Abstract
Mediator is a multi-unit molecular complex that plays a key role in transferring signals from transcriptional regulators to RNA polymerase II in eukaryotes. We have combined biochemical purification of the Saccharomyces cerevisiae Mediator from chromatin with chromatin immunoprecipitation in order to reveal Mediator occupancy on DNA genome-wide, and to identify proteins interacting specifically with Mediator on the chromatin template. Tandem mass spectrometry of proteins in immunoprecipitates of mediator complexes revealed specific interactions between Mediator and the RSC, Arp2/Arp3, CPF, CF 1A and Lsm complexes in chromatin. These factors are primarily involved in chromatin remodeling, actin assembly, mRNA 3′-end processing, gene looping and mRNA decay, but they have also been shown to enter the nucleus and participate in Pol II transcription. Moreover, we have found that Mediator, in addition to binding Pol II promoters, occupies chromosomal interacting domain (CID) boundaries and that Mediator in chromatin associates with proteins that have been shown to interact with CID boundaries, such as Sth1, Ssu72 and histone H4. This suggests that Mediator plays a significant role in higher-order genome organization.
INTRODUCTION
Recent studies of large-scale chromatin organization have identified chromosome territories organized into megabase- to sub-megabase sized interacting domains that have been named ‘topologically associating domains’ (TADs) in mammals (1,2), and ‘chromosomal interacting domains’ (CIDs) in Caulobacter crescentus (3) and Saccharomyces cerevisiae (4). Here, we will use the term ‘CID’ for these domains. CIDs represent a collection of discrete regions, with chromatin within one region interacting preferentially with chromatin in the same CID (5). Boundaries between CIDs are formed by sites that bind architectural proteins (APs), which often function as insulator proteins (6,7). Moreover, tRNA genes have been shown to be involved in barrier functions, acting to restrict the spread of repressive chromatin in human cells (8), S. cerevisiae (9,10) and S. pombe (11,12).
AP positioning and occupancy are dynamic. Heat-shock in Drosophila leads to redistribution of APs from CID boundaries into the body of the CIDs, resulting in weaker boundaries. This redistribution leads to an increase in long-range inter-CID contacts (5). The relative occupancy of APs bound to CID boundaries scales with the strength of the CID boundaries. Stronger boundaries are associated with the CIDs that prefer intra-CID over inter-CID interactions (13). This might correspond to results emerging over the past two decades, which indicate that through DNA looping, multiple active genes transcribed by Pol II cluster into discrete sites in the nucleus and become co-regulated (14,15).
In mammalian cells, binding of CTCF (CCCTC-binding factor) and cohesin is frequently observed at CID boundaries, but these factors are also present inside the CIDs themselves (2). In fact, almost all CID boundaries are occupied by CTCF, but only 15% of all CTCF binding sites are located within CID boundary regions. Thus, additional proteins may be required for the formation of CID boundaries (1). Interestingly, distinct combinations of CTCF, cohesin, and Mediator are involved in chromatin looping in mammalian cells (16). Saccharomyces cerevisiae lack a homolog to CTCF, but the ATP-dependent RSC chromatin remodeling complex and the cohesin loading factor Scc2 are enriched at strong CID boundaries, and mutations in the gene looping factor Ssu72, the Mediator subunits Med1 and Med12, the H3K56 acetyltransferase Rtt109, and the N-terminal part of histone H4 affect chromatin compaction in budding yeast (4).
Transcription regulation of protein coding genes in eukaryotic cells entails an intricate interplay among transcriptional regulators (activators/repressors), co-regulatory factors, general transcription factors (GTFs), and RNA polymerase II (Pol II) on the chromatin template. Mediator is a key co-regulator protein complex, which is required for transcriptional activation and repression (17,18). Depending on the species, it comprises 25–30 evolutionarily conserved subunits, which can be grouped into four modules: Head, Middle, Tail and a distinct Cdk8 kinase module which reversibly associates with Mediator and regulates the Mediator–Pol II interaction in order to control transcription initiation and re-initiation (19,20).
Mediator participates in regulation of gene expression through interactions with promoter-bound transcriptional regulators. Previous work has suggested that Mediator serves as a general component of the Pol II machinery, since mutational inactivation of Med17 or Med22 reduces general transcription to the same extent as the inactivation of Pol II itself (21,22). While initial microarray-based ChIP experiments supported this assumption (23,24), subsequent studies found that Mediator resides at only a few genomic sites in exponentially growing cells, and is recruited to stress-activated genes by a limited number of transcriptional activators (25,26). Thus, the role of Mediator as a pervasive participant in Pol II transcription, particularly in unstressed cells, remains unresolved.
Most reports on the composition of Mediator in S. cerevisiae indicate that it is a well-defined protein complex composed of 21 core subunits and the reversibly associated, four-subunit Cdk8 kinase module. However, these results are based on purifications of the soluble fraction of whole-cell protein extracts, with the proteins interacting strongly with chromatin removed during the initial purification steps (17,18,27). Purification from yeast cells using a procedure that minimized subunit dissociation showed that Mediator was present in at least two different forms, one of which lacked Pol II and several Mediator subunits (28). Another study showed that S. cerevisiae transcriptional activation remained functional even when Tail and Head/Middle modules were artificially separated by mutational inactivation of MED16 (29). Moreover, it was recently reported that Mediator recruitment to SAGA-regulated genes is more dependent on tail module subunits compared to Mediator recruitment to TFIID-regulated genes (30). Thus, the composition of Mediator in yeast seems to vary depending on the purification methods (28) and/or growth conditions (31). Furthermore, Mediator complexes with varying subunit composition have also been identified in mammalian cells (32,33). So far, to our knowledge Mediator has not been purified from chromatin and it is therefore unknown if Mediator bound to DNA through interaction with DNA-binding proteins such as activators or repressors might have a unique and possibly variable subunit composition.
The original characterizations of Mediator identified it as an essential co-regulatory factor that functions by bridging transcriptional activators at promoter-proximal binding sites and Pol II present at the transcription start site (17,18). However, Mediator was also recently shown to be important for formation and maintenance of super-enhancers, which are clusters of enhancers that regulate the expression of genes controlling cell identity and can cause high expression levels of oncogenes in cancer cells (34,35). The distinction between super-enhancers and normal enhancers is vague, but in general super-enhancers are described as a class of regulatory loci that span large genomic regions, which are highly enriched in transcriptional coactivators and specific chromatin modifications, such as H3K27ac, H3K4me1, H3K4me2, and which function in regulation of tissue-specific transcription (36).
In order to examine the role and composition of Mediator in chromatin, we have determined the association of representative Mediator subunits and Pol II across the yeast genome using chromatin immunoprecipitation and DNA sequencing (ChIP-seq). Our results reveal that Mediator is stably associated with strong CID boundaries as well as the promoters of several additional genes. Moreover, we find that the promoters to which Mediator is associated have prominent nucleosome-depleted regions (NDRs). Our co-immunoprecipitation/mass spectrometry experiments revealed that chromatin-bound Mediator is associated with additional proteins and protein complexes which are not found associated with Mediator isolated from the non-chromatin fraction. In line with previous reports in both S. cerevisiae (4) and human cells (16), our results suggest that Mediator in chromatin, in addition to its function as a co-regulator complex involved in regulation of genes transcribed by Pol II, has an important role in the organization of chromatin architecture and higher-order genome structure.
MATERIALS AND METHODS
Yeast strain construction
Occupancy of Mediator in growing yeast was assessed in TAP-tagged Med3, Med7, Med14, Med15, Med17, Med19 and CycC strains (Open Biosystems). Expression of TAP-tagged Mediator subunits was confirmed by western blotting. Strains used in this study are listed in Supplementary Table S1. C-terminally Myc-tagged Mediator subunit strains were constructed by replacing the stop codons of the corresponding ORFs (YGL025C (Med3), YOL135C (Med7), YLR071C (Med14), YOL051W (Med15), YER022W (Med17), YBL093C (Med19) and YNL025C (CycC) with an 8x Gly-13x Myc-KanMx6 construct from the template plasmid pFA6a-13x Myc-kanMX6, using primers listed in Supplementary Table S2. Constructs were transformed into the wild-type prototrophic MATa S288C strain using the standard Li-Ac transformation protocol (37). The G418 positive yeast were sequenced and subjected to western blot analysis to confirm expression of the tagged proteins. All strains were sequenced to confirm the presence of the full Myc-tagged constructs at the correct chromosomal locations. None of the tagged strains showed any growth defects compared to the parent wild-type strain in glucose, glycerol and raffinose media, or in media limited for amino-acids. Since inactivating mutations of Mediator subunits elicit specific growth defects under these conditions (22,38,39), we concluded that the tags did not diminish Mediator function. Nor did expression of any of the Myc-tagged Mediator subunits affect expression of other untagged Mediator subunits, as noted by the consistent levels of endogenous, untagged Med17 in all Myc-tagged strains (Supplementary Figure S1A). Finally, Myc-tagging of Mediator subunits did not impede Mediator complex formation dramatically, since we could co-precipitate Mediator subunits with each of the Myc-tagged versions of Med3, Med7, Med14, Med15, Med 17 and Med19 (Supplementary Figure S1B).
Growth conditions
Yeast cells were grow in 250 ml of SC + 2% glucose medium at 30°C overnight and then diluted to an OD600 of 0.05. The cells were then grown to mid-log phase, filtered (Stericup-GP, 0.22 μm filtering systems, (Millipore Corporation, Billerica, MA, USA)) and cross-linked using a final concentration of 0.6% formaldehyde for 20 min. Cross-linking was quenched by the addition of 2.5 M Glycine and harvested by centrifugation at 5000 × g for 5 min. Cell pellets were frozen in liquid nitrogen and stored at –80°C for subsequent preparation of whole cell extracts (WCE) for ChIP-seq.
