Abstract
We report the synthesis of atom-specifically 13C-modified building blocks that can be incorporated into DNA via solid phase synthesis to facilitate investigations on structural and dynamic features via NMR spectroscopy. In detail, 6-13C-modified pyrimidine and 8-13C purine DNA phosphoramidites were synthesized and incorporated into a polypurine tract DNA/RNA hybrid duplex to showcase the facile resonance assignment using site-specific labeling. We also addressed micro- to millisecond dynamics in the mini-cTAR DNA. This DNA is involved in the HIV replication cycle and our data points toward an exchange process in the lower stem of the hairpin that is up-regulated in the presence of the HIV-1 nucleocapsid protein 7. As another example, we picked a G-quadruplex that was earlier shown to exist in two folds. Using site-specific 8-13C-2′deoxyguanosine labeling we were able to verify the slow exchange between the two forms on the chemical shift time scale. In a real-time NMR experiment the re-equilibration of the fold distribution after a T-jump could be monitored yielding a rate of 0.012 min−1. Finally, we used 13C-ZZ-exchange spectroscopy to characterize the kinetics between two stacked X-conformers of a Holliday junction mimic. At 25°C, the refolding process was found to occur at a forward rate constant of 3.1 s−1 and with a backward rate constant of 10.6 s−1.
INTRODUCTION
The function of nucleic acids is not only encoded in their ground state structure but also in their dynamic properties (1–9). These dynamics can occur over a wide range of timescales depending on the height of the activation barriers separating the alternative folding states (10). Solution NMR spectroscopy has proven to be a valuable tool to study functional dynamics and adequate methods to address kinetics from nanoseconds to hours are available (3,9). A crucial prerequisite to make use of these relaxation-based NMR experiments is the introduction of stable isotopes, such as 13C, 15N or 2H (11,12). For proteins labeling protocols are very advanced and in combination with sophisticated pulse sequences large proteins and even high molecular weight protein complexes can be studied via NMR spectroscopy and methyl group specific 13C-labeling (13–19). In the field of nucleic acid NMR spectroscopy, however, the isotope labeling strategies relying on the enzymatic incorporation of stable isotope labeled (deoxy)ribonucleotides were established in the mid-90s with minor progress since then (20–31). Very recently, the PLOR (position-selective labeling of RNA) method was introduced and holds the promise to site-specifically label RNA using stable isotope labeled ribonucleotide triphosphates and T7 RNA polymerase (32–34). This method will benefit from novel stable isotope labeled ribonucleotide triphosphate, which became recently available making use of a chemo-enzymatic approach (35–38). Concurrently, 13C/15N/2H-labeled RNA phosphoramidite building blocks and their applications for dynamic NMR of RNA were reported (39–43). The chemical labeling strategy proved to be a valuable expansion to the existing enzymatic protocols and will be especially beneficial when studying RNA modifications by NMR spectroscopy (44,45). The most serious limitation of the approach is the size of the RNA under investigation. Using standard tert-butyldimethylsily- (TBDMS) or triisopropylsilyloxy methyl- (TOM) protected RNA building blocks nucleic acids with about 50 nucleotides can be produced in sufficient amounts and the needed quality (>95% purity) for NMR spectroscopic investigations (46). For DNA, however, much larger constructs are accessible through solid phase synthesis as DNA is lacking the 2′-hydroxyl group and no steric interference by the 2′-protecting group during the coupling step is occurring. Thus, the synthesis of 2′-deoxyoligonucleotides has less stringent requirements compared to RNA and DNAs with up to 200 nucleotides can be commercially ordered. Further, 15N and 13C/15N uniformly labeled DNA phosphoramidites are available from companies. This would make solid phase synthesis the method of choice for NMR spectroscopic investigations of DNA. However, the pricing for these building block is considerable. The 13C/15N-DNA phosphoramidites are mainly used to for resonance assignment purposes in G-quadruplex studies on folding and structure with stable isotope dilution for economic reasons (5,47–49). Still, in some cases atom-specific isotope labeling would be advantageous especially for NMR studies on dynamics, for which isolated 1H-X (X = 13C or 15N) spin pairs with simple relaxation behavior and little scalar coupling interactions are important. Surprisingly, there are only scattered reports on the production of site-specifically 13C- or 15N-labeled DNA nucleoside and even a more limited number on the synthesis of stable isotope labeled DNA phosphoramidites so far (25,50–56).
Only recently, a unique feature of B-form DNA was discovered. DNA is able to fluctuate between a ground state Watson–Crick (WC) base paired state, which is well known form textbooks, and an excited state Hoogsteen (HG) base paired conformation (57–60). This high-energy state is populated to only a low extent (0.5–1%) and the exchange between the ground and excited states occurs on the millisecond time scale (kex = ∼3000 s−1 at room temperature). This behavior was revealed by relaxation dispersion NMR methods. The data strongly suggests that the sequence of a DNA double helix codes for the dynamics between WC to HG base pairs. This transient alternative base pairing states can in turn have implications for genome stability and repair or can act as an anchor in protein recognition. These findings underscore, that it would be very beneficial to introduce site- and atom-specific 13C labeled nucleotides into DNA to study such biologically important exchange processes. Furthermore, the synthetic approach can alleviate the problems in the resonance assignment process using site-specific labeling, which can be especially complicated in larger DNAs. The simple relaxation pathways in such atom-specific building blocks also simplify the application of NMR experiments to probe dynamics. Last but not least, the availability of stable isotope labeled DNA phosphoramidites will allow to address structure and dynamics in the presence of modifiers, like naturally occurring methylated residues or nucleotides with heteroatoms replaced with carbon (e.g. deazapurines). Thereby, a deeper understanding of intrinsic DNA dynamics and its function will be amenable.
Here, we present a first comprehensive report on the affordable synthetic access to atom-specifically 13C-modified DNA building blocks that can be incorporated into a target DNA via solid phase synthesis. The approach can be used to facilitate investigations on structural and dynamic features via NMR spectroscopy and can be regarded as an expedient addition to the existing enzymatic methods. The chemical synthesis of 6-13C-modified pyrimidine and 8-13C purine DNA phosphoramidites and their incorporation into DNAs up to 55 nucleotides is described. The approach significantly facilitates the resonance assignment process for a polypurine tract DNA/RNA hybrid with an intrinsically high degree of resonance degeneracy (61–63). We then addressed the applicability of the labeling strategy to study micro- to millisecond dynamics in the mini-cTAR DNA in the absence and presence of the HIV-1 nucleocapsid protein 7 (NCp7) (64,65). With the aid of site-specific 8-13C-guanosine labeling we were further able to verify a slow exchange between two G-quadruplex forms on the NMR chemical shift time scale (66). Using a real-time NMR experiment capitalizing on the Band-selective Excitation Short-Transient (BEST) approach the re-equilibration of the G-quadruplex folds after a temperature jump could be monitored. Finally, we used 13C ZZ exchange spectroscopy to showcase the applicability of the chemical 13C-labeling protocol to kinetically characterize the exchange between two stacked conformations of a four-way junction (67).
MATERIALS AND METHODS
Synthesis of 13C-labeled DNA phosphoramidites
A detailed description of the synthetic procedures together with a high-resolution mass spectrometric analysis of the DNA phosphoramidite building blocks 26, 27, 28 and 29 is given in the supporting information (Supplementary Figure S2). The high-resolution mass data were obtained on a Thermo LTQ Orbitrap XL mass spectrometer.
DNA solid phase synthesis
The stable isotope labeled DNA phosphoramidites were used in combination with labile DNA building blocks (ChemGenes). Custom primer support PS 200 (GE Healthcare) with an average loading of 80 μmol g−1 were used. The sequences were synthesized on an ABI 391 PCR Mate using self-written RNA/DNA synthesis cycles (available on request). Amidite (0.1 M) and activator (5-benzylthio-1H-tetrazole, 0.25 M) solutions were dried over freshly activated molecular sieves overnight. After sequence assembly, the removal of protecting groups and the cleavage from solid support was achieved by treatment with aqueous methylamine (40%, 650 μl) and aqueous ammonia (28%, 650 μl) at 310 K for 1.5 h. After evaporation of the alkaline deprotection solution the crude DNA was dissolved in 1 ml water. The quality of the crude DNA was checked via anion exchange chromatography on an analytical Dionex DNAPac PA-100 column (4 × 250 mm; Eluent A: 25 mM Tris–HCl, 6 M urea, pH 8.0; Eluent B: 25 mM Tris–HCl, 500 mM sodium perchlorate, 6 M urea, pH 8.0) and at elevated temperature (80°C). Purification of the DNA sequences was achieved in a single run by applying the crude nucleic acid on a preparative Dionex DNAPac PA-100 column (22 × 250 mm, eluents as before). The fractions containing the desired RNA or DNA were pooled and loaded on a C18 SepPak catridge (Waters) to remove HPLC buffer salts. The DNA sodium salt form was then eluted from the C18 column with water/acetonitrile (1/1, v/v) and lyophilized. The integrity of the DNAs was further checked by mass spectrometry on a Finnigan LCQ Advantage MAX ion trap instrumentation connected to an Amersham Ettan micro LC (GE Healthcare).
Expression of NCp7 peptide
The expression plasmid NCp7 11–55_pET-11a encoding the NCp7 11–55 peptide was purchased from GeneScript. The protein expression was carried out as earlier described (68). A detailed description of the procedure is given in the supporting information.
NMR spectroscopy
A detailed description is given in the supporting information.
NMR data analysis
A detailed description is given in the supporting information.
Isothermal titration calorimetry
A detailed description is given in the supporting information.
RESULTS AND DISCUSSION
Synthesis of the 13C-modified DNA phosphoramidites
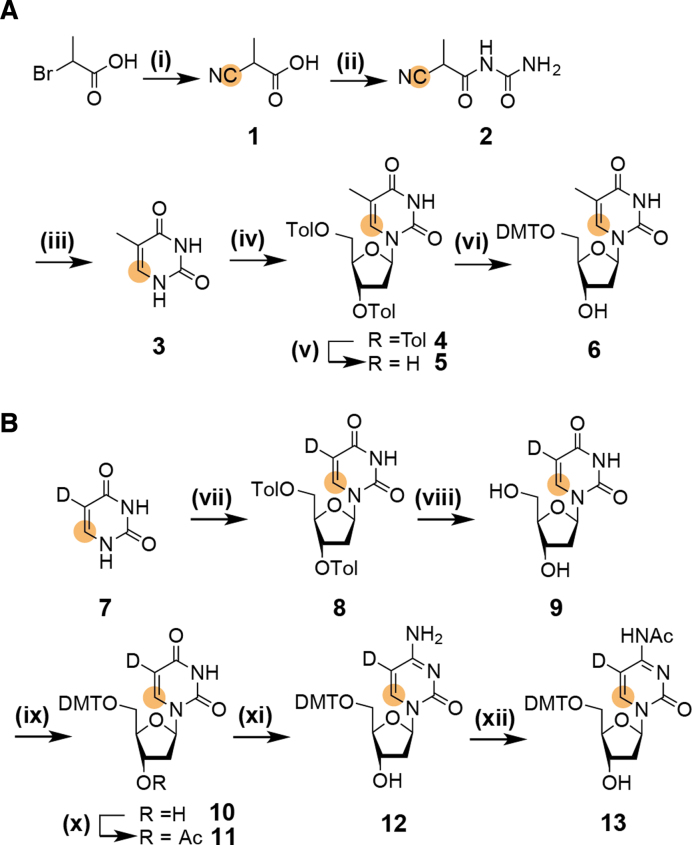
The initial efforts focused on the synthesis of 6-13C-thymidine 5. An early work on the synthesis of a 6-13C-thymidine phosphoramidite building block used a similar synthetic approach, but this synthesis was not optimized and a moderate yield of 10% with respect to 6-13C-potassium cyanide was obtained (52). The presented synthetic route capitalizes on our previous work on 6-13C-pyrimidine RNA phosphoramidites (Scheme 1A and B). Starting from commercially available 2-bromopropionic acid the 13C-tag is introduced via 13C-potassium cyanide, a relatively economic isotope source, in a Kolbe nitrile reaction giving 2-cyanopropionic acid 1 in high yield (97%). The lower temperature (60°C versus 80°C) and the reduced reaction time (3 h versus 16 h) as compared to the synthesis of uracil (see reference (40)) were found to be crucial to avoid hydrolysis of the cyano-function. In the next step urea is introduced via activation as a mixed anhydride and the urea derivative 2 can be isolated from salt cake using diethyl ether, thus by-passing isolation problems mentioned in a previous report (52). Thymine 3 was accessible via a cyclization under reductive conditions. The nucleobase was then fused with a sugar building block. First, we used 1-O-acetyl-(2′,3′,5′-O-tri-benzoyl)-β-d-ribofuranose (ATBR), with which a very high desired β-selectivity can be reached (69). In situ activation of the nucleoside by silylation using bis-trimethylsilyl acetamide and addition of trimethylsilyl trifluoromethanesulfonate gives the benzoyl protected ribothymidine in high yield (86%) and only traces of the undesired α-anomer were detected (Supplementary Figure S1).
