Abstract
Cricket paralysis virus (CrPV) is a dicistrovirus. Its positive-sense single-stranded RNA genome contains two internal ribosomal entry sites (IRESs). The 5′ untranslated region (5′UTR) IRES5′UTR mediates translation of non-structural proteins encoded by ORF1 whereas the well-known intergenic region (IGR) IRESIGR is required for translation of structural proteins from open reading frame 2 in the late phase of infection. Concerted action of both IRES is essential for host translation shut-off and viral translation. IRESIGR has been extensively studied, in contrast the IRES5′UTR remains largely unexplored. Here, we define the minimal IRES element required for efficient translation initiation in drosophila S2 cell-free extracts. We show that IRES5′UTR promotes direct recruitment of the ribosome on the cognate viral AUG start codon without any scanning step, using a Hepatitis-C virus-related translation initiation mechanism. Mass spectrometry analysis revealed that IRES5′UTR recruits eukaryotic initiation factor 3, confirming that it belongs to type III class of IRES elements. Using Selective 2′-hydroxyl acylation analyzed by primer extension and DMS probing, we established a secondary structure model of 5′UTR and of the minimal IRES5′UTR. The IRES5′UTR contains a pseudoknot structure that is essential for proper folding and ribosome recruitment. Overall, our results pave the way for studies addressing the synergy and interplay between the two IRES from CrPV.
INTRODUCTION
Viruses use various strategies to hijack the host cellular translational machinery in order to produce their viral proteins. Among these, positive-stranded RNA viruses downregulate host translation while increasing viral translation (1). For example, during poliovirus infection, an RNA structural element on the viral genome, also named internal ribosome entry site (IRES), is able to recruit the host ribosome while cap-dependent cellular translation is shut-off upon cleavage of essential canonical translation factors such as eukaryotic initiation factor eIF4G and PolyA Binding Protein (2–4). IRES has been classified into four main types according to their structural organization and their eIF requirement (5,6). Type I and II are large IRES that need most of the eIF except cap-binding protein eIF4E. Type I IRES recruits the ribosome upstream of the AUG start codon and then undergoes a scanning step to localize the AUG. In contrast, type II IRES loads the ribosome directly on the start codon without any scanning step. Type III IRES needs only eIF2 and eIF3 in order to bind directly to the 40S ribosomal subunit and to load the ribosome on the start codon without scanning. Finally, type IV IRES is the most compact: they usually contains pseudoknots, do not need any eIFs at all and can initiate translation on a non-AUG start codon.
The genome of Dicistroviridae consists of an ∼9 kb monopartite positive-stranded RNA containing two open reading frames, ORF1 and ORF2, which encode respectively non-structural and structural proteins. Cricket paralysis virus (CrPV) is a prototype member of this family that has been thoroughly investigated. Translation of viral proteins is exclusively driven by two IRES: the 5′UTR contains IRES5′UTR for ORF1 translation, while a type IV IRES is located in the intergenic region (IGR) IRESIGR between the two ORFs and controls expression of structural proteins from ORF2 (7–10). Both IRES act in synergy to produce the viral proteins required for rapid shut-off host protein translation to favor preferential viral protein synthesis. Whereas expression of non-structural proteins driven by IRES5′UTR is constant during the whole infectious process, the expression of structural proteins from the IRESIGR begins during the late phase of infection, with concentrations gradually increasing until reaching supramolar concentration at the end of infection (8–10). The dramatic increase in structural protein expression from the IRESIGR is directly dependent on the expression of non-structural proteins from IRES5′UTR, although the details of the underlying mechanisms remain elusive (11).
Translation initiation mediated by IRESIGR has been extensively studied in the last two decades (12–17). Briefly, a pseudoknot structure named PKI mimics a codon–anticodon interaction in the P-site of the ribosome, therefore allowing direct ribosome recruitment without any translation factors and translation initiation from a non-AUG codon (12,13,18–20). It was recently observed by Cryo-electron microscopy (Cryo-EM) that PKI enters the ribosome in the A-site of the ribosome and is then further translocated into the P-site by eEF2, leaving the A-site free to accept the first aminoacyl-tRNA in order to proceed to elongation (18,19,21). In contrast to IRESIGR, translation mediated by IRES5′UTR has been much less studied. The IRES5′UTR of the related dicistrovirus Rhapdosilum padi virus requires the scanning factors eIF1, eIF2 and eIF3 for efficient translation in a reconstituted cell-free translation extract (22). However, while the IRESIGR sequences of dicistroviruses are highly conserved (23), IRES5′UTR elements are largely variable and do not share any common consensus sequence suggesting the existence of different organizations among the Dicistroviridae family (24). Although it was discovered in early 2000, the CrPV IRES5′UTR remains largely uncharacterized (10). Recently, it was demonstrated that IRES5′UTR-driven translation requires the ribosomal protein RACK1 while the IRESIGR can still promote efficient translation initiation with RACK1-depleted ribosomes (25). Therefore, CrPV IRES5′UTR and IRESIGR are using fundamentally different strategies to initiate translation. In order to better understand the translational events leading to viral propagation, a better understanding of the structure and function of the CrPV IRES5′UTR is needed.
Here, we have characterized structurally and functionally the CrPV IRES5′UTR. We have mapped the IRES to the 760 nt-long 5′UTR and, using selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) and dimethyl sulfate (DMS) probing, established a secondary structure model of the entire 5′UTR. Then, we have determined the minimal IRES element, and shown that it contains a pseudoknot structure. The existence of the pseudoknot was validated by mutational analysis. Finally, we have shown that this structure is essential for proper folding and activity of the IRES5′UTRin vitro in drosophila cell-free translation extracts and in vivo in S2 cells.
MATERIALS AND METHODS
RNA transcription
Renilla reporter mRNAs were synthesized from DNA template by in vitro transcription using recombinant T7 RNA polymerase. After transcription, unincorporated nucleotides were trapped on a G-25 column and RNA transcripts were phenol-extracted and precipitated. RNA pellets were resuspended in water and their concentration was determined by absorbance measurements.
RNA translation in cell-free translation extract
In vitro translation competent extracts from Drosophila melanogaster S2 cells were prepared as previously described (25,26). Cells were lysed in 40 mM HEPES–KOH [pH8], 100 mM potassium acetate, 1 mM magnesium acetate, 1 mM DTT at a density of 109 ml−1 using a Cell Disruption Bomb (Parr Instrument Company). The lysate was then cleared by centrifugations at 4°C and supplemented with creatine kinase at 0, 24°C, aliquoted and stored at −80°C. In vitro translation experiments were performed as previously described under subsaturating conditions to avoid substrates titration (25,26). Translation efficiency was determined by Renilla Luciferase assay. RNA integrity of translated reporter mRNAs was checked by polyacrylamide gel electrophoresis (PAGE) on denaturing 4% acrylamide gels.
RNA translation in S2 cells
The pACT5C–IRES5′CrPV–Renilla Luciferase was mutated by site-directed mutagenesis to obtain the m1–m4 plasmids (25). Drosophila S2 cells were transfected with reporter plasmid DNAs by the CaPO4 precipitation method (adapted from (27)). Twenty-four hours later, medium was changed and copper was added to the culture medium (0.5 mM) to induce the expression of the capped Firefly construct (pMT-Firefly). Forty-eight hours later, cells were lysed and luciferase activity was measured with the Promega Dual-Luciferase assay, using a Berthold luminometer.
