Abstract
Tuberculosis (TB) is one of the leading causes of death worldwide, especially in developing countries. Neonatal BCG vaccination occurs in various regions, but the level of protection varies in different populations. Recently, Mycobacterium vaccae is found to be an immunomodulating therapeutic agent that could confer a significant level of protection against TB. It is the only vaccine in a phase III trial from WHO to assess its efficacy and safety in preventing TB disease in people with latent TB infection. However, the mechanism of immunotherapy of M. vaccae remains poorly understood. In this study, the full genome of M. vaccae was obtained by next-generation sequencing technology, and a proteogenomic approach was successfully applied to further perform genome annotation using high resolution and high accuracy MS data. A total of 3,387 proteins (22,508 unique peptides) were identified, and 581 proteins annotated as hypothetical proteins in the genome database were confirmed. Furthermore, 38 novel protein products not annotated at the genome level were detected and validated. Additionally, the translational start sites of 445 proteins were confirmed, and 98 proteins were validated through extension of their translational start sites based on N terminus-derived peptides. The physicochemical characteristics of the identified proteins were determined. Thirty-five immunogenic proteins of M. vaccae were identified by immunoproteomic analysis, and 20 of them were selected to be expressed and validated by Western blot for immunoreactivity to serum from patients infected with M. tuberculosis. The results revealed that eight of them showed strong specific reactive signals on the immunoblots. Furthermore, cellular immune response was further examined and one protein displayed a higher cellular immune level in pulmonary TB patients. Twelve identified immunogenic proteins have orthologous in H37Rv and BCG. This is the first study to obtain the full genome and annotation of M. vaccae using a proteogenomic approach, and some immunogenic proteins that were validated by immunoproteomic analysis could contribute to the understanding of the mechanism of M. vaccae immunotherapy.
Tuberculosis (TB)1 is an infectious disease caused by the bacillus Mycobacterium tuberculosis, and it is one of the most lethal diseases worldwide. Although over 20 years after WHO declared TB a global public health emergency, the infection remains a major global health problem (1). In 2014, 9.6 million people were estimated to have fallen ill with TB, and 1.5 million people were killed (2). Furthermore, TB now ranks alongside the human immunodeficiency virus (HIV) as a leading cause of death worldwide (3). Although TB is curable and preventable, resistance to medicine and severe adverse drug reactions make the treatment of TB difficult. One of the important tools in the global fight against TB is neonatal vaccination with Mycobacterium bovis bacillus Calmette and Guérin (BCG), which was first used in 1921. In many countries, the immunization of infants with BCG can protect against TB meningitis and other severe forms of TB in children less than five years of age, but the protection is insufficient in various populations (4). It has been suggested that the low levels of protection are likely related to the effectiveness of the vaccine or the absence of complete coverage in these populations (1). The goal of WHO since 2016 has been to end the global TB epidemic by implementing the End TB Strategy (2). Therefore, new vaccination or/and treatment strategies are urgently needed, particularly against primary infection and latent pulmonary infection.
Mycobacterium vaccae (M. vaccae), a heat-killed vaccine derived from a nontuberculosis mycobacterium, could be employed as an immunomodulator to enhance anti-TB mycobacterial infections in patients with cellular immune function. Combined with chemotherapy, the inactivated M. vaccae could also enhance the efficacy for the adjunctive treatment of TB (5). Additionally, M. vaccae could confer a significant level of protection against TB among HIV-infected individuals who were vaccinated with BCG during their childhood (6). Recently, this vaccine was licensed by the China Food and Drug Administration as an immunotherapeutic agent to help shorten TB treatment for patients with drug-susceptible TB (2). It is the only vaccine in a phase III trial to assess its efficacy and safety in preventing TB disease in people with latent TB infection. Despite the growing interest in the therapeutic implications of the vaccine, the exact mechanism by which heat-killed mycobacteria exert their immunomodulatory effects in humans is not fully understood.
The whole-genome sequence and accurate annotations of bacteria are the basis for understanding the immune molecular mechanisms, phylogenetic relationships and the genetic factors that are responsible for their pathogenicity. Conventionally, most genome sequences are annotated with multiple gene prediction algorithms or by manual assignment based on BLAST search results (7). Although considerable emphasis has been placed on accurate analytical tools and updated specialized databases, most genomic data are experimentally uncharacterized (8). Furthermore, the predicted genes exhibit frequent errors, particularly in the false recognition of translational start codons, omission of novel protein coding genes, mis-annotation of pseudogenes, and confusion of overlapping genes (9). Therefore, there is an urgent need for further experimental validation or complementary annotation approaches to conventional genome annotation.
Recently, MS-based proteomic information has been applied to refine genome annotation, which has been termed proteogenomics. This approach can discover previously unidentified genes, and correct and validate the predicted genes in various organisms (10). Because the identification of proteins by MS is more explicit and confident than that from genomic sequence data alone, integrating this protein-level information into the genome annotation process could greatly improve the quality of genome annotation. Proteogenomic analysis has previously been applied to several sequenced prokaryotes and eukaryotes to improve genome annotation, including Mycobacterium smegmatis (11), Mycobacterium tuberculosis (12), Trichophyton rubrum (13), yeast (14), rat (15), and human (16). It has been demonstrated that this approach is likely to become a part of most genome annotation workflows in the future.
Currently, the mechanisms of immunotherapy of M. vaccae are poorly understood and many immune antigens remain unidentified. Among the currently available techniques, immunoproteomics has rendered very good results in the study of the antibody response in experimental infection samples (17). Combined with 2-DE and Western blotting using sera from human patients, an immunoproteomic approach could identify the immunodetected proteins by MS. This strategy has been successfully used for antigen discovery, which is ideally suited to screen and identify potential new candidates for vaccine development as well as biomarkers for infectious diseases (18–22). In this study, we obtained the genome of M. vaccae by next generation sequencing, and applied a proteogenomic approach to perform genome annotation using high resolution and high accuracy MS data. Furthermore, to detect and characterize some immunogenic proteins from M. vaccae, protein profiling by 2-DE and Western blotting were performed, and 35 candidate antigen proteins were identified. Among them, 33 have orthologous in M. tuberculosis H37Rv, BCG or other M. tuberculosis species. Twenty of them were selected to be expressed to validate their immunogenicity by in vitro detection of both humoral and cellular reactivity against TB patients. The results revealed that eight of them showed strong specific reactive signals on the immunoblots of TB+ serum, and one protein displayed a high cellular immune level in pulmonary TB patients. To our knowledge, this is the first study devoted to the identification of M. vaccae immunogenic proteins using immunoproteomics, and the results could provide insight into the immune molecular mechanisms of this newly recognized vaccine strain.
EXPERIMENTAL PROCEDURES
Cell Cultivation and Sample Preparation
The M. vaccae 95051 cells used in this study were grown in 7H9 culture at 30 °C with shaking (100 rpm). When the absorbance of the culture at 600 nm reached 0.8–1.0, the culture supernatant and cells were supplemented with protease inhibitor mixture (Roche, Germany) and separated via filtration first through a 0.45-μm-pore-size membrane and then a 0.22-μm-pore-size membrane (Millipore, Bedford, MA). Genome DNA and protein samples were prepared as previously described with modifications (23). Briefly, total genomic DNA from the cells was extracted using the Qiagen Genomic DNA set (Qiagen, Hilden, Germany). The DNA sequence was determined by the Illumina HiSeq2500 platform using a 100-bp paired-end library with an insert size of 500 bp. For protein preparation, the culture filtrate fraction was treated with sodium deoxycholate and precipitated with trichloroacetic acid as previously described (filtrate fraction) (23). The pelleted cells were probe-sonicated for 30 s with 20 rounds at 30 s interval in ice-cold sonication buffer (50 mm Tris-HCl, 10 mm MgCl2, 0.02% sodium azide, pH 7.4) and the total lysate was roughly divided into three fractions as described in a previous study (24). The lysate was centrifuged at 600 × g for 15 min to collect the cell debris fraction. Upon centrifugation (20,000 × g for 20 min), the resulting supernatant yielded the supernatant (cytoplasmic fraction including plasma membrane) and sediment (cell wall fraction). The protein content from different fractions was quantitated via the bicinchoninic acid protein assay and then subjected to SDS-PAGE.
Two hundred serum samples from patients with pulmonary TB (before treatment) and 50 samples from healthy people were recruited from Shenzhen Third People's Hospital in China and stored in our labs. TB patients were diagnosed and classified according to the 1990 edition of Diagnostic Standards and Classification of Tuberculosis. TB status was also diagnosed based on clinical signs and symptoms, radiographic findings (chest X-ray and/or HRCT) (25). Healthy individuals who had no TB history, no contact with TB patients, and no clinical symptoms of TB and who tested negative for TB in both purified protein derivative test and interferon gamma release assay were recruited as control (26). Written informed consents were obtained from all participates and the study was reviewed and approved by the Ethics Committee of the Shenzhen-Hong Kong Institute of Infectious Disease, Shenzhen Third People's Hospital, China (human subjects research approval number 2016-006).
Genome Annotation and Comparative Analysis
Glimmer3 and tRNAscan-S.E. were used to predict protein-coding sequences (CDS) and tRNA genes, respectively (27, 28). Overlapping and closely clustered CDS were manually inspected. The functional annotation of the predicted CDS was carried out by BLASTP search of the translations products versus GenBank's nonredundant protein database followed by manual curation (29). Ribosomal RNAs were identified by BLASTN search against a database of all publicly available rRNA sequences. Complete sequence of M. vaccae ATCC 95051 has been deposited in GenBank under the accession No. CP011491. Genomic comparisons were carried out by bidirectional BLASTP comparisons of whole genome protein databases.
