Abstract
The 5′ UTR of Coxsackievirus B3 (CVB3) contains internal ribosome entry site (IRES), which allows cap-independent translation of the viral RNA and a 5′-terminal cloverleaf structure that regulates viral replication, translation and stability. Here, we demonstrate that host protein PSF (PTB associated splicing factor) interacts with the cloverleaf RNA as well as the IRES element. PSF was found to be an important IRES trans acting factor (ITAF) for efficient translation of CVB3 RNA. Interestingly, cytoplasmic abundance of PSF protein increased during CVB3 infection and this is regulated by phosphorylation status at two different amino acid positions. Further, PSF protein was up-regulated in CVB3 infection. The expression of CVB3–2A protease alone could also induce increased PSF protein levels. Furthermore, we observed the presence of an IRES element in the 5′UTR of PSF mRNA, which is activated during CVB3 infection and might contribute to the elevated levels of PSF. It appears that PSF IRES is also positively regulated by PTB, which is known to regulate CVB3 IRES. Taken together, the results suggest for the first time a novel mechanism of regulations of ITAFs during viral infection, where an ITAF undergoes IRES mediated translation, sustaining its protein levels under condition of translation shut-off.
INTRODUCTION
Coxsackievirus B3 (CVB3) is an enterovirus and is the most common causative agent for virus induced myocarditis. CVB3 is also known to cause infection in the central nervous system and the pancreas causing meningitis and pancreatitis respectively. Enteroviruses like poliovirus, CVB3 and EV71, have type-1 IRES (1). These IRES are dependent on both canonical translation initiation factors as well as IRES trans acting factors (ITAFs) derived from the host for translation initiation. A complex of host and viral proteins assembles at the 5′UTR to regulate viral replication and translation either positively or negatively (2). Several proteins like PCBP-2, La, PTB, FBP-1 and Srp20 have been reported to interact with the IRES elements of picornaviruses and enhance translation (3–8); while proteins like FBP-2 and AUF-1 bind to the viral 5′UTR and inhibit translation (9,10). These results suggest that outcome of the viral infection depends on the kind of proteins present in the RNP complex assembled at the UTRs. Remodelling of the RNP complex occurs when the virus switches from translation to replication. PCBP-2 plays a central role in both translation and replication while hnRNP K helps only in replication through interactions with the IRES as well as the terminal cloverleaf region of the viral RNA (11). Thus, host proteins interacting with the viral RNA play a critical role in regulating the balance between viral RNA translation and replication.
In the 5′UTR, these host proteins interact with the initial 1–100 nucleotide region in the 5′UTR called the cloverleaf RNA which is a key regulatory region for viral RNA translation, replication and stability (12–14). For example, a complex containing the host protein PCBP-2 and viral protein 3CD is assembled at the cloverleaf RNA and influences viral replication and translation (15). Host protein interactions with the cloverleaf RNA and IRES region plays an important regulatory role in viral positive strand and negative strand synthesis, modulation of the IRES activity and stability of viral genome.
Due to known multifunctional roles of the cloverleaf RNA we aimed to identify the complete host protein profile interacting with the cloverleaf RNA of Coxsackievirus B3 RNA. Using the RNA affinity chromatography approach, we identified 11 host proteins interacting with the cloverleaf RNA. Among these proteins, PTB Associated Splicing Factor (PSF) was further characterized. We demonstrate here that PSF interacts with both the cloverleaf RNA and the IRES and plays a pivotal role in viral RNA translation. It appears that direct interaction of PSF with the IRES is important for viral RNA translation. Interestingly, PSF protein levels in the cell increased upon CVB3 infection, though host mRNA translation is shut off due to cleavage of eIF4G1 by viral protease 2A during CVB3 infection. However, many host proteins still undergo translation during viral infection by unknown mechanisms. Expression of 2A protease induced PSF protein levels in cell. Interestingly, we find that an IRES element is present in the 5′UTR of PSF mRNA. A well characterized viral and cellular ITAF, PTB activates PSF IRES by direct interaction. This is interesting as the cytoplasmic abundance of PTB increases in CVB3 infection, where it could act as ITAF for both cellular mRNAs and viral RNA. Our results suggest involvement of PTB in regulating viral RNA translation by yet another mechanism, acting as an ITAF for an ITAF PSF in the context of CVB3 replication.
MATERIALS AND METHODS
Plasmids and RNAs
CVB3 replicon RNA prepared from pRibCB3-T7-luc vector (kind gift from Prof. Frank van Kuppeveld) was used for viral replication/translation studies.
pCD-CVB3 1–100 and pCD-CVB3 Δ100 constructs were generated from pRibCB3-T7-luc and were used for in vitro transcription to prepare cloverleaf RNA and Δ100 RNA. α-32P uridine triphosphate (Board of Radiation and Isotope Technology, India) body labelled RNA probes were prepared using T7 RNA polymerase enzyme (Promega) and used for UV cross-linking reactions. For preparing non-specific RNAs used in competition UV-crosslinking assays, pGEMt-easy vector was linearized with Sal I enzyme and this was used for in vitro transcription by T7 RNA polymerase. Biotin-11-UTP labelled 1–100nt RNAs were prepared and used for RNA affinity chromatography. pET15b-PSF vector, described earlier (16), was used to express PSF protein, while pET28a-PTB, described earlier (17) was used to express PTB protein.
In the current study, null bicistronic and p53 bicistronic constructs were used as described earlier (18). PSF bicistronic construct was prepared by replacing the p53 5′UTR with PSF 5′UTR in the p53 bicistronic construct. To prepare capped RNAs, all bicistronic constructs were linearized by PmeI enzyme (NEB) and capped RNAs were in vitro prepared using Ribomax (Promega). For generating shRNA targeting PSF, pSUPER vector system was utilized. Targeting sequence utilized is as follows: 5′ GAAGAAGCCTTTAGCCAAT 3′. The non-specific shRNA construct used in this study was described earlier (19). PSF–RFP construct, previously described (20), was a kind gift from Prof. Yossih Shiloh, at Tel-Aviv University. In the same construct, mutations were incorporated to generate Y293F, T687A, Y293D and T687D mutants.
Cell lines, transfections and luciferase assay
HeLaS3 cells, maintained in DMEM (Sigma) containing 10% serum (GIBCO, Invitrogen, Life Technologies) were used for all experiments. For viral replication/translation studies, CVB3 Replicon RNA was transfected in OptiMEM (Invitrogen, Life Technologies) using Lipofectamine-2000 (Invitrogen, Life Technologies). Four hours post-transfection medium was replaced by DMEM containing 10% serum. Cells were harvested at 10 h post-transfection and processed for luciferase assay using dual luciferase reporter assay system (Promega) or in trizol (Sigma) for total RNA isolation using manufacturer's protocol. For partial silencing experiments, cells were transfected with indicated amount of siPSF (Dharmacon) or non-specific siRNA (Dharmacon) or lipofectamine alone, 24 h prior to CVB3 RNA transfection. Luciferase assays was performed using Luciferase assay system (Promega) or Dual luciferase Reagent (DLR, Promega) as per manufacturer's protocol. siRNA sequences used in the pool of siRNAs against PSF are as follows: 5′ GAAGAAGCCTTTAGCCAAT 3′, 5′ GGCAAAGGATTCGGATTTA 3′ and 5′ TGTTCAAGTGGTTCCACAATGACTG 3′.
shRNA constructs were transfected in HeLa S3 cells 24 h prior to either replicon RNA transfection or CVB3 infection. For a 24-well plate, 250 ng per well shRNA plasmid was transfected.
RNA Affinity chromatography and LC/MS analysis
HeLa S3 cells were used to prepare S10 extracts essentially as described in (21). Biotin labelled RNA was incubated with the S10 extracts in RNA binding buffer (1.2 mM MgCl2, 1.65 mM ATP, 30 mM KCl, 0.02% Tween20, 10 mM Tris (pH 7.5), 10 μg/ml tRNA, 10 μg/ml heparin and 5 units of RNasin (Promega)) and Streptavidin coated magna beads (Thermo Scientific) were added to the reaction mixture. RNA–protein complexes bound to magna beads were separated using magnetic separator followed by washes and proteins were eluted at various salt concentrations (300 mM NaCl and 600 mM NaCl). Eluted fractions were run on 12% SDS gel and stained with colloidal coomassie. The lanes were cut and processed for LC/MS. Analysis of the LC/MS data was carried out at C-CAMP, NCBS, Bangalore. For LC/MS analysis, the peptide sequences were matched with protein sequences in SwissProt and TrEMBL databases. Identified proteins, significance score, peptides and query coverage is reported in Supplementary Table S2.
UV-induced crosslinking
α-32P labelled RNA probes were incubated with purified recombinant protein in 1× RNA binding buffer [25 mM HEPES (pH 7.6), 125 mM KCl, 10 mM MgCl2, 19% glycerol, 0.5 mM EDTA, 1.25 mM ATP and 2 mM GTP] for 30 min at 30°C and the RNA–protein complexes were cross-linked in presence of UV light of 254 nm wavelength at 4°C for 20 min followed by RNase A digestion of unprotected RNAs at 37°C. Complexes were resolved by SDS-PAGE and visualized by phosphor-imaging. Indicated amount of molar excess of un-labelled RNAs were added to reaction mixture along with labelled RNA probe for competitive UV-crosslinking assays.