Western blotting and quantification in whole cell extracts
Cells from 10 ml cultures were harvested at an OD600 of 0.3–0.4 and transferred to tubes containing 300 μl acid washed glass beads and 1 ml of 20% trichloroacetic acid containing 2 mM PMSF. Cells were lysed by Fast Prep bead beating (setting 6.5) for 2 × 40 s. Protein pellets were collected by centrifugation and the resulting pellets were washed with 500 μl ice-cold acetone, resuspended in 45 μl SDS loading buffer and separated by 12% SDS-PAGE. Proteins were then transferred to PVDF membranes and stained with Ponceau S to ensure equal loading. The membranes were then subjected to immunoblotting using anti-Myc and anti-Med17 antibodies.
Immunoprecipitation in whole cell extracts
Protein extracts were isolated from yeast grown in YPD media (1% yeast extract, 2% Bacto-Peptone, and 2% glucose) to an OD600 of 4. The cells were harvested by centrifugation at 6000 × g for 5 min and the semi-dry pellet was frozen in liquid nitrogen and broken in a Freezer Mill Model 6850 (SPEX CertiPrep). The resulting cell powder (1 g) was resuspended in 1 ml of 2 × lysis buffer A-100 (40) and centrifuged for 10 min at 10 000 × g to sediment cell debris. Supernatants were used for anti-Myc immunoprecipitation. c-Myc antibodies (monoclonal 9E10, Clontech) were bound to Protein G Dynabeads (Invitrogen) as described in the product manual. 500 μl of the protein extracts were added to 150 μl of the beads and incubated on a rotator at 4°C for 2 h. The flow through was collected and saved and the beads were washed three times with 1 ml lysis buffer. The beads were then dissolved in 100 μl of 1 × SDS buffer. Load and flow through was diluted 10 times and 10 μl was separated on a 7% SDS-PAGE together with 10 μl of beads and immunoblotted with antibodies against Med1, Med5 and Med17.
Isolation of soluble and chromatin-bound protein extracts
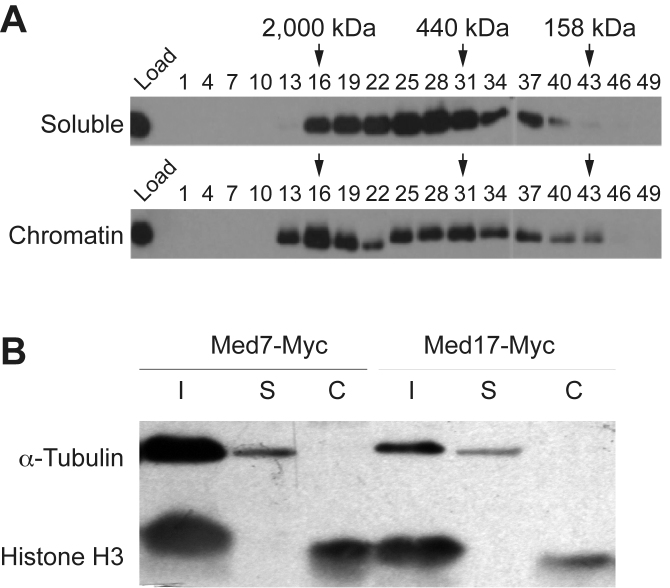
The Med22-TAP (scTAP library, Thermo Scientific), the Med7-Myc, the Med17-Myc (see above) and Arc35-HA (41) strains were grown in YPD media (1% yeast extract, 2% bacto-peptone, and 2% glucose) to an OD600 of 6. Preparation of whole cell extracts and separation of soluble and chromatin-bound protein extracts were performed essentially as described (42), except that 0.5 M instead of 1 M ammonium sulfate was used to release proteins bound to DNA since we found that Mediator started to precipitate at higher concentrations. In order to obtain comparable extracts, also the soluble, or non DNA-bound fraction was treated with 0.5 M ammonium sulfate. After centrifugation for 1 h at 4°C, 42 000 rpm (Beckman Ti45), the supernatants from each of the soluble and chromatin-bound fractions were used for immunoprecipitation using Myc-antibodies coupled to agarose beads (Myc-tagged strains), Calmodulin Sepharose beads (TAP-tagged strains) or HA-antibodies coupled to agarose beads (HA-tagged strains (see below). Separation of soluble and chromatin-bound extracts was verified by Western blotting using antibodies against a-tubulin (soluble protein) and histone H3 (chromatin-bound protein).
Calmodulin purification of proteins from the soluble and chromatin extracts isolated from the Med22-TAP strain
Calmodulin Sepharose 4B (GE Healthcare) was equilibrated with 10 column volumes (CV) of buffer A-100 containing 6 mM CaCl2. 6 mM CaCl2 was also added to both the soluble and the chromatin protein fractions. The two protein fractions were then added to Calmodulin Sepharose and bound in batch for 2 h at 4°C. For the soluble fraction, 50 ml of protein extract (60 mg/ml) was bound to 3 ml of Calmodulin Sepharose. For the chromatin fraction, 18 ml of protein extract (115 mg/ml) was bound to 1.5 ml of Calmodulin Sepharose. The bead slurries were then applied to columns by gravity flow and washed, first with 10 CV of buffer A-100 and then with 125 ml calmodulin wash buffer (10 mM Tris–Cl (pH 8.0), 1 mM magnesium acetate, 1 mM imidazole, 0.1% NP40, 10 mM β-mercaptoethanol, 100 mM potassium acetate, 2 mM CaCl2 and protease inhibitors). Proteins were then eluted by addition of 0.5 CV calmodulin elution buffer (10 mM Tris–Cl (pH 8.0), 10% glycerol, 1 mM Mg-acetate, 1 mM imidazole, 0.1% NP40, 10 mM β-mercaptoethanol, 150 mM potassium acetate, 15 mM EGTA and protease inhibitors). Fractions containing Mediator were pooled (soluble pool: 8 ml (5.2 mg/ml), chromatin pool: 3.5 ml (4.9 mg/ml) and concentrated using a Vivaspin 2 5,000 MWCO HY spin concentrator (Vivaproducts).
Gel filtration of the soluble and chromatin fractions
Gel filtration was performed using a Superose 6 PC 3.2/30 column (GE Healthcare). The columns were equilibrated with 2 column volumes (CV) of buffer A-100 (without protease inhibitors, DTT or NP-40). 50 μl of concentrated soluble (60 mg/ml) or chromatin (26 mg/ml) extracts isolated from the Med22-TAP strain were applied to the columns, which had been equilibrated with buffer A-100. The columns were run at 0.02 ml/min, 50-μl fractions were collected and every third fraction was loaded on to SDS-gels and analyzed by Western blotting using Med22 antibodies.
Anti-Myc immunoprecipitation of proteins from the soluble and chromatin extracts isolated from the Myc-tagged Med7 and Med17 strains
9 mg of proteins from the soluble and chromatin-bound fractions, respectively, were precleared by incubation with 15 μl HA-agarose beads (Abcam) at +4°C for 1 h. The supernatants were transferred to new tubes containing 60 μl c-Myc agarose beads (Abcam) and incubated for 3 h at +4°C. After centrifugation, the beads were washed three times with 750 μl buffer A-100 containing NP-40 and 2 times with 750 μl buffer A-100 without NP-40. The beads were then resuspended in 100 μl 20 mM HEPES–KOH containing 6 M guanidine hydrochloride and stored in –20°C until used for protein extraction and digestion (see below).
Protein extraction and digestion
Samples were placed on a 10 kDa spin-filter, washed twice with 50 mM ammonium bicarbonate (ABC). They were then incubated for 60 min at 95°C in 6 M guanidine hydrochloride with 20 mM DTT, followed by alkylation with iodoacetamide (80 mM final concentration) for 30 min at room temperature in darkness. Samples where then washed twice with 50 mM ABC before overnight digestion in ABC with 100 ng trypsin. The resulting peptides were cleaned-up using a C18 STAGE-tip (43) and the concentration of each sample was measured using a Micro BCA Protein Assay Kit (Thermo Scientific).
Mass spectrometry and data analysis
An amount of 150 ng of the digested proteins were loaded on an HSS T3 C18 analytical column (75 μm i.d. × 200 mm, 1.8 μm particles; Waters, Milford, MA, USA), and separated using a linear 70 min gradient of 5–40% solvent B (3:1 Acetonitrile/2-propanol) balanced with 0.1% aqueous formic acid (solvent A) at a flow rate of 350 nl min−1. The eluate was passed to a nano-ESI equipped SynaptTM G2-Si HDMS mass spectrometer (Waters, Milford, MA) operating in resolution mode. All data were collected using ion-mobility MSe with a scan-time of 0.5 s and mass-corrected using Glu-fibrinopeptide B and Leucine Enkephalin as reference peptides. Data was analyzed using the ProteinLynxGlobalServer v3.0 (Waters, Milford, MA). Databank search parameters were as follows: 10 ppm mass tolerance, <3% FDR, two missed cleavages, carbamidomethylated cysteines as fixed modification, oxidized methionine, deamidation of asparagine and glutamine and protein N-terminal acetylation as variable modifications. A minimum of one unique peptide detected was used as the threshold to call a protein as present in each sample.