Scheme 1.
(A) Synthesis of 5′-DMT-6–13C-thymidine 6. (i) 1 eq. K13CN, sodium carbonate, pH 9, 60°C, 3 h, 97%; (ii) 1.0 eq. urea in 1.1 eq. acetic anhydride, 90°C, 2 h, 85%; (iii) 5% Pd/BaSO4, H2 in 50% aqueous acetic acid, rt, 20h, 70%; (iv) 10 eq. TMS-Cl, 20 eq. HMDS, 120°C, 16 h, then 1 eq. Hoffer's α-chlorosugar in chloroform, 40°C, 4 h, 71% (51% β-anomer); (v) CH3NH2 in ethanol, rt, 16 h, 93%; (vi) 1.3 eq. DMT-Cl in pyridine, rt, 3h, 82%; (B) Synthesis of 5′-DMT-N4-Ac-5-D-6–13C-2′-deoxycytidine 13. (vii) 10 eq. TMS-Cl, 20 eq. HMDS, 120°C, 16 h, then 1 eq. Hoffer's α-chlorosugar in chloroform, 40°C, 4 h, 48% β-anomer; (viii) CH3NH2 in ethanol, rt, 16 h, 95%; (ix) 1.3 eq. DMT-Cl in pyridine, rt, 3 h, 78%; (x) 1.1 eq. Ac2O, DMAP in pyridine, 0°C to rt, 2 h; (xi) 1.5 eq. (TiPS)Cl, 0.2 eq. DMAP, 10 eq. NEt3 in anhydrous methylene chloride, rt, 3 h, then NH3 (aqueous, 28%) and THF, rt, 20 h, then CH3NH2 in ethanol, 2 h; (xii) 1.0 eq. Ac2O in DMF, rt, 16 h, 48% (referred to 11); orange dot = 13C.
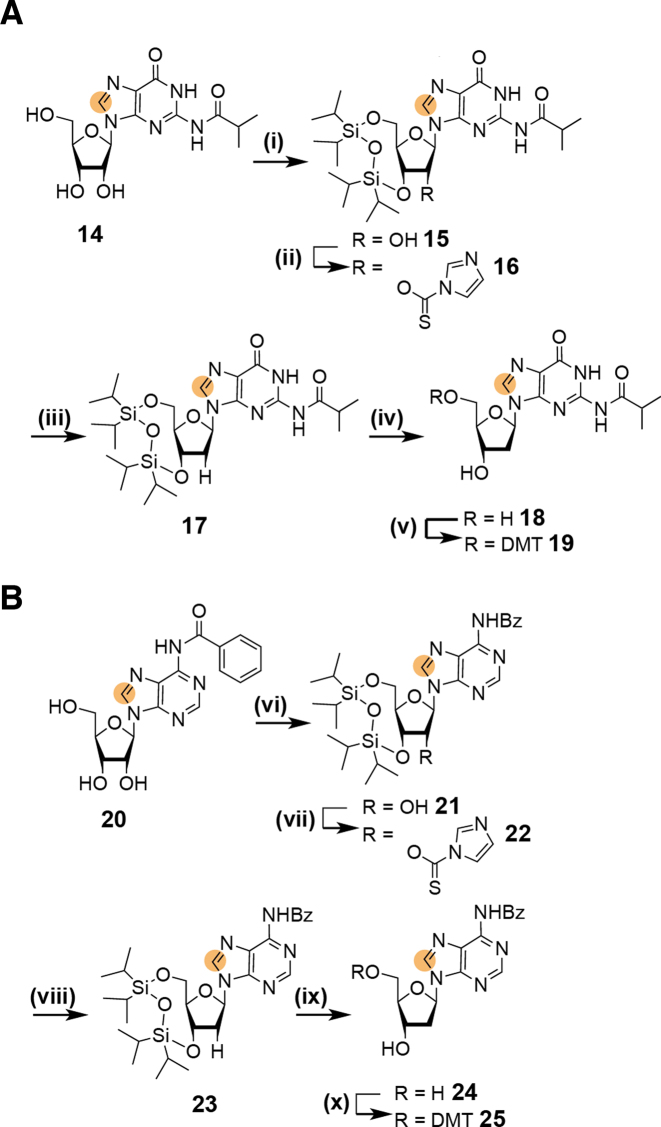
Alternatively, the nucleosidation reaction was carried out using Hoffer's α-chlorosugar (1-chloro-2-deoxy-3,5-di-O-toluoyl-α-d-ribofuranose) (70). The nucleobase is again activated via silylation by treatment with trimethylsilyl chloride and hexadimethylsilazane overnight. Then, the silylated nucleobase is reacted with Hoffer's α-chlorosugar at 40°C in chloroform to maximize the yield of the β-anomer. Thereby, the 2′-deoxynucleoside is directly accessible at the costs of a lower β-selectivity as compared to the silyl-Hilbert–Johnson nucleosidation. A 3/7 mixture of the α/β-nucleoside was obtained at a reasonable yield 71%. After recrystallization of the anomeric mixture in ethanol the desired 5′,3′-bis-toluoyl protected β-6-13C-thymidine nucleoside was isolated with 51% yield. Taken into account, that after the nucleosidation reaction using ATBR six additional steps are necessary to obtain nucleoside 5, the direct access to the 2′-deoxynucleoside using the Hoffer's α-chlorosugar in only two steps proved to be advantageous in terms of yield (46% versus 38%), time and reagent consumption. Thus, the synthetic route using Hoffer's α-chlorosugar is the recommended pathway. The second DNA pyrimidine building block—5-d-6–13C-2′-deoxycytidine—was easily accessible using the direct nucleosidation procedure using Hoffer's α-chlorosugar and 5-D-6-13C-uracil 7. The synthesis of the stable isotope labeled uracil derivative 7 was described in a recent publication (42). In the next steps the 5′,3′-O-bis-toluoyl-2′-deoxyuridine 8 was transformed into the 5′-DMT-N4-Ac-5-d-6-13C-2′-deoxycytidine nucleoside 13 in analogy to the previously described method (40,42). This transformation includes removal of the toluoyl-protection groups, followed by 5′-tritylation and 3′-acetylation. Then, oxygen O4 is activated as a leaving group by trisylation and is replaced by nitrogen using aqueous ammonia. The exocyclic amino group is then selectively acetylated in N,N-dimethylformamide yielding the 13C-labeled 2′-deoxycitidine phosphoramidite precursor 13. To complete the portfolio of 13C-DNA phosphoramidites and to allow a maximum of flexibility on placing the NMR active label the synthesis of 8-13C-purine 2′-deoxy-building blocks was addressed (Scheme 2). Here, the partially protected ribonucleoside derivatives 14 and 20 are used as starting materials. These two compounds are intermediates in the synthesis of 13C-modified RNA phosphoramidite building blocks (42). The selective masking of the 5′- and 3′-hydroxyl groups is achieved using the Markewickz protecting group (71). This makes the 2′-hydroxyl group accessible for the Barton-McCombie deoxygenation yielding 17 and 23, respectively (72). The phosphoramidite precursors 19 and 25 were then obtained by removal of the Markewickz protecting group using triethylamine hydrofluoride in tetrahydrofuran followed by the introduction of the 4,4′-dimethoxytrityl group at the 5′-hydroxyl.
Scheme 2.
(A) Synthesis of 5′-DMT-N2-iBu-8–13C-2′-deoxyguanosine 19. (i) 1.2 eq. TiPDSiCl2 in pyridine, rt, 3 h, 65%; (ii) 1.2 eq. thiocarbonyldiimidazole in DMF, rt, 4 h, 78%; (iii) 0.2 eq. AIBN, 1.5 eq. n-BuSnH, in toluene, 75°C, 1.5 h, 95%; (iv) 3.4 eq. 3 TEA.HF in tetrahydrofuran, rt, 4 h, 100%; (v) 1.2 eq. DMT-Cl in pyridine, rt, 3 h, 70%; (B) Synthesis of 5′-DMT-N6-Bz-8-13C-2′-deoxyadenosine 25. (vi) 1.2 eq. TiPDSiCl2 in pyridine, rt, 3 h, 80%; (vii) 1.2 eq. thiocarbonyldiimidazole in DMF, rt, 4 h; (viii) 0.2 eq. AIBN, 1.5 eq. n-BuSnH, in toluene, 75°C, 1.5 h, 63% over two steps; (ix) 1.3 eq. 3 TEA.HF in tetrahydrofuran, 45°C, 2 h, 72%; (x) 1.2 eq. DMT-Cl in pyridine, rt, 3 h, 80%; orange dot = 13C.
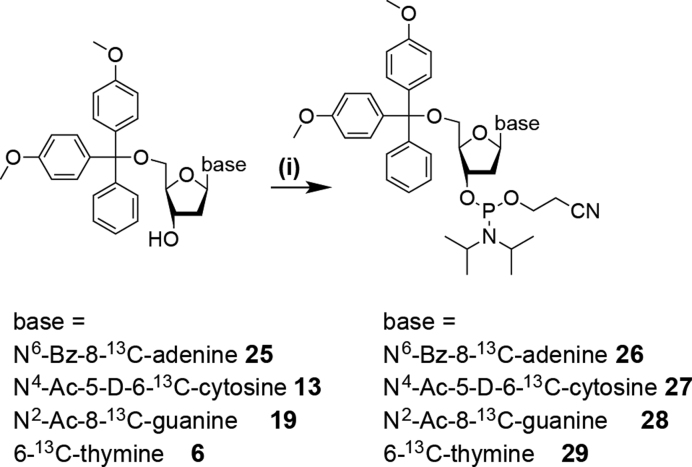
In the final step the phosphoramidite functionality is attached to all four 13C/2H labeled 2′-deoxynucleotides (6, 13, 19 and 25) yielding the desired building blocks (Scheme 3). The identity of the building blocks 26–29 was further confirmed by a high-resolution mass spectrometric analysis (Supplementary Figure S2).
Scheme 3.
Synthesis of stable isotope labeled DNA phosphoramidites 26, 27, 28 and 29. (i) 1.2–3 eq. (2-cyanoethyl)-N,N-diisopropylchlorophosphoramidite (CEP-Cl), 10 eq. DiPEA in anhydrous methylene chloride, rt, 3h, 77% (26), 70% (27), 68% (28) and 77% (29).