Protein purification
Recombinant PSF protein was purified from pET15b-PSF construct. The protocol followed for purification of PSF was essentially as described before (16). Recombinant HuR protein was expressed using pET28a-HuR construct and purified essentially as described as before (22). Recombinant PTB protein was expressed using pET28a-PTB construct and purified essentially as described as before (17).
Western blot analysis
Cell lysates collected in Passive lysis buffer were quantified using Bradford reagent (Bio-Rad) and equal amounts of protein were resolved on SDS-PAGE. Separated proteins were transferred on to nitrocellulose membrane (Pall Biosciences) and analysed using different antibodies. Rabbit raised anti-PSF polyclonal antibody (Santa Cruz Biotechnology), mouse monoclonal anti-PTB antibody (Calnexin), rabbit raised anti-La antibody were used to analyze the samples followed by HRP conjugated secondary antibody (goat raised anti-rabbit and goat raised anti-mouse, Sigma). HRP conjugated monoclonal anti-β-Actin antibody (Sigma) was used to probe β-Actin protein. Protein antibody complexes were analysed by chemiluminescence using Immobilon Western systems (EMD Millipore).
In vitro transcription and translation
RNAs corresponding to different domains of 5′UTR were prepared using templates prepared from T7 promoter sequence tagged primers and reverse primers listed in Supplementary Table S1. DNA templates were resolved on gel and eluted out. Individual RNAs were prepared using T7 polymerase enzyme (Fermentas) as per manufacturer's protocol. In each reaction 50 U of RNAse inhibitor (Promega) was included.
In vitro translation was carried out using Rabbit Reticulocyte Lysate (Promega) supplemented with 10% (v/v) HeLa S10 extract. 1 μg of CVB3 replicon RNA was used per reaction, incubated at 30°C for 90 min and luciferase activity was measured after the reaction using Luciferase assay system (Promega). In some reactions indicated amounts of anti-PTB antibody (Calnexin), anti-PSF antibody (Santa Cruz), anti-rabbit IgG antibody or recombinant PTB and recombinant PSF protein were also included.
RNA immuno-precipitation (RIP)
CVB3 Replicon RNA transfected cells were harvested 10 hours post transfection in lysis buffer containing 100 mM KCl, 5 mM MgCl2, 10 mM HEPES pH 7.0, 0.05% NP-40, 1 mM DTT and 100 U/ml RNasin. Cells were centrifuged and supernatants were quantified and equal amounts of proteins were incubated with PSF antibody saturated Protein G beads, IgG antibody saturated Protein G beads and only Protein G beads overnight. RNP complexes bound to beads were centrifuged, re-suspended in buffer and an aliquot was separated for western blotting. To the remaining lysate, 0.1% SDS and 30 μg Proteinase K was added and incubated at 50°C for 30 min. To this three times volume of TRI reagent was added and RNA was prepared and subjected to RT-PCR and PCR using CVB3 specific primers.
RNA isolation and qPCR
Total cellular RNA was isolated using trizol reagent (Sigma) as per manufacturer's protocol. cDNAs were synthesized from this RNA using gene specific reverse primers and Revert-aid RT enzyme (Fermentas). qPCR was carried out using DyNAmo qPCR kit as per manufacturer's protocol in which 2 μl of cDNA was added per reaction.
For CVB3 positive strand and negative strand determination, strand specific reverse transcription was performed using reverse primer for positive strand RNA and forward primers for negative strand RNA. Following primers sequences were used in the current study. CVB3-F: 5′ GAATGCGGCTAATCCTAACTGC 3′; CVB3-R: 5′ GCTCTATTAGTCACCGGATGGC 3′; for PSF mRNA, PSF-F: 5′ TTGAACGATGCAGTGAAGGTG 3′; PSF-R: 5′ ATACATTGGATTCTTCTGGGC 3′.
Virus titre determination
To determine the copy number of the virus, absolute qPCR approach was used as described by Ertel et al. (23). Standard curve of copy number (log2 scale) and Ct values is presented in Supplementary Figure S4A. To determine virus titre from cells (control or knockdown), RNA was isolated from the supernatant followed by qPCR for positive strand RNA. Absolute copy numbers were determined by comparing the Ct values in the standard curve.
Indirect immunofluorescence
HeLa S3 cells were grown on coverslips for 16 h after which cells were subjected to mock transfection or CVB3 Replicon RNA transfection. At different time-points after transfection, cells were processed for confocal microscopy. Cells were washed with ice cold PBS and fixed using 4% PFA at room temperature for 15 min. 0.1% triton X-100 solution was used to make them permeable and subsequently blocked overnight with 3% BSA. Further, cells were incubated with anti-PSF antibody for 4 h followed by Alexa-488 conjugated anti-rabbit secondary antibody. Nuclei were stained and coverslips were mounted on slides with Prolong Gold Antifade with DAPI (Invitrogen) and images were taken using Zeiss microscope. Image analysis was done using LSM Image Browser software (Zeiss) and Image quantification was carried out using with ImageJ software.
Northern blotting
Total RNA was isolated from HeLa S3 cells transfected with PSF bicistronic RNA and resolved on 0.8% formaldehyde agarose gel. RNAs were blotted onto nitrocellulose membrane by capillary electrophoresis method, followed by probing with α-32P labelled oligos targeting F luc gene.
RESULTS
Profile of host proteins interacting with the CVB3 cloverleaf RNA
In order to identify host proteins interacting with the CVB3 cloverleaf RNA, biotin labelled cloverleaf RNAs were incubated with HeLa S3 S10 extract prepared from cells transfected with CVB3 replicon RNA. RNA protein complexes were pulled down using streptavidin beads and resolved on SDS-10%PAGE (Supplementary Figure S1A). Proteins interacting with cloverleaf RNA were identified by LC/MS and are tabulated in Table 1 and Supplementary Table S2. Beads alone were used as control. To eliminate the bead bound proteins, only the unique proteins present in the cloverleaf RNA pulldown were further considered. As expected, PCBP-2 which is a well characterized protein known to interact with the enteroviral cloverleaf RNA also showed up in the screen. PTB, another well characterized host factor required for viral IRES mediated translation was found to interact with the cloverleaf RNA. PTB associated splicing factor (PSF) was found to be interacting with cloverleaf RNA in our screen. PSF is known to interact with the 5′UTR of cellular mRNAs having IRES and regulate its translation (p53 and c-myc IRES) (16,24). PSF was also identified to interact with poliovirus 5′UTR in a cell based thiouracil cross-linking mass spectrometry approach, the role of PSF in viral life cycle was not studied (24). It was interesting to observe that PSF showed up in our screen with the cloverleaf RNA alone. Interaction of other proteins (PTB and HuR) with cloverleaf was validated by UV-crosslinking assay using radio-labelled cloverleaf RNA (Supplementary Figure S1B). Further, interactions of HuR and PTB proteins were found to be specific to cloverleaf RNA (Supplementary Figure S1C and S1D).
Table 1. Host proteins interacting with the CVB3 cloverleaf RNA.
