Abstract
Microtubule organization and dynamics are critical for key developmental processes such as cell division, elongation, and morphogenesis. Microtubule severing is an essential regulator of microtubules and is exclusively executed by KATANIN 1 in Arabidopsis. In this study, we comparatively studied the proteome-wide effects in two KATANIN 1 mutants. Thus, shotgun proteomic analysis of roots and aerial parts of single nucleotide mutant fra2 and T-DNA insertion mutant ktn1-2 was carried out. We have detected 42 proteins differentially abundant in both fra2 and ktn1-2. KATANIN 1 dysfunction altered the abundance of proteins involved in development, metabolism, and stress responses. The differential regulation of tubulins and microtubule-destabilizing protein MDP25 implied a feedback microtubule control in KATANIN 1 mutants. Furthermore, deregulation of profilin 1, actin-depolymerizing factor 3, and actin 7 was observed. These findings were confirmed by immunoblotting analysis of actin and by microscopic observation of actin filaments using fluorescently labeled phalloidin. Results obtained by quantitative RT-PCR analysis revealed that changed protein abundances were not a consequence of altered expression levels of corresponding genes in the mutants. In conclusion, we show that abundances of several cytoskeletal proteins as well as organization of microtubules and the actin cytoskeleton are amended in accordance with defective microtubule severing.
Microtubules are tubulin filamentous polymers involved in cell division and expansion (1, 2). They are capable of rapid elongation or shortening (polymerization and depolymerization), which is known as dynamic instability. This dynamic instability together with other mechanisms, including nucleation, branching, severing, and bundling, determine the spatiotemporal organization of microtubule arrays, which is crucial for plant growth and development (3–5). Microtubule dynamics and organization are controlled mainly by microtubule-associated proteins (MAPs),1 kinesins, plus-end binding (EB1) proteins, microtubule-severing protein katanin, microtubule-destabilizing protein 25 (MDP25), phospholipase Dα1, and others (6–8). Some of these proteins might be regulated by signaling molecules such as mitogen-activated protein kinases (9, 10), Rop GTPases, calcium, and phosphatidic acid (11–13). Such interactions couple microtubules to the external environment and mediate their developmental or conditional rearrangements.
KATANIN 1 is a microtubule-severing AAA-ATPase assembled from a catalytic subunit of 60 kDa (p60) and a structural 80-kDa subunit (p80 (14)). It is capable of severing microtubules in an ATP-dependent manner (15). At the cellular level, the severing activity of KATANIN 1 was shown to regulate plant microtubule organization (16). Except for microtubule severing, KATANIN 1 activity favors microtubule bundle formation (17) and can be modulated by other microtubule-binding proteins like SPIRAL2 (18). Moreover, KATANIN 1 severing activity is induced by Rho-GTPase signaling, thus connecting hormonal and external stimuli to microtubule dynamics (19).
The importance of KATANIN 1 for plant development is manifested by multiple developmental defects reported in KATANIN 1 mutants such as fra2 or lue1. They exhibit reduced root, hypocotyl, stem, and leaf growth as well as stubby flower organs with reduced anther length (20–22). KATANIN 1 mutants show reduced fertility and defects in ovule and anther development, and they exhibit aberrant embryogenesis and seed formation (23). It is generally accepted that these phenotypes are caused by reduced cell expansion (20, 21). It was also proposed that fra2 and lue1 mutants exhibit some defects in cell division resulting from altered organization of microtubule arrays showing multipolar spindles and disorientation of the cell division plane (24). Advanced live microscopy of microtubules in ktn1-2 uncovered the contribution of KATANIN 1 to dynamic organization of cortical microtubules as well as a new function in the formation and maturation of preprophase band and rectification of cell division plane (25). Moreover, KATANIN 1 mutants displayed altered gibberellic acid (GA) and ethylene responses (22, 26) pointing to the role of KATANIN 1 in controlling microtubule reorganization in response to hormones.
Integrative bioinformatics analyses of Leishmania flagellar genes and proteins revealed that katanin along with profilin and formin are important actin-interacting proteins, which are involved in flagellum assembly, disassembly, and dynamics (27). However, actin-binding properties of katanin have not been experimentally proved so far. In addition, targeted proteomic analysis of mammalian katanin subunits in HeLa cell lines was used for creation of mammalian katanin family interaction network (Katan-ome), which plays an important role in microtubule severing (28).
Although some developmental and cellular roles of KATANIN 1 in plants were relatively well established, a comprehensive proteome-wide study on KATANIN 1 mutants was not performed yet. Therefore, the present proteomic dissection of fra2 and ktn1-2 mutants provides an important survey of new proteins linked to phenotypic and microtubule defects of these mutants.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
Proteomics analyses were carried out with four biological replicates for each of the six biological samples (roots and aerial parts of Col-0, fra2, and ktn1-2). Each replicate contained at least 30 seedlings. Pooling of the specimen was necessary to limit the effects of variations between individual plants. The number of replicates was sufficient to ascertain statistical significance, when analysis of variance (ANOVA) was used to test the differences of protein abundances between biological samples. Because a single factor (wild type and mutants represent one factor) was evaluated and the protein abundance datasets exhibit normal (Gaussian) distribution, it was appropriate to apply one-way ANOVA analysis.
Plant Material
Seeds of ktn1-2, fra2, and wild type Arabidopsis thaliana (ecotype Col-0) were surface-sterilized and placed on half-strength MS culture medium (pH 5.7) containing 1% (w/v) sucrose and 0.8% (w/v) phytagel. Plates with seeds were stored at 4 °C for 48 h to break seed dormancy, and afterward kept vertically in a culture chamber under 16 h light/8 h dark at 22 °C. fra2 is a single nucleotide mutant in the seventh exon of KATANIN 1 (At1g80350), where the A at nucleotide residue 2329 is deleted (21). ktn1-2 is a knockout mutant with T-DNA inserted after the 147th nucleotide in the 5th exon of KATANIN 1 (16). Fourteen-day-old seedlings were used for proteomic and immunoblotting analyses. For whole-mount immunolabeling, 3-day-old seedlings of ktn1-2 and fra2 mutants and Col-0 were used.
Protein Extraction for Proteomic Analysis
Roots and aerial parts of mutant and control plants were subjected to phenol protein extraction, trypsin digestion, and peptide purification as described previously (29).