SHAPE analysis
Full-length RNA was designed to comprise the following elements: a leader sequence (19 nt), the entire CrPV 5′UTR (709 nt), the beginning of the ORF1 (47 nt), a 3′ linker and a generic reverse transcription primer-binding site (28). Mutants comprising the 357–754 sequence did not possess any leader or 3′ linker and generic primer-binding site. RNA was transcribed and purified on 4% acrylamide denaturing gels.
RNA folding assays
For the full-length wt RNA, 18 pmoles RNA in 11 μl of a buffer containing 25 mM NaCl, 15 mM MgCl2, 25 mM Na cacodylate pH 6.5, were incubated at 60°C for 2 min, except for the no-refolding (NR) control, which was not heated. Then, ‘Ra’ samples underwent 1-h long cool down to 37°C. A total of 10 pmoles of NR RNA were used for SHAPE modification (see below). A total of 8 pmoles of NR and refolded RNA were added to an equivalent volume (5 μl) of loading buffer (50% glycerol, 2.5 mM Na cacodylate pH 6.5). Native PAGE was performed at 4°C on 4% acrylamide, 10 mM glycine, 10 mM Tris base. RNA were detected with toluidine blue.
For each wt or mutant RNA (m1–m4), 15 pmoles RNA in 10 μl of translation buffer and incubated at 50°C for 5 min, except for NR controls. Then, ‘Rb’ samples were kept at 25°C for 10 min, while ‘Rc’ samples underwent a 15 min-long cool down to 25°C. A total of 4–8 pmoles of Rb RNA were used for SHAPE modification.
SHAPE
SHAPE was performed in 10 μl containing 2 pmoles RNA (0.5 μM final concentration), 6.5 mM 1-methyl-7-nitroisatoic anhydride (1M7) or 80 mM benzoyl cyanide (BzCN; for full length wt RNA dataset only), 10% dimethylsulfoxyde (DMSO), 90 mM Na HEPES pH 8.0. Modification is complete after 10 min at RT (∼23°C). The modified RNA was precipitated, washed, dried and resuspended in 0.5X TE.
Reverse transcription was performed in 20 μl containing 2 pmoles RNA (except for BzCN dataset: 3 pmoles RNA), 0.9 pmoles of a fluorescently labeled primer (except for BzCN dataset: 6 pmoles; primers used: cr-rev421: 5′-GACCACGCGAGTCGTAATC-3′; cr-rev529: 5′-CAAGGGCTAACTAATCAGGTGTAC-3′; cr-rev769: 5′-GAGTTGATGTTGTTGGTTGCGTTG-3′; 3′gen: 5′-GAACCGGACCGAAGCCCG-3′), 160 U SuperScript III reverse transcriptase, 83 mM KCl, 56 mM Tris–HCl (pH 8.3), 0.56 mM each deoxynucleotides (dNTP), 5.6 mM DTT and 3 mM MgCl2. Denaturing occurred at 95°C for 2 min, followed by annealing at 65°C for 5 min and incubation on ice for 2 min. RT extension parameters were: 42°C for 2 min, 50°C for 30 min and 65°C for 5 min. Sequencing reactions were performed in parallel in similar conditions, but containing 0.5 mM dideoxythymidine triphosphate (ddTTP). Reactions were stopped by the addition of 4 μl 50 mM EDTA pH 8.0, phenol–chloroform extracted, precipitated, washed, dried and resuspended in 10 μl deionized formamide. Samples were loaded on a 96-well plate for sequencing on an Applied Biosystems 3130xl genetic analyzer.
The resulting electropherograms were analyzed using QuSHAPE (29), which aligns signal within and across capillaries, as well as to the dideoxy references and corrects for signal decay. Normalized reactivities range from 0 to ∼2, with 1.0–1.2 being the average reactivity for highly reactive positions. For the sake of simplicity, the final *.shape file contains reactivities for the full length RNA (752 nt) that were combined from reactivities for nucleotides 1–353 from primer cr-rev421, reactivities for nucleotides 354–711 from primer cr-rev769 (three datasets with 1M7 modification) and reactivities for nucleotides 712–752 from primer 3′-gen (two datasets with BzCN modification, for which 75% of the residues have reactivities either >1.2 or undetermined, which suggest that the region is highly unstructured).
Structure prediction
Secondary structure prediction was obtained using the Fold and ShapeKnots algorithms available in RNAStructure v. 5.7 (30). The recommended parameters for pseudoknot and probing-based prediction were used, using the *.shape file as input. A value of −500 was given to nucleotides for which reactivities were not determined. The output 2D models were rendered using VARNA (31). The secondary structure of the region 357–466 region was used as input for automated three-dimensional (3D) modeling using the web-based RNAComposer server (32), using default parameters.
DMS probing
Like the SHAPE experiments, the DMS probing reactions were performed on 2 pmoles of the minimal IRES5′UTR. Briefly, the RNA is incubated for 10 min in dimethylsufate (DMS) buffer (50 mM Na cacodylate, pH 7.5, 5 mM MgCl2 and 100 mM KCl). The RNA was modified in the presence of 1.25% DMS. The reaction is performed for 10 min at 20°C and terminated on ice. Then, the modified RNA are precipitated in ethanol. The modification sites were detected by primer extension with two fluorescent primers complementary to nucleotides 732–755 and 517–707, respectively. The resulting electropherograms were analyzed by the same method as previously described for SHAPE analysis.
CMCT probing
The 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate (CMCT) experiments were performed on 4 pmoles RNA. The RNA is incubated for 10min in CMCT buffer (Na borate 50 mM pH 8, 5; MgCl2 5 mM; KCl 100 mM added with 1 μg of total tRNA). The modifications were done with 10, 5 g/l; final CMCT. The reaction is completed in 30 min at 20°C, then the RNA is precipitated. The primer extension and capillary electrophoresis steps are the same as for the DMS probing.
Sucrose gradients
Pre-initiation complexes were assembled on 3′ 32P-labeled RNA transcript by incubation in S2 cell-free extracts in the presence of 2 mM of a non hydrolysable GTP analog (GMP-PNP). The complexes are then resolved on sucrose gradients 7–47% and centrifuged in a SW41 rotor for 2 h 30 min at 37K at 4°. After centrifugation, the gradient is collected into 45 distinct fractions, then the positions of pre-initiation complexes were monitored by detection of radio-labeled RNA transcripts by Cerenkov counting of each fraction.
Mass spectrometry analysis and data processing.