| Accession | Description | Interacting region | Virus | References |
|---|---|---|---|---|
| P26599 | Polypyrimidine tract-binding protein 1(PTB) | IRES | CVB3, PV | (5,8) |
| F8VXH9 | Poly(rC)-binding protein 2 | CL, IRES | PV, CVB3 | (6,15) |
| E7EQG2 | Eukaryotic initiation factor 4A-II | IRES | PV | (41) |
| P23246 | PTB associated splicing factor (PSF) | 5′UTR | PV | (42) |
| Q15717 | ELAV-like protein 1 (HuR) | — | — | — |
| Q533SS8 | Poly(RC) binding protein 1 | CL, IRES | PV | (43) |
| P61978 | Heterogeneous nuclear ribonucleoprotein K | CL, IRES | EV71 | (11) |
| Q96AE4 | Far upstream element-binding protein 1 | IRES, Linker | EV71 | (44) |
| Q92945 | Far upstream element-binding protein 2 | IRES | EV71 | (9) |
| Q86U42 | Polyadenylate-binding protein 2 | — | — | — |
| Q9PW24 | Human antigen A- HuA | — | — | — |
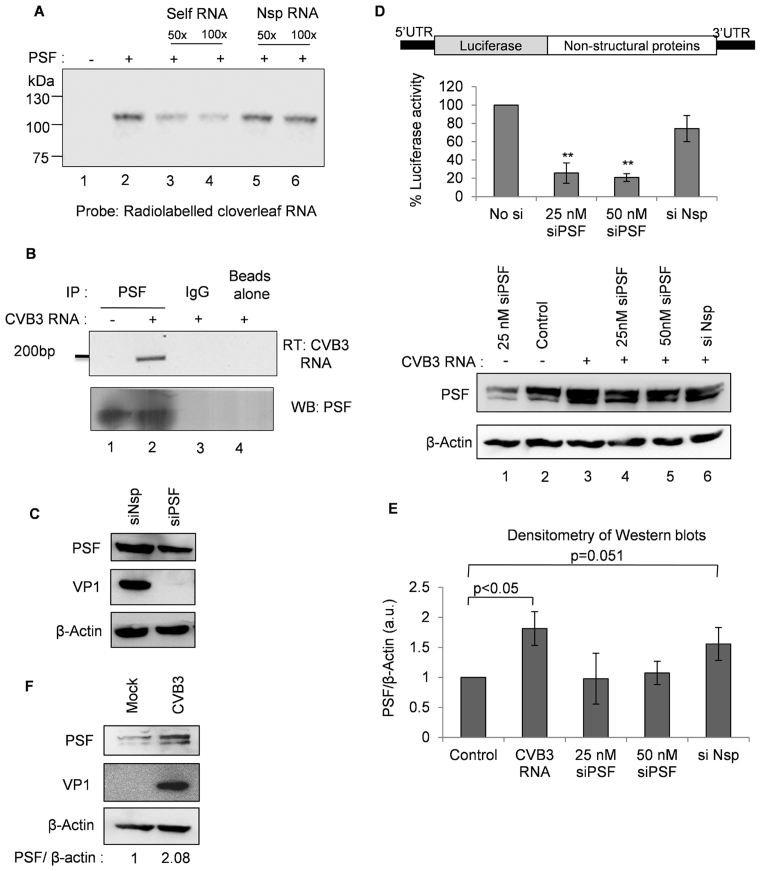
PSF interaction with cloverleaf RNA was further validated through UV cross-linking experiment using recombinant PSF and radio-labelled cloverleaf RNA in the presence of un-labelled competitor RNAs. Results indicate specific interaction between PSF and CVB3 cloverleaf RNA (Figure 1A). Further, this interaction was confirmed in cells by immuno-precipitation of PSF containing RNP complexes followed by RT-PCR for the viral RNA (Figure 1B). To investigate the role of PSF in CVB3 life cycle, PSF was knockdown using siRNAs and viral protein levels were checked by western blot analysis at 8 h post-infection. A drastic reduction in the levels of viral protein VP1 was observed, indicating PSF is important for viral life cycle (Figure 1C, Supplementary Figure S2). Similar experiment was repeated by transfecting CVB3 sub-genomic replicon RNA into the cells. In CVB3 replicon RNA, P1 in viral genome is replaced by firefly luciferase gene, as described earlier (25) (Figure 1D) and the efficiency of viral life cycle can be measured by luciferase activity. Upon partial silencing of PSF, luciferase activity was found to be decreased significantly, consistent with the viral infection experiment (Figure 1D). This suggests that PSF might play a role in viral replication or translation and not in viral entry or assembly as replicon RNA is directly transfected into the cells. Interestingly, we found higher levels of PSF when CVB3 replicon RNA was transfected (lane 3 as compared to lane 2 in Figure 1D). The densitometric analysis of three independent western blots of HeLa S3 cell lysates indicates that PSF levels are significantly higher in presence of CVB3 RNA (Figure 1E). PSF levels were found to be increased during CVB3 infection as well (Figure 1F). Interestingly, we have also observed that PSF knockdown with si-PSF was not very effective in presence of CVB3 infection due to concomitant increase in PSF level (Supplementary Figure S3).
Figure 1.
PSF interacts with cloverleaf RNA and influences CVB3 lifecycle. (A) UV cross-linking assay using recombinant PSF and radio-labelled cloverleaf RNA. Lane 1 corresponds to no protein and only RNA probe and lane 2 shows interaction between RNA and recombinant PSF protein. For competition, 50- and 100-fold molar excess of unlabelled cloverleaf RNA (lanes 3 and 4) or non-specific RNA (lanes 5 and 6) was also included in the reaction. Non-specific RNA of equivalent length was derived from pGEM-T vector MCS region. (B) CVB3 replicon RNA was transfected in HeLa S3 cells, followed by immuno-precipitation of PSF using anti-PSF antibody or IgG isotype antibody (as a control). RNA was purified from the immuno-precipitated complexes and viral positive strand RNA was detected by semi-quantitative PCR. (C) HeLa S3 cells were either transfected with siPSF or siNsp, and 24 h later, CVB3 infection was carried out. Cells were harvested at 8 h post-infection and lysates were subjected to immunoblot analysis. (D) Schematic of CVB3 sub-genomic replicon RNA adapted from Lanke et al. (25).Hela S3 cells were either transfected with siPSF or siNsp, and 24 h later CVB3 sub-genomic replicon RNA was transfected. Cells were harvested at 10 h post-transfection and firefly luciferase activity was measured. Error bars represent standard deviation in three independent experiments. **P value < 0.01. PSF protein levels as determined by western blotting are indicated compared to β-actin protein levels. (E) Densitometry of three independent western blots from (D) are represented in graphical format. Error bars represent standard deviation in three independent experiments. (F) HeLa S3 cells were infected with CVB3 and cells were harvested at 8 h post-infection. Western blot indicating the PSF protein level in presence and absence of CVB3 infection. PSF levels are quantified by densitometric analysis of band intensities and are represented by fold change in PSF/β-actin.
PSF positively influences CVB3 IRES mediated translation
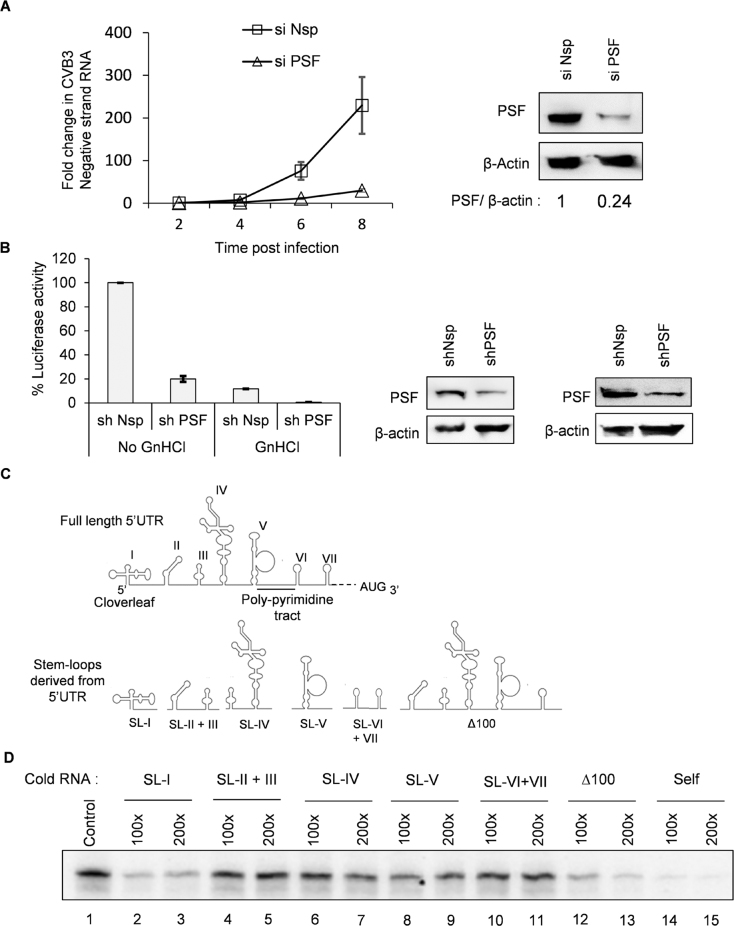
To understand which step in the viral life cycle is influenced by PSF, CVB3 replication was studied in PSF knockdown cells (using siRNAs) as compared to non-specific siRNA transfected cells by measuring negative strand accumulation in cells at different time points using qPCR. CVB3 negative strand synthesis was found to be reduced in siPSF cells as compared to non-specific siRNA transfected cells (Figure 2A). shPSF transfected cells also led to reduced viral titer in the supernatant, as compared to shNsp transfected cells (Supplementary Figure S4A and B). To study the role of PSF in viral RNA translation, CVB3 replication was inhibited by a well characterized inhibitor GnHCl and CVB3 IRES activity was measured in non-specific shRNA transfected or shPSF transfected cells. Since inhibition of viral replication leads to synthesis of very less amount of viral protein, we utilized CVB3 sub-genomic replicon RNA with luciferase reporter for this purpose. Upon siPSF treatment, luciferase activity was further reduced even in presence of GnHCl, indicating that PSF is required for viral RNA translation (Figure 2B).
Figure 2.