Genomic methods
Mononucleosomal DNA was isolated exactly as previously described (44). Chromatin extract production was adapted from (45) with some modifications (44). ChIP assays using anti-Myc and anti-CTD antibodies, reverse crosslinking and purification of DNA were performed as described (44). All anti-Myc ChIPs were performed on three biological replicates. The anti-CTD ChIPs were performed on two biological replicates. ChIP-DNA was amplified (using the LM-PCR method described in Agilent Yeast ChIP-on-chip analysis protocol Version 9.2, May 2007) and subjected to the Illumina TruSeq paired end sequencing protocol and directly used for cluster generation and sequencing using a Illumina HiSeq 2000 Genome Analyzer II (Illumina). Reverse crosslinking and purification of DNA were performed as described (44).
Chromatin immunoprecipitation (ChIP) assays
ChIP was performed using the standard protocol (46). Briefly, the frozen cell pellets were lysed in cold FA lysis buffer with protease inhibitors (Roche) and 0.5 mm glass beads (BioSpec Products) in the Bead Beater (Biospec). The insoluble chromatin obtained from pelleting the WCE was further sheared using the Covaris E220 sonicator. The sheared chromatin was determined to be in the range of 150–600 bp. Immunoprecipitation of TAP- and Myc-bound DNA was performed by incubating the chromatin extracts with 10 μg of polyclonal anti-TAP antibody CAB1001 (Thermo Scientific) and polyclonal anti-rabbit Myc-antibody (Sigma) respectively, bound to magnetic Protein G Dynabeads (Life Technologies) overnight at 4°C. Washes were performed as described in the protocol and the crosslinks were reversed and treated with Proteinase K overnight to obtain ChIP DNA. The DNA was purified using the ChIP DNA Clean and Concentrator (Zymo Research). ChIP assays using anti-CTD antibodies, reverse crosslinking and purification of DNA were performed as described (44).
Library preparation and sequencing
Libraries were prepared using the HiFi KAPA Library prep kit (KAPA Biosystems). ChIP DNA was subjected to end-repair, A-tailing and adapter ligation using NEXTflex Illumina barcode adapters (Bioo Scientific). The adapter-ligated DNA was amplified for 17 cycles after which it was size-selected using AMPure beads (Beckman Coulter) to obtain libraries between 150 and 500 bp. The libraries were pooled to equimolar concentration and 2 × 25 paired-end sequencing was performed on an Illumina MiSeq sequencer.
Bioinformatic analysis
Paired-end sequences were mapped to the S. cerevisiae reference genome sacCer3, using bowtie2 (47) with the default settings. From the resulted alignment files (bam files), we removed the PCR duplicates and the reads with a mapping quality <10, using the samtools software package (48). We size-selected the paired-end reads with the size ≤300 bp, before we computed the raw genome-wide occupancy profiles in MATLAB (using the bioinformatics toolbox), by stacking all paired-end reads and counting the number of fragments that overlapped with every bp. The raw binding profiles were normalized such that the average occupancy for each chromosome is 1. This accounts for the differences in sequencing depths, and allows further comparison among different samples. To visualize the binding profiles, we used igvtools to create tracks (tdf files) that can be loaded in the IGV genome browser (49). ChIP-seq peaks were detected using MACS2 software (50) using the options ‘–gsize 1.2e+7 –qvalue 1e-5 –mfold 3 100’ and the untagged sample as a control. The contact frequency matrix reported previously in a Micro-C experiment (4)was plotted in R using the plotHic function from the Sushi package (51), after we binned the sequencing data in bins of size 100 bp. The heat maps and the average binding figures for the ChIP-seq and ChIP-exo data were plotted in MATLAB using the heatmap function (http://www.mathworks.com/matlabcentral/fileexchange/24253-customizable-heat-maps) and the bioinformatics toolbox.
Data access
Mediator ChIP-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE95051. MNase-seq data and CTD ChIP-seq data were previously published and are available at the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra/) under accession numbers SRX477409 and SRX386369, respectively. Other previously published experimental data used in this study: ChIP-seq data for Sth1, Scc2, Scc4 (52); ChIP-seq data for TFIIIB, TFIIIC (53); ChIP-seq data for Abf1 (54), Med15 and Med17 (55), ChIP-exo data for Rap1, Reb1, Bdf1, TBP (56), DNase-seq data (57), ChIP-chip data for multiple Mediator subunits (58) and ChEC-seq data for Med8 and Med17 (59).
RESULTS
Mediator binds to boundaries of chromosomal interaction domains (CIDs)
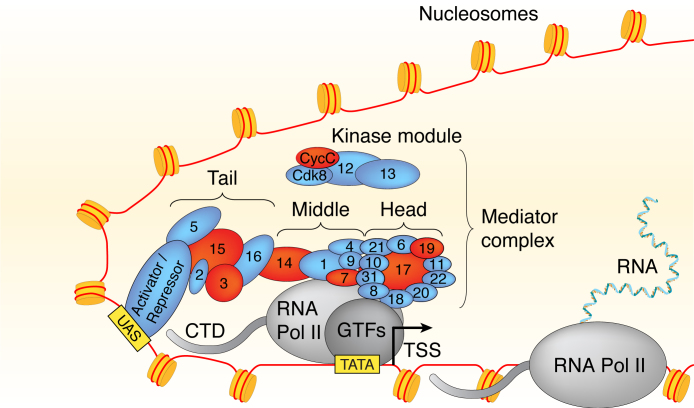
In order to determine the location and composition of Mediator in chromatin of growing cells, we used six strains from the TAP-Fusion Library (Open Biosystems), each of which expressed one individual TAP-tagged mediator subunit. We examined two subunits from each of the Head (Med17, Med19) and Tail (Med3, Med15) Mediator modules, and one subunit each from the Middle (Med14) and the kinase module (CycC) (Figure 1). We also constructed seven strains expressing Myc-epitope-tagged Mediator subunits, including the six above plus a second subunit from the Middle module (Med7). All proteins (both the TAP- and the Myc-tagged) were tagged in their carboxy termini and expressed from their endogenous loci under the control of their native promoters. None of the subunit functions was compromised by the presence of the tag (cf. Materials and Methods).
Figure 1.
Model of the Mediator complex and its subdomains associated with Pol II general transcription factors and a transcriptional activator bound to an UAS. Proteins labeled in red represent those that were used for ChIP-seq assays in this study.
We performed chromatin immunoprecipitation (ChIP) of individual Mediator subunits and analyzed the results by paired-end sequencing (ChIP-seq) using the standard Illumina protocol. For ChIP-seq experiments using the TAP antibodies, we performed control experiments using a non-tagged strain. We also performed ChIP-seq using a monoclonal antibody specific for the C-terminal domain (CTD) of Rpb1, the largest Pol II subunit, in order to detect and quantify transcription of genes by Pol II. Finally, to provide chromatin context for our analysis of Mediator binding, we also mapped genome-wide nucleosome positions by sequencing size-selected DNA fragments following micrococcal nuclease treatment of cross-linked chromatin.
Analysis of our ChIP-seq data using the TAP-tagged strains revealed Mediator subunit occupancy at the expected positions in Pol II promoters, e.g. upstream of YDL055C and YDL047W (Figure 2). ChIP-seq data obtained with the Myc-tagged strains yielded nearly identical results (cf. Figures 2, 3, Supplementary Figures S2 and S3). We also detected Mediator binding at more unexpected locations in the genome, such as the gene bodies of tRNAs, snRNAs, LTR retrotransposons, and autonomously replicating sequences (Supplementary Figure S3A). Mediator binding to most of these latter regions was not associated with detectable transcription of closely located protein-encoding genes, as determined by CTD ChIP-seq experiments. For example, the prominent Mediator peak at tF(GAA)N was associated with only low transcription levels of the surrounding YNL133C or YNL132W, while YNL134C is transcribed at relatively higher levels despite the smaller Mediator subunit peaks in its promoter region (Supplementary Figure S3B). Previous reports suggested that ChIP-seq experiments are affected by artifacts at highly accessible or highly transcribed DNA loci, and reported a list of 238 loci from the yeast genome where ChIP is prone to artifacts (60). To avoid the risk of including false positive peaks of Mediator subunit occupancy, we have eliminated these 238 ‘hyper-ChIP-able’ sites from our subsequent analyses, retaining 638 Mediator binding sites at least 1kb away from these sites.
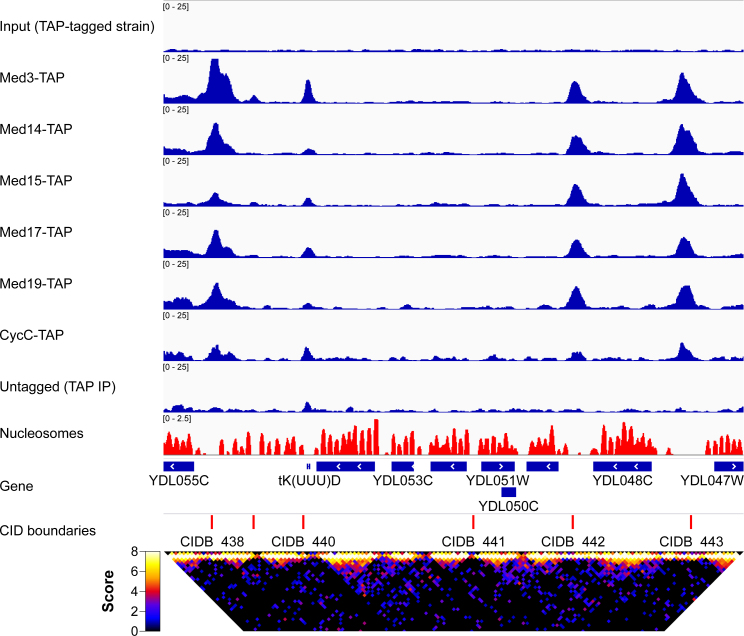
Figure 2.
