Abstract
CRISPR–Cas system provides the adaptive immunity against invading genetic elements in prokaryotes. Recently, we demonstrated that Csa3a regulator mediates spacer acquisition in Sulfolobus islandicus by activating the expression of Type I-A adaptation cas genes. However, links between the activation of spacer adaptation and CRISPR transcription/processing, and the requirement for DNA repair genes during spacer acquisition remained poorly understood. Here, we demonstrated that de novo spacer acquisition required Csa1, Cas1, Cas2 and Cas4 proteins of the Sulfolobus Type I-A system. Disruption of genes implicated in crRNA maturation or DNA interference led to a significant accumulation of acquired spacers, mainly derived from host genomic DNA. Transcriptome and proteome analyses showed that Csa3a activated expression of adaptation cas genes, CRISPR RNAs, and DNA repair genes, including herA helicase, nurA nuclease and DNA polymerase II genes. Importantly, Csa3a specifically bound the promoters of the above DNA repair genes, suggesting that they were directly activated by Csa3a for adaptation. The Csa3a regulator also specifically bound to the leader sequence to activate CRISPR transcription in vivo. Our data indicated that the Csa3a regulator couples transcriptional activation of the CRISPR–Cas system and DNA repair genes for spacer adaptation and efficient interference of invading genetic elements.
INTRODUCTION
CRISPR–Cas constitutes the prokaryotic immune system that defends Bacteria and Archaea against invasive plasmids and viruses (1,2). The CRISPR loci comprise identical repeats (typically 20–50 base pairs in length), interspaced by variable spacer sequences of similar sizes that are derived from invading genetic elements (3). An AT-rich leader sequence is located upstream of the first repeat and promotes transcription of the CRISPR locus (4,5). The cas genes encode diverse families of proteins with RNA-binding, nuclease, and/or helicase activities (6,7). To date, CRISPR–Cas systems have been classified into at least 6 basic types (Types I through VI), which are further divided into subtypes (8). Most Cas proteins have highly divergent sequences. Nevertheless, the Cas proteins implicated in spacer acquisition, i.e. Cas1 and Cas2, are relatively conserved across the major types of CRISPR–Cas systems (2).
CRISPR–Cas adaptive immunity occurs in 3 stages: acquisition of de novo spacers, crRNA biogenesis, and nucleic acid targeting and cleavage (9,10). While crRNA biogenesis and nucleic acid interference have been well characterized for the main types of CRISPR–Cas systems (11,12), the molecular mechanisms involved in spacer acquisition are less well understood. The first successful demonstration of spacer acquisition under laboratory conditions was for the Streptococcus thermophilus Type II-A system (1). Data from subsequent studies have revealed that spacer acquisition occurs for several other types of CRISPR–Cas systems, including Escherichia coli Type I-E (13–15), Pseudomonas aeruginosa Type I-F (16), Sulfolobus solfataricus and Sulfolobus islandicus Type I-A (17–19), Haloarcula hispanica Type I-B (20,21) and Pectobacterium atrosepticum Type I-F (22). A de novo spacer acquisition assay showed that Cas1 and Cas2 of E. coli Type I-E are required for the insertion of new spacers in the host CRISPR locus (14,20,23,24). Furthermore, degradation of DNA into short fragments by CRISPR nucleases via a pre-existing spacer matching the target DNA is capable of triggering primed spacer acquisition by several different CRISPR–Cas systems (20,22,23,25,26). DNA motifs in invasive genetic elements, or in CRISPR loci, also play crucial roles in the spacer-acquisition process. A DNA element known as the protospacer-adjacent motif was found to direct spacer acquisition in E. coli, H. hispanica, S. solfataricus P2 and S. islandicus REY15A (14,15,17,19,21), and the leader-repeat junction was important for acquisition (14,27). Recently, it was found that CRISPR spacer acquisition requires genome-stability proteins (28,29), as well as trans- and cis-acting factors that maintain integration specificity (30,31).
CRISPR–Cas systems have been shown to be regulated at the transcriptional level. In E. coli K12, the heat-stable, nucleoid-structuring protein (H-NS) is involved in silencing CRISPR–Cas promoters and thereby repressing CRISPR-based immunity (32), and H-NS-mediated repression can be relieved by LeuO, a LysR-type transcription factor (33) and by BaeR, an envelope stress-activated regulator (34). Moreover, by competing with the activator LeuO, CRP repressed the Type I-E CRISPR–Cas system in E. coli (33,35). Furthermore, the same factor can serve different functions. For example, CRP activates the transcription of the Type I-F CRISPR–Cas system in P. atrosepticum (36). In a hyperthermophilic archaeon, a dedicated regulator known as Csa3b was found to bind the promoter region of CRISPR-interference cassette genes and facilitated binding of the CRISPR-associated complex for antiviral defence (Cascade) to the promoter region to repress Type I-A cas gene expression in Sulfolobus (37).
Previously, we found that the Sulfolobus Type I-A CRISPR activator Csa3a binds to the promoters of adaptation genes in vitro and in vivo; moreover, overexpression of Csa3a activated expression of the adaptation Cas proteins, thereby triggering de novo spacer acquisition (19). In this study, we demonstrated that the Csa3a regulator activated expression of adaptation cas and DNA repair genes for the spacer-acquisition process and significantly enhanced crRNA biogenesis for efficient targeting of memorized genetic elements.
MATERIALS AND METHODS
Strains, growth, and transformation of Sulfolobus
Sulfolobus islandicus strains, including the genetic host E233S (ΔpyrEFΔlacS) (38) and Δcas3, Δcas3HD, Δcsa5, Δcas5, Δcas6 and Δcas7 mutants constructed previously (39), were cultured in SCVy medium at 78°C. Sulfolobus islandicus cells were transformed by electroporation, and transformants were selected on two-layer phytal gel plates, as described previously (38). Six strains were generated, including the E233S wild-type (wt) strain carrying the empty pSeSD plasmid (wt::p), the wt strain carrying the csa3a-overexpression plasmid pCsa3a (wt::csa3a), a cas1-deletion mutant (Δcas1) constructed with E233S, Δcas1 carrying the empty pSeSD plasmid (Δcas1::p) and pCsa3a (Δcas1::csa3a), a triple mutant strain with all CRISPR-interference activities inactivated (lacking cas3, cmr2α and cmr2β), and the triple mutant carrying the empty pSeSD (Δcas3cmr2αcmr2β::p) plasmid and pCsa3a plasmid (Δcas3cmr2αcmr2β::csa3a). These Sulfolobus strains were grown in SCVy medium (40,41), and growth curves were obtained by measuring their optical densities at 600 nm (OD600). Escherichia coli DH5α cells used for DNA cloning were cultured at 37°C in Luria–Bertani medium, and ampicillin was added to the culture at a final concentration of 100 μg/ml, where required.
Construction of plasmids and gene-deletion mutant strains
The gene-deletion method recently developed for S. islandicus REY15A (42) was employed to delete the cas1 (SiRe_0761), cas2 (SiRe_0762) and cas4 (SiRe_0763) genes, and to prepare the cas3/cmr2α/cmr2β (SiRe_0769/SiRe_0894/SiRe_0598) triple deletion mutant. The left sequence arm (L-arm), right sequence arm (R-arm), and target gene arm (G-arm) were amplified from S. islandicus REY15A genomic DNA using the primer sets listed in Supplementary Table S1. The marker cassette sequence carrying the pyrEF and lacS genes was amplified from the S. islandicus–E. coli shuttle vector pHZ2lacS (38) using the primers M-F-BamHI and M-R-SalI. All fragments were subsequently cloned into vector pUC19 to generate the cas1, cas2, cas4 and cas3/cmr2α/cmr2β gene-deletion plasmids. Selection of deletion mutants using these plasmids was performed as described previously (42).
The csa1 (SiRe_0760) deletion mutant was generated using the endogenous CRISPR–Cas-aided homologous recombination recently developed for Sulfolobus (43). In general, the csa1 gene locus was targeted by the CRISPR interference module with artificial spacers matching these sequences, which were generated from the pSeSD overexpression plasmid carrying the mini-CRISPR cassette with target gene-specific protospacers inserted into the repeat sequences (43). Subsequently, csa1 was deleted by enhanced homologous recombination using two homologous arms upstream and downstream of these genes, which were cloned into the same shuttle vector.
Total RNA extraction
Strains for transcriptome analysis were cultured to log phase (OD600 = 0.3), after which 1 ml of culture of each strain was transferred to 100 ml fresh SCVy medium in 250-ml flasks. Then, total RNA was isolated using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) from exponentially growing Sulfolobus cultures (OD600 = 0.3) in SCVy medium for moderate induction of the csa3a gene under control of the araS promoter, as described previously (19). Genomic DNA in the total RNA sample was removed using DNase I (Roche, Basel, Switzerland). The quality and quantity of purified total RNA were determined by measuring the absorbance at 260 and 280 nm, using a NanoDrop ND-1000 spectrophotometer (Labtech, Wilmington, MA, USA). Total RNA integrity was verified by electrophoresis on a 1.5% agarose gel.
Transcriptome analysis
A total of 3 μg RNA per sample was used as input material for cDNA library preparations. Sequencing libraries were generated using the NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer's recommendations, and index codes were added to assign sequences to each sample. First-strand cDNA synthesis was performed using random hexamer primers and M-MuLV Reverse Transcriptase (RNase H−). Second-strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H, which was followed by 15 cycles of PCR enrichment. Sequencing was performed with an Illumina HiSeq2000 instrument. The raw data were initially processed to obtain clean reads by removing the adapter sequences and low-quality bases. The clean reads were aligned to the reference genome sequence of S. islandicus REY15A (GenBank Accession No. NC_017276). An index of the reference genome was built using Bowtie software v2.0.6, and paired-end clean reads were aligned to the reference genome using TopHat software v2.0.9. HTSeq software v0.6.1 was used to count the number of reads mapped to each gene, following which the reads per kilobase per million mapped reads (RPKM) for each gene was calculated based on the length of the gene and the number of reads mapped to the gene. Each strain was sequenced in duplicate. To investigate the expression level of each gene in different groups, transcript expression levels were expressed as the RPKM. P-values were used to identify differentially expressed genes (DEGs) between two groups using the chi-squared test (2 × 2), and the significance threshold of the P-value in multiple tests was set based on the false discovery rate (FDR). Fold-changes (log2[RPKM1/RPKM2]) were also estimated according to the normalized gene-expression levels. P-values <0.01 and log2 fold-change ≥ 1 (FDR ≤ 0.05) were set as the threshold for DEGs. The transcriptome data were deposited in the GEO database under Accession no. GSE99099.