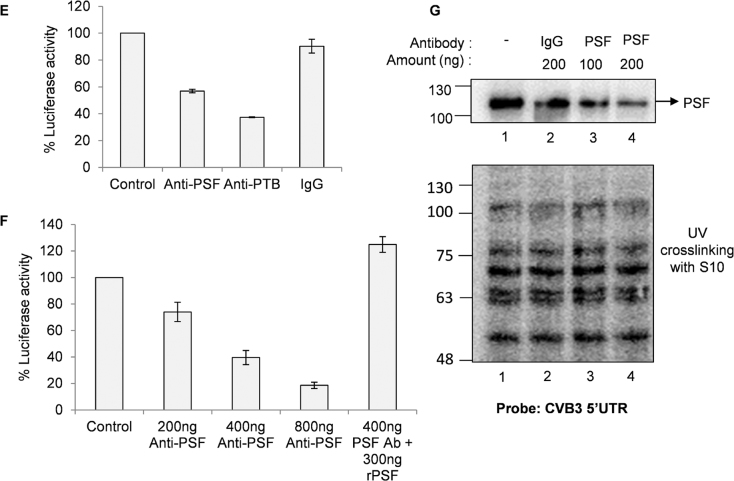
Role of PSF in CVB3 replication and translation. (A) HeLa S3 cells, priorly transfected with siNsp or siPSF, were infected with CVB3 and cells were harvested in trizol at indicated time points post-infection. At each time point, RNA was isolated and negative strand RNA was detected by qPCR. Fold change in negative strand RNA level was calculated in both control cells and PSF knockdown cells and is indicated in the graph. Error bars indicate standard deviation in three independent experiments. (B) Effect of partial knockdown of PSF on CVB3 translation was studied in presence and absence of viral replication inhibitor GnHCl (concentration = 2 mM), using CVB3 sub-genomic replicon RNA as described earlier. Luciferase activity was found to be significantly reduced even in presence of GnHCl. ** indicates P < 0.005. Error bars indicate the standard deviation in three independent experiments. PSF and β-actin protein levels are indicated in the immunoblots. (C) Schematic diagram of CVB3 5′UTR. Different stem-loops derived from the full length 5′UTR for competition UV induced cross-linking assay are indicated. Cloverleaf RNA is indicated by SL-I. (D) UV-crosslinking assay with 32-P labelled full length 5′UTR of CVB3 genomic RNA alone or in presence of indicated un-labelled stem-loop RNAs. Δ1–100 indicates the entire CVB3 5′UTR without the cloverleaf RNA or SL-1. (E) in vitro translation of CVB3 replicon RNA carried out using rabbit reticulocyte system supplemented with S10 extract from HeLa S3 cells and resultant luciferase activities are indicated. Anti-PSF and anti-PTB antibodies were added in the reaction to sequester these proteins. Anti-Rabbit IgG antibody was used as control. Error bars indicate standard deviation in three independent experiments. (F) In vitro translation carried out using rabbit reticulocyte system supplemented with S10 extract from HeLa S3 cells in presence of indicated amounts of anti-PSF antibody. Indicated amount of bacterially expressed recombinant PSF (rPSF) was supplemented in one of the reactions to rescue the inhibitory effects of PSF antibody. Error bars indicate standard deviation in three independent experiments. (G) UV cross-linking assay carried out using radio-labelled CVB3 5′UTR with either recombinant PSF protein or S10 extract from HeLa S3 cells (lane 1). The assay was carried out in presence of PSF antibody (lanes 3 and 4) or IgG antibody (lane 2) as indicated.
Since PSF is involved in CVB3 translation, we investigated the interaction of PSF with CVB3 IRES. For this purpose, un-labelled RNAs corresponding to different stem–loops in 5′UTR were designed (Figure 2C). The secondary structure of the individual stem loops was found to be similar to the full length as observed by prediction of secondary structure using mfold server (data not shown). These un-labelled RNAs were used as competitors with the full length radio-labelled 5′UTR in UV-crosslinking experiments. Results indicate strongest interaction with the cloverleaf RNA (SL-I) and moderate interaction with stem–loop 4 and 5 (SL-IV and V) within the IRES element (lanes 6–9, Figure 2D). A stronger interaction of PSF with full IRES is also observed in Δ1–100 cold RNA which contains both stem–loop IV and V, implying that PSF binding site might be distributed in stem–loop IV and V (lanes 12 and 13, Figure 2D). Hence, PSF can interact with both cloverleaf RNA as well as IRES region. To study the consequence of this interaction, in vitro translation studies using rabbit reticulocyte lysate (RRL) that are supplemented with HeLa S10 extracts were carried out. HeLa S10 extracts are known to provide the necessary factors required by viral IRES for translation. This system is well established to study in vitro translation of picornavirus IRES (26–28). To confirm that direct interaction of PSF with IRES is important for translation, CVB3 IRES mediated translation was studied in vitro in presence of anti-PSF polyclonal antibody (Figure 2E). PTB antibody was used as a positive control and rabbit IgG antibody was used as a non-specific antibody. CVB3 translation was reduced in presence of anti-PSF antibody, while no significant reduction was observed in non-specific antibody. Further, dose dependent reduction in CVB3 IRES activity was observed and this was rescued by supplementation of recombinant PSF protein (Figure 2F). PSF interaction with CVB3 5′UTR was found to be inhibited in presence of PSF antibody, while the global profile of host proteins interacting with 5′UTR was not affected (Figure 2G). These results suggest that PSF interaction to viral RNA is involved in regulating CVB3 IRES activity.
Regulation of CVB3 IRES mediated translation by PSF could be due to the interaction of PSF with CVB3 IRES or cloverleaf RNA or both. To address this question, we utilized the mono-cistronic constructs with full length 5′UTR or cloverleaf-deleted 5′UTR and measured the IRES activity in presence of PSF knockdown condition. In both the full length 5′UTR and cloverleaf deleted 5′UTR constructs, IRES activity was found to be reduced upon knockdown of PSF, suggesting that PSF-cloverleaf interaction might have independent function in CVB3 life-cycle (Supplementary Figure S5A). Role of PSF in cap-dependent translation and CVB3 IRES mediated translation was studied using pRL-CMV construct and CVB3 mono-cistronic RNA respectively. Upon knockdown of PSF using shRNA, CVB3 IRES mediated translation, as measured by Fluc activity, was reduced and cap-dependent translation, as measured by R luc activity, remained unaffected (Supplementary Figure S5B).
Cytoplasmic abundance of PSF increases in presence of CVB3
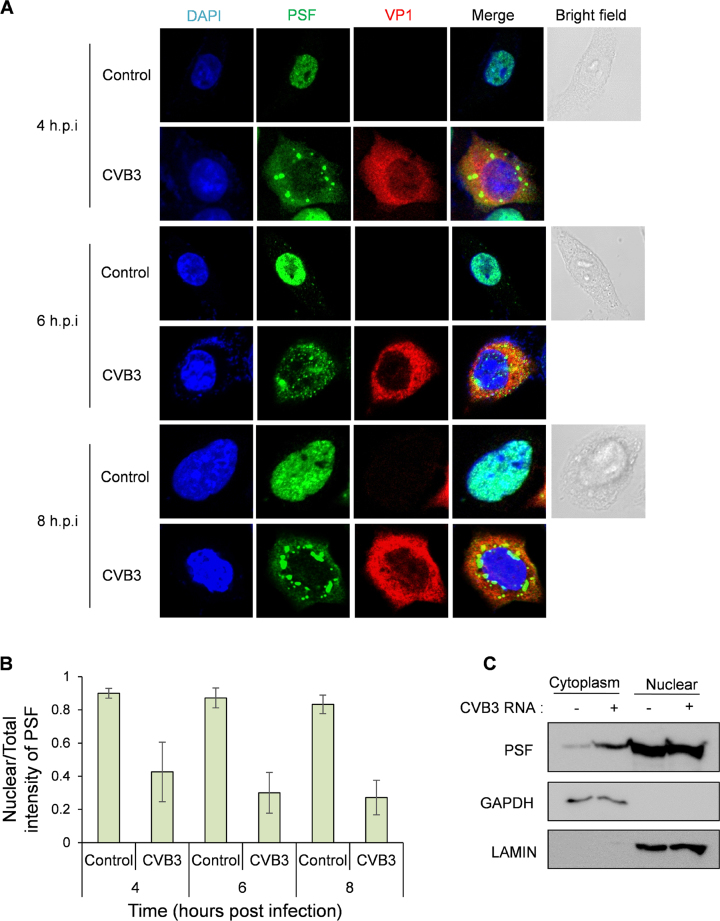
One of the important hallmarks of an IRES transacting factor (ITAF) is its relocalization from nucleus to cytoplasm. Since enteroviral replication and translation occurs in the cytoplasm and PSF being predominantly a nuclear protein, its localization in the cell upon viral infection was investigated by immunofluorescence in HeLa S3 cells. Increase in the cytoplasmic abundance was observed as early as 4 h post-infection in CVB3 infected cells as compared to uninfected cells (Figure 3A). To quantify the increase in cytoplasmic abundance, ratio of intensity of PSF in the nucleus and in total cell is represented in Figure 3B. In CVB3 infection, the nuclear/total intensity of PSF reduced indicating its increase in cytoplasmic abundance (Figure 3B).These results were also confirmed by sub-cellular fractionation, where abundance of PSF was found to be increased in the cytoplasmic fraction (Figure 3C). Interestingly, PSF formed distinct puncta in the cytoplasm. These could be aggresomes formed in the cytoplasm due to high amount of intrinsically disordered proteins in the cytoplasm. In fact, GlobPlot analysis showed that PSF is an intrinsically disordered protein (Supplementary Figure S6) (29).
Figure 3. Cytoplasmic abundance of PSF increases in presence of CVB3. (A) Immunofluorescence microscopy of HeLa S3 cells infected with CVB3 or mock infection at different time points post-infection. Green represents PSF, Red represents viral protein VP1 indicating infected cells, blue represents nucleus of the cell. (B) Quantification of the confocal images. Mean intensities of PSF were calculated in total cell and in nucleus using ImageJ software. Ratio of nuclear intensity by total cell intensity is indicated in graph. For 4 h.p.i., 6 h.p.i. and 8 h.p.i. 20, 25 and 40 cells were quantified from three independent experiments. Error bars represent standard deviation in three independent experiments. (C) Western blot analysis of nuclear and cytoplasmic extracts of HeLa S3 cells with and without CVB3 infection. Cells were harvested at 8 hours post infection processed for fractionation. Lamin and GAPDH were used as nuclear and cytoplasmic markers respectively.