The phosphoramidite functionality was introduced via established methods using (2-cyanoethyl)-N,N-di-iso-propylchlorophosphoramidite (CEP-Cl) and an excess of di-iso-propylethylamine (DiPEA) in dry methylene chloride. For the 2′-deoxyadenosine (26), the 2′-deoxycytidine (27) and thymidine (29) building blocks 1.2–1.5 eq. of phosphitylation reagent were used, for the 2′-deoxyguanosine amidite 3 eq. of reagent were needed. Due to the higher polarity of the DNA phosphoramidites compared to the RNA counterparts carrying an additional apolar 2′-protecting group the removal of the phosphonate side product in the final purification step can be complicated. In our hands, repeated column chromatographic purification steps or alternatively several precipitations of the phosphoramidites in ice cold pentane proved to be the methods of choice to remove this type of impurity.
To sum up, we present a comprehensive description on the synthetic access to atom-specifically 13C/2H labeled DNA phosphoramidites. The described synthetic routes give reproducible yields and can be up-scaled on a gram-scale. In combination with unlabeled commercially available phosphoramidites DNAs tailor-made 13C-isotope labeling patterns become feasible as shown in the next section.
Incorporation of site-specific 13C labels into DNA constructs
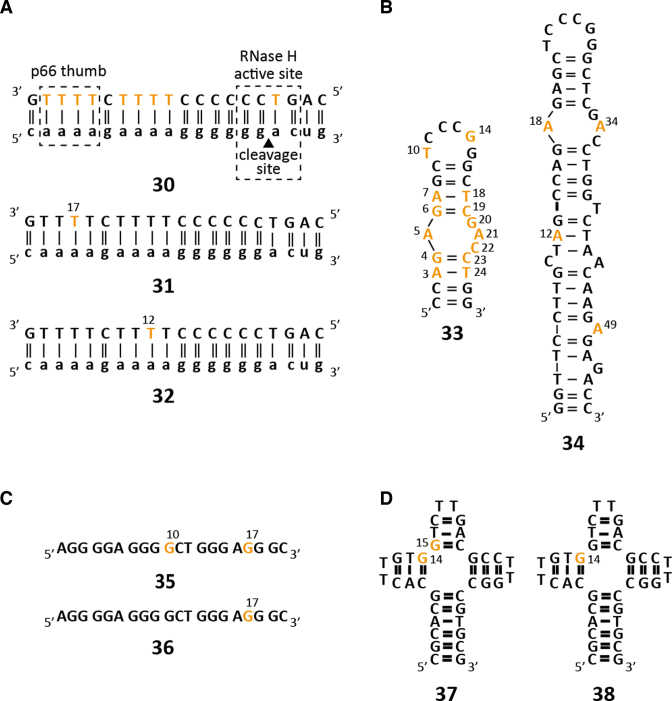
The stable isotope labeled DNA phosphoramidites were then used in the standard solid phase synthesis to obtain DNAs with sizes ranging between 20 and 55 nucleotides (Figure 1). The DNAs were synthesized using the labile building blocks with the phenoxyacetyl (dA, dC) and the dimethylformamidine protection groups (dG). All four 13C-labeled DNA phosphoramidites were incorporated and showed high coupling yields in the synthetic cycle. The number of 13C-labels within the target DNAs varied between one and fourteen and the alternative protecting schemes (benzoyl for 8-13C-dA, acetyl for 6-13C-dC and iso-butyryl for 8-13C-dG) proved to be compatible with the labile protecting groups of the commercially available building blocks. For all syntheses, a major product could be identified using anion-exchange (AIEX) chromatography (Supplementary Figure S3) and mass-spectrometric analyses confirmed the correct assembly of all investigated DNA sequences (Table 1).
Figure 1.
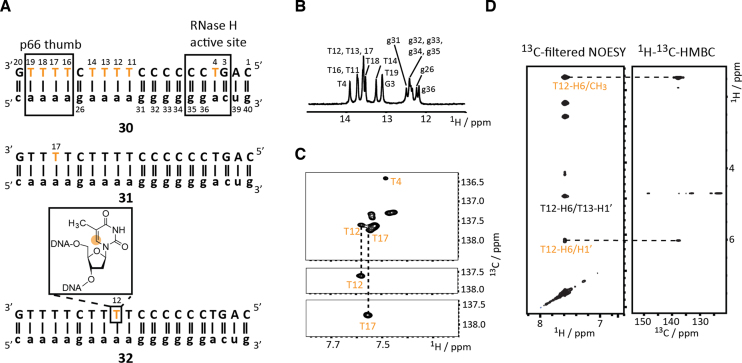
Stable isotope labeled DNA sequences probed by solution NMR spectroscopy. (A) HIV-1 polypurine tract (PPT) DNAs 30–32. (B) HIV-1 cTAR DNA constructs: mini-cTAR DNA 33 and full-length cTAR DNA 34. (C) G-quadruplex DNAs 35 and 36. (D) Holliday fourway-junction mimics 37 and 38. Orange letters indicate 13C labeled residues.
Table 1. Characterization of synthesized stable isotope labeled DNAs 30–38.
| Molecular weightd | ||||
|---|---|---|---|---|
| Sequencea | No. and type of 13C-labeled residuesb | Lengthc | Calculated (amu) | Found (amu) |
| HIV-1 PPT | ||||
| 30 | dT9 | 20 nt | 5969.8 | 5968.8 |
| 31 | dT1 | 20 nt | 5961.8 | 5961.1 |
| 32 | dT1 | 20 nt | 5961.8 | 5961.2 |
| mini cTAR | ||||
| 33 | dA4dC3dG4dT3 | 26 nt | 7975.1 | 7975.4 |
| full length cTAR | ||||
| 34 | dA4 | 55 nt | 16925.9 | 16924.7 |
| G-quadruplex | ||||
| 35 | dG2 | 20 nt | 6371.1 | 6371.4 |
| 36 | dG1 | 20 nt | 6370.1 | 6370.1 |
| J9a-4-way junction | ||||
| 37 | dG2 | 36 nt | 11038.0 | 11038.2 |
| 38 | dG1 | 36 nt | 11037.0 | 11037.4 |
aSequence details and identifier number, b dA, dC, dG, dT denote nucleotide type, c length in nucleotides (nt), d Purified oligonucleotides were characterized by mass spectrometry on a Finnigan LCQ Advantage MAX ion trap instrumentation connected to an Amersham Ettan micro LC system (negative-ion mode with a potential of −4 kV applied to the spray needle). LC: Sample (250 pmol of oligonucleotide dissolved in 20 μl of 20 mM EDTA solution; average injection volume: 10–20 μl); column (Amersham μRPC C2/C18; 2.1 × 100 mm) at 21°C; flow rate: 100 μl min−1; eluant A: 8.6 mM TEA, 100 mM 1,1,1,3,3,3-hexafluoro-2-propanol in H2O (pH 8.0); eluant B: methanol; gradient: 0–100% B in A within 30 min; UV detection at 254 nm.
In detail, the human immunodeficiency virus-1 (HIV) polypurine tract (PPT), a DNA/RNA heteroduplex (30, 31 and 32), was picked as a first target to showcase the potential of site- and atom-specific 13C labeling of DNA (Figure 1A). Three constructs—an all thymidine labeled (30) and two site-specific labeled (T12 (31) and T17 (32)) DNAs—were synthesized. The PPT is a purine-rich region of the plus-strand RNA genome of retroviruses and is resistant to hydrolysis by the RNase H domain of reverse transcriptase (RT) and ultimately serves as a primer for plus-strand DNA synthesis. This RNA/DNA hybrid although relatively small in size (40 nt) represents a challenging target for NMR spectroscopy as discussed in the next section (61).
Then, the labeling strategy was expanded to two constructs of the cTAR DNA (MAL isolate). First, a small mini-cTAR DNA 33 was synthesized with 14 nucleotides out of 26 replaced by their 13C-labeled counterparts (Figure 1B).
This DNA 33 was shown to bind to the nucleocapsid protein NCp 7—a zinc finger protein (73). This 55-amino acid polypeptide acts as a nucleic acid chaperone catalyzing the DNA/RNA hybrid duplex formation step. The μs- to ms dynamics of the mini-cTAR DNA 33 in the presence and absence of NCp 7 was addressed using relaxation dispersion NMR spectroscopy. The other cTAR DNA sequence 34 represents the full-length construct (Figure 1B). This DNA is involved in the viral replication cycle of the HIV-1 and fulfils a priming function via hybridization to the viral genomic TAR RNA (74,75). For this 55 nt DNA construct larger side products eluting after the main product peak were observed in the anion-exchange (AIEX) chromatogram (Supplementary Figure S3). This very likely can be attributed to the use of 5-benzylthiotetrazole for the phosphoramidite activation in the coupling step. Due to the higher acidity of 5-benzylthiotetrazole (pKa 4.08) as compared to 1H-tetrazole (pKa 4.89) it is well possible that DNA amidites can be partially detritylated during the coupling step leading to the addition of multimers. This would lead to later eluting species in the AIEX-chromatogram as observed here. However, a detailed analysis of this undesired side products was not carried out here as it would exceed the scope of the work but we speculate that the use of other more standard and less acidic DNA activator reagents (such as 4,5-dicyanoimidazole or 1H-tetrazole) would reduce the formation of such unwanted larger DNAs.
As a penultimate example for the solid phase synthesis based DNA stable isotope labeling a DNA G-quadruplex sequence 35/36 residing in the human telomerase (hTERT) promoter was picked (Figure 1C). This DNA was previously shown to exist in two folds – a (3+1) and a parallel-stranded G-quadruplex (66). Two constructs 35 and 36 with either one or two 8-13C-2′-deoxyguanosine residues were produced. The assembly of the 20 nt quadruplex bearing either two (36) or one (37) 13C labels was accomplished using the standard synthesis protocol. However, during the analysis using anion-exchange chromatography under denaturing conditions (80°C and 6M urea) some artefacts were observed. With a reported melting temperature of 78°C even the harsh conditions used during analytical anion-exchange chromatography could not fully denature the 20 nt G-quadruplex and two product peaks—one eluting at the expected retention time (32′) and a late eluting species (44.5′), very likely representing a non-denatured quadruplex structure- were observed (Supplementary Figure S3). This effect was not seen during the purification procedure using the preparative anion-exchange column and the desired G-quadruplex sequences 35 and 36 could be isolated in good yields.
Finally, a four-way junction J9a with two (37) or one 8-13C-dG label (38) displaying conformational heterogeneity was picked as a last example (Figure 1D). This DNA was earlier investigated and the exchange parameters between two stacked conformers were determined by proton chemical exchange NMR spectroscopy (67). The 36 nt model Holliday junction was assembled and high quality crude products were obtained (Supplementary Figure S3), which could be purified according to the standard protocol.
Resonance assignment in the HIV-1 polypurine tract RNA/DNA hybrid using site-specific 13C labeling
The stable isotope label protocol was at first used to demonstrate the high suitability of site-specific 13C labeling for resonance assignment purposes in a challenging target, the polypurine tract of the human immunodeficiency virus 1 (HIV-1 PPT, Figure 2A).
Figure 2.
Site-specific 6-13C-thymidine labeling for resonance assignment purposes. (A) HIV-1 polypurine tract DNA/RNA duplex constructs. 6-13C-labeled thymidine residues are highlighted in orange. (B) Imino proton NMR spectrum confirming the RNA/DNA hybrid duplex formation. Assignments are given. (C) 1H–13C-HSQC spectra of the fully 6-13C-thymidine labeled duplex and the selectively T12 and T17 labeled constructs allowing resonance assignment. (D) 13C-filtered NOESY spectrum of T12 labeled construct and HMBC spectrum to differentiate inter- and intraresidue NOEs.