Fresh material (250 mg) was homogenized in liquid nitrogen with 500 μl of cold extraction medium (0.9 m sucrose, 0.1 m Tris-HCl (pH 8.8), 10 mm EDTA, 100 mm KCl, and 0.4% (v/v) 2-mercaptoethanol) and an equal amount of Tris-HCl-buffered phenol (pH 8.1). The mixture was incubated for 30 min at 4 °C. Then, the protein-enriched phenol phase was separated from the aqueous phase by 5 min of centrifugation at 6000 × g at 4 °C. Phenol phase was subjected to ammonium acetate/methanol precipitation at −20 °C overnight. The precipitate was then pelleted by centrifugation at 13,000 × g at 4 °C for 20 min followed by two washes with ice-cold 80% (v/v) acetone and 1 wash in 70% (v/v) ethanol. Precipitate suspensions were stored at −20 °C for 15 min for each washing step. Finally, the pellets were resuspended in 80% (v/v) acetone, centrifuged, and air-dried for 10 min. Subsequently they were dissolved in 6 m urea in Tris-HCl buffer (pH 7.4). After protein content determination (with Bradford assay), equal amounts of proteins were used for in solution digestion. Prior to trypsin application, protein extracts were subjected to a reduction step (by the addition of 50 mm DTT and incubation for 1 h at room temperature), alkylation step (by addition of 50 mm iodoacetamide and incubation at room temperature for 1 h), and the urea concentration was lowered to less than 1 m. The trypsin digestion (1 μg of sequencing grade modified trypsin from Promega per 50 μg of proteins) was performed by permanent gentle shaking at 37 °C overnight. After stopping trypsin digestion by acetic acid, peptides were cleaned on C18 cartridges (Bond Elut C18; Agilent Technologies, Santa Clara, CA) according to manufacturer's instructions. Peptides eluted by 90% (v/v) acetonitrile were dried using SpeedVac and used for LC-MSMS.
Detailed Description of Liquid Chromatography, Mass Spectrometry, Protein Identification, and Relative Quantitative Analysis
Two μg of protein tryptic digest resuspended in 0.1% (v/v) formic acid, 5% (v/v) acetonitrile were loaded on reversed phase fused silica C18 column measuring 75 μm × 150 mm (Thermo Fisher Scientific, Waltham, MA). Peptides were separated and eluted at a constant flow rate of 0.3 μl · min−1 by a 170-min long nonlinear gradient of acetonitrile (in 0.1% formic acid) as follows: 2–55% for 125 min, 95% for 15 min, and 2% for 30 min. The mass spectra were obtained in the data-dependent acquisition mode, with dynamic exclusion being applied, in 18 scan events: one MS scan (m/z range, 300–1700) followed by 17 MSMS scans for the 17 most intense ions detected in MS scan. Other critical parameters were set as given here: normalized collision energy, 35%; automatic gain control “on” with MSn target 4 × 104, isolation width (m/z), 1.5; capillary temperature, 170 °C; spray voltage, 1.97 kV. The method and raw spectral files were created and generated, respectively, by Xcalibur 2.1 (Thermo Fisher Scientific).
The raw files were searched using the SEQUEST algorithm of the Proteome Discoverer 1.1.0 software (Thermo Fisher Scientific) with selection of parameters as follows: minimum and maximum precursor mass, 300 and 6,000 Da, respectively; precursor mass tolerance, 1.5 Da; fragment mass tolerance, 0.8 Da; intensity threshold, 1000; minimum ion count, 7; minimum S/N ratio, 3; enzyme, trypsin; maximum missed cleavages, 2; FDR = 0.01; dynamic (variable) modifications, cysteine carbamidomethylation (+57.021), methionine oxidation (+15.995), and methionine dioxidation (+31.990).
The spectral data were matched against target and decoy databases for more stringent approach to calculating FDR, compared with single search of concatenated database. The NCBI (www.ncbi.nlm.nih.gov) Arabidopsis genus taxonomy referenced protein database (67,924 entries as of November, 2013) served as the target database, and its reversed copy (created automatically by the software) served as a decoy database. The search results were filtered by Xcorr values pertinent to +1, +2, and +3 charged peptides, resulting in FDR <1%. Identified proteins were grouped by default parameters of the software, defining the group as proteins strictly necessary to explain the presence of identified peptides. A representative/master protein of the group is the protein with the highest score, spectral count, and number of matched peptides. If those parameters are equal, the protein with the longest sequence is designated as a master protein. Proteins listed in the supplemental materials are master proteins; however, all groups proteins, their accession numbers, respective peptides, and annotated spectra are included in “.msf” files (see below for how to view them). If the peptide can be attributed to more than one protein, it is indicated by multiple protein accession numbers allocated to a given peptide.
The relative quantitative analysis was based on sums of precursor ion intensities (PII) of filtered peptides attributed to given proteins. PII values were extracted from raw files and exported to spread sheets by Proteome Discoverer software. Even though the peptide-respective experimental PII values are not strictly stoichiometric for a given protein, they are commonly accepted for label-free mass spectrometry-based relative protein quantification. Intensities were summed for each identified master protein in each replicate using in house Xcell script. All data points were considered, and no outliers were excluded. Summed intensities pertinent to proteins in individual replicates were normalized by factors that were calculated to equalize total intensity of all master proteins across all biological samples and replicates. Normalized average protein intensities were used to calculate fold changes when comparing biological samples. The ANOVA analysis of four replicates for each biological sample was performed, and p ≤ 0.05 was used to filter statistically significant results.
Bioinformatic Evaluation of Proteomic Data
Venn diagram was created using Venny 2.1 on-line application (http://bioinfogp.cnb.csic.es/tools/venny/index.html). Differentially abundant proteins were annotated using Gene Ontology annotation analysis by Blast2Go software (30). Blast was performed against Arabidopsis thaliana NCBI database allowing 1 BLAST Hit. The annotation was performed by using these parameters: E value hit filter, 1.0E−6; annotation cutoff: 55; GO weight: 5, GO Slim. For prediction of protein interaction network analysis, STRING (31) applying minimum required interaction score 0.7 was relevant for high confidence prediction.
Immunoblotting Analysis
Roots and aerial parts of wild type and mutant seedlings were crushed in liquid nitrogen to a fine powder. The powder (200 mg) was resuspended and homogenized with 250 μl of 50 mm HEPES (pH 7.5) containing 75 mm NaCl, 1 mm EGTA, 1 mm MgCl2, 10% (v/v) glycerol, 1 mm DTT, and Complete® EDTA-free protease inhibitor mixture (Roche Applied Science) and incubated for 15 min on ice. After centrifugation at 13,000 × g at 4 °C for 15 min, the protein amount was quantitated in supernatants. Extracts were proportionally mixed to give a protein concentration of 1.5 mg of protein/ml with 4-fold concentrated Laemmli buffer (final concentration 62.5 mm Tris-HCl (pH 6.8), 2% (w/v) SDS, 10% (v/v) glycerol, 300 mm 2-mercaptoethanol), heat-denaturated at 95 °C for 5 min, and centrifuged to remove undissolved components. Equal amount of proteins (15 μg) were loaded on 12% TGX Stain-FreeTM (Bio-Rad) gels. After electrophoresis, proteins were transferred to nitrocellulose membranes using TransBlotTM Turbo (Bio-Rad) semidry transfer system. To validate the protein transfer, membranes were documented on ChemiDoc documentation system (Bio-Rad), allowing visualization of proteins transferred from TGX Stain-FreeTM gels. Afterward, they were blocked overnight in 4% (w/v) BSA, 4% (w/v) low-fat dry milk in Tris-HCl-buffered saline with 0.1% (v/v) Tween 20 (TBST). Membranes were then incubated overnight with anti-TSN1/2 (32) (1:750), anti-actin (1:4000; Sigma-Aldrich, Heidelberg, Germany), anti-α-tubulin (clone YOL1/34; 1:2000; ABD Serotec, Raleigh, NC), or anti-β-tubulin (1:2000; Sigma-Aldrich) antibodies, all prepared in TBST with 1% (w/v) BSA. After repeated washings in TBST, membranes were incubated in HRP-conjugated secondary antibody (F(ab′)2 goat anti-rabbit IgG (H+L) secondary antibody, HRP; Thermo Fisher Scientific) diluted to 1:5000 in 1% (w/v) BSA in TBST. The signal was developed after washing by TBST using ClarityTM ECL Western blotting substrate (Bio-Rad) and recorded with ChemiDocTM documentation system (Bio-Rad). Band densities were quantified using ImageLab software (Bio-Rad). All immunoblot analyses were performed at least in three biological replicates. Student's t test was applied to evaluate the statistical significance of differences.