Proteomics analysis
Proteomics analysis by iTRAQ quantification was conducted as reported previously (44). For each sample, 10 mg of cells (OD600 = 0.3) were centrifuged and washed in phosphate buffer (pH 7.2), and immediately frozen in liquid nitrogen. After sonication and centrifugation, supernatants were obtained and precipitated in ice-cold acetone (1:4, v/v) at –20°C overnight, and stored at –80°C prior to sample clean up, if not used immediately. For digestion, protein pellets from the previous step were resuspended in digestion buffer (100 mM triethylammonium bicarbonate and 0.05% [w/v] sodium dodecyl sulfate to a final concentration of 1 mg/ml; total protein concentrations were measured by performing bicinchoninic acid assays [Sigma, St. Louis, MO, USA]). Equal quantities (500 μg) of each lysate were then digested overnight with trypsin at 37°C (Sigma; 1:40 trypsin [w/w] added at 0 and 2 h) and lyophilized. iTRAQ analysis was conducted at BGI Co., Ltd (Beijing, China). Protein identification and quantification were performed using Mascot software, version 2.3.02 (Matrix Science, London, United Kingdom). For iTRAQ quantification, the peptide for quantification was automatically selected with an algorithm used to calculate the reporter peak area, error factor, and P value (using the default parameters of the Mascot Software package). The resulting data sets were automatically bias-corrected to remove any variations imparted because of unequal mixing during combining different labelled samples. The ratio between the csa3a-overexpressing and control strains was obtained directly based on the protein abundance for any given protein. Proteins with a ≥1.2-fold change between the csa3a-overexpressing and control strains, and P < 0.05 were defined as being differentially expressed.
Electrophoretic mobility-shift assay (EMSA) experiments
EMSA probes were generated by PCR or annealing using oligonucleotides with 1 primer biotin-labelled or HEX-labelled at the 5′ end (Supplementary Table S1). Subsequently, the probes were purified from 6% native polyacrylamide gel electrophoresis (PAGE) gels. EMSA binding reactions (10 μl) containing 10 ng/μL of 5′ biotin-labeled probes or 5′ HEX-labelled probes, and different concentrations of Csa3a, were incubated for 20 min at 40°C in binding buffer (20 mM Tris–HCl, pH 8.0, 50 mM KCl, 5% glycerol, 1 mM ethylenediaminetetraacetic acid [EDTA], 1 mM dithiothreitol, and 5 ng/μl poly[dI-dC]). For specific competition reactions, increasing amounts of unlabelled specific competitor DNA (cold probe) were added to the reaction mixture. After the reaction, samples were loaded onto a 6% or 4% native PAGE gel buffered with 0.5× Tris–borate–EDTA (TBE) solution. DNA–protein complexes were separated at 200 V for 30 min, and the resulting fluorescence was detected with a FUJIFILM scanner (FLA-5100). In experiments using biotin-labelled probes, DNA–protein complexes were separated under the same conditions in native PAGE gels and transferred to a polyvinylidene fluoride membrane (Bio-Rad, Hercules, CA, USA) using the Semi-Dry Electrophoretic Transfer Cell system (Bio-Rad, Hercules, CA, USA), and the bands were visualized by chemiluminescence detection using the clarity Western ECL substrate (Bio-Rad, Hercules, CA, USA) and MF-Chemibis 3.2 imaging device (DNR; Jerusalem, Israel).
PCR amplification of the leader-proximal CRISPR regions
Sulfolobus islandicus E233S and the cas-deletion strains harboring an empty expression vector or the csa3a-overexpression plasmids were cultured in 10 ml of SCVy medium at 78°C until the OD600 reached 0.3. Samples of each culture (0.1 ml) were taken, and total DNA from these cells was used as a PCR template. The leader-proximal regions of two CRISPR loci were amplified by PCR using Taq polymerase, the forward primer CRISPR-F, and the reverse primer CRISPR2S5-R for locus 2; the forward primer CRISPR-F and reverse primer CRISPR1S5-R for locus 1. PCR products were separated on a 1.5% agarose gel and visualized by ethidium bromide staining. PCR products from Δcas6::csa3a and Δcas3Δcmr2αΔcmr2β::csa3a were purified using the Axygen Cleanup Kit, and the expanded bands larger than those of the wt control band (parental band) were excised from the gel and purified using the Axygen DNA Extraction Kit. Purified PCR products were cloned into the T-vector (Takara, Dalian, China), following the manufacturer's instructions. Subsequently, the ligation products were transformed into E. coli DH5α cells. Plasmids from single colonies were purified and sequenced at Qingke (Wuhan, China).
RESULTS
Essential cas genes for de novo spacer acquisition in the Sulfolobus Type I-A system
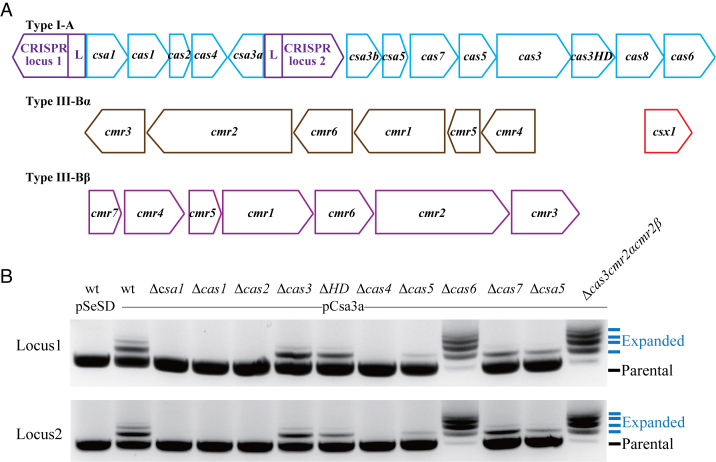
The crenarchaeon S. islandicus strain REY15A contains a I-A adaptation module consisting of csa1, cas1, cas2 and cas4 genes (45) (Figure 1A). Activation of these genes by the transcriptional regulator Csa3a triggers de novo spacer acquisition (19). To investigate whether all four genes could be essential for spacer acquisition, each cas gene was deleted individually from E233S, the genetic host, yielding Δcsa1, Δcas1, Δcas2 and Δcas4 mutants. The overexpression plasmid pCsa3a (19) was introduced into each mutant by electroporation, and the resulting transformants were employed to test de novo spacer acquisition by PCR. As shown in Figure 1B, which represents three independent spacer acquisition analyses, two extended bands were amplified from the proximal regions of the CRISPR loci in the wt::csa3a strain, indicative of the acquisition of new spacers. In contrast, no expanded bands were detected in Δcsa1, Δcas1, Δcas2 and Δcas4 strains transformed with pCsa3a. These results indicated that all genes in this operon were essential for de novo spacer acquisition. In the Cascade complex of the Sulfolobus I-A system, Cas5, Cas7 and Csa5 are implicated in the binding of crRNAs that recognize cognate target DNAs by sequence complementarity (46). Their functions in DNA interference have been demonstrated by genetic analyses of S. islandicus Δcsa5, Δcas5 and Δcas7 mutants (39). Here, their possible roles in de novo spacer acquisition were investigated by overexpression of csa3a gene in each mutant. As shown in Figure 1B, the intensities of the expanded bands were weaker than those obtained with S. islandicus wt::csa3a cells, indicating that the Cascade genes were not essential for spacer acquisition, but might influence spacer acquisition efficiency. Cascade complex along with Csa3b regulator, were found to bind to the promoter sequence of interference and processing cas genes in Sulfolobus (37) and repressed their expression. Deletion of Cascade genes would de-represses the expression of interference genes (cas3 and cas3HD) and cas6 gene, which thereby may enhance DNA targeting activity, resulting in a weaker expanded band.
Figure 1.
The role of cas genes in de novo spacer acquisition. (A) Organization of CRISPR–Cas Type I-A and Type III-B modules in Sulfolobus islandicus REY15A. (B) Detection of spacer acquisition at CRISPR loci 1 and 2 in csa3a-overexpressing strains. Each lane represents an S. islandicus E233S strain (wt carrying the empty vector pSeSD; wt and mutants of the indicated genes carrying the csa3a-overexpression plasmid, pCsa3a). The PCR products of the leader-proximal regions of CRISPR loci 1 and 2 from these strains were separated on a 1.5% agarose gel. The parental and expanded bands are indicated by arrows. This result represents three independent spacer acquisition analyses.
Host genome sampling by the Sulfolobus Type I-A adaptation module
Two mutants, Δcas6 and Δcas3cmr2αcmr2β, which lack DNA-interference activity due to the loss of CRISPR RNA-processing or nuclease activity (39), were also tested for Csa3a-activated spacer acquisition. We found that both mutants were highly active in promoting spacer acquisition since large quantities of expanded bands ranging from 1–4 or more inserted repeat-spacer units were detected in each mutant (Figure 1B). The PCR products, including the expanded bands and parental band, were purified and cloned into the T-vector and transformed into E. coli DH5α cells. Single E. coli colonies were used for sequencing their PCR inserts on plasmids. It was found that >70% of PCR clones carried new spacers with 1–6 de novo repeat-spacer units, whereas the remaining <30% did not possess any new spacers for each mutant strain (Table 1). Notably, >72% of new spacers were derived from the host genomic DNA whereas <28% originated from the plasmid DNA in these mutants. In contrast, we previously found that >90% of adapted spacers were derived from plasmid DNA in the wt::csa3a strain (19). Collectively, these results indicated that the Sulfolobus Type I-A adaptation module acquired spacers from both genomic and plasmid DNA. In the wt cells with new spacers obtained from genomic DNA, Type I-A and/or Type III-B interference complexes cleaved host DNA, resulting in the loss of these cells. However, in Δcas6 and Δcas3cmr2αcmr2β strains, inactivation of DNA interference activity enabled the cells to acquire additional spacers from both plasmid and chromosomal DNA. Furthermore, in S. islandicus cas3 and cas3HD deletion mutants, Csa3a-activated spacer acquisition efficiency did not change (Figure 1B), and this could be due to self-targeting by the type III systems which eliminated cells of cas3 and casHD deletion mutants carrying host-derived spacers.
