Abstract
Preparation of large amount of single-stranded circular DNA in high selectivity is crucial for further developments of nanotechnology and other DNA sciences. Herein, a simple but practically useful methodology to prepare DNA rings has been presented. One of the essential factors is to use highly diluted T4 ligase buffer for ligase reactions. This strategy is based on our unexpected finding that, in diluted T4 buffers, intermolecular polymerization of DNA fragments is greatly suppressed with respect to their intramolecular cyclization. This promotion of cyclization is attributable to abnormally low concentration of Mg2+ ion (0.5–1.0 mM) but not ATP in the media for T4 ligase reactions. The second essential factor is to add DNA substrate intermittently to the mixture and maintain its temporal concentration low. By combining these two factors, single-stranded DNA rings of various sizes (31–74 nt) were obtained in high selectivity (89 mol% for 66-nt DNA) and in satisfactorily high productivity (∼0.2 mg/ml). A linear 72-nt DNA was converted to the corresponding DNA ring in nearly 100% selectivity. The superiority of this new method was further substantiated by the fact that small-sized DNA rings (31–42 nt), which were otherwise hardly obtainable, were successfully prepared in reasonable yields.
INTRODUCTION
Single-stranded circular DNAs possess unique features in both dynamics and topological constraints which can never be fulfilled by linear DNAs. Accordingly, these DNA rings are one of the most fundamental and important parts for DNA nanotechnology (1). By incorporating them to predetermined sites, unprecedentedly sophisticated and functional nanostructures are obtainable. For example, catenanes, in which two or more DNA rings are mechanically interlocked, have been used to construct molecular switches, molecular motors, switchable catalysts and others (1–5). Their conjugation with DNA origami and the relevant technology (6–8) should be promising for advanced nanotechnology. Single-stranded DNA rings are also valuable for molecular biology, medicine and biotechnology. Compared with linear DNAs, they are more resistant against exonucleases in cells and form more stable duplexes and triplexes (9), being advantageous for anti-sense/gene strategies and other therapeutic applications. Furthermore, these DNA rings are employed as templates for rolling circle amplification by DNA polymerase which provides concatemers containing tens to hundreds of tandem repeats and has been widely adopted for various purposes (9,10).
In spite of these remarkable features, the difficulty in large-scale synthesis of single-stranded circular DNAs (c-DNAs) has been hampering their still more versatile applications. Usually, they are produced by ligating two ends of a linear single-stranded DNA (l-DNA) by either chemical or enzymatic means. A short DNA strand is often used as splint additive to place the two ends in a close proximity and facilitates the cyclization (Figure 1A). However, this intramolecular cyclization is inevitably accompanied by intermolecular ligation of several l-DNA strands to form larger polymers (Figure 1B). As we know, T4 DNA ligase is most commonly used for DNA cyclization. When T4 DNA ligase was used, the only strategy to suppress the intermolecular side reactions is to reduce the concentration of DNA substrate in the reaction mixture. In most of previous preparations of single-stranded DNA rings, [l-DNA]0 was thus kept at 0.5 μM or lower (11–15). With this restraint of reaction conditions, however, only a limited amount of c-DNA is obtainable from one reaction vessel. In practical synthesis, these two factors of selectivity and productivity must be somehow compromized. Furthermore, even with this low substrate-concentration strategy, the rings of smaller size are difficult to be produced due to increased steric strain. For example, a linear DNA of 30–45 nt can be hardly cyclized even with this conventional method and is mostly converted to the polymers (see Table 1). Even with linear DNA of larger size, the cyclization is also difficult when it takes a specific secondary structure. An enzyme CircLigase™ from Epicentre Co. (Madison, WS, USA) is available to cyclize single-stranded DNA. However, its dosage is quite large due to the low activity, which only be appropriate for small scale production of DNA rings. Besides, it requires Mn2+ as an essential cofactor, which would be unfavorable especially for in vivo applications of the products.
Figure 1.
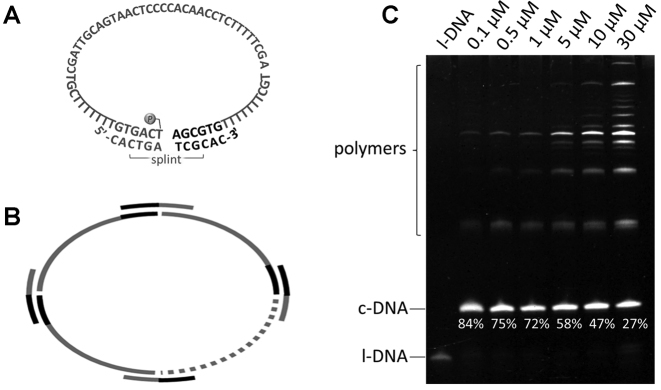
Cyclization of l-DNA66 by T4 DNA ligase. Schematic views of the formation of (A) single-stranded DNA ring (c-DNA66) with the assistance of splint-12 nt and (B) polymers from multiple l-DNA66 strands. The single-stranded DNA substrate (l-DNA66) bears a phosphate at the 5′-terminus. (C) Effects of [l-DNA66]0 on the formation of c-DNA66 and the polymers in 1× T4 ligase buffer (conventional method). The selectivity for the formation of c-DNA66 is presented below the corresponding band. [l-DNA66]0/[splint-12 nt]0 = 1/2 at 20°C for 12 h. All the DNA substrate was added to the mixture all at once at the beginning of the reaction.