Whole-mount Immunolabeling
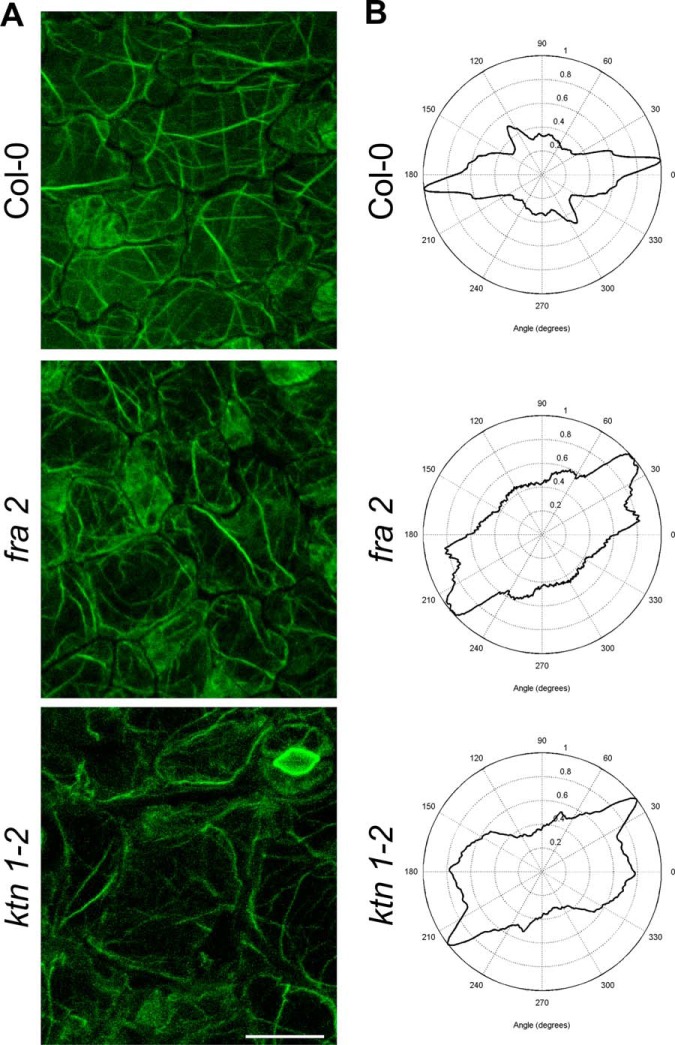
Immunolocalization of microtubules, KNOLLE and TSN1/2, in root whole mounts was done as described previously (33) with a small modification: cell wall digestion enzyme mixture contained 1% (w/v) meicelase, 1% (w/v) cellulase, and 1% (w/v) macerozyme R10 (Desert Biologicals) in PBS. Samples were immunolabeled with rabbit anti-TSN1/2 (29), rat anti-α-tubulin (clone YOL1/34; ABD Serotec), or rabbit anti-KNOLLE (34) primary antibodies diluted 1:75, 1:300, and 1:2000, respectively, in 3% (w/v) BSA in PBS at 4 °C overnight. In the case of KNOLLE and tubulin, co-localization a double immunolabeling was performed. Secondary antibodies included Alexa-Fluor 488 goat anti-rat and Alexa-Fluor 546 goat anti-rabbit IgGs (Thermo Fisher Scientific) and were diluted 1:500 in PBS containing 3% (w/v) BSA for 3 h (1.5 h at 37 °C and 1.5 h at room temperature). Where necessary, nuclei were counterstained with DAPI. Microscopic analyses of immunolabeled samples were examined with a Zeiss 710 CLSM platform (Carl Zeiss, Jena, Germany), using excitation lines at 405, 488, and 561 nm from argon, HeNe, diode, and diode-pumped solid-state lasers. Images were processed using ZEN 2010 software, Photoshop 6.0/CS, and Microsoft PowerPoint. Fluorescence intensity was evaluated using ZEN 2010 software (Carl Zeiss). Maximum intensity projections from Z-stack images (15 μm thick) of root epidermal cells were used for measurements. At least five individual root tips were analyzed. Student's t test was applied to evaluate the statistical significance of differences. Microtubule orientation and degree of isotropy were evaluated using CytoSpectre software (35).
Visualization of Actin Using Alexa-labeled Phalloidin
Actin visualization was performed according Panteris et al. (36) with a small modification: after actin stabilization with 200 μm m-maleimidobenzoyl-N-hydroxysuccinimide ester (BSE), whole seedlings were fixed in a mixture of 2.5% (w/v) paraformaldehyde, 0.5% glutaraldehyde, 10 μm BSE, and 0.1 μm Alexa-Fluor 568 phalloidin in microtubule stabilizing buffer (MTSB; 25 mm K-PIPES (pH 6.8), 2.5 mm EGTA and 2.5 mm MgSO4·7H2O) for 60 min. After washing in MTSB (three times for 2 min), seedlings were extracted in extraction buffer containing 5% (v/v) DMSO, 1% (v/v) Triton X-100 in MTSB for 15 min. Finally, seedlings were stained with 10% (v/v) Alexa-Fluor 488 phalloidin in MTSB for 60 min in the dark. Microscopic analysis of immunolabeled samples was performed using a Zeiss LSM710. Alexa-Fluor 488 phalloidin was excited at 488 nm, and fluorescence was detected between 499 and 566 nm. Images were processed using ZEN 2010 software, Photoshop 6.0/CS, and Microsoft PowerPoint. Actin filament orientation and degree of isotropy were evaluated using CytoSpectre software (35).