Orthologous groups among M. vaccae strain ATCC 95051 and 14 other mycobacterium, including M. tuberculosis H37Rv (NC_000962), M. bovis BCG (NC_008769), M. abscessus ATCC 19977 (NC_010397), M. avium strain 104 (NC_008595), M. chubuense NBB4 (NC_018027), M. gilvum PYR-GCK (NC_009338), M. intracellulare MOTT-02 (NC_016947), M. marinum strain M (NC_010612), M. massiliense GO 06 (NC_018150), M. rhodesiae NBB3 (NC_016604), M. smegmatis MC2 155 (NC_018289), M. ulcerans Agy99 (NC_008611), M. vanbaalenii PYR-1 (NC_008726), and M. yongonense 05–1390 (NC_021715) were identified by the OrthoMCL program with an E-value cutoff of 10−5 (30). Visualization of the M. vaccae genome and comparative results were conducted using the Circular Genome Viewer (31).
Proteogenomic Approach for Genome Annotation
Protein from each fraction was loaded onto a 12% SDS-PAGE gel and each lane was cut into 16 bands and subjected to an in-gel tryptic digestion protocol as previously described (24). All the peptide mixtures were solubilized in 0.1% formic acid and analyzed using a UPLC system (Waters, Milford, MA) coupled to an LTQ Orbitrap Velos mass spectrometer (ThermoFisher Scientific, Waltham, MA) with some modifications (32). Briefly, the LC system is equipped with a C18 reversed-phase microcapillary trapping (nanoAcquity Symmetry C18, 5 μm, 180 μm × 20 mm) and an analytical column (nanoAcquity BEH 300 C18, 1.7 μm, 100 μm ×100 mm). The flow rate was 0.4 μl/min for the nano-column, and peptides were separated in a 160-min chromatography gradient using aqueous solvents A (0.1% HCOOH) and B (0.1% HCOOH, 80% CH3CN). The eluted peptides were electrosprayed with a distally applied spray voltage of 2.0 kV and the analysis was performed in a data-dependent manner with a survey full scan resolution of r = 60,000 at m/z 400 and a scan range of m/z 380 to 2000. Normalized collision energies of 35 and 40% were used for CID and HCD fragmentation, respectively. Following every survey scan, up to 20 and 10 of the most intense precursor ions from the full scan were selected for fragmentation via CID and HCD, respectively. Target ions that had already been picked for MS/MS were dynamically excluded for the next 30 s, and an activation q-Value of 0.25 and an activation time of 10 ms were applied. Lock mass calibration using a background ion from the air (m/z 445.12003) was applied. The LC-MS analysis of all samples was performed in three replicates.
The raw MS/MS data were searched against protein database from sequenced genome of M. vaccae (supplemental Text S1, 6003 entries) and a custom six-frame database (supplemental Text S2). The custom six-frame database was constructed by translating the entire genome in all six reading frame options, three forward and three on the reversed DNA strand (33). The name of the CDSs with the same annotations in the M. vaccae genome were identical to the original names (MYVA_0001 to MYVA_5922), and the CDSs with other reading frame options were replaced with the custom specialized name (ORF00001 to ORF66723). Additionally, sequences for common contaminants (338 unique entries) from two collections (the Max Planck Institute of Biochemistry and the Global Proteome Machine Organization Common Repository of Adventitious Protein) were included in the custom FASTA file. The final database had a total of 72, 983 entries (supplemental Text S2).
The raw data from the proteomic analysis were processed using Proteome Discoverer software (version 1.4.1.12, Thermo Fisher Scientific) with two different search algorithms, MASCOT (version 2.3.02, Matrix Sciences, UK) and SEQUEST (version 1.3, Thermo Fisher Scientific) against two databases above. The search parameters applied in the database searches were: enzyme specificity: trypsin/P; maximum missed cleavages: 2; carboxymethyl (C) as a static modification; oxidation (M) and N-terminal acetylation as dynamic modifications; a precursor mass tolerance of 5 ppm; and a MS/MS mass tolerance of 0.8 Da. The reverse database search option was enabled, and a maximum target decoy-based false discovery rate (FDR) of 1.0% for peptide and protein identification was allowed. The maximal posterior error probability (PEP) must be below or equal to 0.01 and the q-Value could be below 0.01. The IonScore should be above 20 and The XCorr must be no less than 1.9. All of the raw files (.raw) and the merged peak list files (.mgf) generated from Proteome Discoverer software in the present study have been deposited into the publicly accessible database PeptideAtlas with data set Identifier PASS00954 (http://www.peptideatlas.org/PASS/PASS00954).
2-DE and Western Blot Analysis
M. vaccae protein samples were precipitated using the 2D clean-up kit (GE Healthcare, Piscataway, NJ), and the concentration was determined using a PlusOne 2D Quant Kit (GE Healthcare) according to the manufacturer's instruction. Proteins (200 μg) were resuspended in 350 μl rehydration buffer (7 m urea, 2 m thiourea, 4% (w/v) CHAPS, 50 mm DTT, 0.5% IPG Buffer, pH 4–10). IEF was performed using an Ettan IPGphor III system (GE Healthcare). The sample was used to rehydrate the 20-cm IPG strips (pH 4–10) for 10 h at 20 °C. The following focusing parameters were applied: 200 V,1 h; 500 V, 1 h; from 500 V to 1000 V, 1 h; 1000 V to 10000 V, 8000 Vhr; 10000 V, 80000 Vhr; followed by the 1000 V holding step for 3 h at the end of the run (92450 Vhr total).
After focusing was completed, IPG strips were equilibrated with 1% (w/v) DTT in equilibration base buffer (75 mm Tris-HCl (pH 8.8), 6 m urea, 29.3% glycerol, 2% SDS, and 0.002% bromphenol blue) for 15 min, followed by another equilibration with 2.5% (w/v) iodoacetamide in the same buffer for 15 min. Equilibrated IPG strips were placed onto 12% SDS-polyacrylamide gels for the second dimensional separation. Three replicate 2-DE gels for each sample were used: one for Coomassie blue stain, one for Western blotting against immune serum from healthy people (TB−), and the other for Western blotting against immune serum from TB patients (TB+). Serum samples from patients and healthy people were prepared separately for Western blot assay. Briefly, after electrophoresis, one 2D gel was transferred onto nitrocellulose membranes. Pretreated serum was diluted 1:500, and secondary goat anti-human IgG antibody, conjugated with IRDye680 Fluorescence (GENE, Hong Kong, China), was diluted 1:10,000 as the final concentration. The fluorescence signal was analyzed using the LICOR Odyssey Infrared imager and Odyssey software 3.0.xs. A 38-kDa protein (IMMUNO, Woburn, MA) was run in parallel with the positive control and bovine serum albumin was used as the negative control.
The immunoblot profiles against TB− and TB+ were compared, and the Coomassie-stained protein spots on 2-DE gels corresponding to the immunoreactive spots in Western blotting were excised. The in-gel protein digestion was performed and the tryptic peptides were subjected to MALDI-TOF/TOF MS as described previously (34). Briefly, MS measurements were performed in positive ion reflector mode with 20 kV accelerating voltage and 23 kV reflecting voltage, and spectra were calibrated using PeptideCalibStandard II (Bruker, Germany) from mass range 800–3200 Da as external standards. BioTools 3.0 was used for data visualization and peptide mass fingerprinting (PMF) or TOF-TOF MS were analyzed and searched against protein database from sequenced genome of M. vaccae (GenBank accession No. CP011491, supplemental Text S1, 6003 entries) using the Mascot software (version 2.3.02, Matrix Sciences). The following search parameters were applied: trypsin digestion with max missed cleavage: 1; fixed modification: Carbamidomethylation (C); variable modification: Oxidation (M), Carbamyl (N-term), Deamidated (NQ); precursor ion mass tolerance: ± 100 ppm; fragment mass tolerance: ± 1.0 Da. Score greater than 50 was significant (p < 0.05) for a local PMF search and greater than 20 for TOF-TOF MS search. Decoy searches were performed using the automated “Decoy” search option from Mascot. The false discovery rate (FDR) is calculated as follows:
A maximum target decoy-based FDR of 1.0% for peptide identification was allowed. Additionally, proteins identified by a single peptide were validated manually.
Clone, Expression and Validation of Immunogenic Proteins
To validate the immunogenicity of the identified proteins, twenty proteins were chosen for expression and purification. The primer pairs used to amplify the DNA fragments of the recombinant proteins are listed in supplemental Table S1. The gene fragment was PCR amplified, restriction enzyme digested, and linked to the expression vector pET32a. The cloned genes were sequenced to confirm the correct reading frame. The bacterial strain of BL21 (DE3), under two inducible expression temperatures (37 °C and 28 °C), were used for the expression of all target ORF proteins. The bacterial strains were cultured and induced overnight with 1 mm isopropyl β-d-1-thiogalactopyranoside at 37 °C or 28 °C when they grew to OD600 = 0.6–1.0. Extraction of target proteins was performed using the Bugbuster Protein Expression Reagent (Novagen, San Diego, CA) and protein purification was carried out using the His MultiTrap FF purification kit (GE Healthcare) following the manufacturer's instructions. The protein sequences were confirmed by MALDI-TOF/TOF MS analysis as described above, and the raw data is deposited into the publicly accessible database PeptideAtlas with data set Identifier PASS00954.