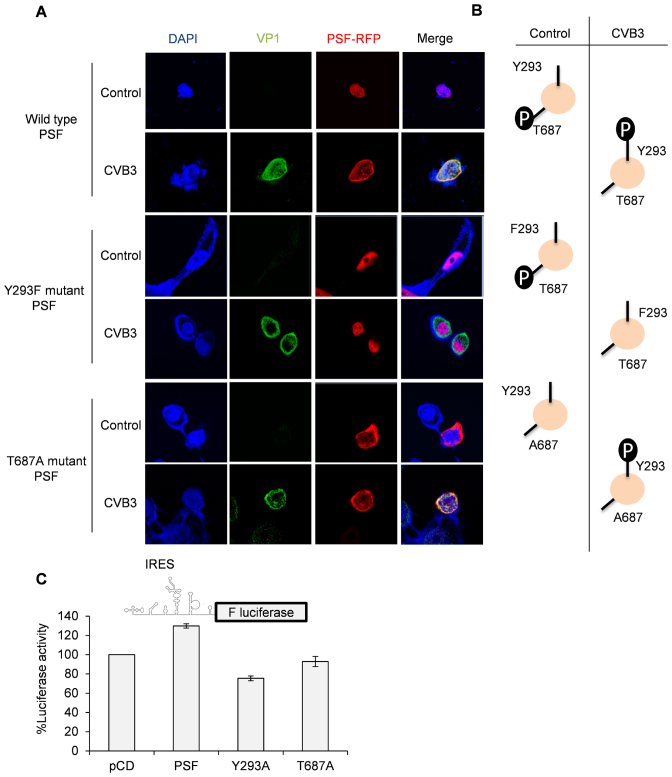
Regulation of PSF localization by phosphorylation upon CVB3 infection
Phosphorylation dependent regulation of PSF localization has been studied earlier in the context of cancer. It is known that PSF is phosphorylated by anaplastic lymphoma kinase (ALK) at Y293 position and this causes relocalization of PSF from nucleus to cytoplasm (30). PSF is also known to be phosphorylated by GSK3 at T687 position upon T-cell activation, but its consequence on PSF localization is not known (31). However, the latter report suggest that T687 phosphorylated PSF interacts with TRAP150, a nuclear protein, and T687A mutant does not interact with TRAP150, suggesting that T687 phosphorylation might have a role in PSF localization. We investigated the role of Y293 and T687 phosphorylation on PSF localization upon CVB3 infection by generating phosphodead mutants in RFP fused PSF expressing construct as described earlier (20). As observed in Figure 4A, wild type PSF relocalizes to the cytoplasm upon CVB3 infection while Y293F phosphodead mutant remained in nucleus in both control cells and CVB3 infected cells. T687A phosphodead mutant was relocalized to the cytoplasm in both control cells as well as upon CVB3 infection. T687D phosphomimic mutant showed exactly opposite phenotype, confirming the phosphorylation dependent regulation of PSF relocalization in CVB3 infection (Supplementary Figure S7). These data suggests that PSF is phosphorylated at T687 position and dephosphorylated at Y293 position in uninfected cells and upon CVB3 infection both phosphorylation of PSF at Y293 position and dephosphorylation at T687 position is required for nuclear to cytoplasmic relocalization (Figure 4B). Next, we checked the effect of over expression of wild-type PSF protein, Y293F mutant and T687A mutant proteins on CVB3 IRES activity using CVB3 monocistronic RNA (Figure 4C). Wild type PSF was able to stimulate CVB3 IRES mediated translation, consistent with earlier results. Interestingly, Y293F mutant that is retained in nucleus inhibited the CVB3 IRES activity. This could be because PSF is known to form dimers and Y293 mutant could form dimers with the endogenous PSF protein, reducing availability of PSF to CVB3 IRES. Interestingly, T687 mutant did not show any change in CVB3 IRES activity and currently we do not have any explanation for this. Y293 phosphorylation has been shown to enhance RNA binding activity of PSF (30). It is possible that T687 phosphorylation could also modulate RNA binding activity of PSF to some extent and hence overexpression of the mutant PSF (T687A) could not stimulate CVB3 IRES activity like the wild type.
Figure 4.
Regulation of PSF localization by phosphorylation. (A) Plasmids harbouring PSF gene fused with RFP were used for immunofluorescence assay. Mutations were incorporated individually in the same plasmid to generate Y293F and T687A phosphodead mutants. HeLa cells were transfected with plasmids expressing wild type PSF fused with RFP or the phosphodead mutants (as described before) and 24 h later cells were infected with CVB3. Eight hours post-infection; cells were processed for immunofluorescence microscopy. (B) A schematic summary of the information obtained from (A). Phosphorylation status of wild type PSF and phosphodead mutants in control cells and upon CVB3 infection is indicated. (C) Luciferase assay carried out using CVB3 monocistronic construct as represented by the schematic. HeLa cells were transfected with plasmids expressing either wild type PSF or Y293F or T687A phosphodead mutants. Twenty four hours post-transfection, cells were transfected with CVB3 monocistronic RNAs and 8 hours post transfection cells were processed for luciferase assay. Graphs indicates the % luciferase activity as compared to pCD vector control. Error bars indicates the standard deviation in three independent experiments.
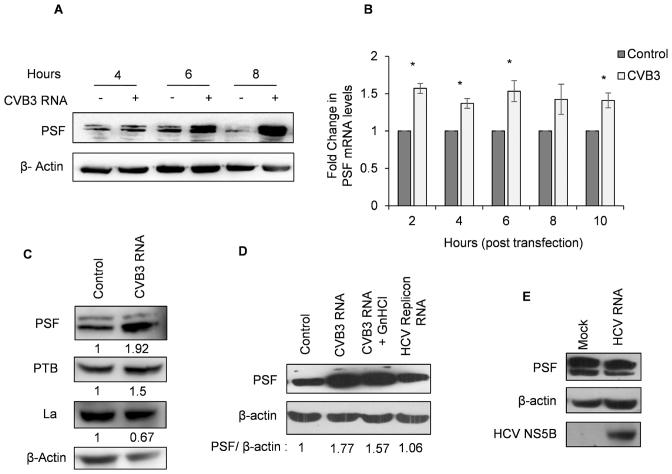
PSF is up-regulated in CVB3 infection
As observed in Figure 1E and F, PSF protein level increased upon CVB3 infection. PSF protein level was found to be increased only at 6 h post-transfection of CVB3 replicon RNA and remained in high level till 8 h (Figure 5A).To understand how PSF is induced in CVB3 infection, PSF mRNA level was determined at different time points post transfection. Marginal but significant increase in mRNA levels was observed as early as 2 h post-transfection (Figure 5B). We investigated whether other known host proteins that are important for CVB3 life cycle are also induced in the presence of CVB3 replicon RNA. Interestingly, PSF and PTB levels were increased in CVB3 infection and La protein which is known to influence CVB3 translation by interacting with the 5′UTR of CVB3 RNA, did not show up-regulation in presence of CVB3 replicon RNA (Figure 5C). In the context of inhibition of CVB3 replication using 2 mM GnHCl, PSF levels were found to be high (Figure 5D), indicating viral genomic RNA or viral proteins are sufficient to induce PSF levels and viral replication event may not be required to cause increase in PSF levels. PSF protein level was not induced during HCV infection (Figure 5E) or during HCV sub-genomic replicon RNA transfection (Figure 5D, lane 4) suggesting that PSF up-regulation could be specific to enterovirus infection.
Figure 5. PSF is specifically up-regulated in CVB3 infection. (A) PSF levels at different time points post CVB3 infection in HeLa S3 cells. (B) PSF mRNA levels at indicated time points post CVB3 RNA transfection as determined by qPCR. At each time point, mRNA level in un-transfected cells were used as control. * indicates P < 0.05. (C) Western blots of HeLa S3 cells transfected with CVB3 replicon RNA indicating levels of PSF, PTB and La. Cells were harvested at 10 h post-transfection. (D) Western blots of HeLa S3 cells transfected with CVB3 replicon RNA in presence or absence of 2 mM GnHCL and HCV replicon RNA. Cells were harvested at 10 h post-transfection. Densitometry of the represented western blot is indicated as PSF/β-actin below the lanes. (E) Western blot to check the levels of PSF upon HCV infection. Huh 7.5 cells were infected with HCV and cells were harvested at 48 h post-infection. NS5B blot is to indicate viral infection.