Table 1. Analysis of the protospacers.
| Protospacer | wt::csa3a* | Δcas6::csa3a | Δcas3cmr2αcmr2β::csa3a |
|---|---|---|---|
| Total | 160 | 218 | 211 |
| genomic DNA | 11 (6.9%) | 159 (72.9%) | 157 (74.4%) |
| plasmid | 149 (93.1%) | 59 (27.1%) | 54 (25.6%) |
| Forward strand | 88 (55%) | 112 (51.4%) | 111 (52.6%) |
| Reverse strand | 72 (45%) | 106 (48.6%) | 100 (47.4%) |
| PAM sequence | |||
| CCN | 115 (71.9%) | 190 (87.2%) | 188 (89.1%) |
| CCA | 47 (29.4%) | 68 (31.2%) | 66 (31.3%) |
| CCT | 32 (20%) | 90 (41.3%) | 83 (39.3%) |
| CCG | 21 (13.1%) | 18 (8.3%) | 20 (9.5%) |
| CCC | 15 (9.4%) | 14 (6.4%) | 19 (9.0%) |
| No PAM | 45 (28.1%) | 28 (12.8%) | 23 (10.9%) |
| Adapted spacer units | |||
| Single | 126 | 57 | 76 |
| two | 15 | 36 | 34 |
| three | 0 | 14 | 16 |
| four | 1 | 9 | 2 |
| five | 1 | 1 | |
| six | 1 | 1 | |
The numbers of protospacers matching the expression plasmid and genomic DNA, as well as the protospacers having an adjacent PAM sequences (5′-PAM-protospacer-3′), are given. Percentages are included in brackets. Cells were cultured in SCVy non-inducible medium. *: data from reference (19).
Overexpression of Csa3a inhibited cell growth
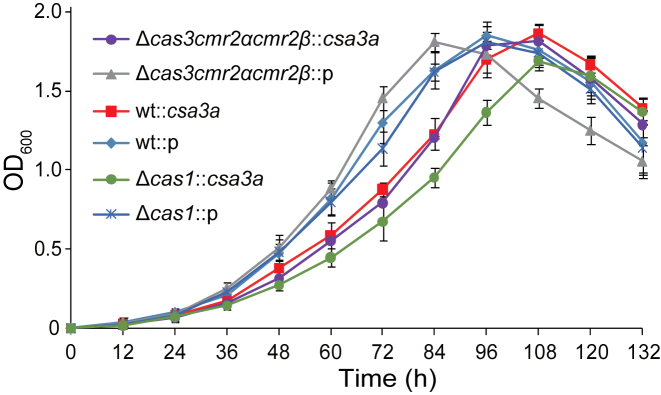
Cas1 plays crucial roles in spacer acquisition (14,24,47), and CRISPR nucleases eliminate cognate target DNA complementary to spacer sequences (9,45,48,49). Therefore, deletion of the cas1 gene results in the loss of spacer acquisition, while deletion of CRISPR nuclease genes results in the accumulation of cells with new adapted spacers. To test the effect of CRISPR spacer acquisition on Sulfolobus cell growth, we compared growth curves of the Sulfolobus wt, Δcas1, and interference-deficient Δcas3cmr2αcmr2β strains carrying the pSeSD control plasmid or pCsa3a in sucrose medium. As shown in Figure 2, all strains carrying the pSeSD vector grew faster than those carrying the csa3a-overexpression plasmid, indicating that overexpression of the csa3a gene may regulate a series of genes involved in CRISPR spacer acquisition and hinder cell growth. It was recently reported that genes in the interference cas and cmr cassettes are expressed to a moderate level in uninfected Sulfolobus cells (29). Interestingly, the interference-deficient mutant carrying pSeSD grew faster than two other strains carrying the same empty vector (Figure 2), suggesting that the moderate, un-induced level of expression of interference nucleases could inhibit Sulfolobus cell growth. Surprisingly, the Δcas3cmr2αcmr2β and wt strains carrying the pCsa3a plasmid showed the same growth curves (Figure 2), although the Δcas3cmr2αcmr2β strain carrying the pCsa3a adapted new spacers much more robustly than did the wt strain carrying pCsa3a (Figure 1B). In contrast, Δcas1::csa3a, a strain that could not acquire new spacers in the CRISPR arrays, grew more slowly than all other strains carrying the pCsa3a plasmid in inducible medium (Figure 2). Taken together, these results indicated that Csa3a overexpression inhibits the growth of S. islandicus.
Figure 2.
Growth curves of S. islandicus wt (E233S), a cas1-deletion mutant, and a cas3/cmr2α/cmr2β triple-deletion mutant carrying the empty pSeSD vector or csa3a-overexpression plasmid in SCVy inducible medium. Growth curves were analysed in triplicate, and average values were calculated.
Transcriptome and proteome analyses revealed differential expression of genes
We found that overexpression of the csa3a gene triggered CRISPR de novo spacer acquisition and hindered cell growth. Therefore, we attempted to identify Csa3a-regulated genes at the genome level. We used S. islandicus wt, Δcas1 and Δcas3cmr2αcmr2β strains carrying the csa3a-overexpression plasmid or empty plasmid for transcriptome and proteome analyses. In the Δcas1::csa3a strain, although Csa3a can possibly regulate a series of genes, the spacer acquisition process is eliminated, while massive spacer acquisition occurred in the Δcas3cmr2αcmr2β::csa3a strain. In this manner, we could identify differentially expressed genes (DEGs) regulated by both the Csa3a regulator and the adaptation process (wt::csa3a vs. wt::p), as well as those that were only regulated by the Csa3a regulator (Δcas1::csa3a vs. Δcas1::p) according to the transcriptome and proteome data. We could also identify DEGs regulated by both Csa3a and massive spacer acquisition processes (Δcas3cmr2αcmr2β::3a vs. Δcas3cmr2αcmr2β::p).
Altered transcript or protein levels identified by transcriptome and proteome analyses were summarized in Supplementary Tables S2 and S3. These data revealed that the adaptation cas genes were significantly up-regulated in all csa3a-overexpression strains, compared to the control strains (Table 2). Furthermore, Cmr4β protein was up-regulated in wt::csa3a and Δcas1::csa3a strains, while Cmr7 protein was up-regulated only in Δcas1::csa3a strain, based on the proteome analysis (Table 2). Because genes of the Cmr-β module clustered into an operon, the up-regulation of cmr7 and cmr4β gene-coding proteins suggests that Csa3a may activate transcription of the whole operon, although other proteins encoded by the operon were not significantly up-regulated according to the proteome data.
Table 2. Differentially expressed genes identified by transcriptome and proteome analyses.
| Gene ID Annotation | wt::csa3a Vs wt::p | Δcas1::csa3a Vs Δcas1::p | Δcas3cmr2αcmr2β::csa3a Vs Δcas3cmr2αcmr2β::p | |||
|---|---|---|---|---|---|---|
| Transcriptome (log2-fold) | Proteome (fold) | Transcriptome (log2-fold) | Proteome (fold) | Transcriptome (log2-fold) | ||
| Cas | ||||||
| SiRe_0602 | Cmr4β | — | 1.22 | — | 1.24 | — |
| SiRe_0603 | Cmr7 | — | — | — | 1.27 | — |
| SiRe_0760 | Csa1 | 9.84 | 2.66 | 12.18 | 4.97 | 10.10 |
| SiRe_0761 | Cas1 | 7.12 | 2.65 | / | / | 6.80 |
| SiRe_0762 | Cas2 | 6.75 | 2.19 | 11.49 | 1.56 | 8.87 |
| SiRe_0763 | Cas4 | 7.74 | 1.52 | 9.81 | 3.24 | 7.57 |
| SiRe_0764 | Csa3a | 8.05 | 3.15 | 10.15 | 2.67 | 8.58 |
| SiRe_1994 | Cas4 | — | — | — | 1.45 | — |
| DNA repair and replication | ||||||
| SiRe_0014 | NurA | 1.70 | — | — | — | 1.08 |
| SiRe_0016 | chromosome segregation protein (SMC)-like protein | — | — | 1.09 | — | — |
| SiRe_0064 | HerA | — | 1.21 | — | 1.21 | — |
| SiRe_0095 | HerA | — | — | — | 1.21 | — |
| SiRe_0236 | DNA Pol 4 | — | — | — | — | (0.93) |
| SiRe_0459 | DNA polymerase beta domain-containing protein | 1.82 | — | 1.92 | 1.22 | 1.16 |
| SiRe_0614 | DNA Pol 2 amino-end | 2.16 | — | 1.60 | — | (0.96) |
| SiRe_0615 | DNA Pol 2 elongation subunit | 1.61 | — | 1.16 | — | 1.14 |
| SiRe_0649 | Chromosome segregation protein (SMC)-like protein | 2.27 | — | 2.29 | — | 2.74 |
| SiRe_1142 | Chromosome segregation protein (SMC)-like protein | 1.14 | — | 2.00 | — | 2.23 |
| SiRe_1231 | Cdc6–2 | 1.56 | — | — | — | 1.01 |
| SiRe_1857 | HerA | — | 1.29 | — | 1.30 | (0.96) |
| Transcription factors | ||||||
| SiRe_1717 | TFB3 | 1.30 | — | — | 1.45 | — |
| Toxin-antitoxin and IS | ||||||
| SiRe_0743 | VapB-type antitoxin | — | — | 1.24 | 1.22 | 1.46 |
| SiRe_0744 | VapC-type toxin | 1.06 | — | 1.00 | — | — |
| SiRe_0856 | IS110 family transposase | (0.96) | — | 1.33 | 1.34 | — |
| SiRe_2296 | IS607 family transposase | 2.84 | — | 3.85 | — | 3.72 |
| Stress response | ||||||
| SiRe_0453 | Ferritin Dps family protein | 3.52 | — | 1.93 | 1.68 | — |
| SiRe_0370 | Flagellar hook-associated protein 2 C-terminus | 1.14 | — | 1.86 | — | — |
‘/’: gene not present in the genome; ‘—’: no change.