Table 1. Selectivities and productivities for the formation of single-stranded DNA rings of various sizes through three cyclization methods.
| Length of l-DNA | Selectivity (%) | Productivity (μg/ml) | ||||
|---|---|---|---|---|---|---|
| 1 × Buf. | 0.1 × Buf. | S-b-S | 1 × Buf. | 0.1 × Buf. | S-b-S | |
| 31 nta | 1.7 | 5.8 | 17.8 | 1.7 | 5.8 | 17.8 |
| 39 nta | 0.7 | 15.0 | 46.7 | 0.9 | 19.3 | 60.1 |
| 42 ntb | 4.5 | 31.8 | 65.5 | 1.2 | 8.8 | 18.2 |
| 66 nta | 55.8 | 75.2d | 89.0d | 121.5 | 163.8d | 193.8d |
| 72 ntb,c | 82.0 | 93.7 | 99.7 | 39.0 | 44.5 | 47.4 |
| 74 ntb,c | 11.0 | 37.8 | 85.3 | 5.4 | 18.5 | 41.7 |
aThe final concentration of l-DNA was 10 μM.
bThe final concentration of l-DNA was 2 μM.
cThe results for 72 nt and 74 nt l-DNA are discussed in terms of their solution structures (see text for details).
dThe buffer concentration used was 0.05 × T4 buffer.
In this paper, we present a simple but very convenient method to efficiently prepare single-stranded DNA rings using T4 DNA ligase. This enzyme is widely available in many laboratories, and employs only non-toxic Mg2+ as cofactor. This new method is primarily based on our unexpected finding that intermolecular ligation of l-DNA strands to form polymers is greatly suppressed by reducing the concentration of T4 ligase buffer. In contrast, the rate of cyclization of l-DNA is only marginally affected, thus the selectivity for target DNA rings is enormously increased. In the present method, the selectivity is further promoted by separating l-DNA substrate to several portions and adding them intermittently to diluted T4 ligase buffers with sufficient time-interval. Throughout the reaction, the concentration of l-DNA in the mixtures is kept small to suppress the polymerization. By this ‘Step by Step’ method, sufficient amount of DNA rings of various sizes, even 30–45 nt rings, can be easily and selectively prepared without using a large-scaled reaction vessel.
MATERIALS AND METHODS
Materials
All DNA oligonucleotides used in this study were ordered from Integrated DNA Technologies, Inc. (Coralville, IA, USA). For the cyclization experiments of linear DNA (l-DNA), the phosphate residue was enzymatically introduced to the 5′-position by using T4 polynucleotide kinase (Thermo Scientific, Pittsburgh, PA, USA). T4 DNA ligase was purchased from Thermo Scientific (Pittsburgh, PA, USA), together with the 10 × T4 ligase buffer. All other chemicals were obtained from Sigma-Aldrich (St. Louis, MO, USA).
Cyclization of single-stranded linear DNA (l-DNA)
Conventional cyclization in 1 × T4 ligase buffer
As control experiments for the new method, the enzymatic cyclization was carried out under conventional conditions. The 1 × T4 DNA ligase buffer ([MgCl2] = 10 mM, [ATP] = 500 μM, [DTT] = 10 mM, and [Tris–HCl] = 40 mM), provided by the manufacturer, was directly employed. A typical cyclization system (20 μl) contained l-DNA (1 μM), splint (2 μM) and T4 DNA ligase (5 U) in 1 × T4 DNA ligase buffer. All these components were added to the mixture at the beginning of the reaction. The ligation was performed at 20°C for 12 h, and terminated by heating the mixture at 65°C for 10 min.
Single-addition cyclization in diluted T4 ligase buffer
The 1 × T4 DNA ligase buffer from the commercial source was diluted by water, and the ligation was achieved in these media. Otherwise, all the procedures were exactly the same as those in conventional cyclization in 1 × T4 ligase buffer.
‘Step by Step’ cyclization involving intermittent addition to diluted T4 ligase buffer
The 1 × T4 DNA ligase buffer was diluted by 10- or 20-fold. Single-stranded linear DNA substrate was separated into several portions, and each portion was step by step added to the mixture with appropriate interval time. Other reaction conditions were the same as the ones described above for the other two methods (20°C for 12 h). Detailed procedures are presented in Results section and in Supplementary materials.
Evaluation of selectivity for the formation of DNA rings
After the reactions, all the products were diluted or concentrated, and loaded on denaturing polyacrylamide gel (dPAGE). For shorter and longer l-DNAs, 10–15% PAGEs were used. The data were analyzed by Image Lab software to quantify the fluorescence emission from each band. The selectivity for the formation of DNA rings was determined from the band intensities of c-DNA and the polymers. The results are averages of three individual experiments.