Mediator binds at the CID boundaries. Mediator binding sites overlap with previously reported CID boundaries (4). Top: binding profiles for the tagged Mediator subunits (CycC, Med3, Med14, Med15, Med17, Med19) and the previously reported nucleosome occupancy (44) on chrIV: 356 000–370 500; bottom: previously reported CID boundaries (4) (red vertical lines) and Micro-C contact frequency heat map for the same locus, on a log2 scale. The bright triangles from the heat map represent the topological domains, or CIDs. In this genomic region (chrIV: 356 000–370 500) four out of six CID boundaries are bound by Mediator as evident from the TAP ChIP-seq data while a fifth site is evident in the Myc ChIP-seq data (Supplementary Figure S2).
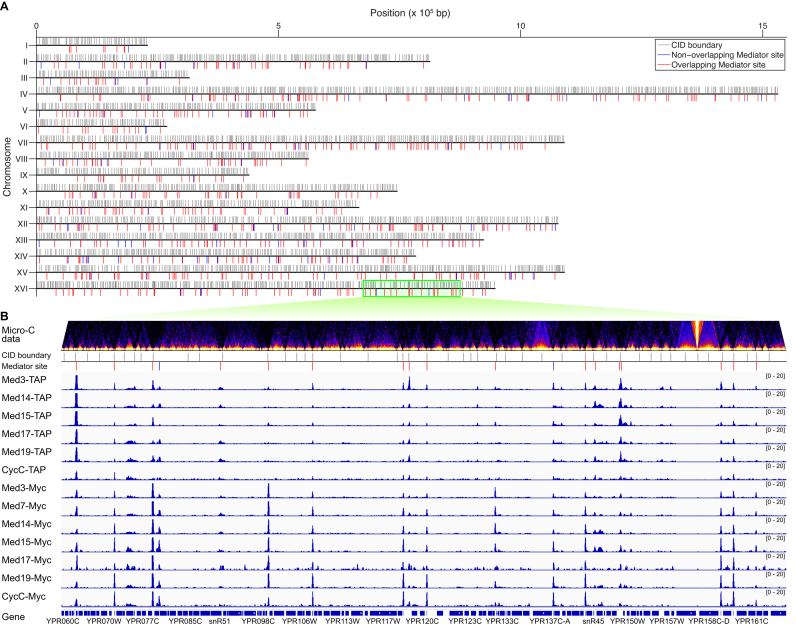
Figure 3.
Significant overlap between the Mediator binding sites and the locations of the CID boundaries. (A) Overview of the locations of CID boundaries (gray lines) and Mediator binding sites that overlap (red lines) or do not overlap (blue lines) with a CID boundary along the 16 yeast chromosomes. (B) Magnified 200 kb region from chrXVI (675 000–875 000), with Micro-C contact frequency represented as a heat map, the locations of previously reported CID boundaries (4) shown by vertical red lines, and the binding profiles of the tagged Mediator subunits shown in blue. The majority of the Mediator peaks are located at CID boundaries.
Among the remaining positions of Mediator binding, we observed significant overlap with CID boundaries previously identified using a chromosome conformation capture method, Micro-C (4) (Figures 2 and 3). The biological significance of this correlation is reinforced by the fact that Mediator participates in chromosome folding (4) and in interactions with cohesin, where it helps connect enhancers with their target promoters through gene looping (61). Overall, 88% of the Mediator peaks that we identify coincided with a CID boundary, and 20% of all CID boundaries were occupied by Mediator (Figure 4A). This contrasts with the results seen with the human CTCF protein, for which only a small fraction (15%) of its binding sites lies within a CID/TAD boundary region, despite the fact that almost all CID/TAD boundaries contain a CTCF binding site (1). Our finding that most Mediator peaks overlap with CID boundaries indicates that participation in chromatin organization is an important function for Mediator, in addition to its well-described role as a transcriptional co-regulator for transcription by Pol II.
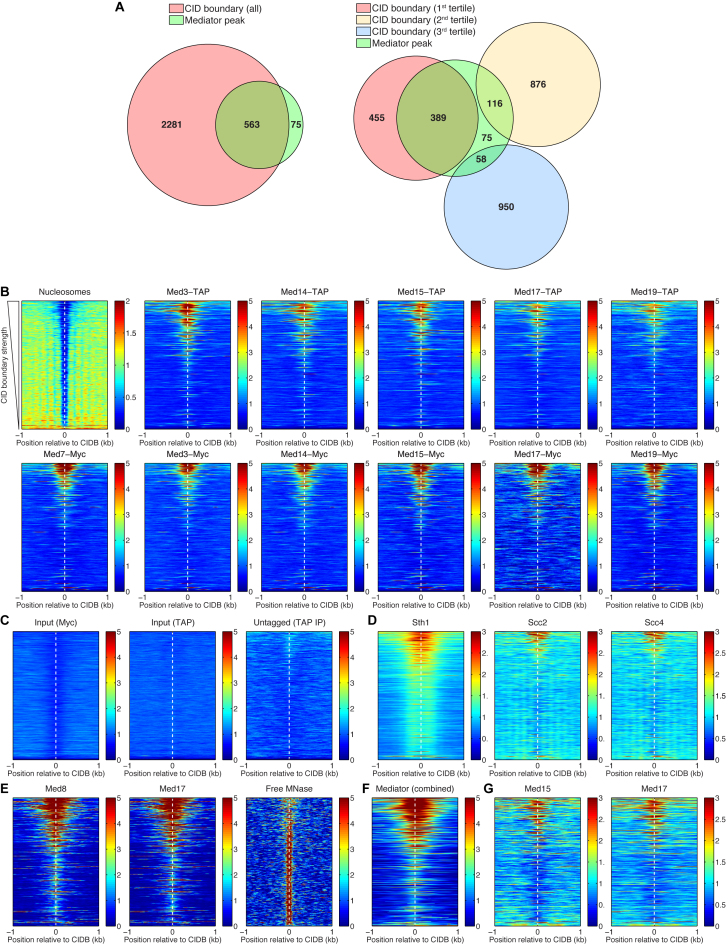
Figure 4.
Mediator, chromatin remodeler RSC, and cohesin loading factors Scc2/Scc4 preferentially bind to the strongest CID boundaries. (A) Venn diagrams representing the overlaps of the CID boundaries and the Mediator binding sites. The CID boundaries are shown as a whole group and split into three tertiles, according to their strength. From the total of 563 Mediator peaks that are found at CID boundaries, about 69% of these are located at the first tertile, containing the strongest CID boundaries. (B) Heat maps representing the nucleosome depletion at all CID boundaries (CIDBs), and the preferential binding of Mediator to the strongest boundaries. The rows of the heat maps are sorted according to strength of the CID boundaries, from the strongest (at the top) to the weakest (at the bottom). (C) Control experiments—sonicated chromatin (input) and IP from an untagged strain—resulted in a clean background, without spurious binding peaks. (D) Sth1 (subunit of the RSC chromatin remodeling complex) and Scc2, Scc4 (cohesin loading factors) also preferentially bind to the strongest CID boundaries. (E) ChEC-seq data for Med8 and Med17 subunits and the control experiment (free MNase cleavage) (59). (F) Combined Mediator ChIP-chip data—MetaMediator Data Set in WT Cells from (58). (G) ChIP-seq data for Med15 and Med17 (55).
Mediator subunits preferentially occupy strong CID boundaries
Analyzing the ChIP-seq data in more detail, we have found that Mediator subunits bind preferentially to the strongest CID boundaries. This is evident from the heat maps in Figure 4B of Mediator binding over the 2 kb regions centered on CID boundaries, which are sorted from top to bottom according to their ‘strength’. The strength of a boundary has been defined as inversely proportional to the number of detected interactions crossing it (4). In fact, we found that Mediator subunits bound 46% of the top third strongest CID boundaries but only 6% of the bottom third (Figure 4A).
Binding of Mediator to CID boundaries could potentially be an artifact caused by non-specific cross-linking of Mediator, since these regions are usually nucleosome-free and therefore in theory more accessible for cross-linking compared to other genomic regions. However, our heat maps show that about two-thirds of the CID boundaries are not bound by Mediator, although these regions are depleted of nucleosomes, and therefore accessible to other DNA-binding proteins (Figure 4B). These conclusions are further supported by calculating the average nucleosome and Mediator subunit occupancies for each of the tertiles (i.e. the top, middle, and bottom thirds of CID boundaries ranked by strength) (Supplementary Figure S4). Finally, the input control experiments and the experiment using a control strain lacking any tags showed essentially no background binding (Figure 4C). As an independent verification of our results, we have examined previously published Mediator binding data: Med8 and Med17 ChEC-seq data (59) (Figure 4E), combined Mediator ChIP-chip data (58) (Figure 4F) and Med15 and Med17 ChIP-seq data (55). Using these data, we have found a similar binding pattern of Mediator to strong CID boundaries. We conclude that Mediator occupancy at strong CID boundaries is not a result of non-specific cross-linking of Mediator subunits to NDRs.
Sth1, Scc2, Scc4, Rap1 and TBP show a pattern of binding to strong CID boundaries similar to that of Mediator
Subunits of other protein complexes such as Sth1 (RSC) and Scc2/Scc4 (cohesin loader complex) are also enriched at yeast CID boundaries and play an important role in their formation and organization (4,52). We therefore used published ChIP-seq data for Sth1, Scc2 and Scc4 (52) to make heat maps corresponding to those shown for the Mediator subunits in order to compare the binding patterns of Mediator to CID boundaries of different strength with the binding patterns of RSC (Sth1) and cohesin loader complex (Scc2, Scc4) (Figure 4D and Supplementary Figure S4). In support of a role for Mediator at CID boundaries, we found that the Mediator, RSC and cohesin loader complex subunits are distributed similarly at these regions.