Up-regulation of DNA repair genes in S. islandicus strains upon csa3a overexpression were also identified (Table 2). The nurA nuclease (SiRe_0014) and herA helicase genes (SiRe_0064, SiRe_0095, and SiRe_1857) involved in DNA double-strand break repair were up-regulated in csa3a-overexpression strains compared to the control strains, based on the transcriptome and proteome data (Table 2). The genes (SiRe_0614 and SiRe_0615) encoding the DNA polymerase II (Dpo2) N-terminal and elongation subunits, which are involved in DNA repair, were significantly up-regulated in all csa3a-overexpression strains according to the transcriptome data (Table 2). However, another DNA repair polymerase gene, DNA polymerase IV gene (dpo4), previously found to be up-regulated during virus-induced spacer acquisition (29), was not significantly up-regulated according to our data, except in the massive spacer-acquisition strain (Δcas3cmr2αcmr2β::csa3a strain) (Table 2). Other transcripts or proteins that were significantly up-regulated by Csa3a, included genes encoding the nucleotidyltransferase domain of DNA polymerase beta (SiRe_0459), the chromosome-segregation protein (SMC)-related ATPase (SiRe_0649), SMC-related protein (SiRe_1142) and Cdc6–2 (SiRe_1231). Furthermore, the DNA damage-inducible general transcription activator TFB3 (50) was significantly up-regulated in the wt::csa3a and Δcas1::csa3a strains compared with the control strains, as revealed by the transcriptome and proteome analyses (Table 2).
In addition, some host toxin and antitoxin genes, and transposase genes were up-regulated in csa3a-overexpression strains and may have contributed to the retarded cell growth (Table 2, Figure 2). These included VapB-type antitoxin (SiRe_0743), VapC-type toxin (SiRe_0744), IS110 family transposase (SiRe_0856), and IS607 family transposase (SiRe_2296) genes. The stress-response genes including the Dps family gene (SiRe_0453) and flagellar hook-associated gene were also significantly up-regulated by Csa3a in both the wt::csa3a and Δcas1::csa3a strains, compared with their respective control strains (Table 2).
Taken together, we found that the DEGs in the wt::csa3a, Δcas1::csa3a and Δcas3cmr2αcmr2β::csa3a strains (relative to the respective control strains) were almost the same (Table 2). This result indicated that all differentially expressed cas genes or DNA repair genes studied were probably regulated by the Csa3a regulator. Therefore, Csa3a may act as a global regulator during CRISPR spacer acquisition.
Csa3a bound to the promoter sequences of up-regulated genes
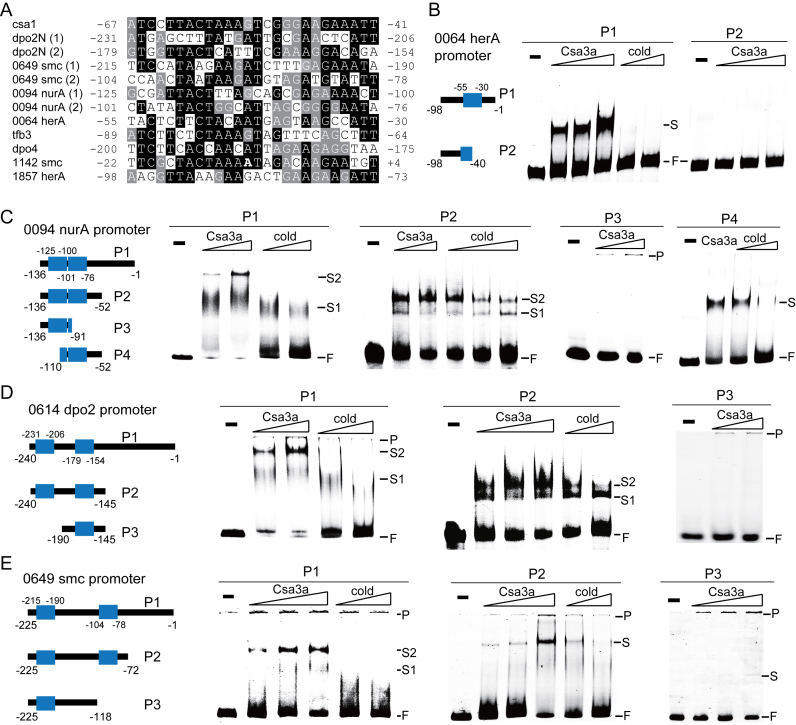
We have shown that adaptation cas genes, DNA repair genes, and stress responsive protein-encoding genes were significantly up-regulated by the Csa3a activator according to the transcriptome and proteome data (Table 2). Previously, we showed that the Csa3a regulator bound to an imperfect palindromic sequence immediately upstream of the promoters of adaptation genes (19). Therefore, we wondered whether other promoters of Csa3a up-regulated genes had similar Csa3a-binding sites. Promoter sequences of the up-regulated DNA repair-related genes were retrieved from the genome sequence (51) and analysed by multiple-sequence alignments. We found several promoters carrying one or two DNA motifs that were very similar to the Csa3a-binding site in the csa1 promoter (Figure 3A), including the promoters of the nurA-mre11-rad50-herA (SiRe_0064, SiRe_0063, SiRe_0062 and SiRe_0061), nurA-herA (SiRe_0094 and SiRe_0095) and dpo2N-dpo2E (SiRe_0614 and SiRe_0615) operons; the dpo4 and tfb3 genes; and 2 SMC-like protein genes (SiRe_0649, and SiRe_1142). Although the SiRe_0094 (NurA) protein was not significantly up-regulated by Csa3a according to the proteome data, the HerA protein encoded by SiRe_0095 gene which clustered in the nurA-herA (SiRe_0094 and SiRe_0095) operon was significantly up-regulated in Δcas1::csa3a strain (Table 2). Therefore, we analysed the SiRe_0094 (nurA) promoter, which controlled transcription of the operon. Each of the nurA-herA, dpo2/4 and smc genes was previously confirmed or proposed to be involved in DNA repair (52–55), suggesting that these DNA repair genes might play an important role in Csa3a-mediated de novo spacer acquisition.
Figure 3.
Csa3a binding to the promoters of up-regulated DNA-repair genes. (A) Alignment of the Csa3a-binding sequence in the csa1 promoter with putative Csa3a-binding sites in the promoter sequences of up-regulated DNA repair genes. The numbers indicate the nucleotide position relative to the translation start codon. Black background: conserved nucleotides; gray background: less-conserved nucleotides; no background: non-conserved nucleotides. B–E, EMSA analysis of Csa3a binding to the SiRe_0064 (herA) promoter, the SiRe_0649 (smc) promoter, SiRe_0094 (nurA) promoter, and the SiRe_0614 (dpo2) promoter. Wt and mutation probes used for EMSA analysis are shown. The putative Csa3a-binding sites are shown in blue boxes. The numbers indicate the nucleotide position relative to the translation start site. The EMSA experiments were conducted using fluorescent wt and mutation probes (10 ng/μl) with Csa3a regulator (10 and 20 ng/μl or 10, 20 and 40 ng/μl for increasing amounts of Csa3a; and 40 ng/μl for the single Csa3a concentration; 10 ng/μl = 0.4 pmol/μl) or unlabelled cold probes (2- and 4-fold excess for the full-length promoter probes, and 1- and 2-fold excess or 1-, 2- and 4-fold excess for the truncated promoter probes).