RESULTS
Formation of single-stranded DNA rings by T4 DNA ligase under conventional reaction conditions
Figure 1 shows the results of cyclization of l-DNA by T4 DNA ligase under conventional conditions. Commercially available 1 × T4 DNA ligase buffer was directly used for the media, and all the reagents were added at the beginning of the reaction. The substrate DNA was a 66-nt single-stranded linear DNA (l-DNA66) bearing a phosphate at the 5′-terminus. In the splint used (12-nt length), 6-nt in the 3′-side is complementary to the 5′-side of l-DNA66, and another 6-nt is complementary to the 3′-side (see Figure 1A). The sequences of l-DNA66 and splint-12 nt were listed in Supplementary Table S1. The intramolecular cyclization of l-DNA66 to c-DNA66 was successful, as shown by the band in the lower part of the denaturing PAGE in Figure 1C. However, a number of other bands assignable to the l-DNA66 polymers of different degrees of polymerization (Figure 1B) were simultaneously formed in the top part of the gel. As [l-DNA66]0 increased (from the left to the right), the intermolecular reactions to yield the polymers became dominant, as expected. When [l-DNA66]0 = 5, 10 and 30 μM, the selectivities for the formation of target cyclization product were only 58, 47 and 27%, respectively.
Optimization of splint length
In Figure 2, the length of splint was optimized for the formation of c-DNA66. In all the cases, the nick site was placed exactly at the center of the splint. With the splint-8 nt, no c-DNA66 was formed, whereas splint-10 nt gave only a little cyclization product with considerable amounts of polymeric products (large amount of l-DNA66 remained unreacted). This is probably because the duplexes between l-DNA66 and the splints (two 4-bp duplexes for splint-8 nt and two 5-bp duplexes for splint-10 nt, respectively) are too short and unstable under the reaction conditions. These results are consistent with previous results that effective ligation by this enzyme requires 11-bp or longer duplex (16). With the splints of 12-, 14-, 16- and 20-nt, the circular DNA ring was successfully formed. However, the polymerization was notable for longer splints, being evident especially for splint-20 nt. According to these results, splint-12 nt was used throughout the following experiments.
Figure 2.
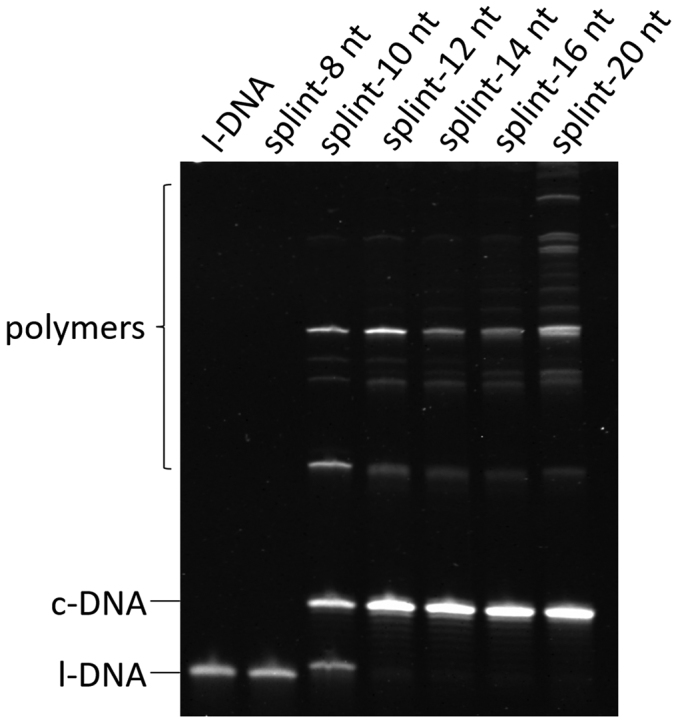
Effects of the length of splints on the efficiencies of cyclization of l-DNA66 and its polymerization under conventional conditions. All the DNA substrate was added to the mixture all at once at the beginning of the reaction. Each of the splints is complementary to equal number of nucleotides in the 5′- and 3′-ends of l-DNA66, respectively (the binding mode of splint-12 nt is presented in Figure 1A). The reaction conditions: [l-DNA66]0 = 5 μM; [splint]0 = 10 μM; 20 U T4 DNA ligase in 1 × T4 ligase buffer at 20°C and 12 h. The sequences of splints were listed in Supplementary Table S1.