In order to assess whether binding to strong CID boundaries is a common feature for transcription-related proteins, we used published ChIP-exo and ChIP-seq data for Brf1 (TFIIIB subunit), Tfc1 (TFIIIC subunit) (53), Abf1 (ARS-binding factor 1) (54), Reb1 (Pol I enhancer binding protein), Rap1 (repressor-activator protein 1), Bdf1 (Bromodomain factor 1) and TBP (TATA-binding protein) (56) to find the occupancies of these factors at CID boundaries (Supplementary Figures S4 and S5). We have found that only TBP and Rap1 preferentially bind to strong CID boundaries with a pattern similar to that of the Mediator, RSC and cohesin loader complex subunits (cf. Figure 4B, D and Supplementary Figure S5).
To check whether Sth1, Scc2, Scc4, Rap1 and TBP co-occupy the same CID boundaries as Mediator and to cross-correlate their DNA-binding patterns, we separated the CID boundaries according to whether they are bound by Mediator or not (Supplementary Figure S6A), keeping each group sorted according to the CID boundary strength. We observed that Sth1, Scc2, Scc4, Rap1 and TBP bind to the same CID boundaries as Mediator (Supplementary Figure S6A). To quantify the binding of each protein to each CID boundary, we computed the average occupancy of the proteins in 100 bp regions centered on each CID boundary. The difference between the occupancy levels of these proteins in the two classes of CID boundaries is extremely significant (Supplementary Figure S6B; two-sample t-test P-values < 10−4 in all cases). The Pearson correlation coefficients for the occupancies of different pairs of proteins are shown in Supplementary Figure S6C.
Mediator is present as a complex in protein extracts isolated both from chromatin and from the soluble fraction
Consistent with our results presented above, several reports have indicated a possible link between Mediator and the proteins that bind to TAD and CID boundaries (e.g. CTCF, cohesin, RSC, Ssu72) (4,16,62–64). These proteins are normally not associated with Mediator in traditional purifications from yeast or mammalian cells, where Mediator is identified as a homogenous complex of ∼25–30 subunits, depending on the species (65). However, previous reports on a possible connection between Mediator and TAD/CID boundaries were based on techniques that involve covalent cross-linking between proteins and nucleic acids (e.g. chromosome conformation capture (3C) and ChIP). In contrast, traditional biochemical purifications of Mediator originate from the non-DNA bound fraction of protein extracts isolated from cells that have not been treated with cross-linking agents. We therefore speculated that Mediator bound to DNA in chromatin might differ in composition compared to the traditional form of Mediator purified from the non-DNA bound fraction.
To assess the native structure of Mediator bound to DNA, we initiated biochemical purification of Mediator from the soluble and chromatin-bound extracts in parallel, without the use of cross-linkers. Whole-cell protein extracts were isolated from cell cultures as previously described (40). Proteins bound to chromatin were then isolated using high-salt extraction. In order to avoid differences between the chromatin and soluble fractions that could be due to differences in treatments, the soluble fractions were treated with the same high-salt extraction. We initially used a yeast strain expressing a TAP-tagged Med22 subunit to facilitate purification of Mediator. Gel filtration of the TAP eluate from the soluble extract indicated that Med22 migrated as one single peak of ∼500 kDa (Figure 5A, top panel), in agreement with previous reports (17). In contrast, Med22 in the chromatin extract eluted in two peaks, one smaller (∼500 kDa), and one larger (∼2 MDa) (Figure 5A, bottom panel). Our results suggest that Mediator in chromatin interacts with additional, and so far unidentified proteins and could potentially represent the form of Mediator which participates in the formation of CID boundaries.
Figure 5.
Chromatin-bound Mediator associates with different sets of proteins. (A) Whole-cell extracts from the Med22-TAP strain were separated into soluble and chromatin-bound extracts as described in Materials and Methods. Proteins from each extract were purified on Calmodulin Sepharose beads. Proteins eluted from the beads were then applied to a Superose 6 PC 3.2/30 gel filtration column. Aliquots from the load and every third fraction were separated on 12% SDS-PAGE and analyzed by Western blotting with Med22 antibodies. (B) Whole cell proteins extracts (I) were isolated from yeast strains expressing Myc-tagged versions of the Med7 and Med17 proteins. The whole cell extracts were separated into a soluble fraction (S) and a chromatin bound fraction (C). The presence of α-tubulin and histone H3 in each fraction was revealed by Western blotting.
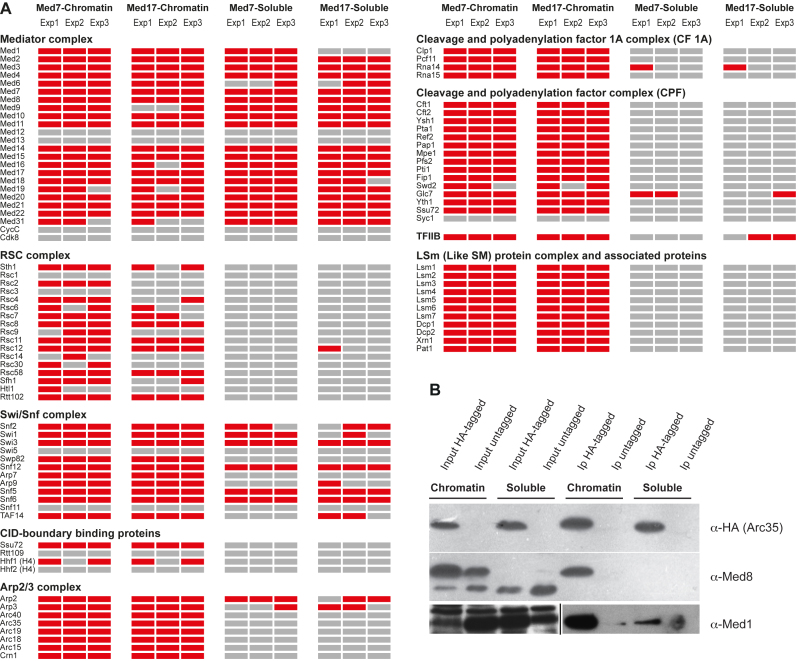
We next used the Myc-tagged Med7 and Med17 strains, which we had used previously in the ChIP-seq experiments described above, in order to affinity purify Mediator and identify additional proteins that might interact specifically with Mediator when it is bound to DNA in the chromatin fractions. Protein extracts eluted from the soluble and chromatin-bound fractions from both strains were isolated as described in Materials and Methods and analyzed by Western blotting using antibodies specific for histone H3 and α-tubulin to verify separation of the soluble and chromatin fractions (Figure 5B). We found histone H3 to be exclusively present in the protein extracts eluted from the chromatin fractions, while α-tubulin was only detected in the soluble fractions, indicating that the two extracts were efficiently separated. Proteins interacting with Med7 or Med17 in each strain and each extract were co-immunoprecipitated using anti-Myc antibodies coupled to agarose beads. Proteins present in each of the four immunoprecipitates were then identified using tandem mass-spectrometry (MS/MS) in three independent experiments. In total, we identified ∼375–600 proteins that were co-precipitated with Mediator isolated from the chromatin fractions of the two Myc-tagged strains (Supplementary Table S3). Similarly, we found ∼400–475 proteins that were co-precipitated with Mediator isolated from the soluble fractions of the two Myc-tagged strains (Supplementary Table S3). We next identified proteins that were present in all three experiments for each strain and extract (Supplementary Table S3). We identified 469 proteins present in all Med7-Myc chromatin experiments, 346 proteins in all Med17-Myc chromatin extracts, 337 proteins in all Med7- Myc soluble extracts and 345 proteins in the Med17-Myc soluble extracts. By combining all proteins detected in each extract, we could identify 332 proteins that interacted with both Med7-Myc and Med17-Myc in the chromatin extracts, and 284 proteins that interacted with both Med7-Myc and Med17-Myc in the soluble extracts.
We found that most Mediator subunits were co-precipitated with Med7-Myc and Med17-Myc from both the soluble and the chromatin fractions (Figure 6A, Supplementary Table S3). This shows that Mediator is present as a complex both in the soluble and the DNA-bound chromatin extracts. The exceptions were Med31, which was not detected in precipitates from the soluble fractions of either of the two strains, and Med1, which was not found in the precipitates from the soluble fractions of the Med17-Myc strain. It is possible that these differences reflect previously reported findings indicating that Mediator complexes with different subunit compositions are present in cells (28–30,32,33). In addition, we were unable to detect the kinase module subunits (CycC, Cdk8, Med12 or Med13) in any of the precipitates from either of the fractions and strains. This is consistent with previous reports showing that the kinase module forms a separate complex in cells, which is only temporarily associated with the other Mediator modules (20). Finally, Tebbji et al. reported that 179 proteins co-precipitate with TAP-tagged Med7 in C. albicans (66). Interestingly, we identified 101 of these 179 proteins in our Med7 pull-down experiments, despite their functional differences; MED7 is an essential gene in S. cerevisiae but not in C. albicans.
Figure 6.
Chromatin-bound Mediator interacts with proteins involved in formation of CID boundaries, mRNA 3′-end processing, gene looping, actin assembly and mRNA decay. (A) Proteins interacting with Mediator in the soluble and chromatin fractions of the Med7-Myc and Med17-Myc strains were immunoprecipitated using Myc-antibodies coupled to agarose beads. Proteins present in each precipitate (red rectangles) were identified using tandem mass-spectrometry (MS/MS). Gray rectangles represent proteins that were not detected in each experiment. (B) Soluble and chromatin fractions of the Arc35-HA strain were immunoprecipitated using HA-antibodies coupled to agarose beads. The presence of Arc35, Med1 and Med8 in each fraction was detected by western blotting using antibodies specific for the HA-tag, Med1 and Med8, respectively. The bottom panel is split in two figures separated by a black line and represent different exposures of the same western blot using anti-Med8 antibodies.