This RNA/DNA hybrid represents a rather demanding target for NMR spectroscopy as the DNA strand comprises almost only pyrimidine nucleotides (nine thymidines, eight 2′-deoxycytidines and only three purine deoxynucleotides) leading to significant resonance overlap. An alternative strategy using fluorine labeling was earlier used to alleviate problems regarding resonance assignment at the costs of introducing a minimally invasive label (61–63). With the atom-specifically labeled 13C-labeled DNA building blocks, however, the native polypurine tract hybrid can be studied. To closely mimic the labeling pattern obtained from enzymatic methods using DNA polymerase and uniformly labeled DNA triphosphates an all 6-13C-thymidine DNA construct 30 was synthesized followed by the synthesis of two site specifically labelled constructs 31 and 32 (Figure 2a). For all constructs the formation of the hybrid duplex was confirmed (exemplarily shown for construct 30) using an unlabeled RNA complementary sequence by observing the imino proton region of the 1H NMR spectrum (Figure 2b). The 1H–13C-HSQC spectrum of the fully 6-13C-thymidine labeled HIV-1 PPT 30 base-paired with RNA showed significant resonance overlap, only one resonance is well resolved (tentatively assigned to T4), whereas the other C6–H6 signals cluster in the region between 7.5 and 7.6 ppm in the proton dimension and in only one ppm between 137 and 138 ppm in the carbon dimension. Noteworthy, the resolution in the indirect dimension of a 13C HSQC spectrum run on a uniformly 13C labeled DNA sample would be even lower as the number of increments is limited (for the atom-specific 13C labeled sample 256 increments were recorded) due to necessity of the constant time element to suppress 13C-13C couplings. In the two HSQC spectra of 31 and 32 with site/atom-specific T12 and T17 labels, respectively, single C6-H6 correlations were observed, which allowed the straightforward assignment of the T12 and T17 resonances in the fully labeled construct (Figure 2C). Furthermore, the site-specific labeling can be exploited to establish the sequential walk in NOE experiments using a combination of a 13C-filtered NOESY and a 1H–13C-HMBC experiment. The latter experiment allows the unambiguous differentiation of intra- and interresidue NOEs in the sequential walk region via long range scalar couplings (Figure 2D). The two experiments were acquired within ∼6 h for a 600 μM sample of 32 on a 600 MHz NMR with a nitrogen cooled prodigy probe. This makes the approach an attractive option to obtain unambiguous assignments, NOE based distance restraints but also other valuable structure restraints, such as residual dipolar coupling information, in highly resonance-degenerated DNAs.
Equilibrium dynamics of the cTAR DNA in absence and presence of NCp7 via CPMG relaxation dispersion NMR
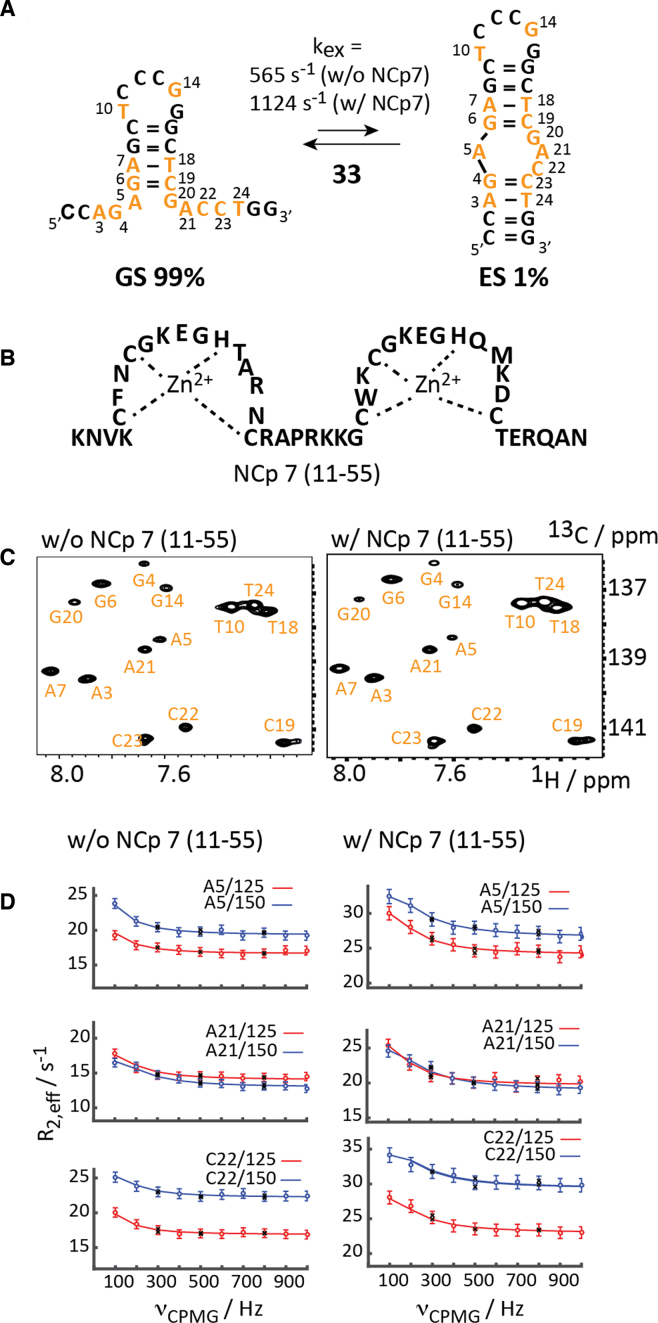
We picked the cTAR DNA (MAL isolate) 33 as a target to study equilibrium μs- to ms-dynamics in DNA using the relaxation dispersion method (Figure 3A).
Figure 3.
13C-CPMG relaxation dispersion NMR of mini-cTAR DNA. (A) Exchange process for the site-specific 13C-modified mini-cTAR DNA. 13C-modified residues are highlighted in orange. (B) Schematic representation of the zinc-finger NCp7 peptide (11–55). (C) 1H-13C-HSQC spectra in the absence and presence of one equivalent NCp7 (11–55). (D) Non-flat carbon relaxation dispersion profiles of residues A5, A21 and C22 with and without NCp7 (11–55). Dots represent experimental data, lines the best fit, black crosses repeat experiments. Residue number and field strengths are specified.
The nucleocapsid protein NCp 7—a zinc finger protein- interacts with the cTAR element and the 55-amino acid polypeptide acts as a nucleic acid chaperone catalyzing the DNA/RNA hybrid duplex formation step (Figure 3B). The mini-cTAR DNA 33 comprises 26 nucleotides and was proposed to fold into a hairpin-bulge conformation. The postulated secondary structure—a stem–loop fold with an internal asymmetric bulge—represents a low populated conformation at ambient temperature, as no observable imino proton resonances were found for the lower stem (Supplementary Figure S5). We recently investigated this DNA using site-specific labeling and proton relaxation dispersion NMR experiments and based on the chemical shift data of the excited state (ES) an equilibrium between a ground state (GS) hairpin structure with 5′- and 3′-dangling termini and a folded state was established (Figure 3A) (42). Fourteen (out of 26) residues were replaced by the respective 13C labeled counterpart making the application of carbon relaxation dispersion experiments feasible. The HSQC spectrum gave a single set of resonances with varying line-widths pointing toward a major conformation in exchange with a minor state in the fast to intermediate NMR chemical shift exchange regime (Figure 3C). This was further corroborated by the application of a carbon relaxation dispersion experiment revealing such a millisecond exchange process. High quality non-flat dispersion profiles were obtained for residues, A5, A21 and C22, all clustered in the bulge region (Figure 3D). A global fit of the proton dispersion data to the Carver-Richards equation gave an excited state population pB of 1.1 ± 0.1% and an exchange rate kex of 565 ± 51 s−1. Thus, the exchange process of the cTAR DNA 33 could be recapitulated using carbon relaxation dispersion methods and the data sets from the earlier proton relaxation dispersion study and the herein described carbon RD study are in striking agreement (Table 2).
Table 2. Summary of exchange parameters and chemical shift difference data obtained from 13C and 1H-CPMG relaxation dispersion profiles of cTAR DNA 33 in absence or presence of NCp7.
| Residue | k ex (s−1)a | p b (%)b | DwES-GS (ppm)c | NMR method and sample information |
|---|---|---|---|---|
| A5 | 0.94 ± 0.14 | |||
| A21 | 565 ± 51 | 1.1 ± 0.1 | 1.09 ± 0.13 | 13C RD |
| C22 | 1.01 ± 0.14 | w/o NCp7 | ||
| A5 | 1.14 ± 0.15 | |||
| A21 | 1124 ± 219 | 1.4 ± 0.2 | 1.16 ± 0.13 | 13C RD |
| C22 | 1.02 ± 0.13 | w/ NCp7 | ||
| G4 | 0.25 ± 0.02 | |||
| A5 | 525 ± 59 | 1.0 ± 0.1 | 0.49 ± 0.03 | |
| C19 | 0.22 ± 0.01 | |||
| G20 | 0.30 ± 0.02 | 1H RD | ||
| A21 | 0.56 ± 0.03 | w/o NCp7[d] | ||
| C22 | 0.34 ± 0.02 | |||
| C23 | 0.24 ± 0.01 |
a Exchange rate constant and b population from global fit of relaxation dispersion data of residues with significant relaxation dispersion at 125/150 MHz 13C Larmor frequency. c Residue specific chemical shift difference between ground and excited state. d For proton relaxation dispersion data and evaluation please refer to (42). Abbreviations: RD relaxation dispersion, NCp7 nucleosidcapsid protein 7.
As aforementioned the nucleocapsid protein NCp7 (Figure 3B) interacts with both the TAR RNA and the cTAR DNA and facilitates the annealing step (65,73). We addressed the binding of the NCp7 peptide to the wildtype mini-cTAR DNA via isothermal titration calorimetry (ITC) and further explored the interaction of NCp7 with two mini-cTAR DNA mutants (Supplementary Figure S4). The data is in qualitative agreement with an earlier study and underscores the importance of the 5′- and 3′ dangling ends for peptide binding but not of the apical 7nt loop for binding of NCp7 (73). We were then interested how the addition of NCp7 modulates the millisecond dynamics of the cTAR DNA construct 33. The addition of one equivalent of NCp7 to the DNA resulted in only minor chemical shift changes for single stranded residues, A5, A21, G4, G14 and G20, indicating a weak, transient and rather unspecific interaction. Significant line broadening effects were found for the single stranded residues G4, A5, G14 and G20 suggesting that NCp7 interacts with easily accessible single stranded residues via electrostatic interactions (Figure 3c). The 13C-CPMG relaxation dispersion experiment revealed subtle changes in the dynamic behavior of the cTAR DNA (Figure 3D and Table 2). The addition of NCp7 leaves the populations of GS and ES almost unchanged—i.e. neither of the both conformations interacts strongly/specifically with NCp7 leading to an energetically favored complex. The most striking effect on the DNA exchange process by the addition of NCp7 is the increase of the exchange rate by a factor of two (kex = 1124 s−1 versus kex = 565 s−1). This means that NCp7 accelerates the forward but also the backward rate to the approximately same extent by lowering the activation barrier either via interactions with folding intermediates or by destabilizing both conformers to the same extent.
Taken together, the chaperon NCp7 speeds up the exchange process of the cTAR DNA 33 in both directions leaving the populations of GS and ES almost untouched. Furthermore, the small chemical shift changes between GS and ES suggest that both the ground and excited state structures are not strongly altered in the presence of NCp7.