Reduction of T4 ligase buffer concentration to promote the cyclization with respect to the polymerization
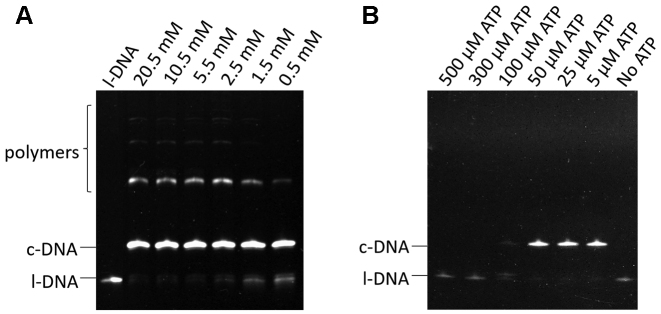
In the course of optimization of the reaction conditions, we unexpectedly found that the formation of polymeric products is drastically suppressed with decreasing concentration of T4 ligase buffer (Figure 3). On the other hand, the rate of cyclization reaction was affected only in much less extent. In 0.05 × T4 ligase buffer (0.05 × Buf., the third lane from the right), for example, the polymeric products were hardly detected, and almost all of l-DNA66 was converted to c-DNA66. In 0.01 × buffer, the selectivity for the cyclization was also very high, but the yield of c-DNA66 was rather low. In this enormously diluted ligation buffer, the enzymatic activity is insufficient.
Figure 3.
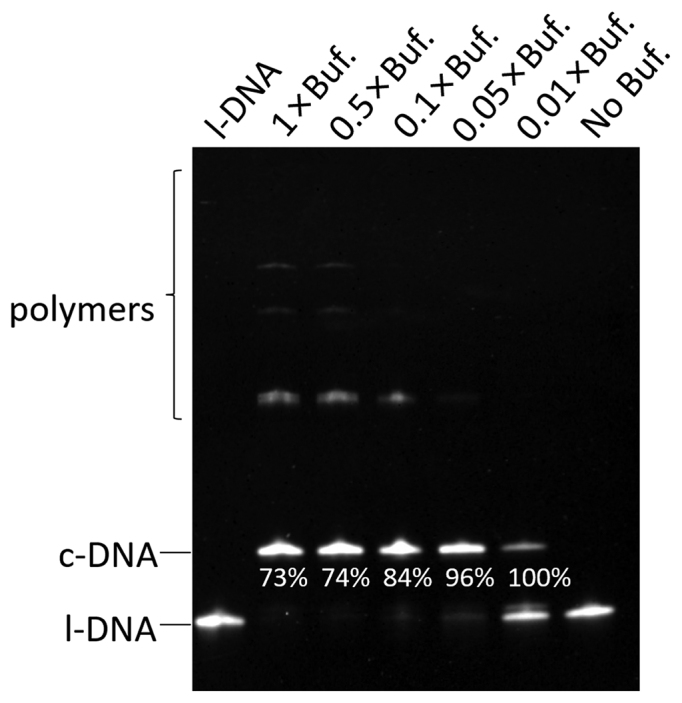
Effects of the concentration of T4 ligase buffer on the efficiencies of cyclization of l-DNA66 and its polymerization. All the DNA substrate was added to the mixture all at once at the beginning of the reaction. The selectivity for the formation of c-DNA66 is presented below the corresponding band. The reaction conditions: [l-DNA66]0 = 1 μM; [splint-12 nt]0 = 2 μM; 5 U T4 DNA ligase at 20°C and 12 h. Note that 1 × T4 ligase buffer contains 10 mM MgCl2, 500 μM ATP, 10 mM DTT and 40 mM Tris-HCl.
The origin of ‘low buffer-concentration effect’ for the T4 DNA ligase-mediated cyclization
Besides Mg2+, ATP is also present in the buffer as the cofactor. In order to figure out the origin of the remarkable effect of buffer concentration, the dependencies of the efficiencies of enzymatic cyclization and polymerization on the concentration of either Mg2+ or ATP were investigated. In Figure 4A, the concentrations of all the components (except for [Mg2+]0) were kept constant at the values in 0.05 × T4 ligase buffer ([ATP]0 = 25 μM, [DTT] = 0.5 mM, and [Tris–HCl] = 2 mM). With decreasing [Mg2+]0, formation of the polymers was monotonously suppressed and got to almost nil when [Mg2+]0 = 0.5 mM (the value in 0.05 × buffer). However, efficiency of the cyclization was not much changed in the range investigated. Apparently, the ‘low buffer-concentration effect’ is primarily ascribed to the decrease in [Mg2+]0 in the media. Although the dilution of T4 ligase buffer can also reduce its pH-controlling ability, the improvement of ligation selectivity is not attributed to the pH change. We found that efficient cyclization could be obtained at a pH range of 7.0–8.5 (data not shown).
Figure 4.
Effects of (A) [Mg2+]0 and (B) [ATP]0 on the cyclization of l-DNA66 by T4 DNA ligase. [l-DNA66]0 = 1 μM, [splint-12 nt]0 = 2 μM, 5 U T4 DNA ligase at 20°C and 12 h. [ATP]0 = 25 μM, [DTT] = 0.5 mM, and [Tris–HCl] = 2 mM in (A), whereas [MgCl2] = 0.5 mM, [DTT] = 0.5 mM, and [Tris–HCl] = 2 mM in (B).