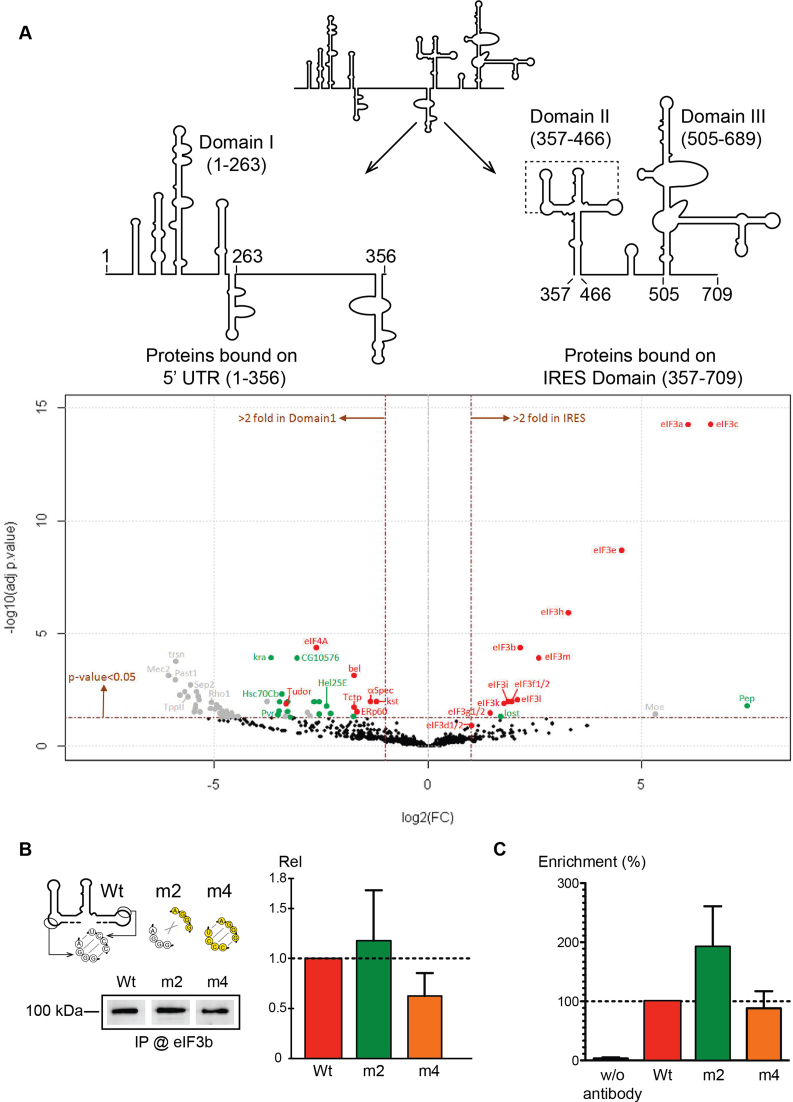
Protein extracts were digested with sequencing-grade trypsin (Promega) as described previously (33). Resulting peptides were analyzed by nanoLC-MS/MS and MS data were searched by the Mascot algorithm against the UniProtKB D. melanogaster database. Identifications were validated with a protein False Discovery Rate (FDR) < 1% using a decoy database strategy. The total number of MS/MS fragmentation spectra was used to quantify each protein from three independent biological replicates. This spectral count was submitted to a negative-binomial test using an edgeR GLM regression through the R-package. For each identified protein, an adjusted P-value corrected by Benjamini–Hochberg was calculated, as well as a protein fold-change (FC = average spectral count in IRES/average spectral count in Domain I). The results are presented in a Volcano plot using protein log2 fold changes and their corresponding adjusted log10P-values highlighted proteins upregulated in each condition (Domain I and IRES).
RNA immunoprecipitation
The reporter plasmids containing the Wt IRES5′UTR, mut2, mut4 upstream of Renilla coding sequence were transfected into drosophila S2 cells (Invitrogen) by the Effectene method following manufacturer’s instructions (Qiagen). Forty-eight hours later, cells were lysed and immunoprecipitated with an eIF3b antibody (gift from M. Hentze) coupled to Dynabeads Protein G (Life technologies). After overnight incubation, RNA and proteins were extracted with TriZol reagent (MRC). For reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR), 500 ng RNA were used to perform reverse transcription (iScript cDNA Synthesis Kit from BioRad) followed by qPCR (BioRad SYBR-Green). Primers qPCR Renilla: Fw 5′-GGATGATAACTGGTCCGCAG-3′, Rev 5′-TTGCCTGATTTGCCCATACC-3′.
RESULTS AND DISCUSSION
The 712-nt long 5′UTR of CrPV contains an IRES that mediates translation of ORF1 polyprotein (7). In order to characterize the minimal IRES element, we inserted nucleotides 1–761 from the 5′UTR or truncated versions upstream of the reporter gene Renilla luciferase which leads to the production of a fusion protein containing the first 17 viral N-terminal amino acids fused to Renilla luciferase peptide sequence. These constructs were in vitro transcribed and used for in vitro translation assays using S2 cell-free translation extracts. Renilla luciferase activity was used to measure the IRES translational activity driven by each RNA construct.
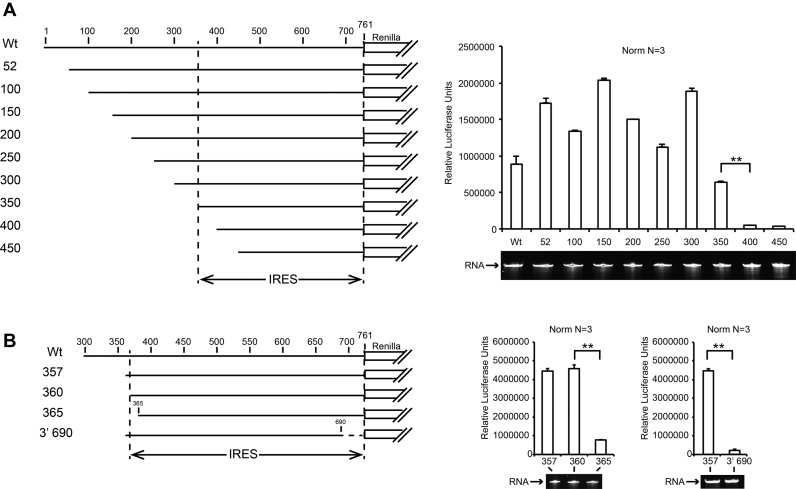
First, we demonstrated that capped and uncapped RNA constructs are stable in S2-cell extracts and have identical translational activity (Supplementary Figure S1). We only used uncapped RNA in subsequent experiments. We then proceeded to map the location of the IRES. Truncations from the 5′ end revealed that the 5′ distal half of the 5′UTR is not required for IRES activity, but that the 5′ proximal half is essential (Figure 1A). More precise 5′ and 3′ truncations revealed that the minimal IRES element is located between nucleotides 357 and 761 (Figure 1B).
Figure 1.
Mapping of the minimal IRES sequence in the 5′UTR of CrPV by 5′ and 3′ truncation. On the left panels, a cartoon representation of Renilla luciferase reporter transcripts used in S2 cell-free translation extracts is shown. On the right panels, translation efficiencies are represented as raw bioluminescence activity (Relative Light Units or RLU) for each transcript. Standard deviations or translational activity for each transcript are shown and calculated from three independent experiments. **P < 0.005 based on Student‘s t-test. RNA integrity was controlled by 4% denaturing polyacrylamide gel electrophoresis (PAGE) and Ethidium Bromide staining. (A) IRES mapping within the entire CrPV 5′UTR and (B) precise mapping of 5′ and 3′ ends of the IRES5′UTR from CrPV.