Site-specific labeling in larger DNAs to probe μs- to ms-dynamics—the 55 nt cTAR DNA as a proof of principle
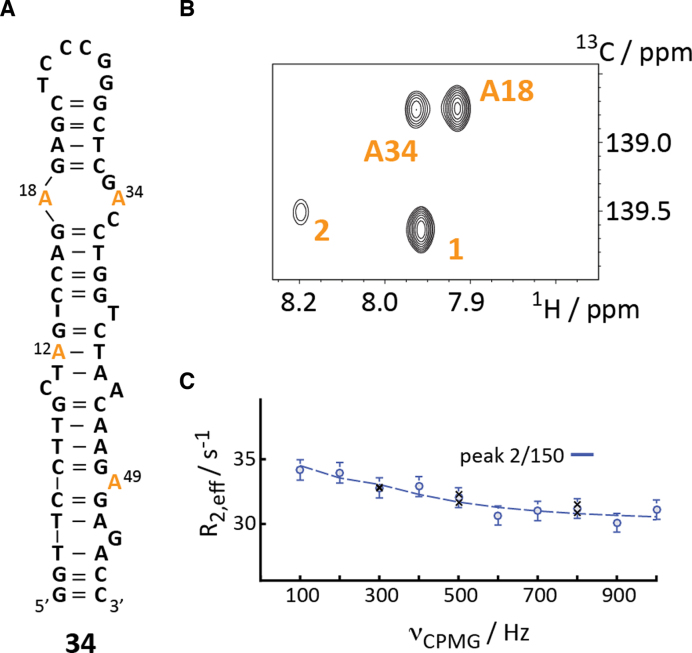
The 26 nt mini-cTAR DNA construct 33 represents a shortened version of the 55 nt full-length cTAR DNA. It is very possible that the observed dynamics in 33 is an artefact resulting from the shortening of the cTAR DNA. To test this hypothesis, a full-length 55 nt cTAR DNA 34 was synthesized with four 8-13C-dA labels (Figure 4A).
Figure 4.
Synthesis of larger DNAs with site-specific 13C labels. (A) Secondary structure representation of full-length cTAR DNA 34 with 8-13C-labeled dA labels highlighted in orange. (B) 1H–13C-HSQC spectrum displaying four C8–H8-resonances. (C) Non-flat carbon relaxation dispersion profile of peak 2 at 150 MHz 13C Larmor frequency. Dots represent experimental data and black crosses repeat experiments. For details see text.
The HSQC spectrum of the 8-13C-dA-labeled DNA 34 gave again a spectrum with a single resonance set and differential line-widths, which is indicative of μs- to ms-dynamics (Figure 4C). The full assignment of the observed resonance is currently pending. Based on the NMR data obtained for the mini cTAR DNA 33 two resonances – A18 (i.e. A5 in 33) and A34 (i.e. A21 in 33) could be tentatively assigned. All C8–H8 cross-peaks are currently assigned using single labeled 8-13C-dA cTAR constructs. We then conducted a preliminary relaxation dispersion study at a single field strength. Interestingly, we did observe only one non-flat dispersion profile (peak 2). The previously detected dynamics for A18 (A5) and A34 (A21) in the mini cTAR DNA 33 could not be reproduced. This impressively illustrates the potential pitfalls when applying the divide-and-conquer approach to study larger nucleic acids. Dynamic features of a nucleic acid can be severely altered cutting out only a certain domain of a large DNA (and also RNA). Studying the folding landscape of an isolated domain can be very misleading as the free energy landscape can have completely different characteristics with ground and excited conformational states not present in the full-size construct. We currently address the μs- to ms-dynamics of the full-length cTAR DNA in presence and absence of NCp7 using several labeling positions. The study will also be expanded to study the cTAR DNA/TAR RNA hybrid duplex formation in real time and the influence of NCp7 on this process. To this end, we will rely on a method recently developed for a RNA/RNA duplex formation study in the presence and absence of the Escherichia coli RNA chaperone CspA (76). For that purpose, we have also produced 15N/13C-labeled NCp7 and assigned the amide resonances to address the role of the peptide during the RNA/DNA duplex formation step (Supplementary Figure S6).
Re-equilibration dynamics of a multi-stable G-quadruplex after temperature jump
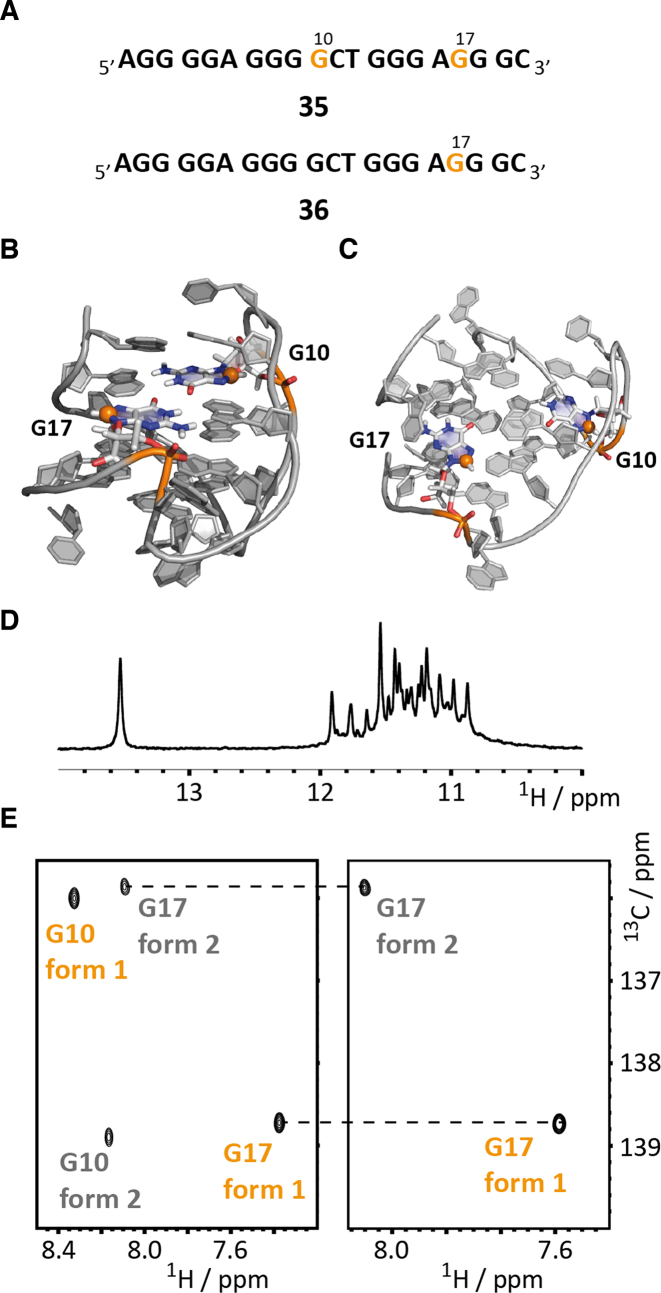
As a penultimate example of the expanded possibilities using the fully chemical stable isotope labeling protocol for DNA we have addressed DNA G-quadruplex folding using site-specifically placed 13C-modified guanosine residues (Figure 5A). The sequence was extracted from the human teleromase (hTERT) promoter region between 20 and 110 nt upstream of the transcription start site with an exceptional high level of guanine rich stretches and thus a high potential for G-quadruplex formation (66). Applying a combination of NMR and circular dichroism (CD) spectroscopy Phan and co-workers could show that a 20 nt four G-tract sequence can fold into two distinct conformations in the presence of potassium ions—in a (3+1) and a parallel stranded G-quadruplex. The introduction of inosine at position 5 replacing a G leads to a preference for the (3+1)-fold and allowed the solution structure determination of this conformational state (form 1) (Figure 5B and C). Alternatively, substitution of G2 and G10 by inosines promotes the parallel quadruplex formation and again NMR spectroscopy could be used to determine the high-resolution structure of this fold (form 2). This fold heterogeneity might be functional as the sequence region contains multiple binding sites for proteins and thus can be a regulator of transcription.
Figure 5.
Studying G-quadruplex folding by site/atom-specific 13C-labeling of DNA. (A) G-quadruplex sequences 35 and 36 with 8-13C-labeled guanosines G10 and G17 highlighted in orange. (B) High-resolution NMR structure of fold 1 (3+1 quadruplex) with G10 and G17 highlighted. The 8-13C-label is shown as an orange sphere. (C) High-resolution NMR structure of fold 2 (parallel quadruplex) with G10 and G17 highlighted. The 8-13C-label is shown as an orange sphere. (D) Imino proton region of 1H NMR spectrum. (E) 1H–13C-HSQC spectra of 35 (left) and 36 (right) with assignments.
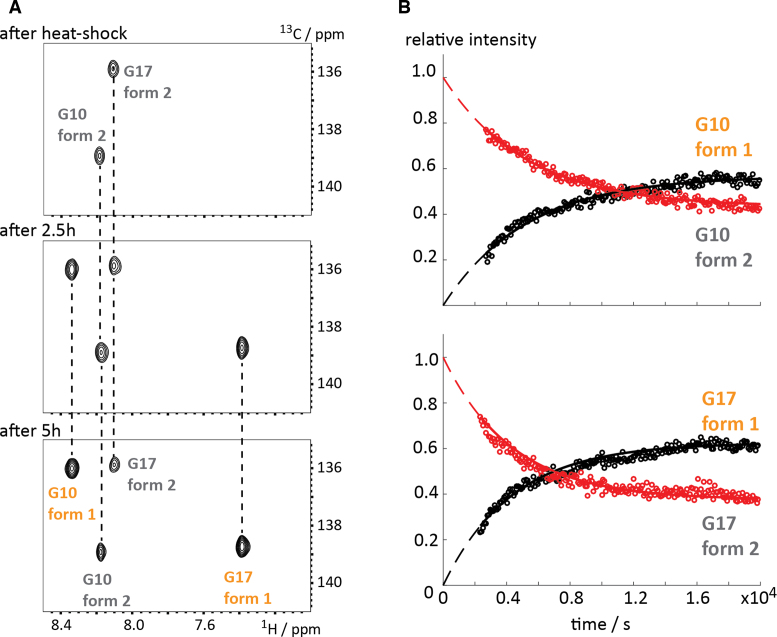
We replaced two guanosines—G10 and G17—by their 8-13C-modified analog and confirmed the correct quadruplex folding of DNA sequence 35 (in potassium phosphate buffer supplemented with potassium chloride) by acquiring a 1D imino proton selective NMR spectrum (Figure 5D) and found a signal pattern in accordance with the published. In the 1H–13C-HSQC spectrum four peaks were found in-line with the fold heterogeneity occurring in the slow chemical shift time regime. The 1H–13C-correlation spectrum of the G17-labeled sequence 36 then allowed the unambiguous assignment of the resonances (Figure 5E). As earlier described under these experimental conditions the (3+1) fold 1 represents the major populated species (∼60%) and the parallel quadruplex fold 2 the minor conformation (∼40%), which could also be confirmed by peak integration. A longitudinal exchange experiment showed that the interconversion between the two quadruplex forms occurs at a very slow rate (>> 1s−1, data not shown). To further demonstrate the potential of the atom/site-specific 13C-labeling protocol for real time NMR applications we monitored the re-equilibration of fold 1 and 2 after a temperature jump. At high temperature fold 2 is the preferred conformational state. This was confirmed by fast real-time NMR data acquisition using the Band-selective Excitation Short-Transient (BEST) TROSY approach after a heat shock of DNA 35 (76–78). In detail, the NMR sample was heated to 90°C for 5 min and the fold re-equilibration from fold 2 to fold 1 was monitored by BEST-TROSY 1H-13C-HSQC spectra with a time resolution of 90 s (Figure 6A and B). After the heat shock the parallel quadruplex form 2 was the almost exclusively populated conformational state and for both residues G10 and G17 a dominating HSQC correlation originating from fold 2 was observed (Figure 6A). The refolding from fold 2 to the (3+1) quadruplex form 1 could be easily followed by acquiring a series of 2D spectra (acquisition time per spectrum: 88 s) (Figure 6A).