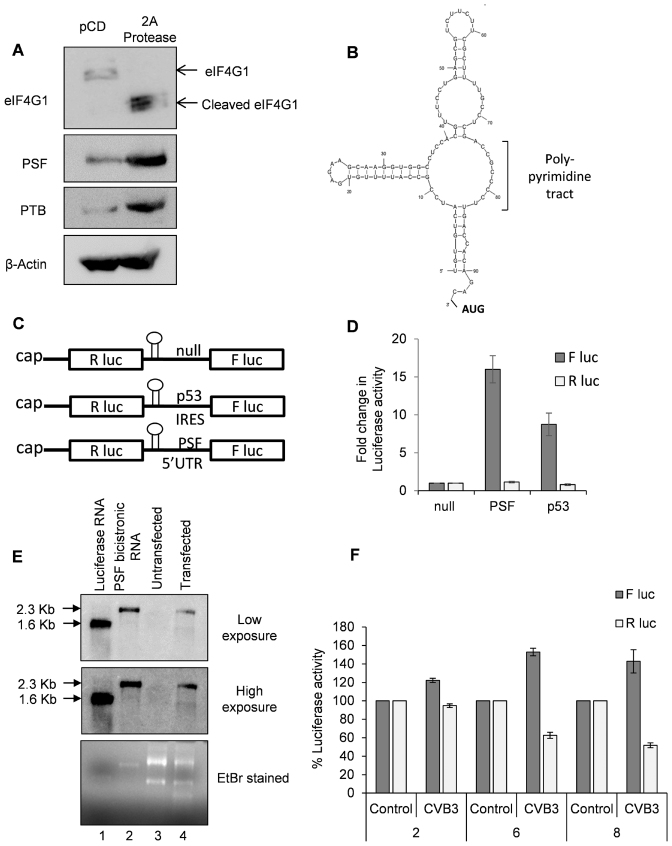
5′UTR of PSF mRNA contains a functional IRES
It is known that cap-dependent translation is reduced during enterovirus infection due to cleavage of eIF4G1 and down-regulation of eIF4E (1,32). Since PSF is up-regulated in CVB3 infection, we investigated the levels of PSF upon 2A protease expression, which inhibits host cell cap-dependent translation. Interestingly, both PSF and PTB were found to be up-regulated upon 2A protease expression (Figure 6A). We further investigated to determine whether PSF mRNA undergoes non-canonical IRES mediated translation or not. 5′UTR structure of PSF mRNA was predicted using mfold server (Figure 6B). The predicted structure showed a hairpin loop followed by poly-pyrimidine stretch, which is a hallmark of PTB dependent IRES (33). To investigate the presence of IRES element in PSF mRNA, the 5′UTR of PSF mRNA was cloned in a bicistronic vector as described earlier (18). A null bicistronic construct, where La ORF sequence was placed up-stream to the firefly luciferase, was used as negative control and p53 IRES was used as positive control. An upstream hairpin loop was introduced just before 5′UTR to prevent initiation by ribosomes that scan through stop codon by leaky scanning (Figure 6C). Capped RNAs were prepared from these constructs and these RNAs were transfected in cell. RNA transfection was chosen over DNA transfection to rule out the possibility of cryptic promoter activity due to which two different RNAs could be formed. Significantly higher amount of Firefly luciferase activity was observed from PSF 5′UTR compared to null bicistronic control (Figure 6D), indicating presence of possible IRES in the 5′UTR of PSF. To further confirm that the capped RNAs are intact in the cells, PSF bicistronic RNAs were transfected in cells and the cells were harvested 8 h post-transfection in trizol for RNA isolation. Total RNA (2 μg) from untransfected cells and PSF bicistronic RNA transfected cells was run on formaldehyde agarose gel along with luciferase control RNA and in vitro transcribed PSF bicistronic RNA and blotted on nitrocellulose membrane followed by probing of firefly luciferase gene to detect the bicistronic RNAs. As observed in Figure 6E, PSF bicistronic RNAs were found to be intact in cells. Similar observation was made by using RT-PCR method where forward and reverse primers encompassing the R luc gene and F luc gene with PSF 5′UTR in between were used, as described earlier (18). A single band of expected length was observed in the PCR amplification indicating the integrity of PSF bicistronic RNA in cell (Supplementary Figure S8). Next, PSF IRES activity in presence of CVB3 infection was studied. Bicistronic construct with PSF IRES was transfected into cells and 24 hours post transfection cell were infected with CVB3 or mock. At 2, 6 and 8 h post-infection, cells were processed for luciferase assay. PSF IRES activity (represented by F luc activity) increased in CVB3 infected cells (as compared to mock) and cap-dependent translation (represented by R luc activity) was reduced as expected (Figure 6F).
Figure 6.
PSF mRNA undergoes IRES mediated translation. (A) Western blots indicating the levels of PSF and PTB upon expression of 2A protease. Both PSF and PTB appear to be up-regulated upon 2A protease expression. 2A protease activity is represented by the cleavage of eIF4G1. (B) Structure of PSF 5′UTR as predicted by mfold server. The presence of hairpin loop followed by poly-pyrimidine tract suggests a potential PTB dependent IRES (33). (C) Schematic of different bicistronic construct used in this study. (D) Capped RNAs were prepared from the constructs described in Figure 5C. These capped RNAs were transfected in cells and cells were harvested 8 h post-transfection, followed by dual luciferase assay. Fold change in luciferase activities from PSF 5′UTR and p53 5′UTR (positive control) compared to La ORF as negative control (null) is represented as firefly luciferase activity while renilla luciferase activity represents the cap-dependent translation. (E) Integrity of capped PSF bicistronic RNA in the cell as assessed by northern blotting. Cells were transfected with in vitro transcribed capped PSF bicistronic RNA and 8 h post-transfection cells were harvested in trizol and RNA was isolated. 2 μg of total RNA from untransfected and transfected cells was used for formaldehyde agarose gel electrophoresis (lanes 3 and 4) along with luciferase RNA (lane 1) and in vitro transcribed PSF bicistronic RNA (lane 2). RNAs were transferred to nitrocellulose membrane by capillary transfer method and the membrane was probed for luciferase gene using complementary radio-labelled primer. Two different exposures after northern blotting are indicated, along with the ethidium bromide stained agarose gel. In both the exposures, PSF bicistronic RNA was found to be intact in the cell with negligible degradation. (F) PSF IRES activity upon CVB3 infection was measured. HeLa S3 cells previously transfected with PSF bicistronic construct were infected with CVB3 or mock. Cells were harvested at 2, 6 and 8 h.p.i. and luciferase activity was measured. Error bars indicate the standard deviation in three independent experiments.
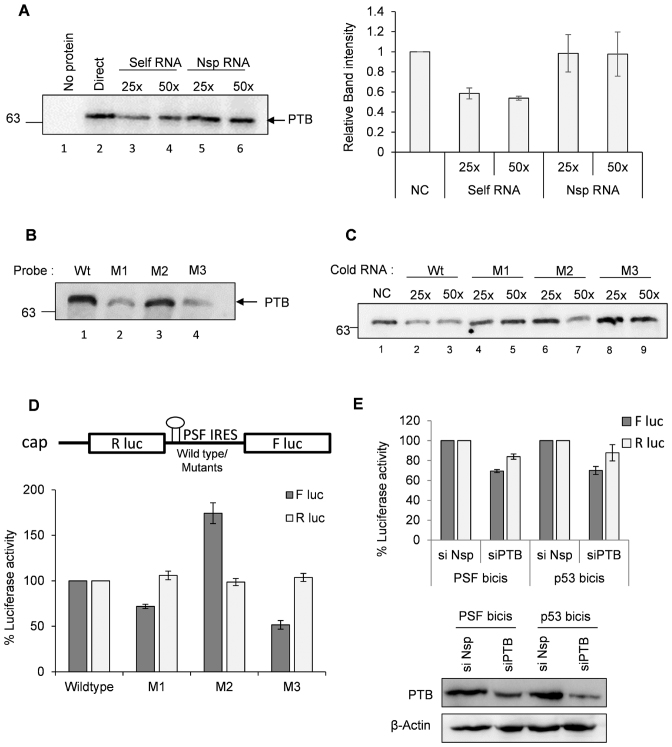
PSF IRES is PTB dependent
PSF IRES contains a poly-pyrimidine tract where PTB could interact. Further, we know that PTB relocalizes from nucleus to the cytoplasm during CVB3 infection, hence its cytoplasmic function could be enhanced. To investigate whether PTB interacts with PSF IRES, UV-crosslinking assay was used where recombinant PTB was incubated with radio-labelled PSF IRES RNA in the presence of un-labelled PSF IRES as competitor RNA or un-labelled non-specific RNA of approximately equal length as non-specific competitor RNA. PTB was found to be interacting with PSF IRES specifically (Figure 7A). The graph represents the densitometry of three independent UV-crosslinking assays. To disrupt the PTB binding on PSF IRES we chose three different pyrimidine rich sites. Two of them were present in the loop region, CCU→GGA at 69–71 nucleotide position (mutant M1) and CCC→GGG at 78–80 nucleotide position (mutant M2). One mutation was present in the stem region at CUU→GAA at 63–65 nucleotide position (mutant M3) (Supplementary Figure S9A). The mfold structures of the mutants are represented in Supplementary Figure S9B–D which shows minor structural changes due to mutation. Interaction of PTB with these mutants was assessed by UV-crosslinking assay using radio-labelled wild type and mutant RNAs and recombinant PTB. Wild type PSF 5′UTR was able to interact with PTB and this interaction was abrogated in M1 and M3 mutants (Figure 7B), however M2 mutant did not show any significant change. Similar observation was found in the competition UV-crosslinking assay where M1 and M3 mutants were not able to compete for interaction with wild type RNA, suggesting abrogated interaction with PTB (Figure 7C). Bicistronic construct harbouring the mutations along with the wild type bicistronic construct were transfected in cells to measure the efficiency of IRES mediated translation ex vivo. Consistent with the binding results, M1 and M3 showed reduced IRES activity as the PTB binding is affected (Figure 7D). Of note, M2 mutant which did not show any change in PTB binding as compared to wild-type showed increased IRES activity. This could be due to change in structure which might make it more efficient IRES or attract other host factors. Upon partial silencing of PTB, PSF IRES (wild type) activity was moderately reduced in cells (Figure 7E). Together these results suggest that PSF IRES is PTB dependent. This is interesting as cytoplasmic abundance of PTB increases during enterovirus infection and also during 2A protease expression (34), possibly enhancing the cytoplasmic functions of PTB, like IRES mediated translation.
Figure 7.