The immunoblotting of the purified proteins against M. tb-specific serum antibody response was performed by Simon-Simple Western (ProteinSimple, San Jose, CA) analysis at room temperature according to the user's manual. Briefly, purified proteins were mixed with a master mix including 40 mm DTT to a final concentration of 0.5 μg/μl. Equal volumes of serum samples from 100 TB patients were collected and pooled for immunoblotting of the expressed antigen proteins. The protein samples, buffers, and pooled serum were dispensed to designated wells in different tubes. The separation electrophoresis and immunodetection steps took place in the capillary system, and they were fully automated. During electrophoresis, proteins were separated based on molecular weight through the stacking and separation matrices at 250 V for 40 min and then immobilized on the capillary wall using proprietary photo activated capture chemistry. The matrices were washed out, and capillaries were incubated with a blocking reagent for 15 min. Target proteins were then immunoprobed with primary antibodies, followed by HRP-conjugated secondary antibodies. Finally, luminol and peroxide (ProteinSimple) were added to generate chemiluminescence, which was captured by a charge-coupled device (CCD) camera. The digital image was analyzed with Compass software (ProteinSimple), and the quantified data of the detected protein were reported. Western analysis was performed in parallel using the commercial 38-kDa protein (IMMUNO, Massachusetts) as the positive control. The ratio between the intensities of the reaction band of the tested protein and the 38-kDa protein was calculated and used to evaluate the humoral response of the tested proteins.
Cellular immune assay of these proteins was further performed using a human IFN-γ cytokine pre-coated ELISPOT kit (Dakewe Biotech Company, Shenzhen, China). Peripheral blood mononuclear cells (PBMC) from participants were obtained from whole blood by centrifugation over Ficoll-Hypaque density gradient (Ficoll-Paque Plus; Amersham Biosciences). Because freshly separated PBMC cells are required for detecting cellular IFN-γ secretion, 23 pulmonary TB patients and 10 nontuberculosis individuals were recruited in order to examine the 20 proteins individually. Characterization of all participants are shown in Table I. A total of 2.5 × 105 cells/well were seeded in duplicate in 96-well plates and each individual sample was tested in parallel with a positive control (phytohemagglutinin, PHA), negative control (blank and buffer for protein purification), the ESAT-6 antigen from the ELISPOT kit as previously described (26). Cells were stimulated with the different antigens with the final concentration of 10 μg/ml and the plates were incubated overnight at 37 °C with 5% CO2 following the manufacturer's instructions. The spot forming cells (SFC) were counted by use of an automated image analysis system the BIOREADER® 4000 PRO-X (Biosys, Karben, Germany) and the individual spots number in the buffer control was subtracted from the number of all test wells. Assays were regarded positive using the criteria for M. tb infection in the commercial kit.
Table I. Characterization of all participants for identification of antigen candidates by ELISPOT.
| Characteristics | E+ pulmonary TB patientsa |
Non-tuberculosis individuals |
||
|---|---|---|---|---|
| ESAT-6 kit | Target protein | ESAT-6 kit | Target protein | |
| Total Number | 23 | 10 | ||
| Age, mean (range, years) | 37 (19–57) | 28 (24–35) | ||
| Male/Female | 14/9 | 1/9 | ||
| Bacteriological test positive/negative | 13/10 | 0/10 | ||
| PCR positive/negative/other | 7/0/16 | 0/10/0 | ||
| ELISPOTb, mean (range) | 153.6 (9–356) | 52.8 (0–343) | 8.2 (1–15) | 14 (3–33) |
a E+ active TB stands for active TB patients with positive response to ESAT-6 antigen by ELISPOT.
b ELISPOT was judged by the spot forming cells (SFC) of the antigen ESAT-6.
Bioinformatics Tools for Protein Analysis
The theoretical Mr and pI value were obtained from Proteome Discoverer software calculation. The subcellular localization of the identified proteins was predicted using the PSORTb v3.0 program (http://www.psort.org/psortb/) and protein subcellular localization prediction with functional gene ontology annotation (CELLO2GO). The gene prediction programs used for prokaryotes were FgeneSB and GeneMark, and homologous proteins were searched using the Blastp program.
RESULTS AND DISCUSSION
The examination of the complete genome and proteome and the identification of the immunoproteins expressed by M. vaccae species during human infection with TB have not been conducted. To address these gaps in our knowledge, we obtained the genome using NGS and proteogenomics and examined the immunoreactive proteins expressed from M. vaccae.
General Features and Comparative Genomics
Previous studies have reported the annotated genome sequence of the M. vaccae type strain, ATCC 25954, and the final assembly has 33 supercontigs (35). However, whole-genome sequencing is important to understand the molecular basis of the strain and to further study the phylogenetic relationships and the genetic factors that are responsible for pathogenicity. Therefore, the complete genome sequence of this microorganism is urgently needed, which could also facilitate a more reliable genetic identification between and within Mycobacterium species.
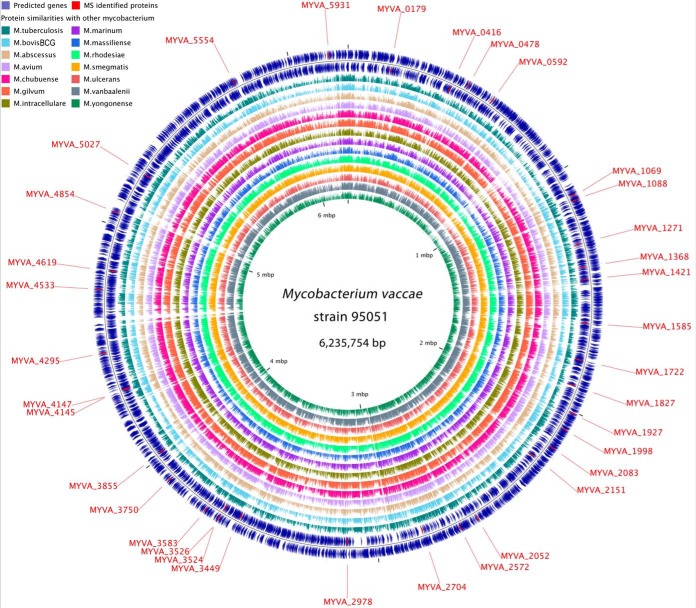
Using NGS and bioinformatic prediction, the genome of the M. vaccae strain ATCC 95051 consists of a single circular chromosome of 6,235,754 bp encoding for 5732 predicted proteins, as well as 48 tRNA genes and two sets of rRNA operons. The overall G+C content of the M. vaccae genome is 68.6%, which is higher than that of M. tuberculosis. Among the predicted proteins, 4535 (79%) are assigned a putative function, whereas 1,187 (21%) are hypothetical proteins. Complete sequence of M. vaccae ATCC 95051 has been deposited in GenBank under the accession CP011491 (https://www.ncbi.nlm.nih.gov/nuccore/CP011491). In addition, comparative genomics with 14 published Mycobacterium genomes revealed that 5177 (90%) proteins in M. vaccae genome have homologous in other mycobacterium (Fig. 1). Among the thirty-five immunogenic proteins identified by immunoproteomic analysis in this study (see below for details), 33 have orthologous in H37Rv, BCG and other M. tuberculosis species (Column H in supplemental Table S2).
Fig. 1.
Circular genome map of M. vaccae strain 95051. From outer to inner, circles 1 and 2 show the predicted genes on leading and lagging strand, respectively; circles 3 to 16 show genome comparisons in amino acids level with 14 other mycobacterium, including M. tuberculosis H37Rv, M. bovis BCG, M. abscessus ATCC 19977, M. avium strain 104, M. chubuense NBB4, M. gilvum PYR-GCK, M. intracellulare MOTT-02, M. marinum strain M, M. massiliense GO 06, M. rhodesiae NBB3, M. smegmatis MC2 155, M. ulcerans Agy99, M. vanbaalenii PYR-1 and M. yongonense 05–1390. Genome location of the 35 proteins identified by MALDI-TOF/TOF MS are highlighted in red and indicated by outside text. The scale is given on the innermost circle.
Proteomic Analysis of M. vaccae
The aims of this study were to obtain a comprehensive experimental catalogue of the genome-wide gene expression in M. vaccae and to use this information to improve the genome annotation. To reduce the complexity of the sample and increase the identification rate of proteins, which results in a greater dynamic range and more comprehensive proteome coverage, the M. vaccae lysate was roughly fractionated into four fractions, including the cell cytoplasm (including membrane), debris, filtrate and wall fractions. In total, 64 gel strips for each replicate were obtained and digested in-gel with trypsin into peptide mixtures. After LC-MS analysis using protein database from sequenced genome of M. vaccae, we obtained a total of 22,508 unique peptide sequences, which corresponded to 3387 proteins with a FDR of less than 1%. On average, more than six unique peptides were used to identify each protein, and the amino acid sequence coverage was ∼39.9%. The highest coverage of the peptides mapping to an identified protein (MYVA_3425), which mapped with 10 unique peptides, was 99.24%. The highest number of unique peptides identified was 113, which mapped to 49.7% of the protein 3-oxoacyl-ACP synthase (MYVA_3919). Other predominant proteins were the DNA-directed RNA polymerase subunit beta′ RpoC (MYVA_1070) with 89 peptides, the polyketide synthase (MYVA_5566) with 74 peptides, and the DNA-directed RNA polymerase subunit beta rpoB (MYVA_1069) with 68 unique peptides. For integral membrane protein (IMP), a total of 607 proteins were identified in this study (excluding the possible signal sequences), representing ∼54% of the total IMPs annotated in the genome. Protein identification based on unique peptide evidence is provided in supplemental Table S3.