Figure 6.
Re-equilibration of G-quadruplex sequence 35 after temperature jump. (A) BEST-TROSY HSQC spectra (88 s time resolution) after heat shock, after 2.5 and 5 h. Assignments are indicated. (B) Decay and build-up curves of form 1 and form 2 for G10 and G17 giving a re-equilibration constant of ∼0.012 min−1. For details of the fitting procedure see the supporting information.
The decay and build-up curves for both residues G10 and G17 could be reliably fitted using a mono-exponential function and yielded a refolding rate of approximately 0.012 min−1 (G10: 0.0115 ± 0.0002 min−1, dead time: 45 ± 1.35 min; G17: 0.0138 ± 0.0002 min−1, dead time: 39 ± 1.5 min). Furthermore, an effective dead time of ∼40–45 min was extracted from the fits, which strongly differs from the real dead time (∼2 min from heat shock until start of NMR measurements). This effective dead time can be easily explained as the refolding rate is accelerated during the cool-down process after the heat shock resulting in this discrepancy. Based on the real-time NMR data the re-equilibration process seems to be a simple monomolecular process as no intermediates could be characterized and the decay/build-up curves can be well reproduced by a mono-exponential function. Taken together, the novel DNA phosphoramidite building blocks proved to be highly useful in DNA G-quadruplex research and the applicability of the atom-specific aromatic 13C-labeling in DNA for real-time NMR applications was showcased.
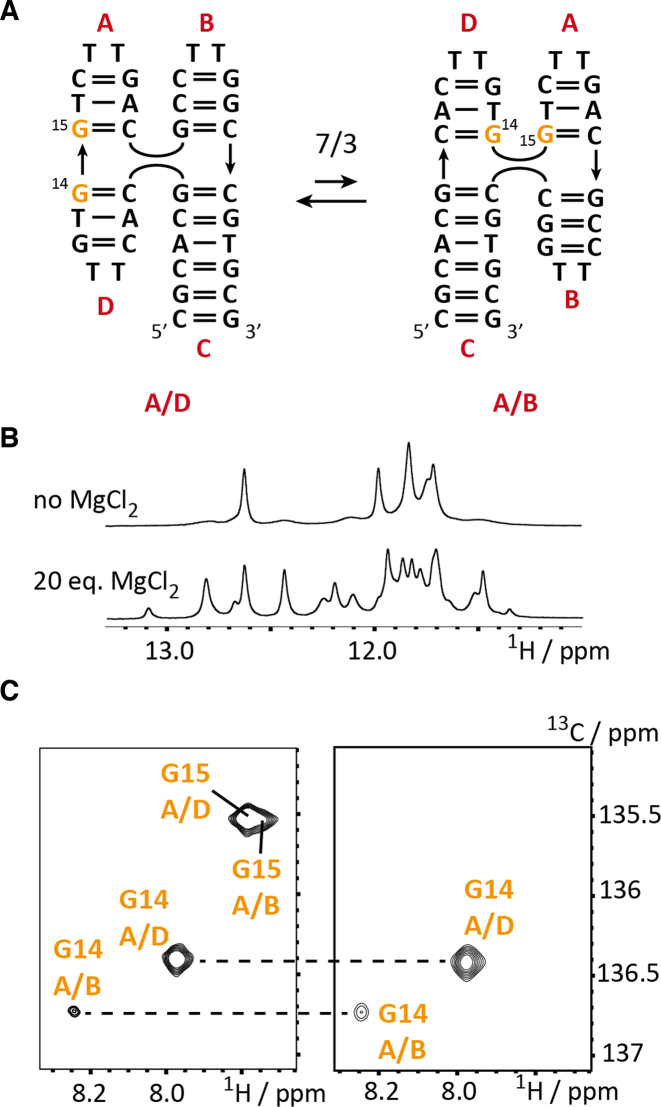
Kinetic characterization of a four-way junction using ZZ exchange spectroscopy
As a last example, we picked the DNA Holliday junction mimic J9a for stable isotope labeling using the synthesized DNA phosphoramdites. The labeling pattern consisted of two 8-13C-guanosine labels, which reside in the center of the four-way junction (Figure 7A). The folding of this model Holliday junction was early described and the formation of the stacked conformers A/D and A/B can be induced by the addition of magnesium chloride (Figure 7B). The four-way junction was previously studied by NMR spectroscopy and Altona et al. could show by proton exchange spectroscopy that J9a exist in a slow dynamic equilibrium of two conformers (67). The nature of the two conformers was deduced from a thorough analysis of Nuclear Overhauser Effect (NOE) patterns and the authors proposed the A/D stacked folding state as the major species (70%) and the minor A/B stacked form is populated to ∼30% (at 21°C). The kinetics of the exchange process was then addressed by ROE/NOE spectroscopy and a forward refolding rate from A/D to A/B of 5.6 s−1 and a backward rate of 2.3 s−1 was determined giving an equilibrium constant of 2.4 at 21°C. To exemplify the suitability of the 13C-labeling protocol for longitudinal exchange NMR spectroscopy the J9a model Holliday junction is ideally suited. The HSQC spectrum at 25°C of the double-labeled DNA 37 confirmed the structural heterogeneity and for G14 two cross-peaks could be identified – one arising from the major fold A/D (76%), the other resonance belonging to minor folding state A/B (24%). Integration of the two peaks also gave an estimate for the equilibrium constant (KA/D-A/B298K = 0.32), which is in good accordance with the previously reported population ratio and the equilibrium constant determined from the kinetic data. Altona et al. also postulated a well resolved exchange cross peak for H8 of G15, which was not reproduced in our investigation. A close inspection of the G15 H8-C correlation reveals strong resonance overlap for the G15 A/B and A/D H8-C signals with the G15 A/B correlation having a slightly different proton chemical shift (Figure 7C).
Figure 7.
NMR characterization of J9a four-way junction 37. (A) Exchange process between the A/D and the A/B coaxial stacked forms with the 8-13C-guanosines labels G14 and G15 highlighted in orange. (B) Imino proton NMR spectrum of 37 in the absence and presence of 20 eq. MgCl2. (C) 1H-13C-HSQC spectra of 37 (G14, G15 labeled) and 38 (G14 labeled) with assignments.
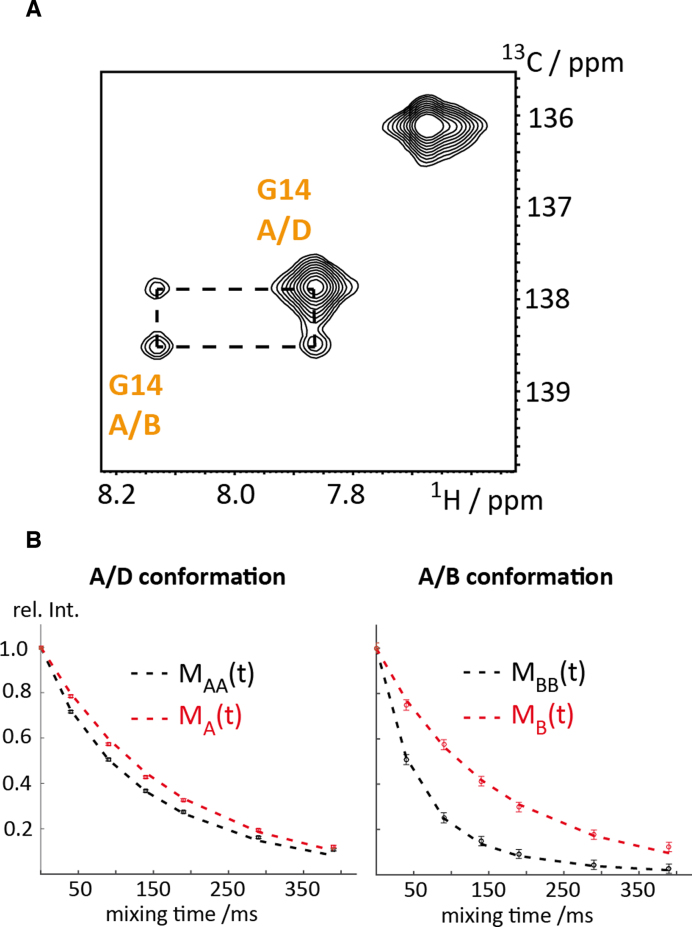
In a final effort, we addressed the refolding kinetics between the two stacked conformational states of the four-way junction J9a 37 by longitudinal exchange NMR spectroscopy (Figure 8A and B). The exchange resides in the slow chemical shift time regime as two separated resonance patterns were observed and thus the 13C ZZ exchange NMR experiment is perfectly suited to address the kinetic signature of the conformational rearrangement between the two stacked X-conformations A/B and A/D. The experimental verification of the interconversion between the A/B and the A/D state is obvious from the ZZ exchange NMR experiment at a mixing time of 50 ms (Figure 8A) displaying exchange cross-peaks. The forward and backward folding rates of the X-stacked conformation equilibrium were determined using a recently introduced approach based on the comparison of the diagonal peak intensities in a T1 and ZZ exchange experiment, respectively (Figure 8B) (79). At 25°C, a forward refolding rate of kA/D→A/B of 3.1 ± 0.3 s–1 and a backward rate kA/B→A/D of 10.6 ± 0.3 s–1 was determined yielding an equilibrium position KA/D-A/B298K of 0.29, which is in very good agreement with previously determined values (from peak integration: KA/D-A/B298K = 0.32; Altona et al.: KA/D-A/B294K = 0.41). The determined refolding rates are also in good agreement with the one reported from Altona and Overmars (kA/D→A/B of 2.3 ± 0.2 s–1 and a backward rate kA/B→A/D of 5.6 ± 0.5 s–1 at 21°C).
Figure 8.
ZZ-exchange NMR spectroscopy of J9a four-way junction 37. (A) ZZ exchange spectrum of 37 at a mixing time of 50 ms and at 25°C. (B) Normalized partial peak volumes of MA(t) and MAA(t) (red and black rectangles) and MB(t) and MBB(t) (red and black circles) corresponding to the diagonal peaks of G14 of the ZZ exchange (MAA/BB(t)) and T1 (MA/B(t)) experiment, respectively. Best fit curves (dashed red and black lines) are shown. Rate constants kA/D→A/B of 3.1 ± 0.3 s–1 and kA/B→A/D of 10.6 ± 0.3 s–1 for the refolding reaction and longitudinal relaxation rates R1A/D = 5.7 ± 0.1 s–1 and R1A/B = 6.7 ± 0.4 s–1 for the 8-13C-label of the G14 residue were obtained.
This final example nicely illustrates that the 13C-DNA labeling protocol can be used to address functional dynamics in biologically relevant DNAs over a large range of timescales from micro-/milliseconds to seconds, but also to monitor very slow structural rearrangements by real-time NMR spectroscopy.
CONCLUSIONS
In this work, we present a 13C-labeling protocol for DNA capitalizing on the synthesis of atom-specific 13C-modified DNA phosphoramidites and site-specific stable isotope labeling using solid phase DNA synthesis. The approach builds upon the previously established synthetic access to atom-specific 13C-modified RNA phosphoramidites and now enables to address functional dynamics in both types of nucleic acids over a wide range of timescales. The chemical syntheses for 6-13C-pyrimdine (27, 29) and 8-13C-purine (26, 28) DNA phosphoramidites starting from affordable stable isotope sources (13C-potassium cyanide 40 €/g and 13C-formic acid 90 €/g) are described taking into account alternative nucleosidation reactions and the most economic synthetic routes are discussed. All four building blocks could be incorporated into target DNAs with high coupling yields (> 98%) and several constructs ranging in size from 20 nt to 55nt were produced in sufficient amounts to conduct high resolution NMR studies.