Quantitative Analysis of mRNA Transcript Levels by Real-time PCR
Total RNA was extracted from roots and aerial parts of 14-day-old seedlings of Col-0, fra2, and ktn1-2 mutants using TRI Reagent® (Sigma-Aldrich) according to the manufacturer's protocol. After DNase I digestion, RNA concentration and purity were determined with NanoDrop Lite spectrophotometer (Thermo Fisher Scientific). Template-primer mix for reverse transcription was composed of 1 μl of oligo(dT) primers (0.5 μg per reaction), 1.5 μg of RNA, and PCR-grade distilled water in total volume of 20 μl. The mixture was denatured at 65 °C for 5 min. The following components were added: 4 μl M-MLV reverse transcriptase 5× reaction buffer (Promega), 2 μl of deoxynucleotide mix (10 mm), 1 μl (40 units) RNasin® Plus RNase inhibitor (Promega), 1 μl (100 units) of M-MLV reverse transcriptase (Promega), and PCR-grade distilled water, in a total volume of 20 μl. PCRs were performed at 42 °C for 1 h followed by inactivation at 70 °C for 10 min. After reverse transcription reaction, the mixture was diluted four times. Quantitative RT-PCRs were performed in a 96-well plate with StepOnePlus Real-time PCR system (Applied Biosystems, Foster City, CA) using SYBR® Green to monitor dsDNA synthesis. Reaction contained 5 μl of Power SYBR® Green PCR master mix (Thermo Fisher Scientific), 0.75 μl of cDNA (corresponds to 140 ng of RNA before reverse transcription), and 0.5 μm gene-specific primers (supplemental Table S1). The following standard thermal profile was used for all PCRs: 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Experiments were run in three biological replicates, and the intra-assay variability was determined with technical triplicates. The expression data were normalized to the expression of EF1α (ELONGATION FACTOR 1-α; At5g60390) as a reference gene, and relative gene expression was calculated by 2−ΔΔCq method. Specificity of the target amplification was further verified by melting curve analysis of reaction products.
RESULTS
Overview of Proteomic Analysis and Functional Classification of the Differential Proteome
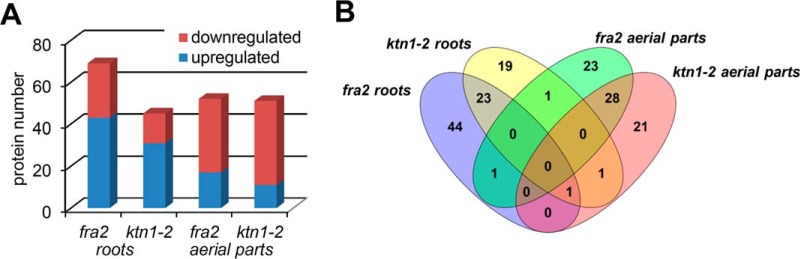
In addition to complete mass spectrometry/proteomics data deposited to PRIDE (see above), the information pertinent to protein identification can be found in the supplemental material in the form of common Excel files given for each individual sample. We compared the proteomes of roots and aerial parts of fra2 and ktn1-2 mutants with Col-0 quantitatively. Only those proteins that differed in abundance more than 1.5-fold between wild type and both mutants have been considered. Forty two proteins, 22 in roots and 20 in aerial parts, were differentially abundant in both mutants and showed a consistent trend in abundance difference (Table I). Twenty six of them were down-regulated, and 16 were up-regulated. Quantification details of all proteins identified in the mutants are presented in supplemental Table S2 (roots) and supplemental Table S3 (aerial parts). We have detected 69 and 45 differentially abundant proteins in roots of fra2 and ktn1-2, respectively. In aerial parts, 53 and 51 differentially regulated proteins were identified in fra2 and ktn1-2 (Fig. 1, A and B). Among them, seven proteins in roots and two proteins in aerial parts were identified only in Col-0, but they were not identified in the mutants. It is likely that differences in proteomes of these two mutants might arise from distinct types of mutations (mentioned under “Experimental Procedures”).
Table I. List of proteins with significantly different abundances consistently found in fra2 and ktn1-2 mutants as compared with wild type (Col-0). NA = not applicable.
| Accession no. | Description | Sample | Normalized intensity average in Col-0 roots | Normalized intensity average in fra2 roots | Normalized intensity average in ktn1-2 roots | Fold change |
p value |
||
|---|---|---|---|---|---|---|---|---|---|
| fra2 vs Col-0 | ktn1-2 vs Col-0 | fra2 vs Col-0 | ktn1-2 vs Col-0 | ||||||
| Amino acid metabolism | |||||||||
| gi15233111 | Cysteine synthase C1 | Roots | 52115.82 | 185447.46 | 136112.56 | 3.56 | 2.61 | 0.05 | 0.01 |
| gi15223910 | Aspartate semialdehyde dehydrogenase | Roots | 55508.12 | 230059.47 | 125986.93 | 4.14 | 2.27 | 0.05 | 0.05 |
| Proteolysis | |||||||||
| gi18402225 | Granulin repeat cysteine protease family protein | Roots | 75658.44 | 343248.11 | 448897.32 | 4.54 | 5.93 | 0.01 | 0.00 |
| Protein synthesis and translation | |||||||||
| gi18410833 | 50S ribosomal protein L31 | Aerial parts | 6417292.09 | 2295801.19 | 4254452.45 | 0.36 | 0.66 | 0.007 | 0.023 |
| gi18399100 | 40S ribosomal protein S18 | Roots | 113741.22 | 227944.46 | 394848.67 | 2.00 | 3.47 | 0.00 | 0.00 |
| gi15221798 | 60S ribosomal protein L6-1 | Roots | 123564.32 | 196333.48 | 383764.34 | 1.59 | 3.11 | 0.00 | 0.01 |
| gi79324564 | 40S ribosomal protein S5-1 | Roots | 219962.29 | 82575.80 | 94830.59 | 0.38 | 0.43 | 0.01 | 0.01 |
| gi79317147 | 50S ribosomal protein L4 | Aerial parts | 7808895.67 | 1985714.83 | 2279779.83 | 0.25 | 0.29 | 0.005 | 0.003 |
| gi30692346 | Small subunit ribosomal protein S1 | Aerial parts | 8943813.19 | 5884181.99 | 5048811.79 | 0.66 | 0.56 | 0.055 | 0.005 |
| gi15232603 | 60S acidic ribosomal protein P0-2 | Aerial parts | 9543945.84 | 4587130.63 | 4871300.09 | 0.48 | 0.51 | 0.001 | 0.009 |
| Energy, metabolism, and photosynthesis | |||||||||
| gi26557005 | ATPase subunit 1 | Aerial parts | 2793480.24 | 7217395.44 | 5089236.13 | 2.58 | 1.82 | 0.007 | 0.030 |