In Figure 4B, [ATP]0 was varied and the concentrations of the other components were kept constant at the values in the 0.05 × T4 ligase buffer. The cyclization efficiency was almost unchanged in the range of 5–50 μM. In 0.05 × T4 buffer, [ATP]0 = 25 μM, so that the decrease in [ATP]0 is irresponsible for the ‘low buffer-concentration effect’. It was also found that too high concentrations of ATP (>100 μM) are unfavourable for the cyclization. Interestingly, in 1 × T4 buffer, the concentration of ATP is as high as 500 μM, and the ligation can be carried out efficiently if the [Mg2+] is as high as 10 mM (Supplementary Figure S1). The concentration of ATP higher than 100 μM decreased the ligation efficiency greatly only when the concentration of Mg2+ is much lower.
‘Step by Step’ method to prepare single-stranded DNA rings in high yield
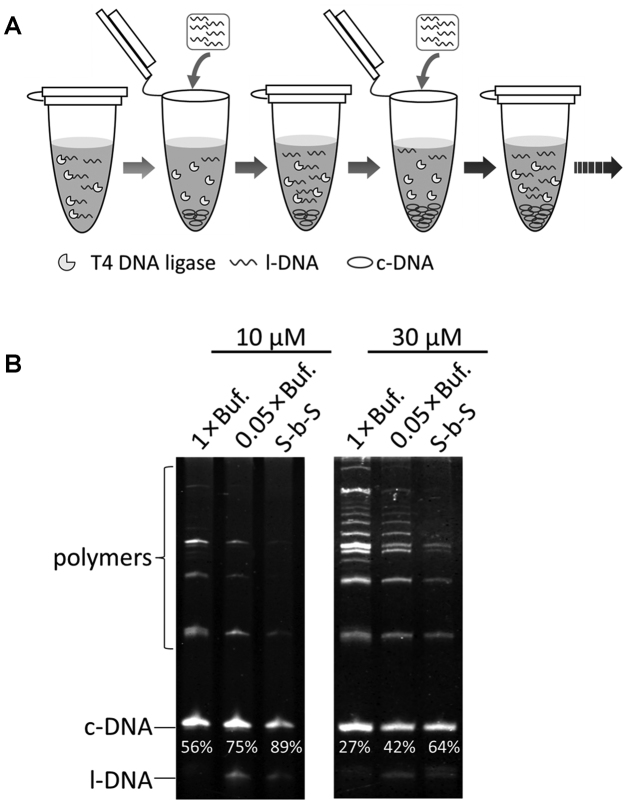
Although the polymerization is notably suppressed in diluted T4 ligase buffers, this side reaction is completely negligible only when [l-DNA]0 is extremely small (Supplementary Figure S2). For practical synthesis, however, [l-DNA]0 cannot be decreased too much, since then the amount of c-DNA obtainable in a reaction vessel should be minimized. In 1999, Prunell et al. reported that the yield of circular DNA duplex was improved when the duplex substrate with sticky ends was added gradually to the ligation solution (17). Accordingly, we developed a new strategy to fulfill both high selectivity and high productivity (Figure 5A). In this method, l-DNA substrates are separated to several portions, and each of them is intermittently added to the diluted T4 buffer. Here, the idea is that the next portion of l-DNA is added only after most of the substrates in reaction mixture are consumed so that its concentration is persistently kept low. With this method, large amount of DNA rings can be obtained in high selectivity and high yield.
Figure 5.
‘Step by Step’ cyclization of l-DNA. (A) Schematic view of the method. (B) Yields of c-DNA66 by this method (S-b-S). The cumulative concentrations of l-DNA66 (the concentrations of all the substrates added to the mixture) = 10 μM (left) and 30 μM (right). Details of the procedure should be referred to the main text. For the purpose of comparison, the results of conventional cyclization in 1 × buffer and single-addition cyclization in 0.05 × buffer are also shown. The selectivity for the formation of c-DNA66 is presented below the corresponding band.
First, the manner of stepwise addition of substrate was optimized. It was found that the selectivity for the cyclization was satisfactorily high (≥89%) as long as [l-DNA]0 < 5 μM (Supplementary Figure S2). Furthermore, under the conditions employed (5 μM of l-DNA in 0.05 × T4 buffer), >80% of l-DNA was converted to c-DNA within 20 min (Supplementary Figure S3). After 20 min, i.e. the concentration of unreacted l-DNA was <1 μM. If new l-DNA substrate was added (e.g. the increased concentration lower than 4 μM), the total concentration of unreacted l-DNA should be under 5 μM, and the by-products will always be kept <10%. Accordingly, hereafter the time interval was determined to be 20 min. A typical protocol is presented in Supplementary Table S2. At the beginning, the reaction mixture (20 μl) contained 2 μM l-DNA66, 25 μM splint and 10 U T4 DNA ligase in 0.05 × T4 DNA ligase buffer. Every 20 min, 3 μl l-DNA (20 μM) was added to the ligation system. After 7 times of addition, the total amount of added l-DNA66 became 10 μM concentration in the final reaction vessel. The T4 DNA ligase and 0.5 × buffer were also separated into two portions, and added to the mixture at the designated timings (see Supplementary Table S2).