Among these identifications in this study, 585 of them were identified by a single tandem MS/MS. The cutoff values used for PEP of the peptide identification is not more than 0.01, which is adopted and accepted by peer experts in the field of proteomics. To ensure the confident identification of these proteins, we further filtered the single peptide with a more stringent threshold of PEP<0.001, which is more suitable to led to a lower error rates of the novel peptides than the known peptides (13). With the stringent criterion (PEP<0.001), 397 out of 585 single peptide-based identifications could pass the threshold and used for annotation improvement. It should be noted that 188 identifications were with PEP < 0.01 and >0.001 for the supporting peptides, which may be somewhat uncertain in confirming those proteins identified by the single tandem MS than those by PEP < 0.001 and multiple unique peptides. However, these single peptide-based identifications (0.001<PEP<0.01) could provide some hints to confirm ORFs, and further experimental validation to confirm the existing of the identifications should be pursued. In addition, the precursor mass tolerance was set as 5 ppm for all the peptides identification when searching the database. So all the peptides qualified the criterion should be ± 5 ppm and we believed the confidence of single peptide could be increased. Furthermore, these proteins identified by a single peptide could not be accepted unless their corresponding MS/MS spectra passed the manual validation, and the annotated spectra of peptides matching to proteins that had a single peptide hit were provided in supplemental Table S4.
Identification of Hypothetical Proteins
Protein identification based on unique peptide evidence was considered to be the existence of the gene products in the genome annotation. These “known genes” whose functions were well-described through a homology search against other organisms or through experimental studies were confirmed at the protein level. In total, 2807 “known gene” products were obtained in our work, representing ∼83% of the identifications in this study.
In the protein database, hypothetical proteins are functionally uncharacterized because of their lack of sequence similarity to any known proteins. Further experimental validation is required to validate the expression of the hypothetical proteins annotated by bioinformatic approaches. One of the most effective methods to validate these hypothetical proteins is to detect these proteins using a proteomic approach. In our study, 581 proteins annotated as hypothetical proteins in the M. vaccae database were identified, representing ∼49% of the predicted hypothetical proteins. These proteomic results provide direct experimental evidence for the expression of these proteins in M. vaccae. These proteins could have some roles in the life cycle of this bacterium, and their functions require further analysis.
Molecular Weight and pI Distributions of the Identified Proteins
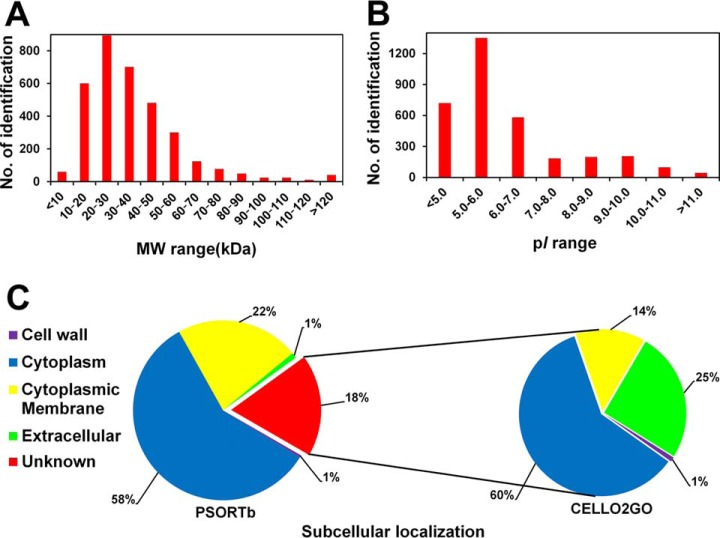
The identified proteins had a wide range of molecular weights (Mr) and pI values. The Mr distribution ranged from 4.3 kDa (MYVA_1195) to 326.8 kDa (MYVA_3919), and the majority of the proteins were between 10 and 60 kDa, representing ∼87.9% (2977 of 3387) of all the identifications. The distribution of the molecular weights of the identified proteins is depicted in Fig. 2A. More than 40 proteins predicted to have molecular weights higher than 140 kDa were identified in our study, indicating that separation by SDS-PAGE could contribute to the identification of high molecular weight proteins.
Fig. 2.
Numbers of proteins identified in this study. The distributions of the identified proteins in terms of different (A) molecular weight (MW) ranges, (B) pI ranges, and (C) subcellular localizations. The identified phosphoproteins are illustrated in the red histogram in (A) and (B). All the identifications were subjected to analysis using the PSORTb v3.0 program to predict their subcellular localizations (left column in C). Those proteins with no localization information by using pSORTb program were further analyzed by a current web server for protein subcellular localization prediction with functional gene ontology annotation (CELLO2GO, right column in C).
The pI values for the identified proteins ranged from 3.5 (MYVA_4311) to 12.3 (MYVA_4664). Approximately 39.9% of the proteins with a pI value between 5 and 6 were identified, which was the majority among the various pI intervals (Fig. 2B). Among them, 2656 out of 3387 proteins were identified with pI value less than 7, which indicated that most of the proteins obtained in this study were acidic.
Subcellular Localization of the Identified Proteins
All the annotated proteins in the M. vaccae database were subjected to analysis using the PSORTb v3.0 program to predict their subcellular localizations. The results showed that 1458 proteins (26%) localized to the cytoplasmic membrane, which is consistent with the average of 20–30% for annotations in various genomes (36). In the present study, 751 proteins (22%) were predicted to localize to the cytoplasmic membrane, which was ∼53% of the cytoplasmic membrane proteins in silico (751 out of 1458). Additionally, 1971 cytoplasmic proteins (58%) and 33 extracellular proteins (1%) were detected, representing 64% (out of 3058) and 57% (33 out of 58) of the annotations in silico, respectively. Several proteins with multiple transmembrane helices were identified. For example, MYVA_0582, which was predicted to have 23 transmembrane helices, was identified as cation: proton antiporter. The relative proportion of the subcellular localization information of the identifications is illustrated in Fig. 2C.
Furthermore, the localization information for ∼18% (620 proteins) of the identifications is unavailable using the current version of the pSORTb program. To address the localization information of these proteins, a current web server for CELLO2GO was used. The results showed that most of the proteins (60%) localized to the cytoplasmic compartment, and 25 and 14% proteins were localized to extracellular and membrane compartments, respectively. Only 1% of the proteins were localized to the cell wall (Fig. 2C). Additionally, some proteins were predicted with two possible localizations. For example, 60 proteins were predicted to localize to cytoplasmic and membrane compartments. The prediction results by CELLO2GO are indicated with blue font and highlighted with yellow boxes in Column G in supplemental Table S3.
Translational Start Site Assignments
The correct translational start site (TSS) is important for the analysis of protein function and transcriptional regulation (37). In genomic annotation, most TSSs are determined by bioinformatics methods and no straightforward experimental methodologies can easily validate the predictions. The TSSs were usually significantly different when predicted by different bioinformatic methods. One of the traditional methods to determine proteins' mature N terminus is N-terminal sequencing. However, this method often requires a large amount of protein and cannot be used when the N termini of proteins are blocked by modifications. Additionally, it is not a high-throughput method for large quantities of proteins. It has been demonstrated that the experimental determination of TSSs using tandem mass spectrometry is a more universal and high-throughput method than N-terminal sequencing (23). In this study, we utilized MS-based proteomic approaches to assign large-scale TSSs by identifying the modified N-terminal peptides. Previous studies have demonstrated that nontryptic nature at the N terminus of the peptide, such as N-terminal peptides with an initiator methionine residue or an initiator methionine cleaved, could indicate the N-terminal of protein (18). Using this approach, 480 unique N-terminal peptides were obtained and 445 existing annotations with predicted TSSs were confirmed. Among these proteins, 348 were confirmed with N-terminal peptides with the initiator methionine cleaved, and 117 were confirmed with initiator methionine residues. Twenty of them were confirmed with both the initiator methionine cleaved and methionine residues (supplemental Table S5).
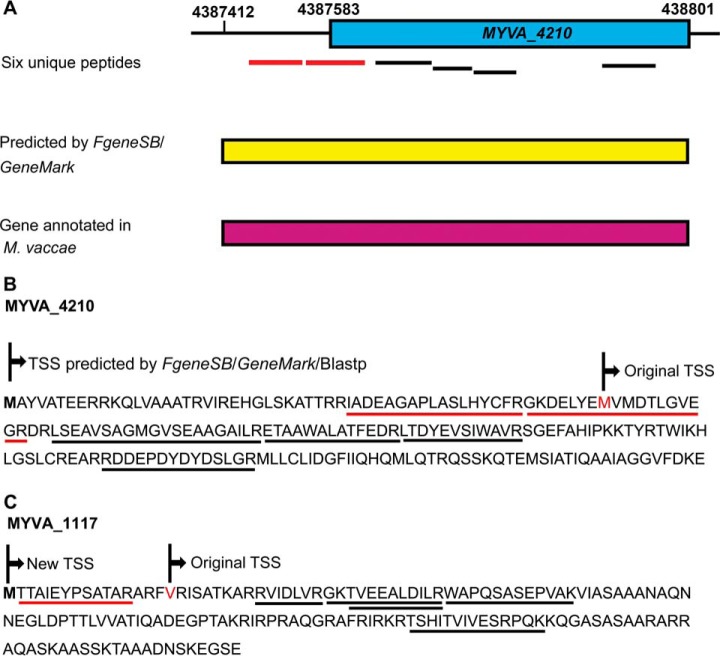
It was fascinating that some TSSs predicted by bioinformatics methods could be experimentally corrected. In the present study, all the MS-derived peptides were screened against a custom 6-frame database, and 123 unique peptides that mapped upstream of the currently annotated TSSs of their corresponding proteins were obtained. These upstream peptide hits suggested that the TSSs of the corresponding proteins should be extended. After manual validation, we were able to identify 98 proteins with extended TSSs (supplemental Table S6), of which 94 contained at least two unique peptides. Fig. 3 depicts an example of a gene model that has an extension of the N terminus. Two unique peptides mapped upstream of the original gene product MYVA_4210 (Fig. 3A and 3B), TetR family transcriptional regulator. Furthermore, another four unique peptides also mapped to MYVA_4210 when we searched against the M. vaccae protein database. The N-terminal extended protein was also validated by performing a Blastp search against the nonredundant protein database. Additionally, the extended gene sequence was searched using the gene prediction programs FgeneSB and GeneMark, and the result indicated that an extension gene model shared higher similarity with its homolog in M. vaccae. Therefore, according to our proteomic results, the length of MYVA_4210 should be extended to 200 aa instead of 143 aa.