To investigate whether Csa3a directly bound the DNA motifs shown in Figure 3A, DNA fragments of the SiRe_0064 (herA), SiRe_0094 (nurA), SiRe_0614 (dpo2) and SiRe_0649 (smc) promoters were used as the probes for EMSA experiments. We first tested the Csa3a-binding ability with the full-length promoters of the herA-nurA operon involved in DNA double-strand break repair. The SiRe_0064 (herA) gene promoter contained 1 putative Csa3a-binding site. The signal intensity of the retarded band increased in parallel with increasing Csa3a amounts in the EMSA experiment, using the full-length SiRe_0064 (herA) promoter DNA as the labelled probe (P1 probe, nucleotide positions –98 to –1, relative to the translation start codon), and the signal of retarded band was completely abolished in the presence of 2- and 4-fold excess of unlabelled specific competitor DNA (cold probe) (Figure 3B). Importantly, deleting half of the predicted site completely abolished the ability of Csa3a to bind the truncated promoter DNA (P2 probe in Figure 3B), indicating that the destroyed motif was crucial for Csa3a binding to the SiRe_0064 (herA) gene promoter. Two Csa3a-binding sites were identified in the other selected promoters (Figure 3C–E). The signal intensity of a sharp retarded band enhanced, and a second smear shift appeared with increased Csa3a amounts using the full-length SiRe_0094 (nurA), SiRe_0614 (dpo2) and SiRe_0649 (smc) promoters as probes (Figure 3C–E). Moreover, increasing the amount of cold probes (2- and 4-fold excess) selectively reduced the signal intensity of the retarded bands (Figure 3C–E). To narrow down the Csa3a-binding regions, truncated promoter sequences carrying both predictive Csa3a-binding sites were used as probes for the EMSA experiments. Two sharp shifts were identified using labelled SiRe_0094 (nurA) P2 probe and SiRe_0614 (dpo2) P2 probe, while 1 sharp retarded band was found using labelled SiRe_0649 (smc) P2 probe (Figure 3C–E). The signal intensity of the retarded bands was enhanced with increasing Csa3a amounts; while increasing the amount of cold probes selectively reduced the signal intensity of the retarded bands (Figure 3C–E). These results restricted the Csa3a-binding region to between nucleotides –136 and –52 for the SiRe_0094 (nurA) promoter, nucleotides –240 and –145 for the SiRe_0614 (dpo2) promoter, and between nucleotides –225 and –72 for the SiRe_0649 (smc) promoter. We further made deletion mutations of the SiRe_0094 (nurA), SiRe_0614 (dpo2) and SiRe_0649 (smc) promoters (Figure 3C–E). In these EMSA experiments, a very weak shift was identified using the SiRe_0649 (smc) P3 probe (Figure 3E), while no retarded bands were detected using SiRe_0094 (nurA) and SiRe_0614 (dpo2) promoter P3 probes (Figure 3C and D), indicating that the deleted motifs are crucial for Csa3a binding. Furthermore, a strong shift was identified in the EMSA experiment using the SiRe_0094 (nurA) P4 probe containing the second putative DNA motif as a probe, and the signal intensity of this shift decreased with increasing amounts of the cold probe (Figure 3C). However, no retarded bands were identified in the EMSA experiments using the probes for the SiRe_0614 (dpo2) and SiRe_0649 (smc) promoters carrying only 1 predicted Csa3a-binding site (data not shown). These data indicated that the Csa3a-binding pattern of the SiRe_0614 (dpo2) and SiRe_0649 (smc) promoters was similar to that of the cas1 promoter: both sites on the promoters were crucial for Csa3a binding, and only one site was insufficient to form a stable complex with the Csa3a regulator (19). Taking the EMSA results and the transcriptome and proteome data together, these results indicated that Csa3a specifically bound the upstream activation motifs of the tested DNA repair gene promoters and activated their transcription.
Csa3a activated CRISPR transcription by binding to a site in the leader sequence
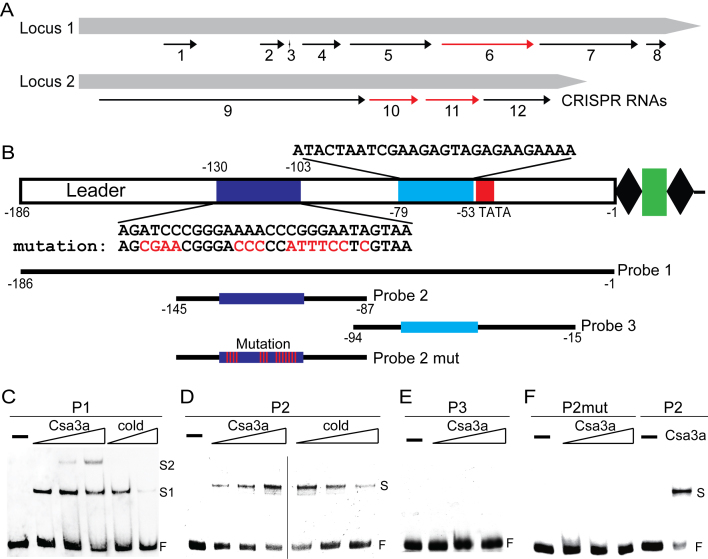
Previously, we found that the Csa3a regulator activated transcription of the adaptation cas genes, thereby triggering CRISPR de novo spacer acquisition in the S. islandicus REY15A strain (19). Interestingly, significantly up-regulated transcription of CRISPR RNAs for spacer regions 69–86 in locus 1, and 51–60 and 62–72 in locus 2 were detected in the wt::csa3a, Δcas1::csa3a and Δcas3cmr2αcmr2β::csa3a strains (Table 3, Figure 4A). In addition, significant up-regulation of CRISPR RNAs for the spacer regions 16–23, 35–39, 40, 43–50, 52–67, 88–106 and 108–111 in locus 1 and the spacer regions 1–50 and 73–85 in locus 2 were found in both the Δcas1::csa3a and Δcas3cmr2αcmr2β::csa3a strains, compared with their control strains carrying the empty pSeSD plasmid (Table 3, Figure 4A). These results indicated that the Csa3a regulator activated transcription of both CRISPR loci in S. islandicus, where the leader sequences are identical. We analysed the leader sequence and found that it contained two DNA motifs similar to the Csa3a-binding site in the csa1 promoter. The DNA motifs were located at nucleotide regions between –130 and –103, and between –79 and –53 in the leader sequence, relative to the first repeat of the CRISPR arrays (Figure 4B).
Table 3. Up-regulation of CRISPR RNAs in Csa3a overexpression strains.
| CRISPR RNA number | Nucleotide range | Spacer range | wt::csa3a Vs wt::p (log2-fold) | Δcas1::csa3a Vs Δcas1::p (log2-fold) | Δcas3cmr2αcmr2β::csa3a Vs Δcas3cmr2αcmr2β::p (log2-fold) |
|---|---|---|---|---|---|
| Locus 1 | |||||
| RNA1 | 726, 244∼726, 652 | 16–23 | — | 2.97 | 1.61 |
| RNA2 | 727, 425–727, 724 | 35–39 | — | 3.27 | 1.88 |
| RNA3 | 727, 777–727, 795 | 40 | — | 2.79 | 1.97 |
| RNA4 | 727, 943–728, 416 | 43–50 | — | 3.16 | 1.89 |
| RNA5 | 728, 527–729, 527 | 52–67 | — | 2.82 | 2.07 |
| RNA6 | 729, 652–730, 779 | 69–86 | 2.44 | 2.86 | 1.71 |
| RNA7 | 730, 850–732, 056 | 88–106 | — | 3.20 | 2.20 |
| RNA8 | 732, 156–732, 401 | 108–111 | — | 3.67 | 2.60 |
| Locus 2 | |||||
| RNA9 | 736, 681–739, 941 | 1–50 | — | 3.25 | 1.27 |
| RNA10 | 739, 992–740, 590 | 51–60 | 2.76 | 3.45 | 1.59 |
| RNA11 | 740, 682–741, 337 | 62–72 | 2.78 | 3.26 | 1.56 |
| RNA12 | 741, 391–742, 212 | 73–85 | — | 3.04 | 1.37 |
‘—’: not detected.
Figure 4.
Csa3a binding to the leader sequence and activating CRISPR transcription. (A) CRISPR RNA activation by Csa3a revealed by transcriptome data. Black arrows: up-regulated CRISPR RNAs in Δcas1::csa3a and Δcas3cmr2αcmr2β::csa3a strains; red arrows: up-regulated CRISPR RNAs in wt::csa3a, Δcas1::csa3a and Δcas3cmr2αcmr2β::csa3a strains, compared with their respective control strains. (B) DNA motifs in leader sequences and probes used for EMSA experiments. Two DNA motifs in leader sequences similar to the Csa3a-binding site are shown in dark and light blue, and their sequences are shown above or below the motifs. A putative TATA box is shown within the red rectangle. Wt and mutation DNA probes for EMSA experiments and their nucleotide ranges relative to the first repeat are indicated. (C) EMSA experiments with increasing amount of Csa3a (10, 20 and 40 ng/μl; 10 ng/μl = 0.4 pmol/μl) or unlabelled cold probe (6- or 12-fold excess) using a biotin-labeled full-leader sequence as the probe (10 ng/μl). (D) EMSA experiments with increasing amounts of Csa3a (10, 20 and 40 ng/μl) or unlabelled cold probe (2-, 6- or 12-fold excess) using HEX-labeled Probe 2 (10 ng/μl). E, EMSA experiments performed with increasing amounts of Csa3a (10, 20 and 40 ng/μl), using HEX-labelled Probe 3 (10 ng/μl). (F) EMSA experiments performed with increasing amounts of Csa3a (10, 20 and 40 ng/μl), using HEX-labeled Probe 2 mut (P2mut) with mutation at the putative Csa3a-binding motif (10 ng/μl). The reaction with 40 ng/μL Csa3a and 10 ng/μl Probe 2 was used as the positive control.
EMSA experiments were then employed to study Csa3a binding to the leader sequence. A strong retarded band was found using the full-length leader (–186 to –1, relative to the first repeat) as the probe, and a second shifted band appeared with increasing amounts of the Csa3a protein (Figure 4C), suggesting that cooperative binding might occur between the two identified DNA motifs (Figure 4B). Moreover, increasing the amount of cold probe (6- or 12-fold excess) significantly reduced the signal intensity of the retarded bands (Figure 4C). These results indicated that Csa3a specifically bound to the full-length leader sequence in vitro. To narrow down the binding region of Csa3a on the leader sequence, two DNA elements carrying the predicted motifs at the regions between –145 and –87 and between –86 and –37, were used as probes to determine the Csa3a-binding ability. The DNA fragment (Probe 2, from –145 to –87) comprising the distal motif (–130 and –103) produced a strongly retarded band with Csa3a, while increasing the cold probe (2-, 6- or 12-fold excess) specifically reduced the signal intensity of the retarded band (Figure 4D). In contrast, no such retarded band was detected when the other fragment (Probe 3, from –94 to –15) comprising the proximal motif (–79 and –53) was incubated with Csa3a (Figure 4E). These results indicated that the Csa3a-binding site was located within the region from –145 to –87, whereas the other DNA motif located in the region from –94 to –15 did not bind Csa3a under these experimental conditions (Figure 4D and E). Based on the results obtained with Probe 2, we made further mutations at conserved nucleotides in the predicted distal motif compared with the Csa3a-binding site of the csa1 promoter, generating Probe 2 mut (P2mut). Using the P2mut probe in EMSA experiments, we found no such retarded bands (Figure 4F), indicating that the Csa3a activator specifically bound the distal motif on the leader sequence. Taken together, these data indicated that Csa3a specifically bound the leader sequence and activated CRISPR transcription. It was previously reported that the crRNA abundance determined the CRISPR–Cas interference efficiency (56) and that the overall crRNA abundance gradually declines from the leader-proximal end to the leader-distal end (57). Therefore, we inferred that transcriptional activation of CRISPR RNA by Csa3a increased the DNA-targeting efficiency against invading viruses and plasmids in Sulfolobus.