By this new ‘Step by Step’ (S-b-S) method, the selectivity for the formation of c-DNA66 was as high as 89%, when the cumulative concentration of l-DNA66 was 10 μM (Figure 5B). This value was much higher than that of conventional cyclization method using 1 × buffer (56% selectivity). The S-b-S method was even better than single-addition cyclization method using 0.05 × buffer (75% selectivity). Note that the amount of l-DNA66 added was exactly the same for all the three methods. When the cumulative concentration of l-DNA66 was 30 μM (the protocol in Supplementary Table S3), the selectivity was 64% and also much higher than the values for the other two methods (42% and 27%). Here, 2 μl l-DNA (100 μM) was added to the ligation system each time, and the concentration of unreacted l-DNA may be more than 5 μM, causing a lower selectivity as compared with the former protocol (Supplementary Table S2).
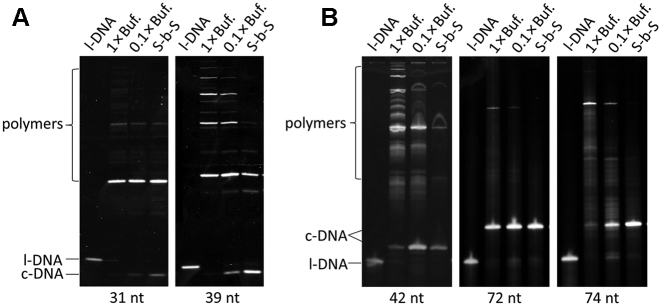
As shown in Figure 6, this new method (S-b-S) was successfully applied to the cyclization of versatile l-DNAs of different lengths (from 31-nt to 74-nt). In Table 1, the selectivities for the formation of c-DNA from these DNA substrates are listed. Compared with conventional cyclization in 1 × T4 ligase buffer and the single-addition cyclization in 0.1 × T4 ligase buffer, S-b-S method always gave the highest selectivity for the formation of single-stranded DNA rings. The productivities of the DNA rings (the amounts obtained from 1 ml reaction vessel) are also the highest for the S-b-S method, showing critical importance of this method for practical preparation of single-stranded DNA rings. The superiority of this method is especially evident, when the ring size is smaller (31- and 39-nt) and the formation of ring structure is less favorable. With conventional ligation, c-DNA31 and c-DNA39 were hardly formed (see the lanes of 1 × Buf. in Figure 6A). Even with the single-addition to 0.1 × T4 buffer, they were marginally formed. Only with S-b-S method, these small rings were successfully prepared in reasonable selectivity and productivity.
Figure 6.
Preparation of DNA rings by ‘Step by Step’ method from l-DNAs of various lengths. For the purpose of comparison, the results of conventional cyclization in 1 × T4 ligase buffer and single-addition cyclization in 0.1 × T4 ligase buffer are also shown. (A) l-DNA31 and l-DNA39. (B) l-DNA42, l-DNA72 and l-DNA74. The concentrations of l-DNA were 10 μM in (A) and 2 μM in (B). Sequences of all the l-DNAs are listed in Supplementary Table S1.
The usefulness of the new method is also very clear, when l-DNA is pretty long but takes a specific secondary structure. For example, l-DNA74 takes a solution structure in Supplementary Figure S4A, and the transition state for the cyclization, formed from five nucleotides, is sterically unfavourable. Accordingly, with conventional method, its cyclization is very inefficient (11.0% selectivity). The productivity is 5.4 μg/ml. Both the selectivity and the productivity are higher in 0.1 × T4 buffer (37.8% and 18.5 μg/ml). With the S-b-S method, however, the selectivity is still more greatly promoted to 85.3%, and the productivity is as high as 41.7 μg/ml. On the other hand, l-DNA72 is intrinsically easy to cyclize due to its favorable solution structure (see Supplementary Figure S4B). Even with the conventional method, the selectivity for the cyclization is pretty high (82.0%). However, the S-b-S method enormously promotes the cyclization still more, providing the DNA ring almost exclusively (99.7% selectivity). The productivity of c-DNA72 is 47.4 μg/ml, which corresponds to nearly quantitative conversion of l-DNA72 to the desired single-stranded DNA ring (the theoretical value is 47.5 μg/ml).
DISCUSSION
With the use of ‘Step by Step’ method, single-stranded DNA rings can be prepared in high selectivity and high productivity. The undesired intermolecular polymerization is enormously suppressed by reducing the concentration of T4 ligase buffer, according to our new finding. Almost all the enzymatic reactions in the literature have been accomplished under the conditions recommended by the manufactures using the buffers in the commercial packages. However, the present results indicate that these enzymes can achieve a variety of unprecedented functions simply by altering the reaction conditions slightly. Furthermore, the concentration of linear DNA substrate is kept low by intermittently addition approach. Obviously, the circular products have almost no effect on the subsequent circularization of linear substrate. As a result, the polymerization is restrained with respect to the intramolecular cyclization. By combining these two factors together, the enzymatic cyclization of l-DNA has been accomplished in enormously high efficiency.