Fig. 3.
N-terminal extension of two gene models using peptide mapping upstream of the annotated translational start sites. A, six unique peptides mapped to the annotated gene MYVA_4210 (blue box), including two (red lines) extended N-terminal peptides. The gene prediction programs FgeneSB and GeneMark predicted the presence of an extension gene model (yellow box). The orthologous protein from M. vaccae also supported this N-terminal extension (purple box). B, the protein sequence of MYVA_4210 was extended by 57 aa at the N terminus. The underlined sequence indicates six unique peptides, including two extended N-terminal peptides (red line).
Furthermore, based on N terminus nontryptic peptides, including peptides with an initiator methionine residue or an initiator methionine cleaved, the accurate TSSs of 16 extension proteins could be confirmed. For example, one peptide with a nontryptic N terminus, M.TTAIEYPSATAR.A, was found to map upstream of the original TSS of an annotated protein MYVA_1117 (Fig. 3C). Another 6 unique peptides were also identified to map this protein when we searched against the M. vaccae protein database. The extension protein was also supported by Blastp search results and FgeneSB and GeneMark predictions.
Discovery of Previously Unidentified Genes
It is interesting that some novel peptides or proteins that were not represented among the predicted proteins of genomes could be identified. An in-house database of M. vaccae that included all possible “gene encoding products” was constructed. This database contains all possible ORFs, including those previously predicted and those that were not predicted. We compared peptide sequences searched against the six-frame translated genome database with those presented in the M. vaccae protein database, and the peptides that map to the M. vaccae database were excluded from those that were mapped to the custom six-reading-frame database. The results provide a list of novel unique peptides. In total, 365 novel unique peptides were obtained. To improve the confidence of the identifications, at least two unique peptides with PEP below 0.01 were required for each novel protein. After manual filtering and validation, 38 novel proteins with 188 supporting unique peptides were obtained. Furthermore, to determine the conservation of these novel proteins across related organisms, all the proteins were searched against the nonredundant protein database by applying performing the Blastp algorithm. Among these proteins, 32 have orthologous in other mycobacteria, and five have orthologous in other organisms. Significantly, one had no homology with proteins from any other organism. The novel proteins along with the supporting unique peptides are listed in supplemental Table S7.
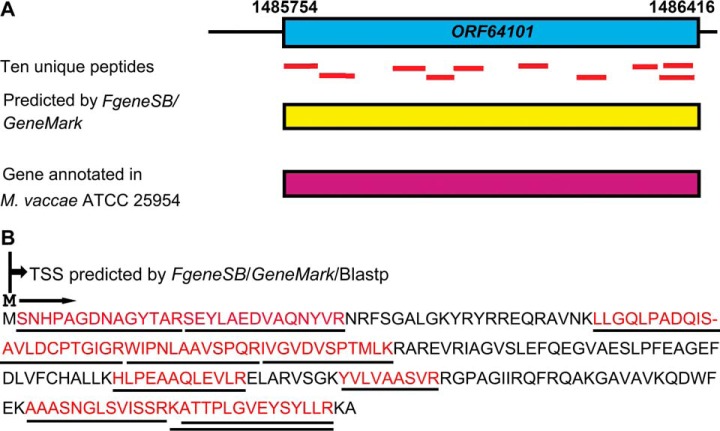
The TSSs of three novel proteins (ORF52985, ORF56032 and ORF64101) were determined by N-terminal peptides with their initiator methionine residues or initiator methionine cleaved. One such example is illustrated in Fig. 4A where a novel protein, ORF64101, was discovered by ten unique peptides. A Blastp search against the nonredundant protein database showed that this “protein” shared 100% identity with the SAM-dependent methyltransferase in M. vaccae ATCC 25954. Additionally, one peptide with a nontryptic N terminus, M.SNHPAGDNAGYTAR.S, was found to map to the “novel” protein (Fig. 4A). The peptide was assumed to have undergone N-terminal methionine excision by methionine aminopeptidase, and could be used as evidence for the correct length of the gene model. Furthermore, the nucleotide sequence of the protein was also supported by FgeneSB and GeneMark predictions. Therefore, our proteomic results could confirm these true novel gene models that have been missed in genome annotations. It is suggested that the approach of using MS-based proteomic data to identify novel proteins could prove to be an essential complementary method for annotating genomes in the future for newly sequenced genomes.
Fig. 4.
Identification of novel gene models based on peptide mapping to the genomic region. A, ten unique peptides (red lines) mapped to the genomic region corresponding to a novel protein, ORF64101. The presence of this novel gene model was also supported by the FgeneSB and GeneMark programs (yellow box). This novel protein was found to share 100% identity with the SAM-dependent methyltransferase in M. vaccae ATCC 25954 (purple box). B, protein sequence of the novel gene product ORF64101. The identified region is underlined with red font. The N terminus nontryptic peptide, M.SNHPAGDNAGYTAR.S, could be used as evidence to determine the N-terminal of the gene model.
Identification and Validation of Immunogenic Proteins by Immunoproteomics
In this study, 2-DE gel map of M. vaccae was provided (Fig. 5), and 2-D immunoblots were performed to investigate the immunogenic proteins using serum from healthy people (TB−) (supplemental Fig. S1A) and TB patients (TB+) (Supplemental Fig. S1B) against M. vaccae whole-cell protein components. The immunoblot profiles against TB− and TB+ were compared, and there was a considerable difference in the number and level of immunoreactive proteins. In total, over forty different strongly immunogenic spots were visible in blots using TB+ serum that were not observed in control blots using TB− serum. The spots on 2-DE gel corresponding to the immunoreactive spots in Western blotting were selected and excised (red arrow in Fig. 5), and thirty-five proteins were unambiguously identified by MALDI-TOF/TOF MS. Among them, seventeen were enzymes, and the others were transcriptional regulators, transporters, chaperone proteins and hypothetical proteins. By using comparative genomics, 33 of them have orthologous in H37Rv, BCG or other M.tuberculosis species (supplemental Table S2). Most of the proteins spanned a broad range of molecular mass from 20 to 80 kDa and pI values between 4.8 and 9.5. Twenty-three were predicted to be cytoplasmic proteins, whereas 10 were predicted to be localized on the cytoplasmic membrane. The remaining two proteins were predicted as “unknown.”
Fig. 5.
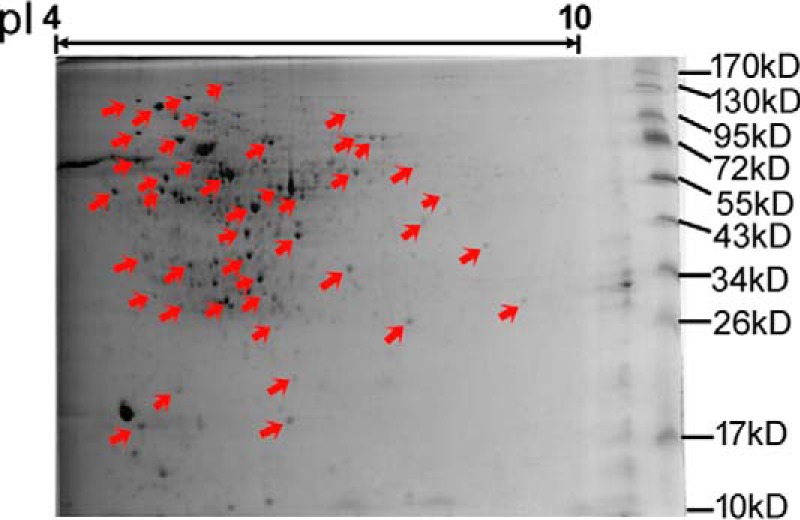
2-DE gel map of M. vaccae. The immunoblot profiles against healthy people (TB−) and TB patients (TB+) were compared, and the Coomassie-stained protein spots on 2-DE gel corresponding to the immunoreactive spots in Western blotting were excised (red arrow).