DISCUSSION
Host DNA sampling in the Sulfolobus Type I-A system
Organisms of Sulfolobales are good models for studying CRISPR–Cas systems in Archaea, since a comprehensive array of genetic tools has been developed for several Sulfolobus species (58). In particular, seminar investigations of CRISPR–Cas mechanisms in S. solfataricus and S. islandicus have recently provided important insights into the adaptation process, the diverse modes of interference, and their modes of regulation (59). The de novo spacer acquisition was first demonstrated for the Sulfolobus Type I-A system using a virus-infected system, and the activation of spacer acquisition coincided with a strong decrease in the growth rate (17,18). Recently, we found that a transcriptional regulator, Csa3a, bound the promoter regions of the csa1 and cas1 genes, activated transcription of the adaptation gene cassette, increased Cas protein levels, and overexpression of Csa3a triggers hyperactive uptake of de novo spacers, primarily from a Csa3a-overexpression vector (19). In this study, we have demonstrated experimentally that Cas proteins essential for de novo spacer acquisition in the Sulfolobus subtype I-A system include Csa1, Cas1, Cas2, and Cas4. This requirement differs from that of the E. coli subtype I-E system where only Cas1 and Cas2 are required for efficient spacer acquisition (14). We have further revealed that, in the absence of DNA interference, the Sulfolobus Type I-A system acquires spacers from both invasive genetic elements (∼25%) and chromosomal DNA (∼75%). These results are in contrast to our previous data obtained in the presence of interference modules, in which 93.1% of adapted spacers are derived from the overexpression plasmid (19). These results indicated that almost all cells acquiring self DNA were destroyed by the CRISPR–Cas system, and no cooperative effect was observed between acquisition of multiple spacers and degradation of chromosomal DNA by the CRISPR–Cas systems. However, a positive-selection mechanism for acquiring invasive genetic elements was present. Although only ∼25% of the spacers were derived from plasmids, considering the size of the plasmid (∼9.0 kb) and its estimated copy number of 3–5, this represents a 14–36-fold enrichment for spacer acquisition from plasmids, compared with what is expected from the DNA content in Sulfolobus cells.
Direct regulation of DNA repair genes by the Csa3a activator
De novo CRISPR spacer acquisition is triggered by Cas1 and Cas2 overexpression in the E. coli Type I-E model system (14,24) and by overexpression of Cas1, Cas2, Cas9 and Csn2 in the S. thermophilus (60) or S. pyogenes (61) Type II-A model systems. Furthermore, it was found that the DNA-repair nuclease provided short DNA fragments for acquisition (62). Indeed, it has been demonstrated in in vitro experiments that the E. coli Cas1–Cas2 complex is capable of integrating oligonucleotide DNA substrates into an acceptor DNA to yield products similar to those generated by retroviral integrases and transposases (63). Therefore, it has been reasoned that, in addition to the adaptation Cas complex, CRISPR spacer acquisition should also requires a DNA-repair nuclease to produce short DNA fragments, a DNA polymerase, and a DNA ligase for gap-filling and nick ligation of the integration intermediate as for other known mechanisms of DNA processing. Recently, it was found that RecB and DNA polymerase I were important for de novo spacer acquisition in E. coli, implying that RecBCD generates short DNA fragments at replication forks for spacer acquisition and that DNA polymerase I fills in the DNA gaps during spacer integration (28). But it remains to be investigated whether DNA-repair proteins are also required for CRISPR spacer acquisition in other model organisms.
In this study, transcriptome and proteome profiles have been generated for wt, Δcas1, and Δcas3cmr2αcmr2β strains carrying csa3a overexpression plasmid versus their control strains, and these analyses have led to the identification of Csa3a as a global regulator during CRISPR spacer acquisition in Sulfolobus. These results are correlated with data from a recent transcriptome study on virus-induced spacer acquisition in Sulfolobus, in which the expression of DNA repair genes, including DNA polymerase 2 and 4, DNA repair HerA helicase, and NurA nuclease protein-encoding genes, are induced (29). Since archaeal NurA nucleases and HurA helicases functioned in homologous recombination together with Rad50 and Mre11 are involved in DNA double strand-break repair (52,64) in analogy to the bacterial RecBCD (53), the archaeal enzymes may also be responsible for production of DNA intermediates for spacer acquisition as demonstrated in E. coli (28,62). Furthermore, it has been suggested that spacer acquisition being dependent on DNA replication in S. islandicus (18) and E. coli (62), and this fits well with the up-regulation of Type B DNA polymerase 2 (Dpo2) and the type Y DNA polymerase 4 (Dpo4), two DNA repair polymerases (54), by Csa3a overexpression. Finally, the expression of TFB3, a DNA damage-inducible general transcriptional activator in Sulfolobus (50) is also activated, suggesting more complex mode of regulation could be involved in Csa3 regulation. More importantly, we found a conserved Csa3a regulator-binding site in the promoter sequences of up-regulated DNA repair genes (Figure 3A) and experimentally demonstrated that the Csa3a regulator specifically bound to these promoters in EMSA experiments (Figure 3). Combining the transcriptome/proteome data and our earlier results (19), we infer that Csa3a directly binds to the promoters of up-regulated DNA-repair genes and adaptation cas genes, sequentially activating their transcription and facilitating de novo CRISPR spacer acquisition in the Sulfolobus Type I-A system.
Conserved regulation model for spacer adaptation and CRISPR RNA transcription in the archaeal genus Sulfolobus
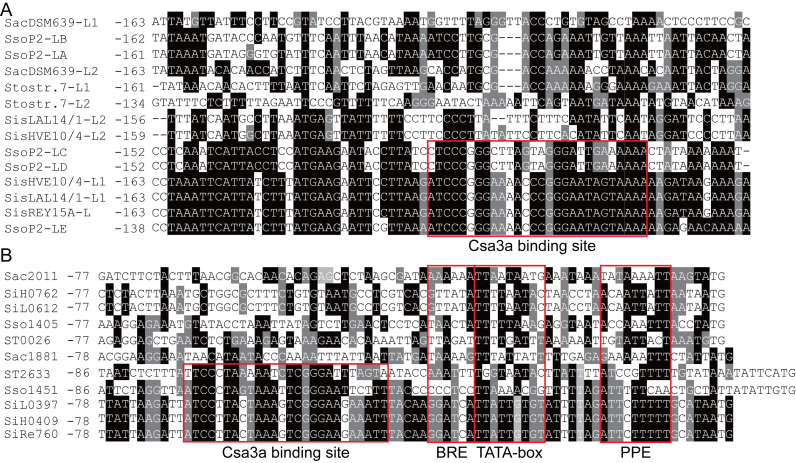
It has been shown that transcriptional-initiation and adaptation signals of CRISPR arrays are located within the leader sequence; however, there is very little knowledge of the structural motifs involved or their possible functions (65). To analyse the regulatory models of spacer adaptation and CRISPR RNA transcription in different Sulfolobus species, we extracted the leader sequences from three S. islandicus strains (REY15A, HVE10/4, and LAL14/1), S. solfataricus P2, S. acidocaldarius DSM639 and S. tokodaii strain 7. Each S. islandicus strain has two identical leader sequences and HVE10/4 and LAL14/1 have two additional identical leader sequences. S. solfataricus P2 has five leader sequences and S. acidocaldarius DSM639 and S. tokodaii strain 7 have two leader sequences. The alignment results are shown in Figure 5A. Three leaders from S. islandicus are identical, and leader E from S. solfataricus P2 is identical to S. islandicus leaders in the region from –138 to –72, relative to the first repeat, indicating that these four leaders all carry the Csa3a-binding site (Figure 5A). Leader C and D from S. solfataricus P2 are identical and are very similar to the leaders of S. islandicus in the region from –152 to –80. However, leader 2 of S. islandicus HVE10/4 and LAL14/1, and leaders A and B from S. solfataricus P2 are nearly identical, but they seem to lack the conserved Csa3a-binding site (Figure 5A). The leaders of S. acidocaldarius DSM639 and S. tokodaii strain 7 also seem to lack the conserved Csa3a-binding site (Figure 5A).
Figure 5.
Alignment of Sulfolobus leader sequences and adaptation cas gene promoter sequences. (A) Leader sequences of S. islandicus strains (REY15A [2 identical leaders], HVE10/4 [two identical leaders] and LAL14/1 [two identical leaders]), S. solfataricus P2 (leaders for CRISPR loci A–E), S. acidocaldarius DSM639 ( leaders), and S. tokodaii strain 7 (leader for CRISPR 1 and 2). (B) Promoter sequences for the csa1–cas1–cas2–cas4 adaptation cas operon (SiRe_0760, SiH_0409, SiL_0397, Sso1451 and ST2633 encoding csa1, the first gene of the adaptation cas operons), single cas1 genes (ST0026, Sso1405, SiL_0612, SiH_0762, and Saci_2011), or the cas1–cas4–cas2 operon (Saci_1881 to Saci_1879). The Csa3a-binding sites, transcription factor B recognition element (BRE), TATA box, and promoter-proximal element (PPE) are boxed. The nucleotide positions are numbered relative to the translation start codon. Black background: conserved nucleotides; gray background: less-conserved nucleotides; no background: non-conserved nucleotides
Adaptation cas genes are usually clustered in operons (66). We further analysed the regulation models of the adaptation genes in the above-mentioned Sulfolobus strains. We found that S. islandicus strains, S. solfataricus P2, S. tokodaii strain 7, and S. acidocaldarius DSM639 encode an adaptation cas gene operon, arranged as csa1 (SiRe_0760, SiH_0409, SiL0397, Sso1451 and ST2633); the cas1, cas2 and cas4 genes; and the cas1–cas4–cas2 operon (Saci_1881 to Saci_1879), or a single cas1 gene (ST0026, Sso1405, SiL0612, SiH_0762 and Saci_2011). All promoters contain a TATA box and AT-rich proximal promoter element (PPE) and transcription factor B-recognition elements (BREs) (Figure 5B). Three S. islandicus strains were found to have identical promoters for the adaptation gene operon (Figure 5B). Significantly, all promoters controlling transcription of the csa1–cas1–cas2–cas4 or cas1–cas4–cas2 adaptation operons contained a conserved Csa3a-binding site (Figure 5B), while the single cas1 gene promoters have no conserved motif for Csa3a binding. In addition, all these organisms encode for a Csa3a protein. The Csa3a protein of S. tokodaii strain 7 (ST2633) and S. acidocaldarius (Saci_1876) showed very low similarity to the Csa3a proteins from S. islandicus and S. solfataricus (Supplementary Figure S1). ST2633 and Saci_1876 potentially recognized distinct DNA motifs present in their leader sequences and adaptation cas promoter, contributing to variation of the Csa3a-binding sites in these organisms (Figure 5B). These results suggest: (1) S. islandicus, S. solfataricus and S. tokodaii strains employ the same regulation model for spacer acquisition and CRISPR transcription using the Csa3a activator and (2) the S. islandicus, S. solfataricus, and S. tokodaii strains have multiple CRISPR arrays that may have evolved to exploit both Csa3a and other regulation models. Importantly, all promoters carrying Csa3a-binding sites have a poorly conserved TATA box. Nucleotides G and C are present in these TATA box sequences, although they should be strictly excluded from a conserved TBP-binding site (Figure 5B). These results also suggest that these organisms may employ Csa3a to recruit TBP to the weak TATA boxes in the promoters of their adaptation-gene operons to activate transcription of these genes.