As evidenced by Figure 4A, the ‘low buffer-concentration effect’ is primarily attributable to the decrease of Mg2+ concentration in the reaction mixture. This could be mainly originated from the effect of stronger electrostatic repulsion between the linear oligonucleotides for intermolecular ligation. The approach of one single-stranded DNA molecule with negative charges to another one should be more difficult when [Mg2+]0 is smaller, as compared with the intramolecular approach. In other word to say, the concentration effect (difference between intermolecular and intramolecular) works well and intramolecular cyclization becomes much more favorable under low Mg2+ concentration.
For further understanding the interesting effect of low Mg2+ concentration, we tried to find some information from the ligation mechanism. It is well known that the nick ligation by T4 DNA ligase proceeds through three steps (18,19). First, the catalytically central Lysine residue of the ligase is adenylated by ATP. Then, this adenylate group is transferred to the 5′-phosphorylated end of a DNA fragment. The resultant ‘highly activated’ 5′-phosphate reacts with the 3′-OH end of another DNA fragment, resulting in the formation of phosphodiester linkage. According to detailed analysis by Cherepanov and de Vries (20,21), there are two parallel pathways in this enzymatic ligation. In a processive ligation, the enzyme transadenylates and seals the nick without dissociating from the DNA. Another is a nonprocessive ligation, in which the enzyme takes part in the abortive adenylation cycle (covalent binding of AMP to the Lysine, transadenylation of the 5′-phosphate, and its dissociation from DNA prior to the formation of phosphodiester linkage). The ligation of this pathway is completed by the nucleophilic attack of the 3′-OH towards the ‘highly activated’ 5′-phosphate to form the phosphodiester. Although the ligation in conventionally employed ligation buffer ([Mg2+] = 10 mM) mostly proceeds via the former pathway, the nonprocessive ligation is dominant when [Mg2+] is small (e.g. <1 mM). These previous studies show that the ligation in 0.05 × T4 buffer ([Mg2+] = 0.5 mM, even lower than the phosphate concentration on DNA under some conditions) overwhelmingly takes the nonprocessive pathway. In other words, the enzyme which has catalyzed the adenylation of 5′-phosphate is removed from the DNA, before the phosphodiester linkage is really formed.
Assuming, the ‘low buffer-concentration effect’ to drastically suppress the intermolecular polymerization, found in this paper, could be also originated from the dynamic characters of this ‘enzyme-free’ nicked double-stranded DNA. In this intermediate, each of the DNA fragment with either 3′-OH or adenylated 5′-phosphate is bound to the splint-12 nt simply by six Watson-Crick base-pairs. Thus, before these DNA fragments are covalently bound by a phosphodiester linkage, either (or both) of them could be removed from the splint and replaced with another equivalent DNA fragment in the solution. Due to entropic difference, the reconstruction of intermediate in the intermolecular polymerization is less efficient and slower than in the intramolecular cyclization. Thus, ‘low buffer-concentration effect’ could be ascribed to this inefficient reconstruction in the polymerization, in which the effect of electrostatic repulsion becomes obvious. Alternatively, the polymerization is more critically retarded, since the activated 5′-phosphate is for a longer time exposed to ambient water to be hydrolyzed to its inactive form. Another explanation is that the efficiency of T4 DNA ligase was too high under normal buffer conditions, and the concentration effect between intramolecular and intermolecular was concealed because the ligation can be carried out efficiently under much low concentrations.
Direct participation of ATP, another cofactor for T4 DNA ligase, to the ‘low buffer-concentration effect’ (increase of the selectivity of circular DNA) is unlikely. Instead, high concentration of ATP strongly inhibited the ligation itself under highly diluted T4 buffers (Figure 4 and Supplementary Figure S1). Yuan et al. (22) reported that ATP (>1 mM) significantly reduces the binding of DNA to the enzyme through the competition in non-covalent complexation with DNA. According to Rossi et al. (23), excess amount of ATP also leads to inability for the T4 DNA ligase to form with DNA a kind of stable complex (S complex), which is crucial for the ligation. Our results showed that the ratio between [ATP] and [Mg2+] is much more important for the competition of ligase binding between DNA and ATP than the concentration itself.