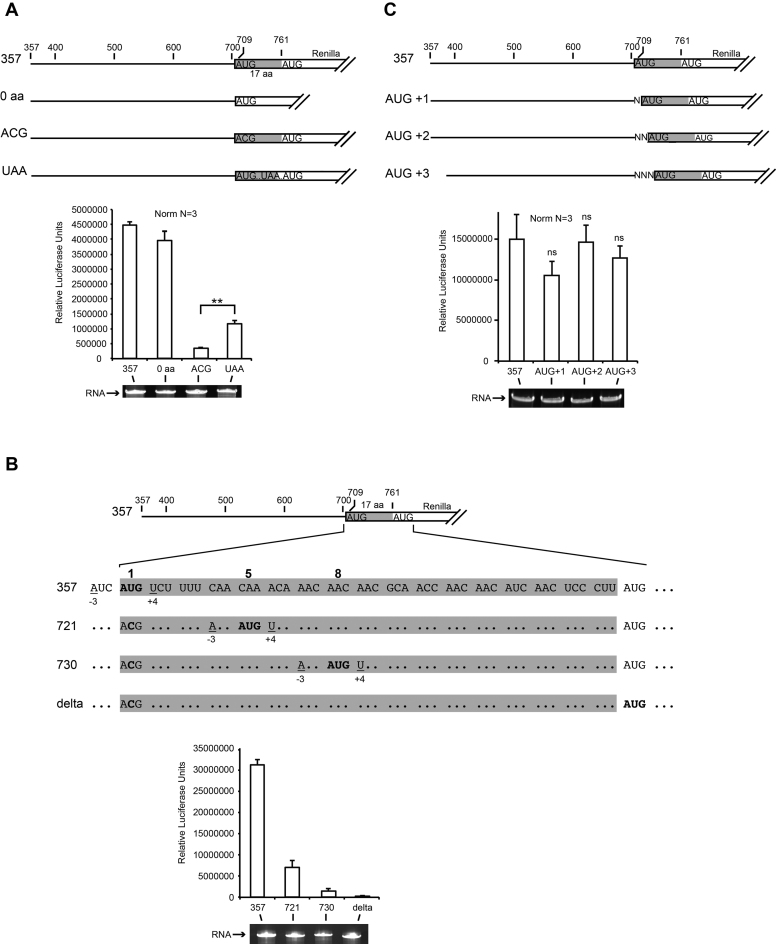
Translation efficiency was not modified when the N-terminal viral sequence coding for the first 17 amino acids was deleted, indicating that the minimal IRES5′UTR is located precisely between nucleotides 357 and 709 (see construct 0 aa in Figure 2A). We next investigated whether the IRES5′UTR is able to recruit the ribosome and promote scanning for a downstream AUG start codon. We mutated the viral AUG start codon at position 709 to ACG and observed a dramatic decrease of translation activity. This indicates that IRES5′UTR is not able to promote scanning to reach the Renilla luciferase AUG codon further downstream and that it needs a genuine AUG start codon in its immediate vicinity (Figure 2A). To further analyze the ability of IRES5′UTR to drive efficient ribosomal scanning, we re-introduced in-frame AUG start codons (at codon number 5 and 8) downstream of the cognate AUG start codon mutated to ACG. In order to avoid AUG context effects, we kept the wt AUG flanking sequence, namely A at position −3 and U at position +4 for these AUGs. (Figure 2B). When the start codon was placed at codon 5, the translation efficiency was already dramatically reduced (∼4-fold reduction), and when it was placed at codon 8 the IRES5′UTR was almost totally inactive.
Figure 2.
AUG start codon recognition during CrPV IRES5′UTR-driven translation initiation Renilla luciferase reporter transcripts used in S2 cell-free translation are represented as raw bioluminescence activity (RLU) for each transcript. Standard deviations or translational activity for each transcript are shown and calculated from three independent experiments. **P < 0.005 based on Student’s t-test; ns, nonsignificant. RNA integrity was controlled by 4% denaturing PAGE and Ethidium Bromide staining. Viral coding sequences are shown in gray and fused to Renilla luciferase coding sequence. (A) IRES5′UTR drives translation initiation on viral cognate AUG start codon but does not promote scanning further downstream to find the Renilla AUG when the viral AUG is mutated to ACG (B) In these transcripts, the cognate viral AUG start codon is mutated to ACG. In addition in-frames AUGs were inserted at codon position 5 and 8 with the same context than the wt viral AUG. The 5′ proximal AUG codons are shown in bold. (C) In these transcripts the cognate viral AUG is shifted by 1, 2 or 3 nts.
Interestingly, when a stop codon is introduced between the two in-frame AUGs, translation is less severely affected. This suggests that the ribosome cannot undergo a shunting mechanism to bypass the stop codon but rather terminates at the UAA codon and then proceed to partial re-initiation on the next Renilla AUG codon. This probably explains the better translation efficiency with the variant containing the UAA codon (Figure 2A). These experiments confirmed that IRES5′UTR drives efficient ribosome assembly on the genuine viral start codon but is not able to promote scanning for another AUG start codon further downstream. The observation that IRES5′UTR was able to drive translation initiation with the same efficiency for three other constructs containing frame shifting of the coding sequence by inserting 1, 2 and 3 nts upstream of the first AUG supports this interpretation (Figure 2C).
Altogether, our data show that IRES5′UTR recruits ribosomes to its cognate viral AUG start codon, and is not able to scan for an alternative AUG start codon further downstream. This is evocative of type III IRES (5), and contrasts with the model proposed for the related discistrovirus Rhapdopsilum padi virus (RhPV). Reconstitution experiments revealed that the IRES5′UTR from RhPV requires the scanning factor eIF1 in addition to eIF2 and eIF3 for translation and belongs to the type I category of IRES (22). Similarly, the IRES5′UTR from another positive-strand unclassified virus, Halastavi arva virus (34), behaves like a type I IRES and allows the recruited ribosome to perform retrograde scanning (35). The cadicivirus IRES5′UTR from the picornavirus family, which shares a similar dicistronic genome structure (36), is also a type I IRES that promotes normal 5′ scanning (37). These differences in the translation mechanism are consistent with the large differences both in size and sequence between the IRES5′UTR from members of this virus family, which contrast with the high conservation of the IRESIGR (10,23,24). They suggest that IRES5′UTR from dicistroviruses might use different strategies to recruit the host translational machinery, reflecting evolution of distinct host adaptation strategies among members of this virus family.