Transcriptional-regulation model for the Sulfolobus Type I-A CRISPR–Cas system
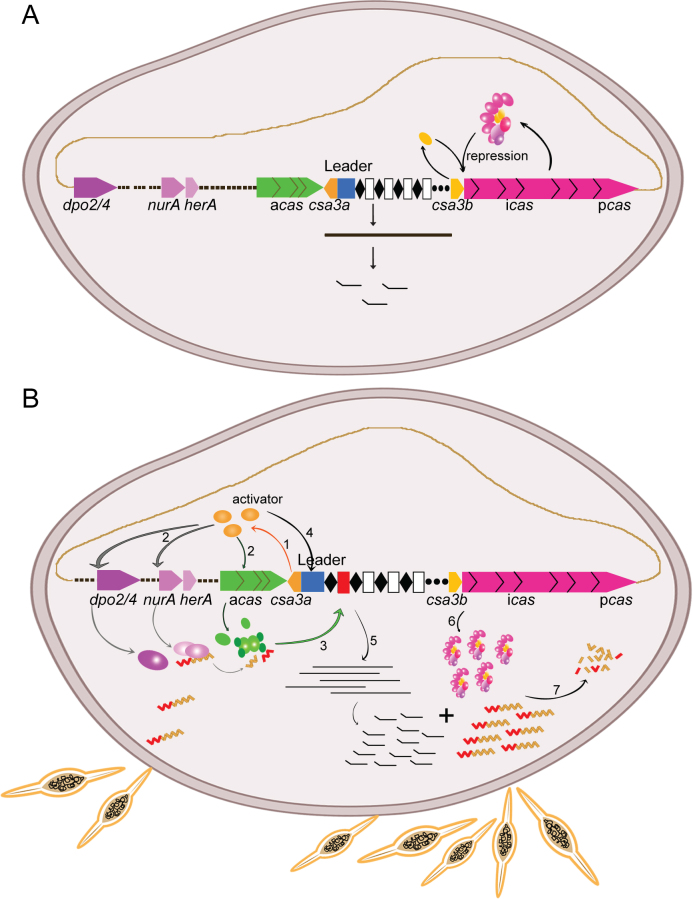
The bacterial CRISPR–Cas systems are well regulated to defend against invading viruses or plasmids. In E. coli K12, the heat-stable H-NS and CRP proteins were shown to repress the Type I-E CRISPR–Cas system (32,35). H-NS-mediated repression of CRISPR-based immunity was relieved by LysR-type transcription factor LeuO (33), while in the P. atrosepticum Type I-F system, CRP activated CRISPR–Cas transcription (36). However, the regulation of archaeal CRISPR–Cas systems is less well understood. By combining the results of our previous work (19) and those from He et al. (37), we generated a transcriptional regulation model for the Sulfolobus Type I-A CRISPR–Cas system, as shown in Figure 6. According to our model, in the absence of invading mobile genetic elements, the Sulfolobus Type I-A adaptation genes (csa1, cas1, cas2 and cas4) are silent, while CRISPR RNAs are transcribed and processed into mature crRNA at a low level. At this stage, the interference genes (cas5, cas7, csa5, cas3 and cas8) are expressed at a low level and form the Cascade complex. The Cascade complex, along with the Csa3b regulator binds to the promoter of interference genes, inhibiting their further transcription (37). Upon invasion by mobile genetic elements, the Csa3a activator is induced by an unknown mechanism. Then, Csa3a directly binds the promoters of the adaptation cas gene, the nurA–herA nuclease–helicase genes, and DNA-repair polymerase genes to activate their transcription. The NurA–HerA complex may act as RecBCD (53) to generate short DNA fragments at the replication forks of mobile genetic elements (28,62), which are integrated into CRISPR arrays by the adaptation complex. The Csa3a regulator simultaneously binds to the CRISPR leader sequences and significantly enhances CRISPR transcription and biogenesis of mature crRNAs. Upon the invasion of genetic elements, the Cascade complex is released from the interference cas gene promoter and binds crRNA to digest genetic elements with cognate protospacers. In this model, a single regulator activates transcription of DNA repair genes, and adaptation cas genes and CRISPR RNAs of Type I-A CRISPR–Cas system to defend against invasive genetic elements in Sulfolobus.
Figure 6.
Transcriptional-regulation model for the Sulfolobus Type I-A CRISPR–Cas system. (A) in the absence of mobile genetic elements, the Sulfolobus Type I-A adaptation genes (csa1, cas1, cas2 and cas4) are silent, while CRISPR RNAs are moderately transcribed and processed into mature crRNAs. At this stage, the interference genes (cas5, cas7, csa5, cas3 and cas8) are transcribed and translated to form the Cascade complex. The Cascade complex, along with the Csa3b regulator binds to the promoter of interference genes, inhibiting their further transcription (37). (B) In the presence of mobile genetic elements, the Csa3a regulator (induced by an unknown mechanism) activates transcription of the adaptation cas, nurA–herA nuclease–helicase, and DNA-repair polymerase genes (step 1). Then, the NurA–HerA protein may generate short DNA fragments at the replication forks of mobile genetic elements (step 2), which are integrated into CRISPR arrays by the adaptation complex (step 3). The Csa3a regulator simultaneously binds to the CRISPR leader sequences (step 4) and significantly enhances CRISPR transcription and mature crRNA biogenesis (step 5). Upon invasion of genetic elements, the Cascade complex is released from interference cas gene promoter and transcription of interference cas genes is de-repressed (step 6). Finally, the Cascade complex binds crRNA to digest the genetic elements with cognate protospacers (step 7).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Roger A Garrett of Danish Archaea Centre in University of Copenhagen for critical reading of the manuscript.
Footnotes
Present address: Wenfang Peng, Hubei Collaborative Innovation Center for Green Transformation of Bioresource, Hubei Key Laboratory of Industrial Biotechnology, College of Life Science, Hubei University, Wuhan 430062, P.R. China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [3167080536 to N.P.]; Fundamental Research Funds for the Central Universities [2662015PX199, 2662015PY038 to N.P.]; Danish Council for Independent Research [DFF-4181-00274, DFF-1323-00330 to Q.S.]. Funding for open access charge: National Natural Science Foundation of China [3167080536].
Conflict of interest statement. None declared.