| gi18412632 | ATP synthase γ chain 1 | Aerial parts | 24872995.56 | 16236642.88 | 17444031.98 | 0.65 | 0.70 | 0.022 | 0.030 |
| gi15230595 | phosphoglycerate kinase 1 | Aerial parts | 99022520.01 | 73663523.69 | 28363047.45 | 0.74 | 0.29 | 0.026 | 0.003 |
| gi79313434 | γ-Hydroxybutyrate dehydrogenase | Roots | 14385.25 | 0 | 0 | Unique in Col-0 | NA | NA | |
| gi145329204 | Triose-phosphate isomerase | Roots | 148158.00 | 366354.23 | 305748.27 | 2.47 | 2.06 | 0.00 | 0.00 |
| gi15235029 | chlorophyll a-b binding protein CP26 | Aerial parts | 4581201.95 | 9236218.52 | 6837980.22 | 2.02 | 1.49 | 0.05 | 0.033 |
| gi15228194 | Sedoheptulose-1,7-bisphosphatase | Aerial parts | 8304146.35 | 13526281.91 | 13754753.35 | 1.63 | 1.66 | 0.010 | 0.045 |
| gi15230358 | Adenylosuccinate synthetase | Roots | 61378.48 | 0 | 0 | Unique in Col-0 | NA | NA | |
| gi7525041 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | Aerial parts | 411927407.76 | 294914235.63 | 277266195.81 | 0.72 | 0.66 | 0.019 | 0.019 |
| gi30684767 | ATP-dependent zinc metalloprotease FTSH 2 (VAR2) | Aerial parts | 10406900.99 | 5403906.75 | 6122905.04 | 0.52 | 0.59 | 0.027 | 0.052 |
| gi15217918 | DNA-damage resistance protein DRT112 | Aerial parts | 13493270.03 | 2986221.23 | 4416446.13 | 0.22 | 0.33 | 0.011 | 0.007 |
| gi15242465 | Soluble inorganic pyrophosphatase 1 | aerial parts | 11022571.57 | 5807403.25 | 5763606.13 | 0.53 | 0.52 | 0.044 | 0.027 |
| gi238479213 | Ribulose bisphosphate carboxylase large chain, catalytic domain | Aerial parts | 15069515.17 | 10777815.24 | 9451326.29 | 0.72 | 0.63 | 0.021 | 0.021 |
| Stress response and redox regulation | |||||||||
| gi15236014 | Lipase/lipooxygenase, PLAT/LH2 family protein | Roots | 38041.31 | 0 | 0 | Unique in Col-0 | NA | NA | |
| gi18413214 | Nucleoside diphosphate kinase 1 | Roots | 226388.99 | 380425.14 | 393491.58 | 1.68 | 1.74 | 0.02 | 0.39 |
| gi15235401 | Glutathione S-transferase F2 | Roots | 644121.71 | 1578139.78 | 2106336.43 | 2.45 | 3.27 | 0.00 | 0.01 |
| gi15230982 | Peroxiredoxin Q | Aerial parts | 9363992.23 | 5543512.31 | 3980676.95 | 0.59 | 0.43 | 0.042 | 0.005 |
| gi79313261 | PYK10-binding protein 1 | Roots | 1166764.61 | 630062.78 | 644475.44 | 0.54 | 0.55 | 0.01 | 0.01 |
| gi18415805 | Ferredoxin/thioredoxin reductase subunit A (variable subunit) 2 | Aerial parts | 2441812.37 | 895399.11 | 1305568.55 | 0.37 | 0.53 | 0.011 | 0.053 |
| gi15236386 | Selenium-binding protein 2 | Aerial parts | 1289730.38 | 0 | 0 | Unique in Col-0 | NA | NA | |
| gi15234648 | Peroxidase 45 | Roots | 447213.47 | 690524.52 | 951861.94 | 1.54 | 2.13 | 0.05 | 0.02 |
| Signaling | |||||||||
| gi18413181 | 14-3-3-like protein GF14 chi | Roots | 797937.21 | 0 | 0 | Unique in Col-0 | NA | NA | |
| Hormonal homeostasis | |||||||||
| gi42570831 | S-Alkyl-thiohydroximate lyase SUR1 | Roots | 6362.74 | 0 | 0 | Unique in Col-0 | NA | NA | |
| gi15221692 | Pyruvate dehydrogenase E1 component subunit α-2 (IAR4) | Roots | 253127.82 | 55389.52 | 66337.07 | 0.22 | 0.26 | 0.01 | 0.02 |
| Protein folding, chaperones | |||||||||
| gi334185828 | Clp ATPase | Roots | 107823.61 | 37454.35 | 48712.02 | 0.35 | 0.45 | 0.02 | 0.03 |
| gi18415982 | Tetratricopeptide repeat protein | Roots | 222702.14 | 427357.63 | 486498.43 | 1.92 | 2.18 | 0.03 | 0.02 |
| gi15237739 | Cyclophilin ROC7 | Aerial parts | 10957142.37 | 3657898.57 | 3546535.06 | 0.33 | 0.32 | 0.002 | 0.004 |
| Unknown function | |||||||||
| gi18404496 | NAD(P)-binding Rossmann-fold-containing protein | Aerial parts | 8607872.32 | 5447058.47 | 5603099.42 | 0.63 | 0.65 | 0.008 | 0.007 |
| gi15237739gi15224648 | Membrane-associated progesterone binding protein 2 | Roots | 203330.29 | 371469.14 | 439583.49 | 1.83 | 2.16 | 0.03 | 0.02 |
| gi15237739gi30682601 | Vacuolar calcium-binding protein-like protein | Roots | 57633.53 | 242968.33 | 103128.61 | 4.22 | 1.79 | 0.01 | 0.05 |
| gi15237739gi334187997 | Uncharacterized protein | Aerial parts | 739698.22 | 0 | 0 | Unique in Col-0 | NA | NA | |
| Membrane transport | |||||||||
| gi15237739gi18397991 | MD-2-related lipid recognition domain-containing protein | Roots | 102011.46 | 0 | 0 | Unique in Col-0 | NA | NA | |
Fig. 1.
Outputs of proteomic analysis of KATANIN 1 mutants. A, graph showing numbers of proteins with increased or decreased abundances in roots and aerial parts of fra2 and ktn1-2 mutants as compared with the Col-0 wild type. B, Venn diagram showing numerical distribution of proteins with significantly changed abundances among roots and aerial parts of fra2 and ktn1-2 mutants.
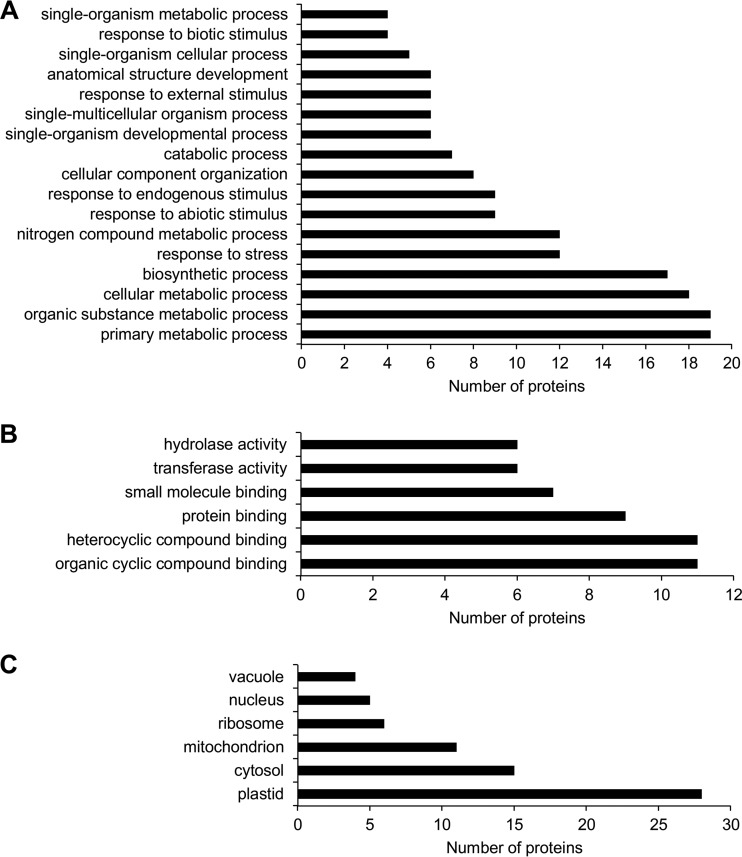
We used gene ontology (GO) annotation to evaluate the impact of defective microtubule severing on the Arabidopsis proteome. According to biological process, the highest number of differentially abundant proteins was annotated to diverse metabolic processes (Fig. 2A). A significant number of proteins was also denoted as involved in the response to biotic and abiotic stimuli. Last but not least, GO annotations connected to cellular component organization and development showed changed abundances in both mutants (Fig. 2A). According to molecular functions, differentially abundant proteins were involved in binding to proteins, organic and inorganic compounds, as well as binding to small molecules (Fig. 2B). GO annotation according to cell compartment showed different abundances of proteins localized to plastids, cytosol, mitochondria, ribosomes, nuclei, and vacuoles (Fig. 2C). A detailed GO annotation performed separately for differentially regulated proteins in roots and aerial parts is provided in supplemental Figs. S1–S6.