PTB interacts with PSF IRES and positively influences its IRES activity. (A) UV-crosslinking assay carried out with recombinant PTB with radio-labelled PSF IRES in presence of un-labelled PSF IRES or non-specific RNA. Graph represents the average intensities of bands visible in the autoradiograph of UV-crosslinking gel from 3 independent experiments. (B) UV-crosslinking assay carried out using recombinant PTB with wild type and mutant radio-labelled PSF IRESs. M1, M2 and M3 mutants were generated as represented in Supplementary Figure S6A. (C) Competition UV crosslinking assay carried out using radio-labelled wild type PSF 5′UTR with recombinant PTB, in presence of un-labelled wild type PSF IRES or mutant PSF IRESs as indicated. (D) Ex vivo IRES activity of wild type and mutant PSF IRESs. Capped RNAs were prepared from bicistronic constructs harbouring mutations in PSF IRES and wild type PSF IRES and were transfected in the cells. Cells were harvested at 8 h post-transfection and firefly luciferase activity indicating the IRES activity of wild type and mutant RNAs and renilla luciferase activity indicating cap-dependent translation are indicated. M1 and M3 mutants showed lower IRES activity as compared to wild type. * indicates P value < 0.05. Error bars indicate standard deviation from three independent experiments. (E) Effect of knockdown of PTB on PSF IRES activity. p53 IRES activity is used as positive control. Cells previously transfected with si-PTB or si-NSP were transfected with the PSF IRES. Luciferase assay was carried out after harvesting the cells in PLB buffer at 8 hours post transfection. * indicates P value < 0.05.
DISCUSSION
In this study, ‘PTB associated splicing factor ‘(PSF) protein was found to be interacting with the cloverleaf RNA, as determined by RNA affinity chromatography followed by LC-MS. PTB is well known IRES trans acting factor for both cellular and viral IRES (8,17,35,36) and hence it was interesting to identify PSF in our screen. In vitro and cell culture interaction studies confirmed direct interaction of PSF protein with CVB3 RNA. Previous reports suggest that PSF is an ITAF for cellular c-myc and p53 mRNAs (16,24), but its role in enteroviral life cycle remains unknown. Host proteins interacting with the viral 5′UTR can be either positive or negative regulators of viral lifecycle, influencing viral replication, translation or both. By uncoupling viral replication and translation using replication inhibitor GnHCl, we have shown that PSF is important for viral RNA translation. To confirm that direct interaction of PSF with viral RNA is important for viral RNA translation, polyclonal antibody against PSF was used to block this interaction in the in vitro translation experiments. A dose dependent reduction in translation was observed, which was successfully rescued upon addition of recombinant PSF. Results clearly indicate that PSF is a positive regulator of CVB3 translation and direct interaction of PSF to the 5′UTR is essential for CVB3 IRES activity.
Knockdown of PSF also reduced viral replication as observed by negative strand accumulation. Defective protein synthesis could lead to lower levels of viral proteins which could in turn affect the replication initiation, and delay the replication kinetics. PSF could also directly participate in the replication process as it interacts with the cloverleaf RNA.Since PSF is involved in viral RNA translation, we investigated whether PSF directly interacts with IRES. PSF was found to be interacting with the IRES region in vitro. Specifically, PSF interaction was observed with domain 4 and domain 5 in the 5′UTR while cloverleaf RNA showed strongest interaction. These results indicate PSF to be a novel IRES trans acting factor for CVB3 IRES. Earlier studies have identified PCBP-2 and hnRNP K in Poliovirus and EV71 respectively to interact with both cloverleaf RNA and IRES. Both these proteins are positive regulators of viral replication, while PCBP-2 is regulator of viral RNA translation as well (11,14). PSF also interacts with both cloverleaf RNA and IRES and this interaction is important for viral RNA translation. However, the consequence of PSF and cloverleaf RNA interaction on viral replication is not clear at this stage and remains to be further elucidated.
PSF is a nuclear protein and CVB3 translation and replication occurs in the cytoplasm. Upon CVB3 infection, PSF abundance in the cytoplasm increases as early as 4 hour post infection. Further, PSF localization was found to be regulated by phosphorylation at Y293 amino acid position and dephosphorylation at T687 amino acid position. Interestingly, GSK3 is known to phosphorylate PSF at T687 amino acid position (30). GSK3 is activated upon CVB3 infection during earlier time points and reaches basal level at 4 hour post CVB3 infection (37). In light of these reports, GSK3 inactivation during CVB3 infection and the subsequent dephosphorylation of PSF protein at T687 could lead to increase in cytoplasmic abundance of PSF. It would be interesting to identify the cognate kinase and phosphatase that are involved in regulation of PSF phosphorylation upon CVB3 infection.
Interestingly, PSF protein was found to be up-regulated in CVB3 replicon harbouring cells which was supported by the observation of up-regulation in PSF RNA levels. PSF up-regulation was not observed in case of HCV infection or HCV replicon RNA transfection suggesting that host response or the viral strategy could be different in case of CVB3 and related viruses. Of note, another ITAF, La protein, did not show up-regulation upon CVB3 infection, but PTB showed up-regulation similar to PSF, suggesting differential regulation of ITAFs during CVB3 infection. Earlier reports showed that many of the IRES trans acting factors like PABP-2, PCBP-2 and PTB are cleaved upon enterovirus infection by viral proteinases (38–40). This further illustrates the complexity in regulation of ITAFs during CVB3 infection. Since PSF is a positive regulator of viral RNA translation, induction in PSF levels might be beneficial for viral RNA translation where other ITAFs are cleaved by viral proteinases.
It is known that cap-dependent translation is shut off in case of enterovirus infection due to the cleavage of eIF4G by viral 2A protease and it has been proposed that cell switches from cap-dependent translation to cap-independent translation mode (32). In this study we observe that expression of 2A protease alone induced both PSF and PTB protein levels. Since PSF levels increased in CVB3 infection, we carried out investigation to determine if PSF mRNA has cap-independent mechanism to initiate translation. Indeed, IRES activity from PSF 5′UTR was detected and upon CVB3 infection, PSF IRES activity is induced. Further, PSF IRES was found to be PTB dependent. Interestingly, PTB is relocalized from nucleus to the cytoplasm upon enterovirus infection as well as expression of 2A protease (34). The increase in the cytoplasmic abundance of PTB upon infection might be responsible for the IRES-mediated translation initiation of PSF mRNA. This might add to the transcriptional up-regulation in PSF mRNA levels and the combined effect leads to higher PSF levels during CVB3 translation.
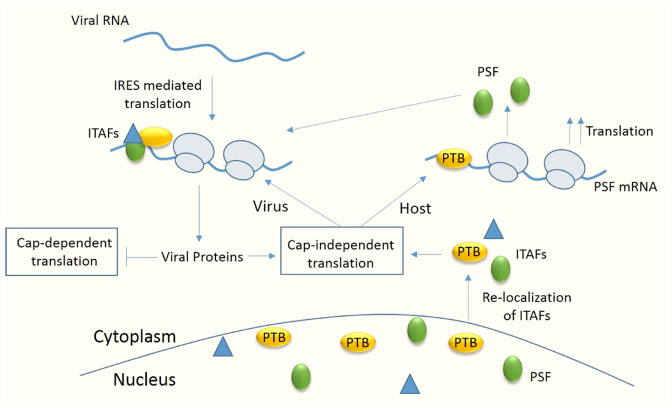
Together, our results suggest that PSF is a novel host factor for CVB3, an enterovirus. Upon viral infection, viral genomic RNA undergoes IRES mediated translation with the help of canonical translation initiation factors as well as ITAFs. This leads to production of viral proteins which further induces shut-off in host cap-dependent translation by cleavage of eIF4G1. However, 2A protease activity also leads to relocalization of ITAFs like PTB and SRp20 which leads to increase in the cytoplasmic abundance of these proteins, favouring the viral IRES mediated transaction. Additionally, we report for the first time the presence of PTB dependent IRES on PSF 5′UTR. Relocalization of other ITAFs like PTB, from nucleus to cytoplasm during viral infection favour IRES mediated translation of PSF mRNA, increasing PSF protein levels in the cell. This further helps in CVB3 IRES mediated translation (Figure 8). With the available knowledge from the literature and with the current study, it appears that there are two important mechanisms that are exploited by virus to their advantage during shut down of cap-dependent translation. First, viral infection induces relocalization of nuclear proteins to cytoplasm which increases the availability of the protein for viral RNA translation. Second, ITAFs like PSF also undergo IRES mediated translation and are able to sustain their levels in cell. The results highlight the complex interplay between the host factors orchestrated by the viruses to their advantage.
Figure 8.