REFERENCES
- 1. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P.. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007; 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- 2. Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F. et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 2011; 9:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Soria E.. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005; 60:174–182. [DOI] [PubMed] [Google Scholar]
- 4. Agari Y., Sakamoto K., Tamakoshi M., Oshima T., Kuramitsu S., Shinkai A.. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J. Mol. Biol. 2010; 395:270–281. [DOI] [PubMed] [Google Scholar]
- 5. Pougach K., Semenova E., Bogdanova E., Datsenko K.A., Djordjevic M., Wanner B.L., Severinov K.. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 2010; 77:1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haft D.H., Selengut J., Mongodin E.F., Nelson K.E.. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005; 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Makarova K.S., Grishin N.V., Shabalina S.A., Wolf Y.I., Koonin E.V.. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006; 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koonin E.V., Makarova K.S.. CRISPR–Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013; 10:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J.. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008; 321:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Westra E.R., Swarts D.C., Staals R.H., Jore M.M., Brouns S.J., van der Oost J.. The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 2012; 46:311–339. [DOI] [PubMed] [Google Scholar]
- 11. Plagens A., Richter H., Charpentier E., Randau L.. DNA and RNA interference mechanisms by CRISPR–Cas surveillance complexes. FEMS Microbiol. Rev. 2015; 39:442–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Charpentier E., Richter H., van der Oost J., White M.F.. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR–Cas adaptive immunity. FEMS Microbiol. Rev. 2015; 39:428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swarts D.C., Mosterd C., van Passel M.W., Brouns S.J.. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012; 7:e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yosef I., Goren M.G., Qimron U.. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012; 40:5569–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yosef I., Shitrit D., Goren M.G., Burstein D., Pupko T., Qimron U.. DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:14396–14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cady K.C., Bondy-Denomy J., Heussler G.E., Davidson A.R., O’Toole G.A.. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J. Bacteriol. 2012; 194:5728–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Erdmann S., Garrett R.A.. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 2012; 85:1044–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Erdmann S., Le Moine Bauer S., Garrett R.A.. Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol. Microbiol. 2014; 91:900–917. [DOI] [PubMed] [Google Scholar]
- 19. Liu T., Li Y., Wang X., Ye Q., Li H., Liang Y., She Q., Peng N.. Transcriptional regulator-mediated activation of adaptation genes triggers CRISPR de novo spacer acquisition. Nucleic Acids Res. 2015; 43:1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li M., Wang R., Zhao D., Xiang H.. Adaptation of the Haloarcula hispanica CRISPR–Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014; 42:2483–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li M., Wang R., Xiang H.. Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res. 2014; 42:7226–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richter C., Dy R.L., McKenzie R.E., Watson B.N., Taylor C., Chang J.T., McNeil M.B., Staals R.H., Fineran P.C.. Priming in the Type I-F CRISPR–Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014; 42:8516–8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E.. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012; 3:945. [DOI] [PubMed] [Google Scholar]
- 24. Arslan Z., Hermanns V., Wurm R., Wagner R., Pul U.. Detection and characterization of spacer integration intermediates in type I-E CRISPR–Cas system. Nucleic Acids Res. 2014; 42:7884–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Diez-Villasenor C., Guzman N.M., Almendros C., Garcia-Martinez J., Mojica F.J.. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR–Cas I-E variants of Escherichia coli. RNA Biol. 2013; 10:792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fineran P.C., Gerritzen M.J., Suarez-Diez M., Kunne T., Boekhorst J., van Hijum S.A., Staals R.H., Brouns S.J.. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E1629–E1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wei Y., Chesne M.T., Terns R.M., Terns M.P.. Sequences spanning the leader-repeat junction mediate CRISPR adaptation to phage in Streptococcus thermophilus. Nucleic Acids Res. 2015; 43:1749–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ivancic-Bace I., Cass S.D., Wearne S.J., Bolt E.L.. Different genome stability proteins underpin primed and naive adaptation in E. coli CRISPR–Cas immunity. Nucleic Acids Res. 2015; 43:10821–10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leon-Sobrino C., Kot W.P., Garrett R.A.. Transcriptome changes in STSV2-infected Sulfolobus islandicus REY15A undergoing continuous CRISPR spacer acquisition. Mol. Microbiol. 2016; 99:719–728. [DOI] [PubMed] [Google Scholar]
- 30. Nunez J.K., Bai L., Harrington L.B., Hinder T.L., Doudna J.A.. CRISPR immunological memory requires a host factor for specificity. Mol. Cell. 2016; 62:824–833. [DOI] [PubMed] [Google Scholar]
- 31. McGinn J., Marraffini L.A.. CRISPR–Cas systems optimize their immune response by specifying the site of spacer integration. Mol. Cell. 2016; 64:616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pul Ü., Wurm R., Arslan Z., Geißen R., Hofmann N., Wagner R.. Identification and characterization of E. coli CRISPR–Cas promoters and their silencing by H-NS. Mol. Microbiol. 2010; 75:1495–1512. [DOI] [PubMed] [Google Scholar]
- 33. Westra E.R., Pul U., Heidrich N., Jore M.M., Lundgren M., Stratmann T., Wurm R., Raine A., Mescher M., Van Heereveld L. et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 2010; 77:1380–1393. [DOI] [PubMed] [Google Scholar]
- 34. Perez-Rodriguez R., Haitjema C., Huang Q., Nam K.H., Bernardis S., Ke A., DeLisa M.P.. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol. Microbiol. 2011; 79:584–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang C.-D., Chen Y.-H., Huang H.-Y., Huang H.-D., Tseng C.-P.. CRP represses the CRISPR/Cas system in Escherichia coli: evidence that endogenous CRISPR spacers impede phage P1 replication. Mol. Microbiol. 2014; 92:1072–1091. [DOI] [PubMed] [Google Scholar]
- 36. Patterson A.G., Chang J.T., Taylor C., Fineran P.C.. Regulation of the Type I-F CRISPR–Cas system by CRP-cAMP and GalM controls spacer acquisition and interference. Nucleic Acids Res. 2015; 43:6038–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He F., Vestergaard G., Peng W., She Q., Peng X.. CRISPR–Cas type I-A Cascade complex couples viral infection surveillance to host transcriptional regulation in the dependence of Csa3b. Nucleic Acids Res. 2016; 45:1902–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deng L., Zhu H., Chen Z., Liang Y.X., She Q.. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles. 2009; 13:735–746. [DOI] [PubMed] [Google Scholar]
- 39. Peng W., Li H., Hallstrom S., Peng N., Liang Y.X., She Q.. Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus. RNA Biol. 2013; 10:738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peng N., Xia Q., Chen Z., Liang Y.X., She Q.. An upstream activation element exerting differential transcriptional activation on an archaeal promoter. Mol. Microbiol. 2009; 74:928–939. [DOI] [PubMed] [Google Scholar]
- 41. Peng N., Deng L., Mei Y., Jiang D., Hu Y., Awayez M., Liang Y., She Q.. A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 2012; 78:5630–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang C., Guo L., Deng L., Wu Y., Liang Y., Huang L., She Q.. Revealing the essentiality of multiple archaeal pcna genes using a mutant propagation assay based on an improved knockout method. Microbiology. 2010; 156:3386–3397. [DOI] [PubMed] [Google Scholar]
- 43. Li Y., Pan S., Zhang Y., Ren M., Feng M., Peng N., Chen L., Liang Y.X., She Q.. Harnessing Type I and Type III CRISPR–Cas systems for genome editing. Nucleic Acids Res. 2016; 44:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Song Z., Chen L., Wang J., Lu Y., Jiang W., Zhang W.. A transcriptional regulator Sll0794 regulates tolerance to biofuel ethanol in photosynthetic Synechocystis sp. PCC 6803. Mol. Cell Proteomics. 2014; 13:3519–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deng L., Garrett R.A., Shah S.A., Peng X., She Q.. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 2013; 87:1088–1099. [DOI] [PubMed] [Google Scholar]
- 46. Lintner N.G., Kerou M., Brumfield S.K., Graham S., Liu H., Naismith J.H., Sdano M., Peng N., She Q., Copie V. et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J. Biol. Chem. 2011; 286:21643–21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rollie C., Schneider S., Brinkmann A.S., Bolt E.L., White M.F.. Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. eLife. 2015; 4:e08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peng W., Feng M., Feng X., Liang Y.X., She Q.. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015; 43:406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gasiunas G., Barrangou R., Horvath P., Siksnys V.. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Paytubi S., White M.F.. The crenarchaeal DNA damage-inducible transcription factor B paralogue TFB3 is a general activator of transcription. Mol. Microbiol. 2009; 72:1487–1499. [DOI] [PubMed] [Google Scholar]
- 51. Guo L., Brugger K., Liu C., Shah S.A., Zheng H., Zhu Y., Wang S., Lillestol R.K., Chen L., Frank J. et al. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J. Bacteriol. 2011; 193:1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang S., Wei T., Hou G., Zhang C., Liang P., Ni J., Sheng D., Shen Y.. Archaeal DNA helicase HerA interacts with Mre11 homologue and unwinds blunt-ended double-stranded DNA and recombination intermediates. DNA Repair (Amst.). 2008; 7:380–391. [DOI] [PubMed] [Google Scholar]
- 53. Blackwood J.K., Rzechorzek N.J., Abrams A.S., Maman J.D., Pellegrini L., Robinson N.P.. Structural and functional insights into DNA-end processing by the archaeal HerA helicase-NurA nuclease complex. Nucleic Acids Res. 2012; 40:3183–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Choi J.Y., Eoff R.L., Pence M.G., Wang J., Martin M.V., Kim E.J., Folkmann L.M., Guengerich F.P.. Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication. J. Biol. Chem. 2011; 286:31180–31193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Herrmann U., Soppa J.. Cell cycle-dependent expression of an essential SMC-like protein and dynamic chromosome localization in the archaeon Halobacterium salinarum. Mol. Microbiol. 2002; 46:395–409. [DOI] [PubMed] [Google Scholar]
- 56. Dwarakanath S., Brenzinger S., Gleditzsch D., Plagens A., Klingl A., Thormann K., Randau L.. Interference activity of a minimal Type I CRISPR–Cas system from Shewanella putrefaciens. Nucleic Acids Res. 2015; 43:8913–8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Richter H., Zoephel J., Schermuly J., Maticzka D., Backofen R., Randau L.. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012; 40:9887–9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Peng N., Han W., Li Y., Liang Y., She Q.. Genetic technologies for extremely thermophilic microorganisms of Sulfolobus, the only genetically tractable genus of crenarchaea. Sci. China Life Sci. 2017; 60:1–16. [DOI] [PubMed] [Google Scholar]
- 59. Garrett R.A., Shah S.A., Erdmann S., Liu G., Mousaei M., Leon-Sobrino C., Peng W., Gudbergsdottir S., Deng L., Vestergaard G. et al. CRISPR–Cas adaptive immune systems of the Sulfolobales: unravelling their complexity and diversity. Life (Basel, Switzerland). 2015; 5:783–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wei Y., Terns R.M., Terns M.P.. Cas9 function and host genome sampling in Type II-A CRISPR–Cas adaptation. Genes Dev. 2015; 29:356–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Heler R., Samai P., Modell J.W., Weiner C., Goldberg G.W., Bikard D., Marraffini L.A.. Cas9 specifies functional viral targets during CRISPR–Cas adaptation. Nature. 2015; 519:199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Levy A., Goren M.G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., Sorek R.. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015; 520:505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nunez J.K., Lee A.S., Engelman A., Doudna J.A.. Integrase-mediated spacer acquisition during CRISPR–Cas adaptive immunity. Nature. 2015; 519:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Constantinesco F., Forterre P., Koonin E.V., Aravind L., Elie C.. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Res. 2004; 32:1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Alkhnbashi O.S., Shah S.A., Garrett R.A., Saunders S.J., Costa F., Backofen R.. Characterizing leader sequences of CRISPR loci. Bioinformatics. 2016; 32:i576–i585. [DOI] [PubMed] [Google Scholar]
- 66. Vestergaard G., Garrett R.A., Shah S.A.. CRISPR adaptive immune systems of Archaea. RNA Biol. 2014; 11:156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.