Fig. 2.
Graphs showing GO annotations of differentially abundant proteins found consistently in fra2 and ktn1-2 mutants (roots and aerial plants collectively) according to biological process (A), molecular function (B), and cellular compartments (C).
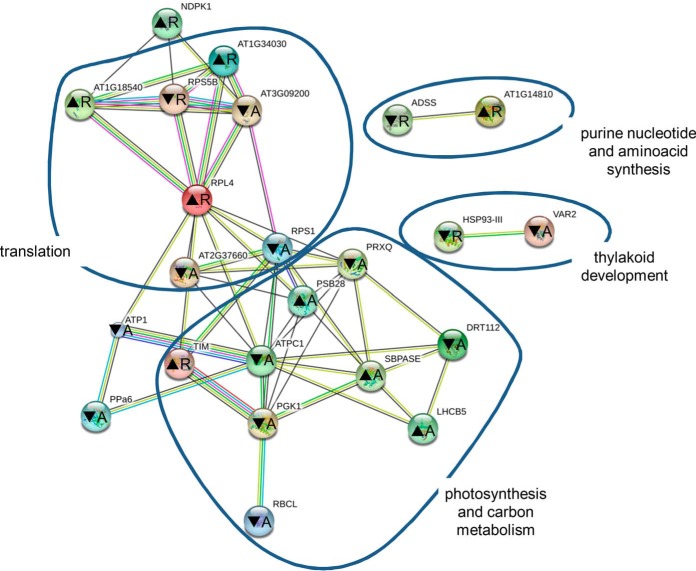
Analysis of protein interaction networks occurring among these proteins using STRING application showed deregulation of proteins involved in photosynthesis, carbon metabolism, and translation in both mutants (Fig. 3). Other less abundant networks were annotated to purine nucleotide and amino acid synthesis and chloroplast thylakoid development. The majority of proteins involved in photosynthesis showed decreased abundance in the mutants, whereas carbon metabolism appears to be enhanced. Two proteins contributing to thylakoid development showed unequivocal decreased abundance. Abundances of proteins involved in translation did not show uniform changes in the mutants. Similar protein networks were predicted when both mutants were analyzed separately (supplemental Fig. S7).
Fig. 3.
Representation of protein interaction networks in differential proteome consistently found in both fra2 and ktn1-2 roots and aerial parts and as generated by STRING web-based application. Letters in nodes mean roots (R) and aerial (A) parts. ▴ means increased abundance, and ▾ means decreased abundance. Different line colors represent the types of evidence used in predicting the associations: gene fusion (red), neighborhood (green), co-occurrence across genomes (blue), co-expression (black), experimental (purple), association in curated databases (light blue), or co-mentioned in PubMed abstracts (yellow). RPL4 is 50S ribosomal protein L4 (gi79317147); At1g14810 is aspartate semialdehyde dehydrogenase (gi15223910); ADSS is adenylosuccinate synthetase (gi15230358); At1g18540 is 60S ribosomal protein L6-1 (gi15221798); DRT112 is DNA-damage resistance protein DRT112 (gi15217918); At1g34030 is 40S ribosomal protein S18 (gi18399100); TIM is triose-phosphate isomerase (gi145329204); VAR2 is ATP-dependent zinc metalloprotease FTSH 2 (VAR2) (gi30684767); RPS5B is 40S ribosomal protein S5-1 (gi79324564); At2g37660 is NAD(P)-binding Rossmann fold-containing protein (gi18404496); At3g09200 is 60S acidic ribosomal protein P0-2 (gi15232603); PGK1 is phosphoglycerate kinase 1 (gi15230595); PRXQ is peroxiredoxin Q (gi15230982); HSP93-III is Clp ATPase (gi334185828); SBPASE is sedoheptulose-1,7-bisphosphatase (gi15228194); ATPC1 is ATP synthase γ chain 1 (gi18412632); NDPK1 is nucleoside diphosphate kinase 1 (gi18413214); LHCB5 is chlorophyll a-b-binding protein CP26 (gi15235029); PSB28 is photosystem II reaction center PSB28 protein (gi18417239); PPa6 is soluble inorganic pyrophosphatase 1 (gi15242465); RPS1 is small subunit ribosomal protein S1 (gi30692346); RBCL is ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (gi7525041); and ATP1 is ATPase subunit 1 (gi26557005).
KATANIN1 Mutants Exert Altered Abundances of MAPs, Disturbed Microtubule Organization, and Abnormal Nuclear Shape
Disrupted microtubule severing resulted in severe developmental defects of fra2 and ktn1-2 mutants (21, 23, 26). Several proteins related to microtubule severing may contribute to these developmental phenotypes. Therefore, by performing differential proteomics on fra2 and ktn1-2 mutants, we focused on proteins involved in plant development as classified by gene ontology annotation (supplemental Tables S4–S7) or experimentally (Table II). These proteins are mostly involved in embryogenesis, germination, root growth, and hypocotyl elongation.
Table II. Differentially abundant proteins having developmental roles in fra2 and ktn1-2 mutants.
Proteins annotated in GO cellular component organization (GO:0016043), single organism developmental process (GO:0044767), anatomical structure development (GO:0048856), and proteins reported to have developmental roles are listed.