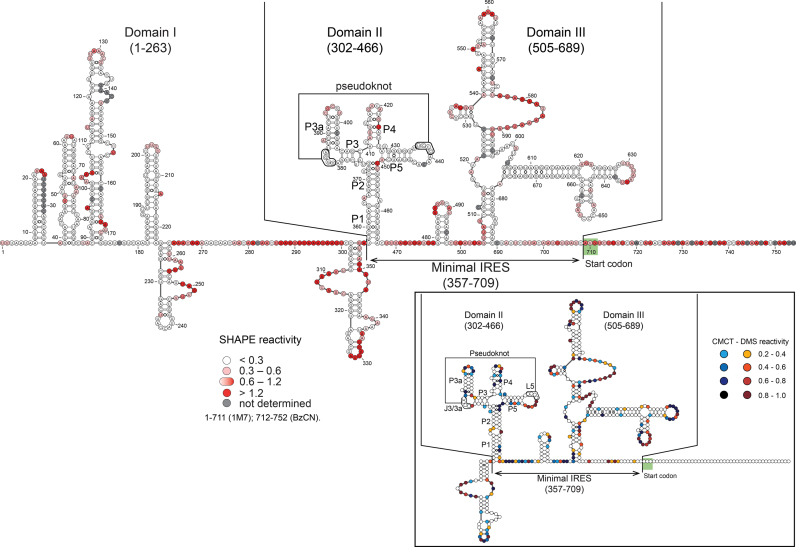
We next determined the secondary structure of the whole 5′UTR using the SHAPE method (28), after checking that the 709-nt long RNA was homogeneously folded (Supplementary Figure S2A). The resulting predicted 2D model shows that the overall 5′UTR contains three highly structured domains that are separated by flexible linkers (Figure 3). Domain I encompasses nucleotides 1–263 and contains five stem-loops. Domain II (302–466) and III (505–689) present a more sophisticated secondary structure with hairpins and three- and four-way junctions. Based on our deletion experiments (Figures 1 and 2), we mapped the minimal IRES5′UTR to nucleotides 357–709, which is in good agreement with our predicted secondary structure model. Indeed, construct 365 in which the first-half of P1 in domain II is deleted has a 4-fold reduction of its translation activity compared with constructs 360 and 357. Therefore, the minimal IRES5′UTR requires domains II and III to be fully active.
Figure 3.
The predicted secondary structure of the IRES5′UTR of CrPV reveals three highly structured domains named I, II and III separated by flexible linkers. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) data are overlaid on the structure prediction for the full-length 5′UTR RNA. Reactivities are shown as averages from three independent experiments (except for region 711–754: average from two experiments). Position of the minimal IRES is indicated (375–709). DMS and CMCT reactivity data for the minimal IRES domain (357–710). Reactivities are represented as averages from three independent experiments in the box of the right part of the figure. Nucleotides from J3/3a and L5 involved in long-distance interaction to form the pseudoknot are boxed. Reactivity values with standard deviations are listed in Supplementary Tables S1–4.
Domains II and III from IRES5′UTR are separated by a flexible region, which contains a 7-nt loop and 6 bp stem. Since domains II and III are sufficient for IRES5′UTR activity, we also determined the secondary structure of the minimal IRES5′UTR construct isolated from the full-length 5′UTR. The SHAPE analysis revealed highly similar reactivities between the full length and minimal IRES construct (Supplementary Figure S3) indicating that domain II and III retain their structure in an RNA fragment containing residues 357–754 (Pearson correlation coefficient RPearson = 0.8). Therefore, domains II and III can fold independently from domain I.
A closer look at the SHAPE reactivity in domain II revealed that loops J3/3a (382GGGA385) and L5 (436UCCC439) are completely inaccessible to the SHAPE reagent (Figure 4A; Supplementary Figure S2B and C). Moreover, the J3/3a and L5 loops contain complementary sequences suggesting long-range base pairs mediated by a pseudoknot structure, as the ShapeKnots algorithm helped us pinpoint (30). To strengthen this observation, we performed DMS and CMCT probing. The pattern of DMS/CMCT reactivities supports the 2D structure predicted on the basis of the SHAPE data (Figure 3, see insert). The absence of DMS/CMCT reactivity for the nucleotides involved in the putative pseudoknot structure confirms the SHAPE analysis and further supports the existence of a long-range interaction between these residues.
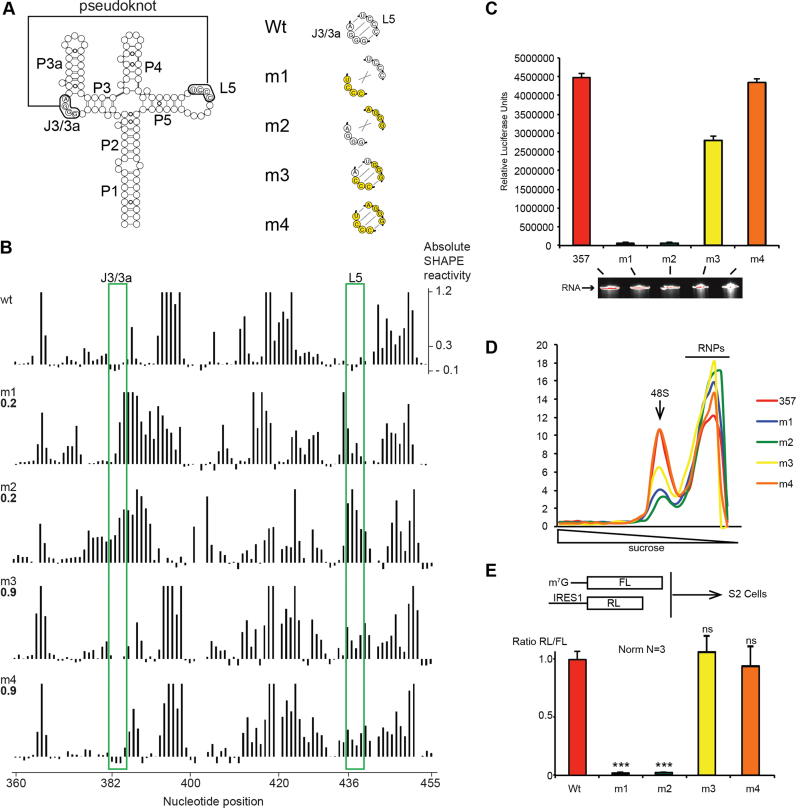
Figure 4.
A pseudoknot is required for correct folding and efficient IRES5′UTR -driven translation. (A) Mutants in the J3/3a and/or L5 regions used in this study, mutated nucleotides are shown in yellow. (B) Absolute SHAPE reactivities for 357–754 RNA transcripts with either wt or m1–m4 mutant sequence. Reactivities are shown as averages from two independent experiments (reactivity values with standard deviation are given in Supplementary Table S2). The Pearson correlation coefficients (RPearson) with the 357–754 wt dataset are shown on the left of each histogram. J3/3a and L5 are boxed in green. (C) Translation activity of IRES 357–754 wt and mutants m1–m4 when placed upstream of Renilla luciferase coding region. RNA integrity control by denaturing 4% sodium dodecyl sulphate-PAGE is shown under the histogram. (D) Pre-initiation complex assembly and analysis on 7–47% sucrose gradient with P32 radio-labeled wt and mutant m1–m4 IRES. (E) In vivo translation assay using monocistronic reporters transfected in S2 cells. The mutants m1–m4 are compared with wt IRES5′UTR activity. *P < 0.05 and **P < 0.005, ***P < 0.0005 based on Student’s t-test.