In detail, the labeling approach was used to facilitate or even allow the resonance assignment process in DNAs displaying a high degree signal overlap as DNA 30 forming the HIV-1 polypurine tract RNA–DNA hybrid duplex This DNA 30 comprises almost only pyrimidine nucleotides (nine thymidines, eight 2′-deoxycytidines and only three purine 2′-deoxynucleotides) and site-specific 6-13C-thymidine labeling showed the high potential of the presented approach to resolve assignment ambiguities even in such small constructs. We then started to address functional dynamics in DNA using the relaxation dispersion NMR experiment. We have previously investigated the mini-cTAR DNA 33 using proton relaxation dispersion NMR spectroscopy and now used the labeling scheme to conduct 13C-Carr Purcell Meiboom Gill (CPMG) relaxation dispersion experiments in the absence and presence of the nucleocapsid 7 peptide (NCp 7), a RNA/DNA chaperone assisting in the formation of the TAR/cTAR RNA/DNA hybrid duplex in the viral replication cycle. We were able to obtain high quality data and the dispersion experiment captured a subtle effect of the addition of NCp 7, namely an up-regulation of the dynamic process also found for the mini cTAR DNA 33 alone. This is in-line with mode of action of RNA/DNA chaperones to lower the activation barriers on the folding landscape allowing the formation of the thermodynamic most stable species. Furthermore, the labeling strategy was used to address millisecond dynamics in a larger DNA construct—the full-length HIV-1 cTAR DNA 34 comprising 55 nucleotides. Here, a potential pitfall of the divide and conquer approach used in the NMR spectroscopy of nucleic acids became obvious. We were not able to recapitulate the dynamic behavior of shared labeled residues (A18/A5 and A34/A21) of the full-length DNA 35 and the mini cTAR DNA 33, but instead found dynamics for another adenosine residue. Thus, the exchange process of the 5′- and 3′-dangling ends of the mini-cTAR DNA 33 very likely is an artefact from the truncation of the sequence and such effects should be kept in mind when applying the divide and conquer approach to large DNAs but also RNAs. DNA quadruplexes play vital roles in the cellular environment. Solution NMR spectroscopy has proven to be well suited to address structural and functional features of this species capitalizing on site-specific labeling using high-priced uniformly 13C/15N-labeled DNA phosphoramidites. We introduced the novel 13C-labeled DNA building blocks in the challenging G-quadruplex 35 displaying structural heterogeneity between a (3+1)- and a parallel quadruplex form. The resonance assignment process for DNA quadruplexes is very challenging as this species provides inherently a low signal dispersion due to the predominance of G residues. The resonance assignment process for DNA 35 is even more complicate as the fold heterogeneity (60% for (3+1)-fold and 40% for parallel fold at room temperature) leads to a doubling of resonances and to severe overlap. The here proposed building blocks can be produced at significantly lower costs and the high potential of atom- and site-specific 13C-labeling for G-quadruplex studies is illustrated by a real-time NMR re-equilibration study of 35 after a temperature jump. As a final example, the four-way Holliday junction mimic J9a 37 was chosen to showcase the applicability of the building blocks for longitudinal exchange NMR experiments. We were able to detect and quantify the exchange process between two stacked X-conformers in unperturbed equilibrium.
Taken together, for the first time a comprehensive synthetic access to atom-specific 13C-labeled DNA phosphoramidites is presented along with solution NMR spectroscopic applications for resonance assignment purposes and studying functional dynamics of DNA covering timescales from micro-/milliseconds, to seconds and even slower processes with a time resolution on the minute time scale can be studied using fast NMR methods and the labeling protocol. Furthermore, as the solid phase synthesis of DNA is very advanced constructs with approximately 100 nucleotides should be directly accessible for site-specific stable isotope labeling pushing the limit currently encountered in the field of nucleic acid NMR spectroscopy (∼50 nt).
Supplementary Material
ACKNOWLEDGEMENTS
CK thanks Ronald Micura (Innsbruck) for scientific discussions. CK thanks Bernhard Brutscher (Grenoble) for providing the 1H–13C-BEST TROSY pulse sequence.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Austrian Science Fund (FWF) [P26550, P28725 to CK]. Funding for open access charge: FWF.
Conflict of interest statement. None declared.
REFERENCES
- 1. Shajani Z., Varani G.. NMR studies of dynamics in RNA and DNA by 13C relaxation. Biopolymers. 2007; 86:348–359. [DOI] [PubMed] [Google Scholar]
- 2. Latham M.P., Brown D.J., McCallum S.A., Pardi A.. NMR methods for studying the structure and dynamics of RNA. ChemBioChem. 2005; 6:1492–1505. [DOI] [PubMed] [Google Scholar]
- 3. Zhao B., Zhang Q.. Characterizing excited conformational states of RNA by NMR spectroscopy. Curr. Opin. Struct. Biol. 2015; 30:134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Torchia D.A. NMR studies of dynamic biomolecular conformational ensembles. Prog. Nucl. Magn. Reson. Spectrosc. 2015; 84–85:14–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adrian M., Heddi B., Phan A.T.. NMR spectroscopy of G-quadruplexes. Methods. 2012; 57:11–24. [DOI] [PubMed] [Google Scholar]
- 6. Dominguez C., Schubert M., Duss O., Ravindranathan S., Allain F.H.T.. Structure determination and dynamics of protein–RNA complexes by NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2011; 58:1–61. [DOI] [PubMed] [Google Scholar]
- 7. Al-Hashimi H.M., Walter N.G.. RNA dynamics: it is about time. Curr. Opin. Struct. Biol. 2008; 18:321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bardaro M.F., Varani G.. Examining the relationship between RNA function and motion using nuclear magnetic resonance. Wiley Interdiscipl. Rev.: RNA. 2012; 3:122–132. [DOI] [PubMed] [Google Scholar]
- 9. Bothe J.R., Nikolova E.N., Eichhorn C.D., Chugh J., Hansen A.L., Al-Hashimi H.M.. Characterizing RNA dynamics at atomic resolution using solution-state NMR spectroscopy. Nat. Methods. 2011; 8:919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mustoe A.M., Brooks C.L., Al-Hashimi H.M.. Hierarchy of RNA functional dynamics. Annu. Rev. Biochem. 2014; 83:441–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu K., Miyazaki Y., Summers M.F.. Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR. 2009; 46:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nelissen F.H.T., Tessari M., Wijmenga S.S., Heus H.A.. Stable isotope labeling methods for DNA. Prog. Nucl. Magn. Reson. Spectrosc. 2016; 96:89–108. [DOI] [PubMed] [Google Scholar]
- 13. Weininger U. Site-selective 13C labeling of proteins using erythrose. J. Biomol. NMR. 2017; 67:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang H., van Ingen H.. Isotope-labeling strategies for solution NMR studies of macromolecular assemblies. Curr. Opin. Struct. Biol. 2016; 38:75–82. [DOI] [PubMed] [Google Scholar]
- 15. Kurzbach D., Kontaxis G., Coudevylle N., Konrat R.. Felli IC, Pierattelli R. Intrinsically Disordered Proteins Studied by NMR Spectroscopy. 2015; Cham: Springer International Publishing; 149–185. [Google Scholar]
- 16. Kerfah R., Plevin M.J., Sounier R., Gans P., Boisbouvier J.. Methyl-specific isotopic labeling: a molecular tool box for solution NMR studies of large proteins. Curr. Opin. Struct. Biol. 2015; 32:113–122. [DOI] [PubMed] [Google Scholar]
- 17. Rosenzweig R., Kay L.E.. Bringing dynamic molecular machines into focus by methyl-TROSY NMR. Annu. Rev. Biochem. 2014; 83:291–315. [DOI] [PubMed] [Google Scholar]
- 18. Gossert A.D., Jahnke W.. Atreya HS. Isotope Labeling in Biomolecular NMR. 2012; Netherlands, Dordrecht: Springer; 179–196. [Google Scholar]
- 19. Wiesner S., Sprangers R.. Methyl groups as NMR probes for biomolecular interactions. Curr. Opin. Struct. Biol. 2015; 35:60–67. [DOI] [PubMed] [Google Scholar]
- 20. Pardi A. [15]Multidimensional heteronuclear NMR experiments for structure determination of isotopically labeled RNA. Methods Enzymol. 1995; 261:350–380. [DOI] [PubMed] [Google Scholar]
- 21. Masse J.E., Bortmann P., Dieckmann T., Feigon J.. Simple, efficient protocol for enzymatic synthesis of uniformly 13C,15N-labeled DNA for heteronuclearNMR studies. Nucleic Acids Res. 1998; 26:2618–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith D.E., Su J.-Y., Jucker F.M.. Efficient enzymatic synthesis of 13 C,15N-labeled DNA for NMR studies. J. Biomol. NMR. 1997; 10:245–253. [DOI] [PubMed] [Google Scholar]
- 23. Zimmer D.P., Crothers D.M.. NMR of enzymatically synthesized uniformly 13C15N-labeled DNA oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:3091–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kojima C., Ono A.M., Ono A., Kainosho M.. Thomas L, James VD, Uli S. Methods in Enzymology. 2002; 338:Academic Press; 261–283. [DOI] [PubMed] [Google Scholar]
- 25. Quant S., Wechselberger R.W., Wolter M.A., Wörner K.H., Schell P., Engels J.W., Griesinger C., Schwalbe H.. Chemical synthesis of 13C-labelled monomers for the solid-phase and template controlled enzymatic synthesis of DNA and RNA oligomers. Tetrahedron Lett. 1994; 35:6649–6651. [Google Scholar]
- 26. Földesi A., Yamakage S.I., Nilsson F.P., Maltseva T.V., Chattopadhyaya J.. The use of non-uniform deuterium labelling [‘NMR-window’] to study the NMR structure of a 21mer RNA hairpin. Nucleic Acids Res. 1996; 24:1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saito Y., Nyilas A., Agrofoglio L.A.. Efficient synthesis of D-[1′-13C]-ribonucleosides for incorporation into oligo-RNA. Nucleosides. Nucleotides. Nucleic. Acids. 2001; 20:937–940. [DOI] [PubMed] [Google Scholar]
- 28. Agrofoglio L.A., Jacquinet J.-C., Lancelot G.. A multigram, stereoselective synthesis of d-[13C5]ribose from d-[13C6]glucose and its conversion into [13C5]nucleosides. Tetrahedron Lett. 1997; 38:1411–1412. [Google Scholar]
- 29. Milecki J., Földesi A., Fischer A., Adamiak R.W., Chattopadhyaya J.. Synthesis of multiply labelled ribonucleosides for sequence-specific labelling of oligo-RNA. J. Labelled Compd. Radiopharm. 2001; 44:763–783. [Google Scholar]
- 30. Shallop A.J., Gaffney B.L., Jones R.A.. Use of both direct and indirect 13C tags for probing nitrogen interactions in hairpin ribozyme models by 15N NMR. Nucleosides. Nucleotides. Nucleic. Acids. 2004; 23:273–280. [DOI] [PubMed] [Google Scholar]
- 31. Shallop A.J., Gaffney B.L., Jones R.A.. Use of 13C as an indirect tag in 15N specifically labeled nucleosides. Syntheses of [8-13C-1,7,NH2-15N3] adenosine, -guanosine, and their deoxy analogues. J. Org. Chem. 2003; 68:8657–8661. [DOI] [PubMed] [Google Scholar]