| Accession no. | Description | Sample | Fold change |
GO annotation | Experimental evidence | |
|---|---|---|---|---|---|---|
| fra2 vs Col-0 | ktn1-2 vs Col-0 | |||||
| gi15221692 | Pyruvate dehydrogenase E1 component subunit α-2 (IAR4) | Roots | 0.22 | 0.26 | Auxin homeostasis (67, 68) | |
| gi42570831 | S-Alkyl-thiohydroximate lyase SUR1 | Roots | Unique in Col-0 | GO:0016043 | Auxin homeostasis (66) | |
| GO:0044767 | ||||||
| GO:0048856 | ||||||
| gi18397991 | MD-2-related lipid recognition domain-containing protein | Roots | Unique in Col-0 | Gravitropism, auxin transport (72) | ||
| gi18413181 | 14-3-3-like protein GF14 χ | Roots | Unique in Col-0 | Regulation of ethylene synthesis (62) | ||
| gi18402225 | Granulin repeat cysteine protease family protein | Roots | 4.54 | 5.93 | ABA signaling (73) | |
| gi334185828 | Clp ATPase | Roots | 0.35 | 0.45 | GO:0016043 | Photosystem biogenesis, thylakoid membrane biogenesis, leaf development (74) |
| gi79324564 | 40S ribosomal protein S5-1 | Roots | 0.38 | 0.43 | GO:0016043 | Delayed embryo development, abnormal venation (75) |
| gi79317147 | 50S ribosomal protein L4 | Aerial parts | 0.25 | 0.29 | Embryo development (76) | |
| gi22326646 | TUDOR-SN protein 1 | Roots | 1.50 | Embryogenesis and fertility (78) | ||
| gi15240352 | TUDOR-SN protein 2 | Roots | 4.48 | 3.12 | Embryogenesis and fertility (78) | |
| gi15237739 | Cyclophilin ROC7 | Aerial parts | 0.33 | 0.32 | GO:0044767 | |
| GO:0048856 | ||||||
| gi30684767 | ATP-dependent zinc metalloprotease FTSH 2 (VAR2) | Aerial parts | 0.52 | 0.59 | GO:0016043 | Photosystem biogenesis, thylakoid membrane biogenesis, leaf development (74) |
| GO:0044767 | ||||||
| GO:0048856 | ||||||
| gi334187997 | Uncharacterized protein | Aerial parts | Unique in Col-0 | GO:0016043 | ||
| gi15236386 | Selenium-binding protein 2 | Aerial parts | Unique in Col-0 | GO:0016043 | ||
| GO:0044767 | ||||||
| GO:0048856 | ||||||
| gi145329204 | Triose-phosphate isomerase | Roots | 2.47 | 2.06 | GO:0016043 | |
| GO:0044767 | ||||||
| GO:0048856 | ||||||
| gi15233111 | cysteine synthase C1 | Roots | 3.56 | 2.61 | GO:0044767 | |
| GO:0048856 | ||||||
| gi15235029 | Chlorophyll a-b-binding protein CP26 | Aerial parts | 2.02 | 1.49 | GO:0016043 | |
| gi79325183 | Plasma membrane-associated cation-binding protein 1 (MDP25) | Roots | 1.61 | Hypocotyl elongation (37, 38) | ||
| gi15241472 | Tubulin β-4 chain | Roots | 0.20 | Cell division and elongation (5), seed imbibition (41, 77) | ||
| gi18401618 | WPP domain-containing protein 2 | Roots | 0.46 | Root growth, cell division (39) | ||
| gi15224838 | Profilin 1 | Roots | 1.09 | 1.48 | Cell elongation (79), embryo development and germination (80) | |
| gi30697298 | Actin-depolymerizing factor 3 | Aerial parts | 0.30 | Tip growth (81) | ||
| gi15242516 | Actin 7 | Aerial parts | 2.09 | Root growth (82), seed imbibition (41, 83) | ||
| gi15222929 | V-type proton ATPase subunit B1 | Aerial parts | 1.23 | Actin stabilizing (84) | ||
| gi15219901 | Patellin 2 | Roots | 0.22 | Cell plate formation (54) | ||
| gi18391066 | 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase 1 | Roots | 1.78 | Stomatal movements, pollen development (85) | ||
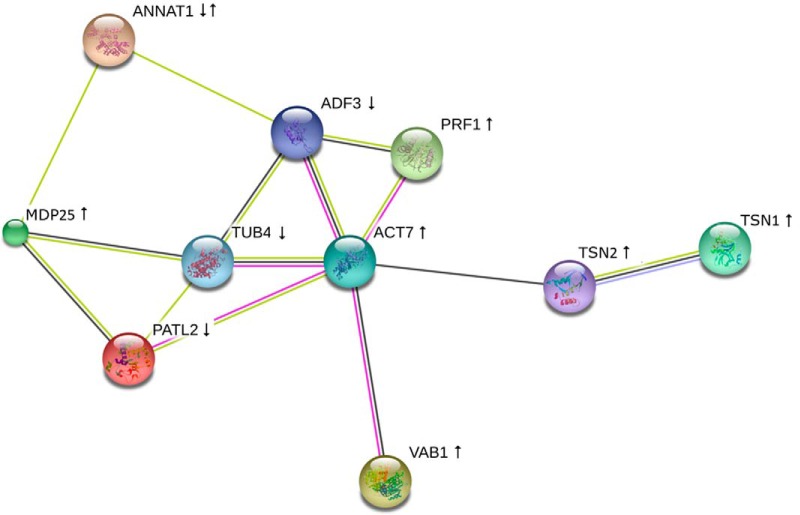
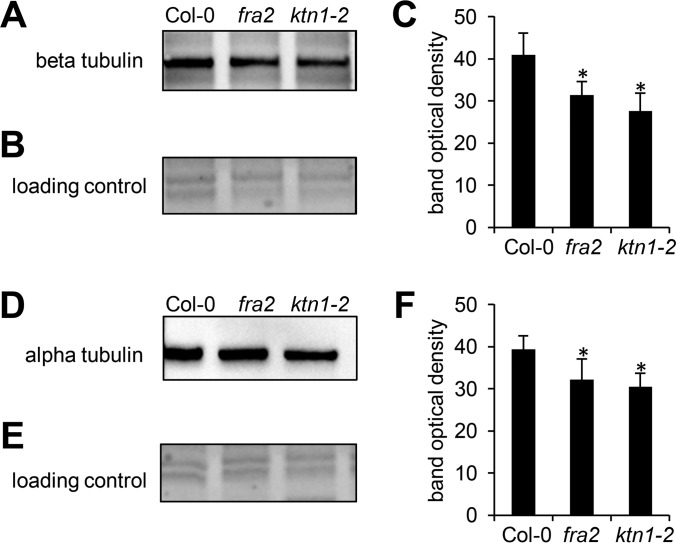
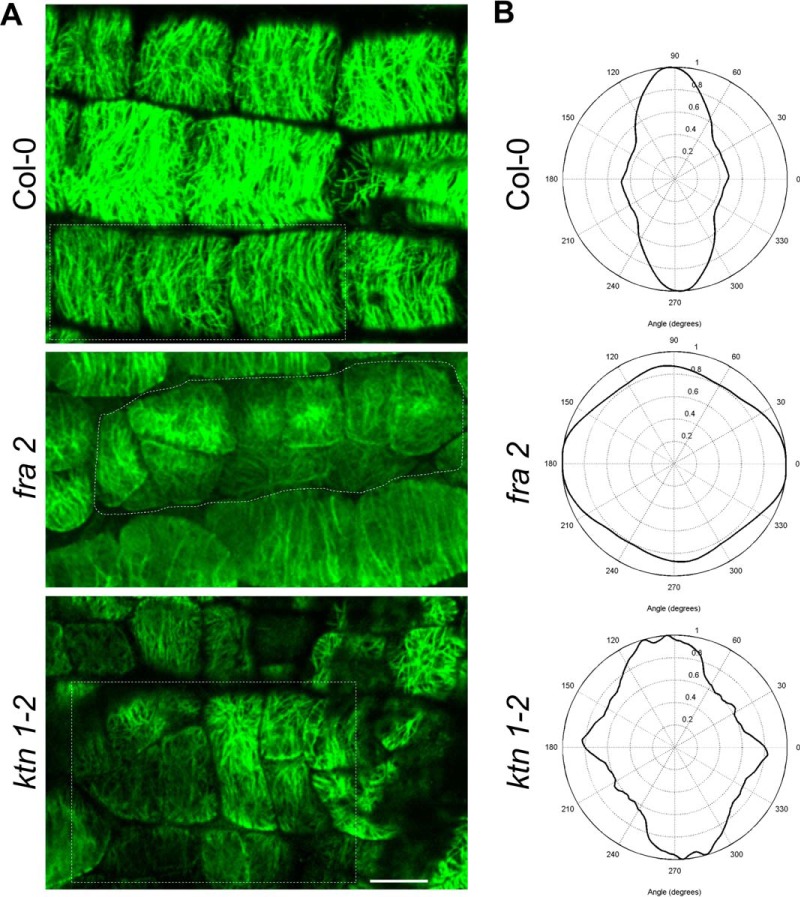
Our proteomic analysis revealed several cytoskeletal and cytoskeleton-related proteins. Interestingly, these proteins form a functionally interconnected network consisting of both microtubule-binding as well as actin-binding and regulatory proteins, as predicted by STRING (Fig. 4). Among them, tubulin β-4 was down-regulated in aerial parts of fra2 (Table II). To validate such down-regulation, we carried out an immunoblotting analysis of fra2 and ktn1-2 roots using anti-β-tubulin antibody. In agreement with proteomic data, β-tubulin showed decreased abundances in both mutants (Fig. 5, A–C). As expected, a similar trend was observed in the case of α-tubulin (Fig. 5, D–F). Whole-mount immunofluorescence labeling of microtubules performed on root cells revealed reorganization and randomization of microtubules in root epidermal cells of fra2 and ktn1-2 mutants (Fig. 6). Furthermore, plasma membrane-associated cation-binding protein 1, also called microtubule-depolymerizing protein 25 (MDP25) (37, 38), showed an increased abundance in the fra2 mutant (Table II). In addition, we have found proteins