To demonstrate the presence of the pseudoknot structure, we constructed minimal IRES5′UTR fragments containing mutations in J3/3a and L5. Mutants m1 and m2 contained non-complementary sequences in the two loops, whereas mutants m3 and m4 contained compensatory mutations that are complementary but different from the wt sequence (Figure 4A). In compensatory mutant m3, 3 nts in the loops of the putative pseudoknot have been swapped whereas in m4, the 4 nts have been swapped. To assess the impact of these mutations on the IRES5′UTR secondary structure, we determined the SHAPE reactivity profiles of the four mutants in the buffer used for translation assays (see ‘Materials and Methods’ section; Supplementary Figure S4) and compared it with the wt IRES5′UTR. As expected, the loops J3/3a and L5 are more reactive to the SHAPE reagent in the non-complementary mutants m1 and m2 than in the wt sequence indicating an increased flexibility (Figure 4B, green boxes). On the contrary, compensatory mutants m3 and m4 have reduced accessibility to the SHAPE reagent in loops J3/J3a and L5, as observed for the wt IRES5′UTR, suggesting that these loops are indeed involved in a pseudoknot interaction. Moreover, mutations m1 and m2 not only destabilize the long-range interaction but also affect the overall folding of domain II when compared with the wt IRES5′UTR as shown by the Pearson correlation coefficients for both m1 and m2 mutants with the wt structure (RPearson = 0.2) (Alternative folds for m1 and m2 are presented in Supplementary Figure S5). Both compensatory mutants m3 and m4 present an overall folding of domains II and III very close to that of the wt IRES5′UTR (RPearson = 0.9) (Figure 4B). Altogether, these data demonstrate that loops J3/3a and L5 are involved in a pseudoknot structure and that this long-range interaction is essential to reach the native folding of domain II.
We next addressed the requirement of the pseudoknot structure for the IRES5′UTR translation activity. Mutants m1–m4 were inserted upstream of Renilla luciferase coding sequences in reporter constructs and the IRES activity was tested in in vitro translation assays with S2 cell-free extracts. The mutants m1 and m2 were inactive and unable to drive efficient translation, whereas mutants m3 and m4 showed partial or even fully restored IRES5′UTR activity (60 and 90% respectively) (Figure 4C). We conclude that the pseudoknot structure is essential for the IRES5′UTR activity, indicating that these long-range distances are required for proper folding and translation initiation. In addition, a 3 bp inversion in mutant m3 is less efficient than a full 4 bp inversion as shown by the near wt IRES activity driven by mutant m4.
In order to determine the precise role of the pseudoknot structure, we performed translation initiation complex assembly and analyzed these complexes by sucrose gradient analysis. We first checked that the mutations did not affect RNA stability in S2-cell extracts (Supplementary Figure S6). Then, in the presence of GMP-PNP (a non-hydrolysable GTP-analog), the wt IRES showed accumulation of the 48S pre-initiation complexes (Figure 4D and Supplementary Figure S7). As expected the null mutants m1 and m2 showed a significant decrease in the 48S complex formation. The compensatory mutant m3 and m4 restored 48S complex assembly although with different efficiencies. The most efficient mutant was m4 as already observed in previous in vitro translation experiments with S2 cell-free extracts (Figure 4C). Taken together, these experiments indicate that the pseudoknot structure in domain II is essential for 48S pre-initiation complex formation and is therefore essential at an early step of translation initiation.
Finally, mutants m1–m4 were tested for IRES activity in vivo using monocistronic reporters transfected in S2 cells (Figure 4E). Here again, m1 and m2 mutants were totally inactive, whereas the IRES activity was restored to the same extent for m3 and m4 mutants. Taken together, this demonstrates that the pseudoknot structure is essential for IRES5′UTR activity in vivo as well.
To further characterize the CrPV IRES5′UTR, we performed mass spectrometry analysis on translation initiation complexes in order to determine the factors that bind specifically to the IRES. To do so, translation initiation complexes were assembled on a biotinylated CrPV IRES5′UTR in S2-cell extracts. Translation initiation complexes were then first immobilized on magnetic streptavidin beads and subsequently eluted by DNAse treatment as previously described (33,38). The composition of the purified translation initiation complex was then determined by mass spectrometry. To determine the factors required at the initial step of the translational process, translation initiation was blocked by addition of edeine, which prevents initiator tRNAMet codon–anticodon interaction in the P-site of the ribosome (39–41). As a negative control, we performed the same experiments with the 5′ proximal part of the 5′UTR that is not essential for IRES activity. Interestingly, 12 of the 13 subunits of eIF3 are specifically found on the IRES fragment, indicating that IRES5′UTR recruits this factor (Figure 5A and Supplementary Table S5), the only missing factor being eIF3j. However, we have previously shown that eIF3j is required for the activity of IRES5′UTR (25). In yeast, eIF3j/hcr1 has been shown to participate in efficient AUG recognition during the scanning process by directly interacting with eIF1A for canonical cap-dependent translation (42). A similar function has also been proposed for mammalian eIF3j (43,44). Since we have shown that IRES5′UTR does not promote scanning for initiation on the viral AUG, our data show that eIF3j has a different function for IRES5′UTR. We propose that eIF3j is essential for ribosome recruitment to the IRES5′UTR and that it dissociates after complex formation prior to AUG recognition.
Figure 5.
Eukaryotic initiation factor (eIF)3 is recruited specifically by the IRES5′UTR in a pseudoknot-independent manner. (A) Mass spectrometry analysis of edeine-blocked translation initiation complexes on the 5′ proximal fragment (1–356) and the minimal IRES (357–709). Graphical representation of proteomics data: protein log2 spectral count fold changes (on the x-axis) and the corresponding adjusted log10P-values (on the y-axis) are plotted in a pairwise volcano plot. The significance thresholds are represented by a horizontal dashed line (P-value = 0.05, negative-binomial test with Benjamini–Hochberg adjustment) and two vertical dashed lines (−2.0-fold on the left and +2.0-fold on the right). Data points in the upper left and upper right quadrants indicate significant negative and positive changes in protein abundance. Protein names are labeled next to the off-centered spots and they are depicted according to the following color code: proteins represented by a red spot are identified by more than 30 MS/MS spectra, by a green spot when identified by 11–30 spectra and by a gray spot when identified by <10 spectra. Data points are plotted on the basis of average spectral counts from triplicate analysis. For an exhaustive list of hits, see Supplementary Table S5. (B) Western blots analysis with drosophila eIF3b antibody of edeine-blocked translation initiation complexes programmed with Wt and m2 and m4 mutants. The complexes were assembled in vitro in S2-cell extracts. The histogram represents the quantification of three independent experiments. (C) In vivo RNA immunoprecipitation of IRES5′UTR-Renilla reporter mRNA in S2 cells. Wt, m2 and m4 mRNA were immunoprecipitated using drosophila eIF3b antibody and quantified by RT-qPCR. The histogram represents the quantification of three independent experiments. The enrichment fold was calculated by the ratio between immunoprecipitated and the input and was set to 100% for the Wt.