Mediator isolated from chromatin interacts with architectural proteins involved in formation of CID boundaries, mRNA 3′-end processing, gene looping, actin assembly and mRNA decay
By comparing the mass spectrometry results from all experiments, we could identify 88 proteins that interacted with both Med7-Myc and Med17-Myc in all chromatin extracts, but were absent from all experiments using soluble extracts (Supplementary Table S3). These 88 proteins therefore constitute a set that interacts uniquely with Mediator purified from chromatin. Gene ontology (GO) analysis showed that the two top categories were ‘mRNA cleavage factor complex’ and ‘macromolecular complex’ (Supplementary Table S4). Specifically, we have found that 38 of the 88 identified proteins (see proteins in bold style in Supplementary Table S3) represent subunits of six protein complexes involved in chromatin remodeling (RSC, SWI/SNF), RNA metabolism (CFT, CF 1A, Lsm-Pat1), and actin assembly (Arp2/Arp3 complex). Regardless of the functions that are normally attributed to these protein complexes, they have all been shown to have functions in Pol II transcription, as described in the following sections.
The Arp2/3 complex
Arp2/3 is a complex composed of seven protein subunits. We have found that all Arp2/3 subunits co-precipitate with both Med7 and Med17 from the chromatin extracts. In addition, the Arp2, and to some extent Arp3 were also found in the pull-downs from the soluble extracts of both strains, which might indicate that the Arp2/3 complex is present in several forms in yeast. In line with this, the Arp2/3 complex is well known to function in the cytoplasm as one of the most important mediators for actin assembly (67), but it has also been shown that the Arp2/3 complex has a function in the nucleus where it interacts with Pol II and is involved in transcriptional regulation (68).
In an attempt to confirm that Mediator and the Arp2/3 complex interact specifically in protein extracts isolated from the chromatin fraction, we used a strain expressing an HA-tagged Arc35 subunit (41). As shown in Figure 6B, we have found that the HA-tagged Arc35 protein was present in extracts isolated from both the chromatin and soluble fractions isolated from the strain that express HA-tagged Arc35, but not present in either extract isolated from a corresponding untagged strain (top panel). After immunoprecipitation using HA-antibodies, we found that Arc35 was precipitated to the same level from extracts isolated from both the soluble and chromatin fractions. Western blotting of the same protein extracts using antibodies specific for the Med8 Mediator subunit showed that it was present in both extracts and both strains. We found that the extracts isolated from the chromatin fractions of both strains predominantly contain a slower migrating form of Med8, whereas the extracts isolated from the soluble fractions of both strains only contain a faster migrating form of Med8 (Figure 6B, middle panel). Interestingly however, only the slower migrating form of Med8 is present in extracts isolated from the chromatin fraction that was co-immunoprecipitated with Arc35 (middle panel). We found no Med8 precipitation in either extract isolated from the un-tagged control strain. Similar results were obtained using the same extracts and antibodies specific for the Med1 Mediator subunit (lower panel). Previous reports have shown that Med8 is present in two forms, which differ in their phosphorylation states, with the slower-migrating form representing phosphorylated Med8 (69,70). Furthermore, Med8 was also identified as a phosphoprotein by the Phosphopep project (http://www.phosphopep.org/index.php) and it was reported that the phosphorylation site is located within the C-terminal amino acids (amino acids 205–222). Our results showing that Med8 is present in two migration forms in proteins isolated from the soluble and chromatin fractions might reflect differences in Med8 phosphorylation, but further experiments are required in order to confirm these results.
RSC, SWI/SNF and proteins previously identified to interact with CID boundaries
We found that four subunits of the RSC chromatin remodeling complex (Rsc8, Arp7/Rsc11, Rsc58 and Rtt102) exclusively interact with Mediator isolated from the chromatin fractions of both strains (Supplementary Table S3). By analyzing all 12 experiments from the MS, we found that the Sth1, Rsc2, Rsc4, Rsc6, Rsc7, Rsc8, Rsc9, Rsc11, Rsc12, Rsc14, Rsc30, Rsc58, Sfh1, Htl1 and Rtt102 RSC subunits also interacted with Mediator isolated from chromatin, albeit each of these RSC subunits was not detected in all six precipitates from the chromatin extracts (Figure 6A). This finding is in line with our ChIP-seq results described above, which show co-occupancy of Mediator and RSC at strong CID boundaries (Figure 4B, D), and with results reported by other groups showing that RSC is enriched at CID boundaries (4,16,52). As described above (Figure 4D), we found that both Sth1, the catalytic subunit of RSC, and the Scc2 and Scc4 cohesin loader complex subunits preferentially bind to strong CID boundaries, but that the binding pattern of Sth1 was most similar to the pattern that we found for Mediator subunit occupancies at CID boundaries. The RSC complex likely acts upstream of cohesin in order to recruit the Scc2 and Scc4 subunits to the CID boundaries (52). We did not find any cohesin loader complex subunits in our pull-downs of Mediator, indicating that Mediator might act upstream of RSC in the recruitment of factors to the CID boundaries.
We also detected interaction between Mediator and the SWI/SNF chromatin remodeling complex. However, only two SWI/SNF subunits, Arp7 and Swp82, showed specific interactions with Mediator isolated from chromatin and did not interact with Mediator isolated from the soluble fraction. The Snf2, Swi1, Arp9 and Taf14 subunits were co-precipitated with Mediator from both extracts and both strains, but showed preferential interaction with Mediator in the chromatin extracts. Finally, Swi3, Snf5, Snf6 and Snf12 interacted equally frequently with Mediator in both the soluble and chromatin extracts, while no interaction was observed between Mediator and Swi5 or Snf11 in any of the precipitates from either the soluble or chromatin extracts. We conclude that Mediator appears to interact with the SWI/SNF complex, but that this interaction is more specific for Mediator bound to DNA.
Another interesting finding was that the Ssu72 and histone H4 proteins were only co-precipitated with Mediator in protein extracts isolated from the chromatin fractions, but not from the soluble extracts isolated from either of the Med7-Myc or Med17-Myc strains. Mutations in the genes encoding either Ssu72 or histone H4 resulted in a decrease in global chromosome compaction, similarly to mutations in MED1 or MED14 (4).
The cleavage-polyadenylation-factor (CPF)
The Ssu72 protein mentioned above is also a subunit of the CPF RNA 3′-end-processing machinery, which has been shown to interact with the C-terminal domain (CTD) of the largest subunit of Pol II (71). CPF has been implicated in transcription by acting as a Ser5 phosphatase on the Pol II CTD during transcription termination (72). Ssu72 also functionally interacts with other components of the transcription pre-initiation machinery (e.g. TFIIB) (73) and may facilitate interactions between the 5′- and 3′-ends of genes to promote gene looping (74). We found that 12 of the 15 CPF subunits (75) were uniquely present in both the Med7 and Med17 pull-downs from the chromatin fractions, but completely absent from either pull-down from the soluble fractions (Figure 6B, Supplementary Table S3). In addition, the thirteenth subunit, Swd2, was also identified in four out of six experiments using the chromatin fractions, but was not found in any of the experiments with the soluble extracts. The fourteenth subunit, Glc7, was present in all experiments using the chromatin extracts, but was also detected in two of the experiments using the soluble fractions. It has been reported that TFIIB (SUA7) interacts with CPF through Ssu72 (71,76). In agreement with this, we find that TFIIB was the only GTF present in pull-downs from the chromatin fractions of both the Med7-Myc and Med17-Myc strains.
The cleavage and polyadenylation factor 1A (CF 1A) complex
The CF 1A complex is composed of the Clp1, Pcf11, Rna14 and Rna15 protein subunits (77). Together with Hrp1, it forms a larger CF 1 complex, which is involved in mRNA 3′-end processing. However, several reports demonstrate that CF 1A also interacts functionally with promoters, again through interactions with TFIIB (78,79). We found that three of the CF 1A complex subunits (Clp1, Pcf11 and Rna15) were co-precipitated with both Med7 and Med17 specifically in the chromatin extracts, but not in the soluble extracts, while Rna14 was detected in all experiments using the chromatin extracts, but also in two of the experiments using the soluble extracts.
The Lsm-Pat1 protein complex
The cytoplasmic mRNA decay pathway in eukaryotes is initiated by shortening of the poly A-tail by the Ccr4/Not and Pan2/3 complexes, followed by further degradation via two pathways; by the exosome, which degrade mRNAs from the 3′-end, and by Xrn1 exonuclease, which degrades from the 5′-end (for a review, see (80)). The Xrn1 pathway involves prior removal of the 5-cap, a process catalyzed by the Lsm-Pat1 complex which includes the Lsm1–7, Dcp1, Dcp2, Xrn1, Pat1, Dhh1 and Edc1/2/3 proteins. These proteins have also been shown to interact in two-hybrid assays (81). We found that all the subunits of the Lsm-Pat1 complex co-precipitated specifically with both Med7 and Med17 in the chromatin extracts, except Dhh1, which was also pulled down from the soluble extracts. Interestingly, all subunits of the Lsm-Pat1 complex have been shown to shuttle between the cytoplasm and the nucleus (82). In the nucleus, the Lsm-Pat1 complex apparently associates with transcription start sites and is known to stimulate transcription initiation and elongation (82).