Model showing the regulation of host cell translation under CVB3 infection. Upon entry of CVB3 RNA in cells, host factors known as ITAFs relocalize from nucleus to cytoplasm and these proteins stimulate viral IRES mediated translation. Viral proteins inhibit cap-dependent translation and favour cap-independent translation. ITAFs like PSF also have an IRES in their mRNA and are stimulated by PTB, also an ITAF for viral RNA. Thus, level of PSF in cell is maintained even under condition where cap-dependent translation is shut off and this is beneficial for viral IRES mediated translation.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to the Mass Spectrometry facilities at C-camp, NCBS, Bangalore and Molecular Biophysics Unit, IISc. Prof. Frank van Kuppeveld and Nora Chapman are acknowledged for various constructs. BSL3 facility at Centre for Infectious Disease, IISc is acknowledged. Prof. Yossih Shiloh, Tel-Aviv University, is acknowledged for providing PSF-RFP construct. We thank Prof. Umesh Varshney, Department of Microbiology and Cell Biology, Indian Institute of Science, and his lab members, Riyaz Ahmad Shah and V Rajagopal, for the help in northern blotting experiment. We are grateful to S. Shwetha, Stanford University, for critical reading of the manuscript and useful suggestions. DST Fund for Improvement of Science and Technology Infrastructure (FIST) level II infrastructure and University Grants Commission Centre of Advanced Studies support to the department is acknowledged.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology (DBT), Government of India; Council of Scientific and Industrial Research (CSIR) (to P.D.); D.S. Kothari fellowship from UGC, Government of India (to B.G.); J.C. Bose fellowship from Department of Science and Technology (DST), Govt. of India (to S.D.). Funding for open access charge: Research grant from Department of Biotechnology, Govt. of India. This work was also supported by the Department of Biotechnology, New Delhi through its partnership program with the Indian Institute of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jackson R.J., Hellen C.U.T., Pestova T.V.. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010; 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andino R., Rieckhof G.E., Baltimore D.. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990; 63:369–380. [DOI] [PubMed] [Google Scholar]
- 3. Ray P.S., Das S.. La autoantigen is required for the internal ribosome entry site-mediated translation of Coxsackievirus B3 RNA. Nucleic Acids Res. 2002; 30:4500–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bedard K.M., Daijogo S., Semler B.L.. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007; 26:459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Verma B., Bhattacharyya S., Das S.. Polypyrimidine tract-binding protein interacts with coxsackievirus B3 RNA and influences its translation. J. Gen. Virol. 2010; 91:1245–1255. [DOI] [PubMed] [Google Scholar]
- 6. Walter B.L., Nguyen J.H., Ehrenfeld E., Semler B.L.. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999; 5:1570–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Verma B., Ponnuswamy A., Gnanasundram S.V., Das S.. Cryptic AUG is important for 48S ribosomal assembly during internal initiation of translation of coxsackievirus B3 RNA. J. Gen. Virol. 2011; 92:2310–2319. [DOI] [PubMed] [Google Scholar]
- 8. Hunt S.L., Jackson R.J.. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999; 5:344–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin J.-Y., Li M.-L., Shih S.-R.. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 2009; 37:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cathcart A.L., Rozovics J.M., Semler B.L.. Cellular mRNA decay protein AUF1 negatively regulates enterovirus and human rhinovirus infections. J. Virol. 2013; 87:10423–10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shih S.-R., Stollar V., Li M.-L.. Host factors in enterovirus 71 replication. J. Virol. 2011; 85:9658–9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vogt D.A., Andino R.. An RNA element at the 5′-end of the poliovirus genome functions as a general promoter for RNA synthesis. PLoS Pathog. 2010; 6:e1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murray K.E., Roberts A.W., Barton D.J.. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA. 2001; 7:1126–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kempf B.J., Barton D.J.. Poly(rC) binding proteins and the 5′ cloverleaf of uncapped poliovirus mRNA function during de novo assembly of polysomes. J. Virol. 2008; 82:5835–5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parsley T.B., Towner J.S., Blyn L.B., Ehrenfeld E., Semler B.L.. Poly (rC) binding protein 2 forms a ternary complex with the 5΄-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997; 3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 16. Sharathchandra A., Lal R., Khan D., Das S.. Annexin A2 and PSF proteins interact with p53 IRES and regulate translation of p53 mRNA. RNA Biol. 2012; 9:1429–1439. [DOI] [PubMed] [Google Scholar]
- 17. Grover R., Ray P.S., Das S.. Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle. 2008; 7:2189–2198. [DOI] [PubMed] [Google Scholar]
- 18. Ray P.S., Grover R., Das S.. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 2006; 7:404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Subramanian N., Mani P., Roy S., Gnanasundram S.V., Sarkar D.P., Das S.. Targeted delivery of hepatitis C virus-specific short hairpin RNA in mouse liver using Sendai virosomes. J. Gen. Virol. 2009; 90:1812–1819. [DOI] [PubMed] [Google Scholar]
- 20. Salton M., Lerenthal Y., Wang S.-Y., Chen D.J., Shiloh Y.. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010; 9:1568–1576. [DOI] [PubMed] [Google Scholar]
- 21. Walter B.L., Parsley T.B., Ehrenfeld E., Semler B.L.. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 2002; 76:12008–12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shwetha S., Kumar A., Mullick R., Vasudevan D., Mukherjee N., Das S.. HuR displaces polypyrimidine tract binding protein to facilitate La binding to the 3′untranslated region and enhances hepatitis C virus replication. J. Virol. 2015; 89:11356–11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ertel K.J., Brunner J.E., Semler B.L.. Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative-strand RNA intermediates. J. Virol. 2010; 84:4229–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cobbold L.C., Spriggs K.A., Haines S.J., Dobbyn H.C., Hayes C., de Moor C.H., Lilley K.S., Bushell M., Willis A.E.. Identification of internal ribosome ntry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell. Biol. 2008; 28:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lanke K.H.W., van der Schaar H.M., Belov G.A., Feng Q., Duijsings D., Jackson C.L., Ehrenfeld E., van Kuppeveld F.J.M.. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J. Virol. 2009; 83:11940–11949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sweeney T.R., Abaeva I.S., Pestova T.V., Hellen C.U.T.. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J. 2014; 33:76–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jünemann C., Song Y., Bassili G., Goergen D., Henke J., Niepmann M.. Picornavirus internal ribosome entry site elements can stimulate translation of upstream genes. J. Biol. Chem. 2007; 282:132–141. [DOI] [PubMed] [Google Scholar]
- 28. de Breyne S., Yu Y., Pestova T.V., Hellen C.U.T.. Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site. RNA. 2008; 14:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Linding R., Russell R.B., Neduva V., Gibson T.J.. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003; 31:3701–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Galietta A., Gunby R.H., Redaelli S., Stano P., Carniti C., Bachi A., Tucker P.W., Tartari C.J., Huang C.-J., Colombo E. et al. . NPM/ALK binds and phosphorylates the RNA/DNA-binding protein PSF in anaplastic large-cell lymphoma. Blood. 2007; 110:2600–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heyd F., Lynch K.W.. Phosphorylation-dependent regulation of PSF by GSK3 controls CD45 alternative splicing. Mol. Cell. 2010; 40:126–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ho B.-C., Yu S.-L., Chen J.J.W., Chang S.-Y., Yan B.-S., Hong Q.-S., Singh S., Kao C.-L., Chen H.-Y., Su K.-Y. et al. . Enterovirus-Induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe. 2011; 9:58–69. [DOI] [PubMed] [Google Scholar]
- 33. Mitchell S.A., Spriggs K.A., Bushell M., Evans J.R., Stoneley M., Le Quesne J.P.C., Spriggs R.V., Willis A.E.. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev. 2005; 19:1556–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Álvarez E., Castelló A., Carrasco L., Izquierdo J.M.. Poliovirus 2A protease triggers a selective nucleo-cytoplasmic redistribution of splicing factors to regulate alternative pre-mRNA splicing. PLoS ONE. 2013; 8:e73723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pickering B.M., Mitchell S.A., Evans J.R., Willis A.E.. Polypyrimidine tract binding protein and poly r(C) binding protein 1 interact with the BAG‐1 IRES and stimulate its activity in vitro and in vivo. Nucleic Acids Res. 2003; 31:639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schepens B., Tinton S.A., Bruynooghe Y., Beyaert R., Cornelis S.. The polypyrimidine tract-binding protein stimulates HIF-1α IRES-mediated translation during hypoxia. Nucleic Acids Res. 2005; 33:6884–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yuan J., Zhang J., Wong B.W., Si X., Wong J., Yang D., Luo H.. Inhibition of glycogen synthase kinase 3[beta] suppresses coxsackievirus-induced cytopathic effect and apoptosis via stabilization of [beta]-catenin. Cell Death Differ. 2005; 12:1097–1106. [DOI] [PubMed] [Google Scholar]
- 38. Bonderoff J.M., LaRey J.L., Lloyd R.E.. Cleavage of poly(A)-binding protein by poliovirus 3C proteinase inhibits viral internal ribosome entry site-mediated translation. J. Virol. 2008; 82:9389–9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perera R., Daijogo S., Walter B.L., Nguyen J.H.C., Semler B.L.. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J. Virol. 2007; 81:8919–8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Back S.H., Kim Y.K., Kim W.J., Cho S., Oh H.R., Kim J.-E., Jang S.K.. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro). J. Virol. 2002; 76:2529–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Belsham G.J., Sonenberg N.. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 1996; 60:499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lenarcic E.M., Landry D.M., Greco T.M., Cristea I.M., Thompson S.R.. Thiouracil cross-linking mass spectrometry: a cell-based method to identify host factors involved in viral amplification. J. Virol. 2013; 87:8697–8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sean P., Nguyen J.H.C., Semler B.L.. The linker domain of poly(rC) binding protein 2 is a major determinant in poliovirus cap-independent translation. Virology. 2008; 378:243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang P.-N., Lin J.-Y., Locker N., Kung Y.-A., Hung C.-T., Lin J.-Y., Huang H.-I., Li M.-L., Shih S.-R.. Far upstream element binding protein 1 binds the internal ribosomal entry site of enterovirus 71 and enhances viral translation and viral growth. Nucleic Acids Res. 2011; 39:9633–9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.