In order to determine whether the pseudoknot is necessary for eIF3 recruitment, pre-initation complexes were programmed in vitro with S2-cell extracts with Wt, m2 and m4 and were purified as previously described. Western blot analysis with an antibody specific for drosophila eIF3b subunit showed that mutant m2 (which has no pseudoknot) is also able to recruit eIF3 suggesting that eIF3 interaction does not require the pseudoknot (Figure 5B). To further corroborate this statement in vivo, Renilla reporter mRNAs containing the Wt IRES5′UTR and mutants m2 and m4 were transfected in S2 cells. Then we performed RNA immunoprecipication with an antibody raised against drosophila eIF3b subunit, we quantified the Renilla reporter mRNA by RT-qPCR. Indeed, mutant m2 is also able to interact with eIF3 in vivo confirming that the pseudoknot is not required for eIF3 recruitment (Figure 5C). The interaction between the IRES5′UTR and eIF3 is transient during the translation initiation process. In both experiments, mutant m2 seems to interact more efficiently than Wt and m4, this might be explained by the fact that mutant m2 is inactive and therefore accumulates more complexes with eIF3 than Wt and m4.
The IRES5′UTR is preceded by domain I which is dispensable for translational activity. A few other proteins bind specifically to this domain (Figure 5A and Supplementary Table S5). These secondary structures might be involved in other steps required for viral propagation such as initiation of replication or encapsidation. Our data are in good agreement with previous observations made while constructing an infectious molecular clone of CrPV (45). The authors isolated a clone containing a duplication of fragment 75–271, which inhibits viral infectivity. In our secondary structure model, this duplication is located in domain I, away from the minimal IRES sequence. The duplication however has no effect on viral translation and RNA accumulation during CrPV infection suggesting it impacts viral entry and/or viral packaging (46).
In addition, the structured arrangements of domains II and III with several helices connected by three- and four-way junctions and loops is reminiscent of those in the classical swine fever virus (CSFV), the Hepatitis-C virus (HCV), the foot-and-mouth disease virus (FMDV) and other IRESes (6). These structured domains were shown to be important for IRES function, through binding to eIF3 (47), promoting long-range interactions (48,49), or binding to IRES-trans acting factors (50,51). The secondary structure of the apical domain II in CrPV IRES5′UTR that harbors a pseudoknot bears some resemblance to that of residues 153–255 in CSFV and residues 134–249 in FMDV (Supplementary Figure S8). Automatic 2D-based 3D-RNA modeling suggests that, because of the pseudoknot, the four-way junction would adopt a topology closer to that of the L-shaped CSFV (52), than to the topology modeled for FMDV (49). Future studies using X-ray crystallography or cryo-EM will be needed to pinpoint the similarities and differences in how type III IRES domains fold, and what role they play in IRES function.
In summary, the IRES5′UTR contains a three-way junction structure with a pseudoknot that recruits the ribosome on the cognate viral AUG start codon without any scanning step. Moreover IRES5′UTR specifically recruits eIF3. This is reminiscent of translation driven by the HCV–IRES, the prototypic type III IRES (5). This similarity between CrPV IRES5′UTR and HCV–IRES is in agreement with the fact that both IRES are strictly dependent on the ribosomal protein RACK1. On the contrary IRESIGR-driven translation efficiency does not require the presence of RACK1 on the ribosome, suggesting fundamentally distinct molecular mechanisms for ribosome recruitment (53). The structural characterization of the IRES5′UTR from CrPV will facilitate the investigation of the role played by RACK1 in cap-independent translation. During CrPV infection, the IRES5′UTR promotes translation from ORF1 in a constitutive manner during the whole infectious process, whereas IRESIGR-driven translation starts in the second half of infection and is boosted in the late phase of infection (11). The molecular basis for this differential expression pattern, which is essential for the progression of the viral infection, remains unexplored. Our data on the IRES5′UTR represent a first step toward a better understanding of the concerted action of the two structurally and functionally different IRES active in dicistroviruses.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Claire Batisse and Adeline Renaud for S2-cell extracts preparation. We are thankful to Matthias Hentze for drosophila eIF3 antibodies. We also thank Redmond P. Smyth for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique (CNRS); Université de Strasbourg; Investissement d’Avenir program [NetRNA ANR-10-LABX-36, ANR ANR-11-SVSE802501]. Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
REFERENCES
- 1. Walsh D., Mohr I.. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011; 9:860–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Joachims M., Van Breugel P.C., Lloyd R.E.. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 1999; 73:718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Etchison D., Milburn S.C., Edery I., Sonenberg N., Hershey J.W.. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220, 000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 1982; 257:14806–14810. [PubMed] [Google Scholar]
- 4. Kuyumcu-Martinez N.M., Joachims M., Lloyd R.E.. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J. Virol. 2002; 76:2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jackson R.J., Hellen C.U.T., Pestova T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010; 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Filbin M.E., Kieft J.S.. Toward a structural understanding of IRES RNA function. Curr. Opin. Struct. Biol. 2009; 19:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bonning B.C., Miller W.A.. Dicistroviruses. Annu. Rev. Entomol. 2010; 55:129–150. [DOI] [PubMed] [Google Scholar]
- 8. Garrey J.L., Lee Y.-Y., Au H.H.T., Bushell M., Jan E.. Host and viral translational mechanisms during cricket paralysis virus infection. J. Virol. 2010; 84:1124–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moore N.F., Kearns A., Pullin J.S.. Characterization of cricket paralysis virus-induced polypeptides in Drosophila cells. J. Virol. 1980; 33:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilson J.E., Powell M.J., Hoover S.E., Sarnow P.. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000; 20:4990–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khong A., Bonderoff J.M., Spriggs R. V, Tammpere E., Kerr C.H., Jackson T.J., Willis A.E., Jan E.. Temporal regulation of distinct internal ribosome Entry sites of the dicistroviridae cricket paralysis virus. Viruses. 2016; 8, doi:10.3390/v8010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilson J.E., Pestova T. V, Hellen C.U., Sarnow P.. Initiation of protein synthesis from the A Site of the Ribosome. Cell. 2000; 102:511–520. [DOI] [PubMed] [Google Scholar]
- 13. Costantino D.A, Pfingsten J.S., Rambo R.P., Kieft J.S.. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol. 2008; 15:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spahn C.M.T., Jan E., Mulder A., Grassucci R.A., Sarnow P., Frank J.. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell. 2004; 118:465–475. [DOI] [PubMed] [Google Scholar]
- 15. Pestova T. V., Hellen C.U.T.. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003; 17:181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jan E., Kinzy T.G., Sarnow P.. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:15410–15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jan E., Sarnow P.. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002; 324:889–902. [DOI] [PubMed] [Google Scholar]