- 32. Liu Y., Yu P., Dyba M., Sousa R., Stagno J.R., Wang Y.-X.. Applications of PLOR in labeling large RNAs at specific sites. Methods. 2016; 103:4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y., Sousa R., Wang Y.-X.. Specific labeling: an effective tool to explore the RNA world. Bioessays. 2016; 38:192–200. [DOI] [PubMed] [Google Scholar]
- 34. Liu Y., Holmstrom E., Zhang J., Yu P., Wang J., Dyba M.A., De C., Ying J., Lockett S., Nesbitt D.J. et al. . Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature. 2015; 522:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Longhini A.P., LeBlanc R.M., Dayie T.K.. Chemo-enzymatic labeling for rapid assignment of RNA molecules. Methods. 2016; 103:11–17. [DOI] [PubMed] [Google Scholar]
- 36. Longhini A.P., LeBlanc R.M., Becette O., Salguero C., Wunderlich C.H., Johnson B.A., D'Souza V.M., Kreutz C., Dayie T.K.. Chemo-enzymatic synthesis of site-specific isotopically labeled nucleotides for use in NMR resonance assignment, dynamics and structural characterizations. Nucleic Acids Res. 2016; 44:e52–e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alvarado L.J., Longhini A.P., LeBlanc R.M., Chen B., Kreutz C., Dayie T.K.. Donald HB-A. Methods in Enzymology. 2014; 549:Academic Press; 133–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alvarado L.J., LeBlanc R.M., Longhini A.P., Keane S.C., Jain N., Yildiz Z.F., Tolbert B.S., D'Souza V.M., Summers M.F., Kreutz C. et al. . Regio-selective chemical-enzymatic synthesis of pyrimidine nucleotides facilitates rna structure and dynamics studies. ChemBioChem. 2014; 15:1573–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wunderlich C.H., Huber R.G., Spitzer R., Liedl K.R., Kloiber K., Kreutz C.. A novel paramagnetic relaxation enhancement tag for nucleic acids: a tool to study structure and dynamics of RNA. ACS Chem. Biol. 2013; 8:2697–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wunderlich C.H., Spitzer R., Santner T., Fauster K., Tollinger M., Kreutz C.. Synthesis of (6-13C)Pyrimidine nucleotides as spin-labels for RNA dynamics. J. Am. Chem. Soc. 2012; 134:7558–7569. [DOI] [PubMed] [Google Scholar]
- 41. Neuner S., Kreutz C., Micura R.. The synthesis of 15N(7)-Hoogsteen face-labeled adenosine phosphoramidite for solid-phase RNA synthesis. Monatsh. Chemie - Chem. Monthly. 2017; 148:149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Juen M.A., Wunderlich C.H., Nußbaumer F., Tollinger M., Kontaxis G., Konrat R., Hansen D.F., Kreutz C.. Excited states of nucleic acids probed by proton relaxation dispersion NMR spectroscopy. Angew. Chem. Int. Ed. 2016; 55:12008–12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Neuner S., Santner T., Kreutz C., Micura R.. The “Speedy” synthesis of atom-specific 15N Imino/Amido-labeled RNA. Chemistry. 2015; 21:11634–11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Väre Y.V., Eruysal R.E., Narendran A., Sarachan L.K., Agris F.P.. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules. 2017; 7:E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vendeix F.A.P., Murphy Iv F.V., Cantara W.A., Leszczyńska G., Gustilo E.M., Sproat B., Malkiewicz A., Agris P.F.. Human tRNALys3UUU Is pre-structured by natural modifications for cognate and wobble codon binding through Keto–Enol tautomerism. J. Mol. Biol. 2012; 416:467–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Micura R. Small interfering RNAs and their chemical synthesis. Angew. Chem. Int. Ed. 2002; 41:2265–2269. [DOI] [PubMed] [Google Scholar]
- 47. Phan A.T., Modi Y.S., Patel D.J.. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004; 126:8710–8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Phan A.T., Patel D.J.. A site-specific low-enrichment 15N,13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002; 124:1160–1161. [DOI] [PubMed] [Google Scholar]
- 49. Phan A.T., Patel D.J.. Differentiation between unlabeled and very-low-level fully 15N,13C-labeled nucleotides for resonance assignments in nucleic acids. J. Biomol. NMR. 2002; 23:257–262. [DOI] [PubMed] [Google Scholar]
- 50. Jones R. Agrawal S. Protocols for Oligonucleotide Conjugates. 1994; 26:Humana Press; 207–231. [Google Scholar]
- 51. Jones R. Stabel Isotope Applications in Biomolecular Structure and Mechnism. 1994; Los Alamos: Los Alamos National Laboratory. [Google Scholar]
- 52. Williamson J.R., Boxer S.G.. Synthesis of a thymidine phospboramidite labelled with 13C at C6: relaxation studies of the loop region in a 13C labelled DNA hairpin. Nucleic Acids Res. 1988; 16:1529–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lafrancois C.J., Fujimoto J., Sowers L.C.. Synthesis and utilization of 13C(8)-enriched purines. Nucleosides. Nucleotides. 1999; 18:23–37. [DOI] [PubMed] [Google Scholar]
- 54. Gmeiner W.H., Poulter C.D.. An efficient synthesis of [8-13C]adenine. J. Org. Chem. 1988; 53:1322–1323. [Google Scholar]
- 55. Orji C.C., Michalczyk R., Silks L.A.P.. Synthesis of 2′β-Deoxy-[8-13C;amino,9-15N2]adenosine: unusual annulation conditions to assemble the purine core. J. Org. Chem. 1999; 64:4685–4689. [DOI] [PubMed] [Google Scholar]
- 56. Yamada H., Hirobe M., Higashiyama K., Takahashi H., Suzuki K.T.. Detection of carbon-13-nitrogen-15 coupled units in adenine derived from doubly labeled hydrogen cyanide or formamide. J. Am. Chem. Soc. 1978; 100:4617–4618. [Google Scholar]
- 57. Szymanski E.S., Kimsey I.J., Al-Hashimi H.M.. Direct NMR evidence that transient tautomeric and anionic states in dG·dT form Watson–Crick-like base pairs. J. Am. Chem. Soc. 2017; 139:4326–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou H., Kimsey I.J., Nikolova E.N., Sathyamoorthy B., Grazioli G., McSally J., Bai T., Wunderlich C.H., Kreutz C., Andricioaei I. et al. . m1A and m1G disrupt A-RNA structure through the intrinsic instability of Hoogsteen base pairs. Nat. Struct. Mol. Biol. 2016; 23:803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kimsey I.J., Petzold K., Sathyamoorthy B., Stein Z.W., Al-Hashimi H.M.. Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature. 2015; 519:315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nikolova E.N., Kim E., Wise A.A., O/'Brien P.J., Andricioaei I., Al-Hashimi H.M.. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011; 470:498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yi-Brunozzi H.Y., Brinson R.G., Brabazon D.M., Lener D., Le Grice S.F.J., Marino J.P.. High-resolution NMR analysis of the conformations of native and base analog substituted retroviral and LTR-retrotransposon PPT primers. Chem. Biol. 2008; 15:254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brinson R.G., Turner K.B., Yi-Brunozzi H.Y., Le Grice S.F.J., Fabris D., Marino J.P.. Probing anomalous structural features in polypurine tract-containing RNA–DNA hybrids with neomycin B. Biochemistry. 2009; 48:6988–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brinson R.G., Miller J.T., Kahn J.D., Le Grice S.F.J., Marino J.P.. Zvi K. Methods in Enzymology. 2016; 566:Academic Press; 89–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zargarian L., Kanevsky I., Bazzi A., Boynard J., Chaminade F., Fossé P., Mauffret O.. Structural and dynamic characterization of the upper part of the HIV-1 cTAR DNA hairpin. Nucleic Acids Res. 2009; 37:4043–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zargarian L., Tisné C., Barraud P., Xu X., Morellet N., René B., Mély Y., Fossé P., Mauffret O.. Dynamics of linker residues modulate the nucleic acid binding properties of the HIV-1 nucleocapsid protein zinc fingers. PLoS One. 2014; 9:e102150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lim K.W., Lacroix L., Yue D.J.E., Lim J.K.C., Lim J.M.W., Phan A.T.. Coexistence of two distinct G-Quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010; 132:12331–12342. [DOI] [PubMed] [Google Scholar]
- 67. Overmars F.J.J., Altona C.. NMR study of the exchange rate between two stacked conformers of a model holliday junction1. J. Mol. Biol. 1997; 273:519–524. [DOI] [PubMed] [Google Scholar]
- 68. Lee B.M., De Guzman R.N., Turner B.G., Tjandra N., Summers M.F.. Dynamical behavior of the HIV-1 nucleocapsid protein1. J. Mol. Biol. 1998; 279:633–649. [DOI] [PubMed] [Google Scholar]
- 69. Vorbrüggen H., Krolikiewicz K., Bennua B.. Nucleoside syntheses, XXII1. Nucleoside synthesis with trimethylsilyl triflate and perchlorate as catalysts. Chem. Ber. 1981; 114:1234–1255. [Google Scholar]
- 70. Hodge R.P., Brush C.K., Harris C.M., Harris T.M.. Synthesis of 1- and 1,2,2′-deuteriated deoxyribose and incorporation into deoxyribonucleosides. J. Org. Chem. 1991; 56:1553–1564. [Google Scholar]
- 71. Markiewicz W.T., Wiewiórowski M.. A new type of silyl protecting groups in nucleoside chemistry. Nucleic Acids Res. 1978; 1:s185–s190. [Google Scholar]
- 72. Barton D.H.R., McCombie S.W.. A new method for the deoxygenation of secondary alcohols. J. Chem. Soc., Perkin Trans. 1. 1975; 1574–1585. [PubMed] [Google Scholar]
- 73. Bazzi A., Zargarian L., Chaminade F., Boudier C., De Rocquigny H., René B., Mély Y., Fossé P., Mauffret O.. Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC. Nucleic Acids Res. 2011; 39:3903–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Boudier C., Storchak R., Sharma K.K., Didier P., Follenius-Wund A., Muller S., Darlix J.L., Mély Y.. The mechanism of HIV-1 tat-directed nucleic acid annealing supports its role in reverse transcription. J. Mol. Biol. 2010; 400:487–501. [DOI] [PubMed] [Google Scholar]
- 75. Godet J., de Rocquigny H., Raja C., Glasser N., Ficheux D., Darlix J.-L., Mély Y.. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J. Mol. Biol. 2006; 356:1180–1192. [DOI] [PubMed] [Google Scholar]
- 76. Brutscher B., Felli I.C., Gil-Caballero S., Hošek T., Kümmerle R., Piai A., Pierattelli R., Sólyom Z.. Felli IC, Pierattelli R. Intrinsically Disordered Proteins Studied by NMR Spectroscopy. 2015; Cham: Springer International Publishing; 49–122. [Google Scholar]
- 77. Lescop E., Schanda P., Brutscher B.. A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J. Magn. Reson. 2007; 187:163–169. [DOI] [PubMed] [Google Scholar]
- 78. Lescop E., Kern T., Brutscher B.. Guidelines for the use of band-selective radiofrequency pulses in hetero-nuclear NMR: example of longitudinal-relaxation-enhanced BEST-type 1H–15N correlation experiments. J. Magn. Reson. 2010; 203:190–198. [DOI] [PubMed] [Google Scholar]
- 79. Kloiber K., Spitzer R., Grutsch S., Kreutz C., Tollinger M.. Longitudinal exchange: an alternative strategy towards quantification of dynamics parameters in ZZ exchange spectroscopy. J. Biomol. NMR. 2011; 51:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.