CONCLUSION
A new practical method (‘Step by Step’ cyclization) has been developed to synthesize single-stranded DNA rings efficiently for various sizes and sequences. Here, linear DNA substrate (bearing 5′-phosphate) is added intermittently to highly diluted T4 ligase buffer. Both high selectivity and high productivity are fulfilled, and large amount of DNA ring can be obtained by one-batch reaction. This is a successful solution to the long-lasting problem that a high selectivity for the cyclization is attainable only at very low substrate concentration and thus the productivity cannot be satisfactorily high. Simply by magnifying the size of the experiments in this paper (e.g. 100 ml), large amounts of single-stranded DNA rings (e.g. 10–20 mg), which would be sufficient for versatile applications, should be easily prepared. The purification process of the products should be easy because of the high selectivity of cyclization reactions. Further improvements of the reaction conditions should be possible, if necessary. The present new method could greatly facilitate the studies using single-stranded DNA rings and lead to still more remarkable progresses in the relevant fields.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31571937 to X.L., 31301420 to J.L.]; China Postdoctoral Science Foundation Funded Project [2015M572086 to X.P.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Qi X.J., Lu C.H., Cecconello A., Yang H.H., Willner I.. A two-ring interlocked DNA catenane rotor undergoing switchable transitions across three states. Chem. Commun. 2014; 50:4717–4720. [DOI] [PubMed] [Google Scholar]
- 2. Lu C.H., Cecconello A., Qi X.J., Wu N., Jester S.S., Famulok M., Matthies M., Schmidt T.L., Willner I.. Switchable reconfiguration of a seven-ring interlocked DNA catenane nanostructure. Nano Lett. 2015; 15:7133–7137. [DOI] [PubMed] [Google Scholar]
- 3. Lu C.H., Cecconello A., Willner I.. Recent advances in the synthesis and functions of reconfigurable interlocked DNA nanostructures. J. Am. Chem. Soc. 2016; 138:5172–5185. [DOI] [PubMed] [Google Scholar]
- 4. Gaillard C., Strauss F.. Construction of DNA hemicatenanes from two small circular DNA molecules. PLoS One. 2015; 10:e0119368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu L., Lu C.H., Willner I.. Switchable catalytic DNA catenanes. Nano Lett. 2015; 15:2099–2103. [DOI] [PubMed] [Google Scholar]
- 6. Seeman N.C. DNA in a material world. Nature. 2003; 421:427–431. [DOI] [PubMed] [Google Scholar]
- 7. Rothemund P. DNA curvature and flexibility in vitro and in vivo. Nature. 2006; 440:297–302.16541064 [Google Scholar]
- 8. Douglas S.M., Chou J.J., Shih W.M.. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:6644–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kool E.T. Circular oligonucleotides: new concepts in oligonucleotide design. Biophysics. 1996; 25:1–28. [DOI] [PubMed] [Google Scholar]
- 10. Wang X., Li C., Gao X., Jing W., Liang X.. Preparation of small RNAs using rolling circle transcription and site-specific RNA disconnection. Mol. Ther.-Nucleic Acids. 2015; 4:e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y., Kuzuya A., Sha R., Guillaume J., Wang R., Canary J.W., Seeman N.C.. Coupling across a DNA helical turn yields a hybrid DNA/organic catenane doubly tailed with functional termini. J. Am. Chem. Soc. 2008; 130:10882–10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang X., Kuhn H., Frank-Kamenetskii M.D.. Monitoring single-stranded DNA secondary structure formation by determining the topological state of DNA catenanes. Biophys. J. 2006; 90:2877–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Billen L.P., Li Y.. Synthesis and characterization of topologically linked single-stranded DNA rings. Bioorg. Chem. 2004; 32:582–598. [DOI] [PubMed] [Google Scholar]
- 14. Wu Z.S., Shen Z., Tram K., Li Y.. Engineering interlocking DNA rings with weak physical interactions. Nat. Commun. 2014; 5:4279. [DOI] [PubMed] [Google Scholar]
- 15. Liu M., Zhang Q., Li Z., Gu J., Brennan J.D., Li Y.. Programming a topologically constrained DNA nanostructure into a sensor. Nat. Commun. 2016; 7:12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ng P.S., Bergstrom D.E.. Protein–DNA footprinting by endcapped duplex oligodeoxyribonucleotides. Nucleic Acids Res. 2004; 32:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prunell A., Alilat M., De L.. Nucleosome structure and dynamics. The DNA minicircle approach. Methods Mol. Biol. 1999; 119:79–101. [DOI] [PubMed] [Google Scholar]
- 18. Lehman I.R. DNA ligase: structure, mechanism, and function. Science. 1974; 186:790–797. [DOI] [PubMed] [Google Scholar]
- 19. Subramanya H.S., Doherty A.J., Ashford S.R., Wigley D.B.. Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell. 1996; 85:607–615. [DOI] [PubMed] [Google Scholar]
- 20. Cherepanov A.V., De V.S.. Kinetics and thermodynamics of nick sealing by T4 DNA ligase. Eur. J. Biochem. 2003; 270:4315–4325. [DOI] [PubMed] [Google Scholar]
- 21. Cherepanov A.V., De V.S.. Kinetic mechanism of the Mg2+-dependent nucleotidyl transfer catalyzed by T4 DNA and RNA ligases. J. Biol. Chem. 2002; 277:1695–1704. [DOI] [PubMed] [Google Scholar]
- 22. Yuan C., Lou X.W., Rhoades E., Chen H., Archer L.A.. T4 DNA ligase is more than an effective trap of cyclized dsDNA. Nucleic Acids Res. 2007; 35:5294–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rossi R., Montecucco A., Ciarrocchi G., Biamonti G.. Functional characterization of the T4 DNA ligase: a new insight into the mechanism of action. Nucleic Acids Res. 1997; 25:2106–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.