- 18. Fernàndez I.S., Bai X.C., Murshudov G., Scheres S.H.W., Ramakrishnan V.. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014; 157:823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muhs M., Hilal T., Mielke T., Skabkin M.A., Sanbonmatsu K.Y., Pestova T. V., Spahn C.M.T.. Cryo-EM of ribosomal 80s complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol. Cell. 2015; 57:422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pfingsten J.S., Costantino D.A., Kieft J.S.. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006; 314:1450–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murray J., Savva C.G., Shin B.-S., Dever T.E., Ramakrishnan V., Fernández I.S.. Structural characterization of ribosome recruitment and translocation by type IV IRES. Elife. 2016; 5, doi:10.7554/eLife.13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Terenin I.M., Dmitriev S.E., Andreev D.E., Royall E., Belsham G.J., Roberts L.O., Shatsky I.N.. A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry. Mol. Cell. Biol. 2005; 25:7879–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanamori Y., Nakashima N.. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA. 2001; 7:266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roberts L.O., Groppelli E.. An atypical IRES within the 5′ UTR of a dicistrovirus genome. Virus Res. 2009; 139:157–165. [DOI] [PubMed] [Google Scholar]
- 25. Majzoub K., Hafirassou M.L., Meignin C., Goto A., Marzi S., Fedorova A., Verdier Y., Vinh J., Hoffmann J.A., Martin F. et al. RACK1 controls IRES-mediated translation of viruses. Cell. 2014; 159:1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wakiyama M., Kaitsu Y., Yokoyama S.. Cell-free translation system from Drosophila S2 cells that recapitulates RNAi. Biochem. Biophys. Res. Commun. 2006; 343:1067–1071. [DOI] [PubMed] [Google Scholar]
- 27. Green M.R., Sambrook J.. Molecular cloning: a laboratory manual. 2012; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 28. Wilkinson K.A., Merino E.J., Weeks K.M.. Selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006; 1:1610–1616. [DOI] [PubMed] [Google Scholar]
- 29. Karabiber F., McGinnis J.L., Favorov O.V, Weeks K.M.. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA. 2013; 19:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hajdin C.E., Bellaousov S., Huggins W., Leonard C.W., Mathews D.H., Weeks K.M.. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:5498–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Darty K., Denise A., Ponty Y.. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Popenda M., Szachniuk M., Antczak M., Purzycka K.J., Lukasiak P., Bartol N., Blazewicz J., Adamiak R.W.. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012; 40:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chicher J., Simonetti A., Kuhn L., Schaeffer L., Hammann P., Eriani G., Martin F.. Purification of mRNA-programmed translation initiation complexes suitable for mass spectrometry analysis. Proteomics. 2015; 15:2417–2425. [DOI] [PubMed] [Google Scholar]
- 34. Boros Á., Pankovics P., Simmonds P., Reuter G.. Novel positive-sense, single-stranded RNA (+ssRNA) virus with di-cistronic genome from intestinal content of freshwater carp (Cyprinus carpio). PLoS One. 2011; 6:e29145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abaeva I.S., Pestova T.V, Hellen C.U.T.. Attachment of ribosomal complexes and retrograde scanning during initiation on the Halastavi árva virus IRES. Nucleic Acids Res. 2016; 44:2362–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Woo P.C.Y., Lau S.K.P., Choi G.K.Y., Huang Y., Teng J.L.L., Tsoi H.-W., Tse H., Yeung M.L., Chan K.-H., Jin D.-Y. et al. Natural occurrence and characterization of two internal ribosome entry site elements in a novel virus, Canine Picodicistrovirus, in the Picornavirus-like superfamily. J. Virol. 2012; 86:2797–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Asnani M., Pestova T.V, Hellen C.U.T.. Initiation on the divergent Type I cadicivirus IRES: factor requirements and interactions with the translation apparatus. Nucleic Acids Res. 2016; 44:3390–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Prongidi-Fix L., Schaeffer L., Simonetti A., Barends S., Ménétret J.-F., Klaholz B.P., Eriani G., Martin F.. Rapid purification of ribosomal particles assembled on histone H4 mRNA: a new method based on mRNA-DNA chimaeras. Biochem. J. 2013; 449:719–728. [DOI] [PubMed] [Google Scholar]
- 39. Odon O.W., Kramer G., Henderson A.B., Pinphanichakarn P., Hardesty B.. GTP hydrolysis during methionyl-tRNAf binding to 40 S ribosomal subunits and the site of edeine inhibition. J. Biol. Chem. 1978; 253:1807–1816. [PubMed] [Google Scholar]
- 40. Garreau de Loubresse N., Prokhorova I., Holtkamp W., Rodnina M.V., Yusupova G., Yusupov M.. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014; 513:517–522. [DOI] [PubMed] [Google Scholar]
- 41. Kozak M., Shatkin A.J.. Migration of 40 S ribosomal subunits on messenger RNA in the presence of edeine. J. Biol. Chem. 1978; 253:6568–6577. [PubMed] [Google Scholar]
- 42. Aylett C.H.S., Boehringer D., Erzberger J.P., Schaefer T., Ban N.. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat. Struct. Mol. Biol. 2015; 22:269–271. [DOI] [PubMed] [Google Scholar]
- 43. ElAntak L., Wagner S., Herrmannova A., Karaskova M., Rutkai E., Lukavsky P.J., Valasek L.. The indispensable n-terminal half of eIF3j/hcr1 cooperates with its structurally conserved binding partner eIF3b/prt1-rrm and with eiF1A in stringent aug selection. J. Mol. Biol. 2010; 396:1097–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sokabe M., Fraser C.S.. Human eukaryotic initiation factor 2 (eIF2)-GTP-Met-tRNAi ternary complex and eIF3 stabilize the 43 S preinitiation complex. J. Biol. Chem. 2014; 289:31827–31836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kerr C.H., Wang Q.S., Keatings K., Khong A., Allan D., Yip C.K., Foster L.J., Jan E.. The 5′ untranslated region of a novel infectious molecular clone of the dicistrovirus cricket paralysis virus modulates infection. J. Virol. 2015; 89:5919–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kerr C.H., Wang Q.S., Keatings K., Khong A., Allan D., Yip C.K., Foster L.J., Jan E.. The 5′ untranslated region of a novel infectious molecular clone of the dicistrovirus cricket paralysis virus modulates infection. J. Virol. 2015; 89:5919–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hashem Y., des Georges A., Fu J., Buss S.N., Jossinet F., Jobe A., Zhang Q., Liao H.Y., Grassucci R.A., Bajaj C. et al. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature. 2013; 494:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fernández-Miragall O., Martínez-Salas E.. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003; 9:1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jung S., Schlick T.. Candidate RNA structures for domain 3 of the foot-and-mouth-disease virus internal ribosome entry site. Nucleic Acids Res. 2013; 41:1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pacheco A., Martinez-Salas E.. Insights into the biology of IRES Elements through riboproteomic approaches. J. Biomed. Biotechnol. 2010; 2010:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lozano G., Martínez-Salas E.. Structural insights into viral IRES-dependent translation mechanisms. Curr. Opin. Virol. 2015; 12:113–120. [DOI] [PubMed] [Google Scholar]
- 52. Hashem Y., des Georges A., Dhote V., Langlois R., Liao H.Y., Grassucci R.A., Pestova T. V, Hellen C.U.T., Frank J.. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013; 503:539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Majzoub K., Hafirassou M.L., Meignin C., Goto A., Marzi S., Fedorova A., Verdier Y., Vinh J., Hoffmann J.A., Martin F. et al. Ribosomal protein RACK1 is a specific host factor required for IRES-mediated translation of fly and human viruses. Cell. 2014; 159:1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.