To validate the immunogenicity of the candidate antigen proteins identified in this study, 20 genes were selected to be expressed and reacted with TB+ serum and peripheral blood mononuclear cells (PBMC) from participants. Twelve of the expressed proteins have orthologous in H37Rv and BCG genomes (supplemental Table S8).Immunoblotting of these proteins against M. tb-specific antibody from the mixture of serum samples from 100 TB patients showed a clear difference in the pattern and intensity of the immunogenic proteins blots. The results revealed that eight of them showed a strong specific reactive band, and seven exhibited faints signals. The other five presented no obvious blotting signals (supplemental Fig. S2). The raw data of the Simon-Simple Western analysis against serum antibody response were provided in supplemental Table S9. Some of these serology findings were also identified in previous studies by comparing responses in TB patients and non-TB patients. For example, in 2010, Kunnath-Velayudhan et al. reported 198 out of 484 mycobacterial antigens were recognized by serum from more than one TB patient (38). Among them, three proteins, Rv0350, Rv1837c and Rv2215, which are orthologous in this study (MYVA_0478, MYVA_2978 and MYVA_3526, respectively) could react to sera from TB patients but not to sera of uninfected individuals. Li et al. tested for antigenicity of expressed mycobacterial proteins and found 249 proteins had significant reactions with the mixture of serum samples from 15 TB patients. Protein Rv3071, which is orthologous of MYVA_1827, was identified as distinct reactive (39). In 2014, Deng et al. used the M. tuberculosis proteome microarray to identify 14 serum biomarkers that can together differentiate between TB patients with active disease and recovered individuals (40). However, no orthologous protein in M. vaccae was discovered in their study. My colleagues, Liu et al. previously have identified 514 out of the 1250 expressed mycobacterial proteins which had positive reactions with pooled serum from 200 TB patients (26). Among them, three proteins, Rv1013, Rv2214c and Rv2578c, which are orthologous in M. vaccae (MYVA_4619, MYVA_3524 and MYVA_2502, respectively), had significant reactions with the serum samples from TB patients in this study. The identification of immune antigens in previous studies supports the reliable serology findings in this study.
Cellular antigen-stimulated IFN-γ secretion was further examined to screen the purified proteins to identify novel antigens by using a cell-based ELISPOT assay. Under a unified standard, the ratio of SFCs of examined proteins to SFCs of the commercial ESAT-6 protein (positive control) was calculated and analyzed by using three pulmonary TB patients. During the first-round of screening, most of proteins had an average SFC ratio less than 10%, including three proteins in which the reaction to PBMCs from TB patients was not detectable (supplemental Table S10A). MYVA_1927, which was an aminoglycoside phosphotransferase, had an average SFC ratio of 66.8% compared with the SFC of commercial ESAT-6, indicating potential cellular antigenicity. This protein was further examined for the second-round of screening in 20 pulmonary TB patients and 10 nontuberculosis individuals (supplemental Table S10B and S10C). Although the sensitivity of such tests is dependent on the immune status of the patients, the complexity of the infection and the contribution of suppressive cytokines related to TB, results showed that MYVA_1927 had the average SFC ratio of 51%. Therefore, after two-rounds of screening, MYVA_1927 protein displayed a high cellular immune level in pulmonary TB patients. Further study on the antigenicity of the protein should be pursued.
Understanding the functional information of the identified mycobacterial antigens, especially those for which the homologues have been identified in Mycobacterium species, could provide insight into the physiology of production and the likely roles of these detected antigen homologues in this newly recognized vaccine strain. For example, DNA-directed RNA polymerase subunit beta (MYVA_1069, RpoB), has been demonstrated to have a strong association between specific rpoB mutations and differential resistance to rifampin and rifambutin (RFB) (41). It has orthologous in H37Rv (Rv0667) and BCG (BCG_0716) genomes. It was also suggested that the detection of some of rpoB mutations could be used to rapidly and reliably identify RFB-susceptible MDR-TB isolates that could benefit from RFB treatment (42).
It has been demonstrated DnaK, a chaperone protein, is essential for cell growth and required for native protein folding in Mycobacterium species, and its loss is accompanied by proteotoxic collapse characterized by the accumulation of insoluble newly synthesized proteins (43). In this study, this protein showed great signal intensities on the immunoblots probed with TB+ serum, which indicated that a high concentration of specific antibodies were produced in serum specifically in response to TB. Additionally, orthologous of the chaperone protein in H37Rv (Rv0350) and BCG (BCG_0389) were also found. Previous studies suggested that DnaK could constitute a virulence factor to play roles as an important immunomodulator of macrophage responses (44).
It is reported that S-adenosylmethionine synthase (SAMS) is a key enzyme that produces SAM and it is related to ethylene and polyamine synthesis (45). It has orthologous in H37Rv (Rv1392) and BCG (BCG_1453). SAM plays a major role in the regulation of methionine and threonine synthesis and it affects methionine accumulation. Therefore, SAMS complexes play important roles in the organism growth and development of the organized tissues and/or organs (46). MoeA is required for molybdenum cofactor biosynthesis and it functions in the addition of molybdenum to the dithiolene of molybdopterin to form molybdenum cofactor (47). It is also found to be necessary for nitrate reductase activity and heterocyst suppression on nitrate, suggesting that the moeA gene is necessary for cellular growth in nitrate-based medium (48). Additionally, flavoprotein (MYVA_3583) was suggested to contribute to the mycobacterial resistance to oxidative stress and to affect global gene expression (49).
Oxidative stress resistance is one of the key properties that enable pathogenic bacteria to survive the toxic reactive oxygen species released by the host. It has been shown that NADPH quinone reductase could play an important role in managing oxidative stress and that it contributes to the successful colonization of the host (50). Because the reductase homologs are present in the genomes of many pathogenic bacteria, it will likely have broad implications for our understanding of bacterial infection and pathogenesis by investigating the function and mechanism of this protein in oxidative stress resistance (51). Six aminoglycoside phosphotransferases were identified in this study and one of them showed strong immunoreactivity in blots probed with TB+ serum. It has been demonstrated these proteins likely play a role in the normal function of the cell and they exhibit a potential for mycobacteria to act as reservoirs for antibiotic resistance genes (52). For example, previous studies on the crystal structure of aminoglycoside phosphotransferase of M. tuberculosis have suggested this protein responsible for the acquisition of aminoglycoside resistance (53). Mutations in the protein might increase its aminoglycoside phosphotransferase activity, thereby conferring greater aminoglycoside resistance to M. tuberculosis. Diol dehydratase could catalyze the conversion of 1,2-propanediol, glycerol, and 1,2-ethanediol to the corresponding aldehydes, but it undergoes inactivation by glycerol or by O2 in the absence of substrate (54). Specific protein factors, such as diol dehydratase reactivation protein identified in this study, were involved in the reactivation of inactivated diol dehydratase (55). Therefore, this reactivation protein is important for the anaerobic metabolism of 1,2-propanediol and glycerol.
The GntR family is one of the most abundant and widely distributed groups of helix-turn-helix transcriptional regulators in bacteria. GntR regulators are associated with diverse metabolic pathways and the regulation of virulence in pathogenic bacteria (56). ABC transporter ATPases are a superfamily of enzyme pumps that hydrolyze ATP in exchange for the translocation of chemically diverse substrates across the lipid bilayers of cellular membranes, and they are essential for cell survival through their role in nutrient uptake and osmoregulation, and they are also involved in virulence (57). An understanding of the molecular mechanism of ABC transporters is critical to the development of new diagnostics and treatments for a wide range of pathological conditions affecting human health (58).
Additionally, it has been demonstrated that Acyl-CoA dehydrogenase (MYVA_3750) plays an important role in the oxidation of fatty acyl-CoA esters, which could bring about catalysis, promote specificity and determine the selective transfer of electrons to electron transferring flavoprotein (59). The long chain fatty acid CoA-ligase (MYVA_4619) plays a pivotal role in the transport and activation of exogenous fatty acids prior to their subsequent degradation, which could serve as a potential target candidate for the development of selective inhibitors against some diseases (60). The von Willebrand factor type A has a widespread localization and its functions are varied in many different proteins of the immune system and the extracellular matrix, as well as in blood coagulation (61).
CONCLUSIONS
In this study, a proteogenomic approach was successfully applied to perform genome annotation using high resolution and high accuracy MS data. We validated the existence of 3,387 proteins predicted by genome software and confirmed 445 existing annotations with predicted TSSs based on N-terminal peptides. Furthermore, 98 proteins were validated through extension of the translational start sites based on N terminus-derived peptides, and 38 novel protein products not annotated in the M. vaccae database were detected and validated. By comparative immunoproteomic analysis, 35 candidate antigen proteins were unambiguously identified, and 20 of them were selected to be expressed to validate their immunogenicity by in vitro detection of both humoral and cellular reactivity. The results revealed that eight of them showed strong specific reactive signals on the immunoblots of TB+ serum, and one protein displayed a high cellular immune level in pulmonary TB patients. These detected antigens will likely provide insight into the physiology and play important roles in this newly recognized vaccine strain.
DATA AVAILABILITY
Complete sequence of M. vaccae ATCC 95051 has been deposited in GenBank under the accession CP011491 (https://www.ncbi.nlm.nih.gov/nuccore/CP011491). All of the raw files (.raw) and the merged peak list files (.mgf) generated from Proteome Discoverer software in the present study have been deposited into the publicly accessible database PeptideAtlas with data set Identifier PASS00954 (http://www.peptideatlas.org/PASS/PASS00954).
Supplementary Material
Acknowledgments
We thank Professor Guozhi Wang and Miao Xu (Institute for Biological Products Control, National Institutes for Food and Drug Control, Beijing, China) and Dr. Haiying Liu (Institute of Pathogen Biology, and Centre for Tuberculosis, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China) for providing strain Mycobacterium vaccae 95051.
