M.D., Barbosa-Morais N.L., Schmidt D., Stark R., Pan Q., Schwalie P.C., Menon S., Lukk M., Watt S. et al. . Latent regulatory potential of human-specific repetitive elements. Mol. Cell. 2013; 49:262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rose N.R., Klose R.J.. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim. Biophys. Acta. 2014; 1839:1362–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Suto R.K., Clarkson M.J., Tremethick D.J., Luger K.. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Bio.l. 2000; 7:1121–1124. [DOI] [PubMed] [Google Scholar]
- 91. Coleman-Derr D., Zilberman D.. DNA methylation, H2A.Z, and the regulation of constitutive expression. Cold Spring Harb. Symp. Quant. Biol. 2012; 77:147–154. [DOI] [PubMed] [Google Scholar]
- 92. Ruggero D. Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 2013; 5:a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kang N., Koo J.. Olfactory receptors in non-chemosensory tissues. BMB Rep. 2012; 45:612–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Muranen T.A., Greco D., Fagerholm R., Kilpivaara O., Kampjarvi K., Aittomaki K., Blomqvist C., Heikkila P., Borg A., Nevanlinna H.. Breast tumors from CHEK2 1100delC-mutation carriers: genomic landscape and clinical implications. Breast Cancer Res. 2011; 13:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Weber L., Al-Refae K., Ebbert J., Jagers P., Altmuller J., Becker C., Hahn S., Gisselmann G., Hatt H.. Activation of odorant receptor in colorectal cancer cells leads to inhibition of cell proliferation and apoptosis. PLoS One. 2017; 12:e0172491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Giandomenico V., Cui T., Grimelius L., Oberg K., Pelosi G., Tsolakis A.V.. Olfactory receptor 51E1 as a novel target for diagnosis in somatostatin receptor-negative lung carcinoids. J. Mol. Endocrinol. 2013; 51:277–286. [DOI] [PubMed] [Google Scholar]
- 97. Weng J., Wang J., Hu X., Wang F., Ittmann M., Liu M.. PSGR2, a novel G-protein coupled receptor, is overexpressed in human prostate cancer. Int. J. Cancer. 2006; 118:1471–1480. [DOI] [PubMed] [Google Scholar]
- 98. Chedotal A., Kerjan G., Moreau-Fauvarque C.. The brain within the tumor: new roles for axon guidance molecules in cancers. Cell Death Differ. 2005; 12:1044–1056. [DOI] [PubMed] [Google Scholar]
- 99. Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G.. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014; 26:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhang Z., Zheng Y., Zhang X., Liu C., Joyce B.T., Kibbe W.A., Hou L., Zhang W.. Linking short tandem repeat polymorphisms with cytosine modifications in human lymphoblastoid cell lines. Hum. Genet. 2016; 135:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.