DISCUSSION
Here, we describe ChIP-seq and co-immunoprecipitation experiments aimed at disclosing the composition of Mediator bound to DNA in chromatin. Our results have revealed both expected and unexpected findings. As anticipated, we identified Mediator occupancy at Pol II promoters, consistent with its proposed role as a co-regulator complex that bridges promoter-bound regulatory transcription factors (activators/repressors) and the general RNA polymerase II transcriptional machinery. More unexpectedly however, we also detected enrichment of Mediator at the strongest CID boundaries (4). CID boundaries were originally perceived as insulators, mainly based on their interaction with the CTCF insulator protein in vertebrates. However, more recent reports have suggested that the insulator activity is a minor, context-dependent function of CTCF, and that its primary role is facilitating long-range DNA interactions between distant chromosomal regions (83–85). In mammalian cells, CTCF has been found to co-localize with cohesin and Mediator in different combinations, which correlate with different scales of chromatin interactions; CTCF and cohesin together bridge long-range interactions further than 1 Mb apart, while Mediator and cohesin together bridge shorter-range interactions (<100 kb apart); all three proteins were detected at chromatin loops of the intermediate (100 kb–1 Mb) size (16). In general, long-range CTCF/Cohesin interactions appear to be more static, while medium- to short-range Mediator/Cohesin and Mediator/CTCF/Cohesin-dependent interactions are more dynamic and change in response to ES cell differentiation (16).
Saccharomyces cerevisiae lacks a CTCF-like regulator, but other features of boundaries between CIDs, such as enrichment for active promoters and binding sites for the cohesin loading complex, appear to be conserved (4). Yeast CIDs are two orders of magnitude smaller compared to the corresponding domains in mammalian cells and constitute self-associating domains that cover 1–5 genes; these genes are often co-regulated by the factors that bind to the boundaries that surround them. Strong CID boundaries overlap with promoters of highly transcribed genes and regions that are bound by the RSC chromatin remodeling complex and the cohesin loading complex. Our ChIP-seq results described here show that Mediator is enriched at the same strong CID boundaries as those bound by RSC and the cohesin loading complex, suggesting that these boundaries encompass CIDs that include highly transcribed genes. These results are further supported by our findings that Mediator purified in the absence of cross-linking from the chromatin fraction, but not from the soluble, non-DNA bound fraction, interacts with RSC, which has been shown to bind to CID boundaries in yeast, and with Ssu72 and histone H4, both of which have been implicated in folding of the yeast genome (4). Hsieh et al. (4) also reported that mutation of MED1 affects chromatin compaction. This provides functional validation of the hypothesis supported by the results presented here, which suggest involvement of Mediator in establishment and/or maintenance of CID boundaries. Transcription does not occur diffusely throughout the nucleus, but rather takes place at sub-nuclear sites enriched in RNA polymerase II and other components of the transcription and RNA-processing machinery (86–88). Thus, binding of Mediator at the CID boundaries could be involved in the formation of such ‘transcription factories’ by connecting distantly located enhancers with their target promoters.
In addition to identifying Mediator as an architectural protein, our purification of Mediator from chromatin using non-crosslinking methods revealed unique interactions with a set of protein complexes (CPF, CF 1A, Arp2/3 and Lsm) that traditionally have been linked to gene looping, actin assembly, mRNA decay, and cytoplasmic processes such as mRNA 3′-end processing. However, all these complexes shuttle between the cytoplasm and the nucleus and have specific nuclear functions in different transcriptional processes such as phosphorylation of, and interaction with, Pol II (CPF, Arp2/3), interaction with TFIIB (Ssu72 and CF 1A), association with transcription start sites and stimulation of transcription initiation and elongation (68,72,78,79,82). It is interesting to note that, in their traditionally described activities, several of these factors function by looping DNA in a way that physically connects the 3′- and 5′-ends of the gene. These results, combined with the well-known function of Mediator as a complex required to connect enhancers with promoters, and our results presented here showing interaction of Mediator with strong chromatin-organizing CID boundaries, point to a common function for Mediator in different processes that require DNA looping, such as regulation of gene expression by super-enhancers, long-range chromatin rearrangements, and formation of physical contacts between the 5′- and 3′-end of genes during transcription.
ACCESSION NUMBER
Mediator ChIP-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE95051.
Supplementary Material
ACKNOWLEDGEMENTS
R.V.C. thanks Ho Sung Rhee and Bongsoo Park for helpful discussions regarding the ChIP-exo experiments. We are grateful to the staff at the KBC Proteomics Facility, Umeå University for assistance with the LC–MS analysis. Bioinformatic support by BILS (Bioinformatics Infrastructure for Life Sciences) is gratefully acknowledged. We also thank Dr Bruce Goode for providing us with the HA-tagged Arp35 strain. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). The Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Cancer Society (to to S.B.); Swedish Research Council; Knut and Alice Wallenberg Foundation; National Institutes of Health [GM076562]; Pennsylvania Department of Health using Tobacco CURE funds (to J.R.B., in part); Intramural Research Program of the NIH (to R.V.C.). Funding for open access charge: The Swedish Cancer Society and the Swedish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J. et al. . Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012; 485:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Le T.B., Imakaev M.V., Mirny L.A., Laub M.T.. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013; 342:731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hsieh T.H., Weiner A., Lajoie B., Dekker J., Friedman N., Rando O.J.. Mapping nucleosome resolution chromosome folding in yeast by micro-C. Cell. 2015; 162:108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nguyen H.Q., Bosco G.. Gene positioning effects on expression in eukaryotes. Annu. Rev. Genet. 2015; 49:627–646. [DOI] [PubMed] [Google Scholar]
- 6. Hou C., Li L., Qin Z.S., Corces V.G.. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell. 2012; 48:471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li L., Lyu X., Hou C., Takenaka N., Nguyen H.Q., Ong C.T., Cubenas-Potts C., Hu M., Lei E.P., Bosco G. et al. . Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell. 2015; 58:216–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raab J.R., Chiu J., Zhu J., Katzman S., Kurukuti S., Wade P.A., Haussler D., Kamakaka R.T.. Human tRNA genes function as chromatin insulators. EMBO J. 2012; 31:330–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Valenzuela L., Gangadharan S., Kamakaka R.T.. Analyses of SUM1-1-mediated long-range repression. Genetics. 2006; 172:99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Bortle K., Corces V.G.. tDNA insulators and the emerging role of TFIIIC in genome organization. Transcription. 2012; 3:277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haldar D., Kamakaka R.T.. tRNA genes as chromatin barriers. Nat. Struct. Mol. Biol. 2006; 13:192–193. [DOI] [PubMed] [Google Scholar]
- 12. Scott K.C., Merrett S.L., Willard H.F.. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol.: CB. 2006; 16:119–129. [DOI] [PubMed] [Google Scholar]
- 13. Van Bortle K., Nichols M.H., Li L., Ong C.T., Takenaka N., Qin Z.S., Corces V.G.. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014; 15:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cook P.R. The organization of replication and transcription. Science. 1999; 284:1790–1795. [DOI] [PubMed] [Google Scholar]
- 15. Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J. et al. . Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012; 148:84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phillips-Cremins J.E., Sauria M.E., Sanyal A., Gerasimova T.I., Lajoie B.R., Bell J.S., Ong C.T., Hookway T.A., Guo C., Sun Y. et al. . Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013; 153:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim Y.J., Bjorklund S., Li Y., Sayre M.H., Kornberg R.D.. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994; 77:599–608. [DOI] [PubMed] [Google Scholar]
- 18. Koleske A.J., Young R.A.. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994; 368:466–469. [DOI] [PubMed] [Google Scholar]
- 19. Dotson M.R., Yuan C.X., Roeder R.G., Myers L.C., Gustafsson C.M., Jiang Y.W., Li Y., Kornberg R.D., Asturias F.J.. Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:14307–14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Knuesel M.T., Meyer K.D., Donner A.J., Espinosa J.M., Taatjes D.J.. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol. Cell. Biol. 2009; 29:650–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takagi Y., Kornberg R.D.. Mediator as a general transcription factor. J. Biol. Chem. 2006; 281:80–89. [DOI] [PubMed] [Google Scholar]
- 22. Thompson C.M., Young R.A.. General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:4587–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Andrau J.C., van de Pasch L., Lijnzaad P., Bijma T., Koerkamp M.G., van de Peppel J., Werner M., Holstege F.C.. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol. Cell. 2006; 22:179–192. [DOI] [PubMed] [Google Scholar]
- 24. Zhu X., Wiren M., Sinha I., Rasmussen N.N., Linder T., Holmberg S., Ekwall K., Gustafsson C.M.. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol. Cell. 2006; 22:169–178. [DOI] [PubMed] [Google Scholar]
- 25. Fan X., Chou D.M., Struhl K.. Activator-specific recruitment of Mediator in vivo. Nat. Struct. Mol. Biol. 2006; 13:117–120. [DOI] [PubMed] [Google Scholar]
- 26. Fan X., Struhl K.. Where does mediator bind in vivo. PLoS One. 2009; 4:e5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Myers L.C., Kornberg R.D.. Mediator of transcriptional regulation. Annu. Rev. Biochem. 2000; 69:729–749. [DOI] [PubMed] [Google Scholar]
- 28. Liu Y., Ranish J.A., Aebersold R., Hahn S.. Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. J. Biol. Chem. 2001; 276:7169–7175. [PubMed] [Google Scholar]