Footnotes
Author contributions: J.Z., L.C., L.L., J.Y., X.Z., and Q.J. designed research; J.Z., L.C., L.L., H.L., J.D., L.S., Y.Z., X.Z., and Q.J. performed research; J.Z., L.C., L.L., H.L., B.L., D.Z., T.L., J.D., L.S., Y.Z., J.Y., X.Z., and Q.J. contributed new reagents or analytic tools; J.Z., L.C., L.L., B.L., D.Z., T.L., J.Y., X.Z., and Q.J. analyzed data; J.Z., J.Y., and Q.J. wrote the paper.
* This work was supported by the National Twelve-Fifth Mega-Scientific Project on the “prevention and treatment of AIDS, viral hepatitis and other infectious diseases” (2012ZX10003002) (http://www.nmp.gov.cn/zxjs/crb/201012/t20101208_2127.htm), CAMS Innovation Fund for Medical Sciences (2016-I2M-1-013) (http://www.pumc.edu.cn/blog/), and the Fundamental Research Funds for Central Public-interest Scientific Institution (Centre for Tuberculosis) (2016ZX310183) (http://www.gov.cn/xinwen/2016-07/27/content_5095236.htm).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- TB
- Tuberculosis
- BCG
- Mycobacterium bovis bacillus Calmette and Guérin
- CDS
- protein-coding sequences
- CELLO2GO
- protein subCELlular LOcalization prediction with functional Gene Ontology annotation
- FDR
- false discovery rate
- HIV
- human immunodeficiency virus
- IMP
- integral membrane protein
- PEP
- posterior error probability
- RFB
- rifambutin
- SAMS
- S-adenosylmethionine synthase
- TSS
- translational start site.
REFERENCES
- 1. Gao L., Lu W., Bai L., Wang X., Xu J., Catanzaro A., Cardenas V., Li X., Yang Y., Du J., Sui H., Xia Y., Li M., Feng B., Li Z., Xin H., Zhao R., Liu J., Pan S., Shen F., He J., Yang S., Si H., Wang Y., Xu Z., Tan Y., Chen T., Xu W., Peng H., Wang Z., Zhu T., Zhou F., Liu H., Zhao Y., Cheng S., and Jin Q. (2015) Latent tuberculosis infection in rural China: baseline results of a population-based, multicentre, prospective cohort study. Lancet Infect. Dis. 15, 310–319 [DOI] [PubMed] [Google Scholar]
- 2. WHO. (2016) Global tuberculosis report 2015. Geneva: WHO Press [Google Scholar]
- 3. Wallis R. S., Maeurer M., Mwaba P., Chakaya J., Rustomjee R., Migliori G. B., Marais B., Schito M., Churchyard G., Swaminathan S., Hoelscher M., and Zumla A. (2016) Tuberculosis-advances in development of new drugs, treatment regimens, host-directed therapies, and biomarkers. Lancet Infect. Dis. 16, e34–e46 [DOI] [PubMed] [Google Scholar]
- 4. Andersen P., and Doherty T. M. (2005) The success and failure of BCG - implications for a novel tuberculosis vaccine. Nat. Rev. Microbiol. 3, 656–662 [DOI] [PubMed] [Google Scholar]
- 5. Weng H., Huang J. Y., Meng X. Y., Li S., and Zhang G. Q. (2016) Adjunctive therapy of vaccine in the treatment of multidrug-resistant tuberculosis: A systematic review and meta-analysis. Biomed. Rep. 4, 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. von Reyn C. F., Mtei L., Arbeit R. D., Waddell R., Cole B., Mackenzie T., Matee M., Bakari M., Tvaroha S., Adams L. V., Horsburgh C. R., and Pallangyo K. (2010) Prevention of tuberculosis in Bacille Calmette-Guerin-primed, HIV-infected adults boosted with an inactivated whole-cell mycobacterial vaccine. AIDS 24, 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nielsen P., and Krogh A. (2005) Large-scale prokaryotic gene prediction and comparison to genome annotation. Bioinformatics 21, 4322–4329 [DOI] [PubMed] [Google Scholar]
- 8. Reeves G. A., Talavera D., and Thornton J. M. (2009) Genome and proteome annotation: organization, interpretation and integration. J. R. Soc. Interface 6, 129–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao L., Liu L., Leng W., Wei C., and Jin Q. (2011) A proteogenomic analysis of Shigella flexneri using 2D LC-MALDI TOF/TOF. BMC Genomics 12, 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang M. K., Yang Y. H., Chen Z., Zhang J., Lin Y., Wang Y., Xiong Q., Li T., Ge F., Bryant D. A., and Zhao J. D. (2014) Proteogenomic analysis and global discovery of posttranslational modifications in prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 111, E5633–E5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Potgieter M. G., Nakedi K. C., Ambler J. M., Nel A. J., Garnett S., Soares N. C., Mulder N., and Blackburn J. M. (2016) Proteogenomic Analysis of Mycobacterium smegmatis Using High Resolution Mass Spectrometry. Front. Microbiol. 7, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kelkar D. S., Kumar D., Kumar P., Balakrishnan L., Muthusamy B., Yadav A. K., Shrivastava P., Marimuthu A., Anand S., Sundaram H., Kingsbury R., Harsha H. C., Nair B., Prasad T. S., Chauhan D. S., Katoch K., Katoch V. M., Chaerkady R., Ramachandran S., Dash D., and Pandey A. (2011) Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol. Cell. Proteomics 10, M111.011627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu X., Liu T., Ren X., Liu B., Yang J., Chen L., Wei C., Zheng J., Dong J., Sun L., Zhu Y., and Jin Q. (2015) Proteogenomic Analysis of Trichophyton rubrum Aided by RNA Sequencing. J. Proteome Res. 14, 2207–2218 [DOI] [PubMed] [Google Scholar]
- 14. Yagoub D., Tay A. P., Chen Z., Hamey J. J., Cai C., Chia S. Z., Hart-Smith G., and Wilkins M. R. (2015) Proteogenomic discovery of a small, novel protein in yeast reveals a strategy for the detection of unannotated short open reading frames. J. Proteome Res. 14, 5038–5047 [DOI] [PubMed] [Google Scholar]
- 15. Kumar D., Yadav A. K., Jia X., Mulvenna J., and Dash D. (2016) Integrated transcriptomic-proteomic analysis using a proteogenomic workflow refines rat genome annotation. Mol. Cell. Proteomics 15, 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilhelm M., Schlegl J., Hahne H., Moghaddas Gholami A., Lieberenz M., Savitski M. M., Ziegler E., Butzmann L., Gessulat S., Marx H., Mathieson T., Lemeer S., Schnatbaum K., Reimer U., Wenschuh H., Mollenhauer M., Slotta-Huspenina J., Boese J. H., Bantscheff M., Gerstmair A., Faerber F., and Kuster B. (2014) Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587 [DOI] [PubMed] [Google Scholar]
- 17. Hernandez-Haro C., Llopis S., Molina M., Monteoliva L., and Gil C. (2015) Immunoproteomic profiling of Saccharomyces cerevisiae systemic infection in a murine model. J. Proteomics 112, 14–26 [DOI] [PubMed] [Google Scholar]
- 18. Shinoy M., Dennehy R., Coleman L., Carberry S., Schaffer K., Callaghan M., Doyle S., and McClean S. (2013) Immunoproteomic analysis of proteins expressed by two related pathogens, Burkholderia multivorans and Burkholderia cenocepacia, during human infection. PLoS ONE 8, e80796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rodrigues A. M., Kubitschek-Barreira P. H., Fernandes G. F., de Almeida S. R., Lopes-Bezerra L. M., and de Camargo Z. P. (2015) Immunoproteomic analysis reveals a convergent humoral response signature in the Sporothrix schenckii complex. J. Proteomics 115, 8–22 [DOI] [PubMed] [Google Scholar]
- 20. Pan J., Li C., and Ye Z. (2016) Immunoproteomic approach for screening vaccine candidates from bacterial outer membrane proteins. Methods Mol. Biol. 1404, 519–528 [DOI] [PubMed] [Google Scholar]
- 21. Pang H., Chen L., Hoare R., Huang Y., Zaohe Wu, and Jian J. (2016) Identification of DLD, by immunoproteomic analysis and evaluation as a potential vaccine antigen against three Vibrio species in Epinephelus coioides. Vaccine 34, 1225–1231 [DOI] [PubMed] [Google Scholar]
- 22. Wareth G., Eravci M., Weise C., Roesler U., Melzer F., Sprague L. D., Neubauer H., and Murugaiyan J. (2016) Comprehensive identification of immunodominant proteins of Brucella abortus and Brucella melitensis using antibodies in the sera from naturally infected hosts. Int. J. Mol. Sci. 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng J., Ren X., Wei C., Yang J., Hu Y., Liu L., Xu X., Wang J., and Jin Q. (2013) Analysis of the secretome and identification of novel constituents from culture filtrate of bacillus Calmette-Guerin using high-resolution mass spectrometry. Mol. Cell. Proteomics 12, 2081–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng J., Liu L., Wei C., Leng W., Yang J., Li W., Wang J., and Jin Q. (2012) A comprehensive proteomic analysis of Mycobacterium bovis bacillus Calmette-Guerin using high resolution Fourier transform mass spectrometry. J. Proteomics 77, 357–371 [DOI] [PubMed] [Google Scholar]
- 25. Chen X., Zhang M., Zhu X., Deng Q., Liu H., Larmonier N., Graner M. W., and Zhou B. (2009) Engagement of Toll-like receptor 2 on CD4(+) T cells facilitates local immune responses in patients with tuberculous pleurisy. J. Infect. Dis. 200, 399–408 [DOI] [PubMed] [Google Scholar]