- 29. Zhang F., Sumibcay L., Hinnebusch A.G., Swanson M.J.. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol. Cell. Biol. 2004; 24:6871–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paul E., Zhu Z.I., Landsman D., Morse R.H.. Genome-wide association of mediator and RNA polymerase II in wild-type and mediator mutant yeast. Mol. Cell. Biol. 2015; 35:331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petrenko N., Jin Y., Wong K.H., Struhl K.. Mediator undergoes a compositional change during transcriptional activation. Mol. Cell. 2016; 64:443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malik S., Guermah M., Yuan C.X., Wu W., Yamamura S., Roeder R.G.. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol. Cell. Biol. 2004; 24:8244–8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taatjes D.J., Tjian R.. Structure and function of CRSP/Med2; a promoter-selective transcriptional coactivator complex. Mol. Cell. 2004; 14:675–683. [DOI] [PubMed] [Google Scholar]
- 34. Loven J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A.. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013; 153:320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pott S., Lieb J.D.. What are super-enhancers. Nat. Genet. 2015; 47:8–12. [DOI] [PubMed] [Google Scholar]
- 37. Gietz R.D., Schiestl R.H.. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007; 2:31–34. [DOI] [PubMed] [Google Scholar]
- 38. Piruat J.I., Chavez S., Aguilera A.. The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics. 1997; 147:1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sakai A., Shimizu Y., Kondou S., Chibazakura T., Hishinuma F.. Structure and molecular analysis of RGR1, a gene required for glucose repression of Saccharomyces cerevisiae. Mol. Cell. Biol. 1990; 10:4130–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Y., Bjorklund S., Kim Y.J., Kornberg R.D.. Yeast RNA polymerase II holoenzyme. Methods Enzymol. 1996; 273:172–175. [DOI] [PubMed] [Google Scholar]
- 41. Daugherty K.M., Goode B.L.. Functional surfaces on the p35/ARPC2 subunit of Arp2/3 complex required for cell growth, actin nucleation, and endocytosis. J. Biol. Chem. 2008; 283:16950–16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Svejstrup J.Q., Petrakis T.G., Fellows J.. Purification of elongating RNA polymerase II and other factors from yeast chromatin. Methods Enzymol. 2003; 371:491–498. [DOI] [PubMed] [Google Scholar]
- 43. Rappsilber J., Ishihama Y., Mann M.. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003; 75:663–670. [DOI] [PubMed] [Google Scholar]
- 44. Elfving N., Chereji R.V., Bharatula V., Bjorklund S., Morozov A.V., Broach J.R.. A dynamic interplay of nucleosome and Msn2 binding regulates kinetics of gene activation and repression following stress. Nucleic Acids Res. 2014; 42:5468–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fisher T.S., Taggart A.K., Zakian V.A.. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004; 11:1198–1205. [DOI] [PubMed] [Google Scholar]
- 46. Lefrancois P., Zheng W., Snyder M.. ChIP-Seq using high-throughput DNA sequencing for genome-wide identification of transcription factor binding sites. Methods Enzymol. 2010; 470:77–104. [DOI] [PubMed] [Google Scholar]
- 47. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Phanstiel D.H., Boyle A.P., Araya C.L., Snyder M.P.. Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics. 2014; 30:2808–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lopez-Serra L., Kelly G., Patel H., Stewart A., Uhlmann F.. The Scc2-Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome-free regions. Nat. Genet. 2014; 46:1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nagarajavel V., Iben J.R., Howard B.H., Maraia R.J., Clark D.J.. Global ‘bootprinting’ reveals the elastic architecture of the yeast TFIIIB-TFIIIC transcription complex in vivo. Nucleic Acids Res. 2013; 41:8135–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Paul E., Tirosh I., Lai W., Buck M.J., Palumbo M.J., Morse R.H.. Chromatin mediation of a transcriptional memory effect in yeast. G3 (Bethesda). 2015; 5:829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Eyboulet F., Wydau-Dematteis S., Eychenne T., Alibert O., Neil H., Boschiero C., Nevers M.C., Volland H., Cornu D., Redeker V. et al. . Mediator independently orchestrates multiple steps of preinitiation complex assembly in vivo. Nucleic Acids Res. 2015; 43:9214–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rhee H.S., Pugh B.F.. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012; 483:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hesselberth J.R., Chen X., Zhang Z., Sabo P.J., Sandstrom R., Reynolds A.P., Thurman R.E., Neph S., Kuehn M.S., Noble W.S. et al. . Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods. 2009; 6:283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jeronimo C., Langelier M.F., Bataille A.R., Pascal J.M., Pugh B.F., Robert F.. Tail and kinase modules differently regulate core mediator recruitment and function in vivo. Mol. Cell. 2016; 64:455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grunberg S., Henikoff S., Hahn S., Zentner G.E.. Mediator binding to UASs is broadly uncoupled from transcription and cooperative with TFIID recruitment to promoters. EMBO J. 2016; 35:2435–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Teytelman L., Thurtle D.M., Rine J., van Oudenaarden A.. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:18602–18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S. et al. . Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010; 467:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ebmeier C.C., Taatjes D.J.. Activator-Mediator binding regulates Mediator-cofactor interactions. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:11283–11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lupien M., Eeckhoute J., Meyer C.A., Wang Q., Zhang Y., Li W., Carroll J.S., Liu X.S., Brown M.. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008; 132:958–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang D., Garcia-Bassets I., Benner C., Li W., Su X., Zhou Y., Qiu J., Liu W., Kaikkonen M.U., Ohgi K.A. et al. . Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011; 474:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bourbon H.M., Aguilera A., Ansari A.Z., Asturias F.J., Berk A.J., Bjorklund S., Blackwell T.K., Borggrefe T., Carey M., Carlson M. et al. . A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell. 2004; 14:553–557. [DOI] [PubMed] [Google Scholar]
- 66. Tebbji F., Chen Y., Richard Albert J., Gunsalus K.T., Kumamoto C.A., Nantel A., Sellam A., Whiteway M.. A functional portrait of Med7 and the mediator complex in Candida albicans. PLoS Genet. 2014; 10:e1004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pollard T.D., Borisy G.G.. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003; 112:453–465. [DOI] [PubMed] [Google Scholar]
- 68. Yoo Y., Wu X., Guan J.L.. A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J. Biol. Chem. 2007; 282:7616–7623. [DOI] [PubMed] [Google Scholar]
- 69. Balciunas D., Hallberg M., Bjorklund S., Ronne H.. Functional interactions within yeast mediator and evidence of differential subunit modifications. J. Biol. Chem. 2003; 278:3831–3839. [DOI] [PubMed] [Google Scholar]
- 70. Willger S.D., Liu Z., Olarte R.A., Adamo M.E., Stajich J.E., Myers L.C., Kettenbach A.N., Hogan D.A.. Analysis of the Candida albicans phosphoproteome. Eukaryotic Cell. 2015; 14:474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dichtl B., Blank D., Ohnacker M., Friedlein A., Roeder D., Langen H., Keller W.. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell. 2002; 10:1139–1150. [DOI] [PubMed] [Google Scholar]
- 72. Krishnamurthy S., He X., Reyes-Reyes M., Moore C., Hampsey M.. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol. Cell. 2004; 14:387–394. [DOI] [PubMed] [Google Scholar]
- 73. Pappas D.L. Jr, Hampsey M.. Functional interaction between Ssu72 and the Rpb2 subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000; 20:8343–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tan-Wong S.M., Zaugg J.B., Camblong J., Xu Z., Zhang D.W., Mischo H.E., Ansari A.Z., Luscombe N.M., Steinmetz L.M., Proudfoot N.J.. Gene loops enhance transcriptional directionality. Science. 2012; 338:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ansari A., Hampsey M.. A role for the CPF 3΄-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005; 19:2969–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wu W.H., Pinto I., Chen B.S., Hampsey M.. Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics. 1999; 153:643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kessler M.M., Zhao J., Moore C.L.. Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. Separation into two components that are required for both cleavage and polyadenylation of mRNA 3΄ ends. J. Biol. Chem. 1996; 271:27167–27175. [DOI] [PubMed] [Google Scholar]
- 78. El Kaderi B., Medler S., Raghunayakula S., Ansari A.. Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J. Biol. Chem. 2009; 284:25015–25025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Medler S., Al Husini N., Raghunayakula S., Mukundan B., Aldea A., Ansari A.. Evidence for a complex of transcription factor IIB with poly(A) polymerase and cleavage factor 1 subunits required for gene looping. J. Biol. Chem. 2011; 286:33709–33718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012; 191:671–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fromont-Racine M., Rain J.C., Legrain P.. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 1997; 16:277–282. [DOI] [PubMed] [Google Scholar]
- 82. Haimovich G., Medina D.A., Causse S.Z., Garber M., Millan-Zambrano G., Barkai O., Chavez S., Perez-Ortin J.E., Darzacq X., Choder M.. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013; 153:1000–1011. [DOI] [PubMed] [Google Scholar]
- 83. Handoko L., Xu H., Li G., Ngan C.Y., Chew E., Schnapp M., Lee C.W., Ye C., Ping J.L., Mulawadi F. et al. . CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011; 43:630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Phillips J.E., Corces V.G.. CTCF: master weaver of the genome. Cell. 2009; 137:1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sanyal A., Lajoie B.R., Jain G., Dekker J.. The long-range interaction landscape of gene promoters. Nature. 2012; 489:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Brown J.M., Green J., das Neves R.P., Wallace H.A., Smith A.J., Hughes J., Gray N., Taylor S., Wood W.G., Higgs D.R. et al. . Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J. Cell Biol. 2008; 182:1083–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fraser P., Bickmore W.. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007; 447:413–417. [DOI] [PubMed] [Google Scholar]
- 88. Iborra F.J., Pombo A., Jackson D.A., Cook P.R.. Active RNA polymerases are localized within discrete transcription ‘factories’ in human nuclei. J. Cell Sci. 1996; 109:1427–1436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.