- 26. Liu L., Zhang W. J., Zheng J., Fu H., Chen Q., Zhang Z., Chen X., Zhou B., Feng L., Liu H., and Jin Q. (2014) Exploration of novel cellular and serological antigen biomarkers in the ORFeome of Mycobacterium tuberculosis. Mol. Cell. Proteomics 13, 897–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delcher A. L., Harmon D., Kasif S., White O., and Salzberg S. L. (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27, 4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lowe T. M., and Eddy S. R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clark K., Karsch-Mizrachi I., Lipman D. J., Ostell J., and Sayers E. W. (2016) GenBank. Nucleic Acids Res. 44, D67–D72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li L., Stoeckert C. J. Jr, and Roos D. S. (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stothard P., and Wishart D. S. (2005) Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539 [DOI] [PubMed] [Google Scholar]
- 32. Zheng J., Liu L., Liu B., and Jin Q. (2015) Phosphoproteomic analysis of bacillus Calmette-Guerin using gel-based and gel-free approaches. J. Proteomics 126, 189–199 [DOI] [PubMed] [Google Scholar]
- 33. de Souza G. A., Softeland T., Koehler C. J., Thiede B., and Wiker H. G. (2009) Validating divergent ORF annotation of the Mycobacterium leprae genome through a full translation data set and peptide identification by tandem mass spectrometry. Proteomics 9, 3233–3243 [DOI] [PubMed] [Google Scholar]
- 34. Liu X., Wang D., Ren J., Tong C., Feng E., Wang X., Zhu L., and Wang H. (2013) Identification of the immunogenic spore and vegetative proteins of Bacillus anthracis vaccine strain A16R. PLoS ONE 8, e57959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ho Y. S., Adroub S. A., Abadi M., Al Alwan B., Alkhateeb R., Gao G., Ragab A., Ali S., van Soolingen D., Bitter W., Pain A., and Abdallah A. M. (2012) Complete genome sequence of Mycobacterium vaccae type strain ATCC 25954. J. Bacteriol. 194, 6339–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wallin E., and von Heijne G. (1998) Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 7, 1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rison S. C., Mattow J., Jungblut P. R., and Stoker N. G. (2007) Experimental determination of translational starts using peptide mass mapping and tandem mass spectrometry within the proteome of Mycobacterium tuberculosis. Microbiology 153, 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kunnath-Velayudhan S., Salamon H., Wang H. Y., Davidow A. L., Molina D. M., Huynh V. T., Cirillo D. M., Michel G., Talbot E. A., Perkins M. D., Felgner P. L., Liang X., and Gennaro M. L. (2010) Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc. Natl. Acad. Sci. U.S.A. 107, 14703–14708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Y., Zeng J., Shi J., Wang M., Rao M., Xue C., Du Y., and He Z. G. (2010) A proteome-scale identification of novel antigenic proteins in Mycobacterium tuberculosis toward diagnostic and vaccine development. J. Proteome Res. 9, 4812–4822 [DOI] [PubMed] [Google Scholar]
- 40. Deng J., Bi L., Zhou L., Guo S. J., Fleming J., Jiang H. W., Zhou Y., Gu J., Zhong Q., Wang Z. X., Liu Z., Deng R. P., Gao J., Chen T., Li W., Wang J. F., Wang X., Li H., Ge F., Zhu G., Zhang H. N., Wu F. L., Zhang Z., Wang D., Hang H., Li Y., Cheng L., He X., Tao S. C., and Zhang X. E. (2014) Mycobacterium tuberculosis proteome microarray for global studies of protein function and immunogenicity. Cell Rep. 9, 2317–2329 [DOI] [PubMed] [Google Scholar]
- 41. ElMaraachli W., Slater M., Berrada Z. L., Lin S. Y., Catanzaro A., Desmond E., Rodrigues C., Victor T. C., Crudu V., Gler M. T., and Rodwell T. C. (2015) Predicting differential rifamycin resistance in clinical Mycobacterium tuberculosis isolates by specific rpoB mutations. Int. J. Tuberc. Lung Dis. 19, 1222–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Berrada Z. L., Lin S. Y., Rodwell T. C., Nguyen D., Schecter G. F., Pham L., Janda J. M., Elmaraachli W., Catanzaro A., and Desmond E. (2016) Rifabutin and rifampin resistance levels and associated rpoB mutations in clinical isolates of Mycobacterium tuberculosis complex. Diagn. Microbiol. Infect. Dis. 85, 177–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fay A., and Glickman M. S. (2014) An essential nonredundant role for mycobacterial DnaK in native protein folding. PLoS Genet. 10, e1004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lopes R. L., Borges T. J., Araujo J. F., Pinho N. G., Bergamin L. S., Battastini A. M., Muraro S. P., Souza A. P., Zanin R. F., and Bonorino C. (2014) Extracellular mycobacterial DnaK polarizes macrophages to the M2-like phenotype. PLoS ONE 9, e113441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hacham Y., Song L., Schuster G., and Amir R. (2007) Lysine enhances methionine content by modulating the expression of S-adenosylmethionine synthase. Plant J. 51, 850–861 [DOI] [PubMed] [Google Scholar]
- 46. He L., Ban Y., Miyata S., Kitashiba H., and Moriguchi T. (2008) Apple aminopropyl transferase, MdACL5 interacts with putative elongation factor 1-alpha and S-adenosylmethionine synthase [corrected]. Biochem. Biophys. Res. Commun. 366, 162–167 [DOI] [PubMed] [Google Scholar]
- 47. Nichols J., and Rajagopalan K. V. (2002) Escherichia coli MoeA and MogA. Function in metal incorporation step of molybdenum cofactor biosynthesis. J. Biol. Chem. 277, 24995–25000 [DOI] [PubMed] [Google Scholar]
- 48. Sandu C., and Brandsch R. (2002) Evidence for MoeA-dependent formation of the molybdenum cofactor from molybdate and molybdopterin in Escherichia coli. Arch. Microbiol. 178, 465–470 [DOI] [PubMed] [Google Scholar]
- 49. Du Y., Zhang H., He Y., Huang F., and He Z. G. (2012) Mycobacterium smegmatis Lsr2 physically and functionally interacts with a new flavoprotein involved in bacterial resistance to oxidative stress. J. Biochem. 152, 479–486 [DOI] [PubMed] [Google Scholar]
- 50. Hong Y., Wang G., and Maier R. J. (2008) The NADPH quinone reductase MdaB confers oxidative stress resistance to Helicobacter hepaticus. Microb. Pathog. 44, 169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang G., and Maier R. J. (2004) An NADPH quinone reductase of Helicobacter pylori plays an important role in oxidative stress resistance and host colonization. Infect. Immun. 72, 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wright G. D., and Thompson P. R. (1999) Aminoglycoside phosphotransferases: proteins, structure, and mechanism. Front. Biosci. 4, D9–D21 [DOI] [PubMed] [Google Scholar]
- 53. Ahn J. W., and Kim K. J. (2013) Rv3168 phosphotransferase activity mediates kanamycin resistance in Mycobacterium tuberculosis. J. Microbiol. Biotechnol. 23, 1529–1535 [DOI] [PubMed] [Google Scholar]
- 54. Toraya T., Tanokuchi A., Yamasaki A., Nakamura T., Ogura K., and Tobimatsu T. (2016) Diol dehydratase-reactivase is essential for recycling of coenzyme B12 in diol dehydratase. Biochemistry 55, 69–78 [DOI] [PubMed] [Google Scholar]
- 55. Mori K., Tobimatsu T., Hara T., and Toraya T. (1997) Characterization, sequencing, and expression of the genes encoding a reactivating factor for glycerol-inactivated adenosylcobalamin-dependent diol dehydratase. J. Biol. Chem. 272, 32034–32041 [DOI] [PubMed] [Google Scholar]
- 56. An S. Q., Lu G. T., Su H. Z., Li R. F., He Y. Q., Jiang B. L., Tang D. J., and Tang J. L. (2011) Systematic mutagenesis of all predicted gntR genes in Xanthomonas campestris pv. campestris reveals a GntR family transcriptional regulator controlling hypersensitive response and virulence. Mol. Plant Microbe Interact. 24, 1027–1039 [DOI] [PubMed] [Google Scholar]
- 57. Davidson A. L., Dassa E., Orelle C., and Chen J. (2008) Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. George A. M., and Jones P. M. (2013) An asymmetric post-hydrolysis state of the ABC transporter ATPase dimer. PLoS ONE 8, e59854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wipperman M. F., Yang M., Thomas S. T., and Sampson N. S. (2013) Shrinking the FadE proteome of Mycobacterium tuberculosis: insights into cholesterol metabolism through identification of an alpha2beta2 heterotetrameric acyl coenzyme A dehydrogenase family. J. Bacteriol. 195, 4331–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kaur J., Tiwari R., Kumar A., and Singh N. (2011) Bioinformatic analysis of leishmania donovani long-chain fatty acid-CoA ligase as a novel drug target. Mol. Biol. Int. 2011, 278051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Perkins S. J., Hinshelwood J., Edwards Y. J., and Jenkins P. V. (1999) Structural and functional modelling of von Willebrand factor type A domains in complement and coagulation. Biochem. Soc. Trans. 27, 815–820 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complete sequence of M. vaccae ATCC 95051 has been deposited in GenBank under the accession CP011491 (https://www.ncbi.nlm.nih.gov/nuccore/CP011491). All of the raw files (.raw) and the merged peak list files (.mgf) generated from Proteome Discoverer software in the present study have been deposited into the publicly accessible database PeptideAtlas with data set Identifier PASS00954 (http://www.peptideatlas.org/PASS/PASS00954).