controlling microtubule-dependent processes such as cell division and cell plate formation (patellin 2 and WPP domain-containing protein 2; Table II). This is in agreement with the obliquely oriented cell plates in fra2 and lue1 mutants as it was previously proposed due to the aberrantly oriented phragmoplasts (24). Our more detailed whole-mount immunolabeling of cell plates using specific antibody recognizing cell plate marker protein KNOLLE (34) showed obliquely oriented and misaligned cell plates in the ktn1-2 mutant (supplemental Fig. S8), and misaligned phragmoplasts were observed by simultaneous co-visualization of microtubules by using anti-α-tubulin antibody (supplemental Fig. S8). It seems that patellin 2 and WPP2 are co-regulated with KATANIN 1 to control phragmoplast and cell plate formation.
Fig. 4.
Depiction of functional protein association networks predicted by STRING among cytoskeletal proteins found to be differentially abundant in fra2 and ktn1-2 as compared with Col-0. Different line colors represent types of evidence used in predicting associations: gene fusion (red); neighborhood (green); co-occurrence across genomes (blue); co-expression (black); experimental (purple); association in curated databases (light blue); or co-mentioned in PubMed abstracts (yellow). ↑ means increased abundance; ↓ means decreased abundance. TUB4 is tubulin β-4 chain; TSN1 is TUDOR-staphylococcal nuclease protein 1, TSN2; MDP25 is microtubule-destabilizing protein 25; PATL2 is patellin 2; PRF1 is profilin 1; ACT7 is actin 7; ADF3 is actin-depolymerizing factor 3; ANNAT1 is annexin 1; and VAB1 is V-type proton ATPase subunit B1.
Fig. 5.
Immunoblotting analysis of β-tubulin and α-tubulin in roots of Arabidopsis wild type Col-0 and KATANIN 1 mutants fra2 and ktn1-2. A and D, immunoblots probed with anti-β-tubulin (A) and anti-α-tubulin (D) antibodies. B and E, visualization of proteins transferred on nitrocellulose membranes. C and F, optical density quantifications of respective bands in A and D. Asterisks indicate significant differences between mutants and wild type at p ≤ 0.05 according to Student's t test. Error bars represent standard deviations.
Fig. 6.
A, immunolocalization of cortical microtubules in root epidermal cells of Arabidopsis wild type Col-0 and KATANIN 1 mutants fra2 and ktn1-2. B, microtubule orientation and degree of isotropy analyzed by CytoSpectre software. Note distorted microtubule orientation and anisotropy in both mutants in contrast to mostly parallel microtubule orientation in Col-0. Bar, 10 μm.
Because WPP2 is also a nuclear envelope localized protein (39), its down-regulation might indicate altered nuclear shape in KATANIN 1 mutants. Consistent with this assumption, visualization of nuclei by DAPI revealed that nuclei of fra2 and ktn1-2 root epidermal cells showed aberrant shapes (supplemental Fig. S9).
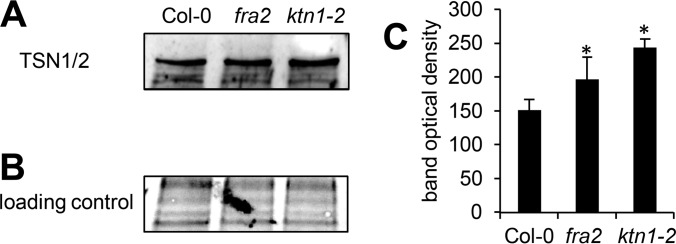
We have also detected increased abundances of TUDOR-staphylococcal nuclease protein 1 (TSN1) and TSN2 in the fra2 mutant (Table II). These proteins are co-localizing with and move along cortical microtubules (32). They are involved in the formation of stress granules, which is dependent on microtubule dynamics (32). Thus, our results support a tight link between TSN proteins and microtubules. Immunoblotting analysis using primary antibody recognizing both TSN1 and TSN2 verified increased abundances of these two isoforms in fra2 and ktn1-2 roots (Fig. 7). This was further confirmed by immunolocalization of TSN proteins in intact roots of fra2 and ktn1-2 mutants (Fig. 8). Root cells of both mutants showed prominent accumulation of TSN proteins preferentially at cell peripheries and in the whole cytoplasm that was not so prominent and was quantitatively less abundant in the control Col-0. Altogether, these data indicate a feedback mechanism controlling microtubule organization in KATANIN 1 mutants.
Fig. 7.
Immunoblotting analysis of TSN1/2 (TUDOR-staphylococcal nuclease protein 1/2) abundances in roots of Arabidopsis wild type Col-0 and KATANIN 1 mutants fra2 and ktn1-2. A, immunoblots probed with anti-TSN1/2 antibody. B, visualization of proteins transferred on nitrocellulose membranes. C, optical density quantification of band in A. Asterisks indicate significant differences between mutants and wild type at p ≤ 0.05 according to the Student's t test. Error bars represent standard deviations.
Fig. 8.
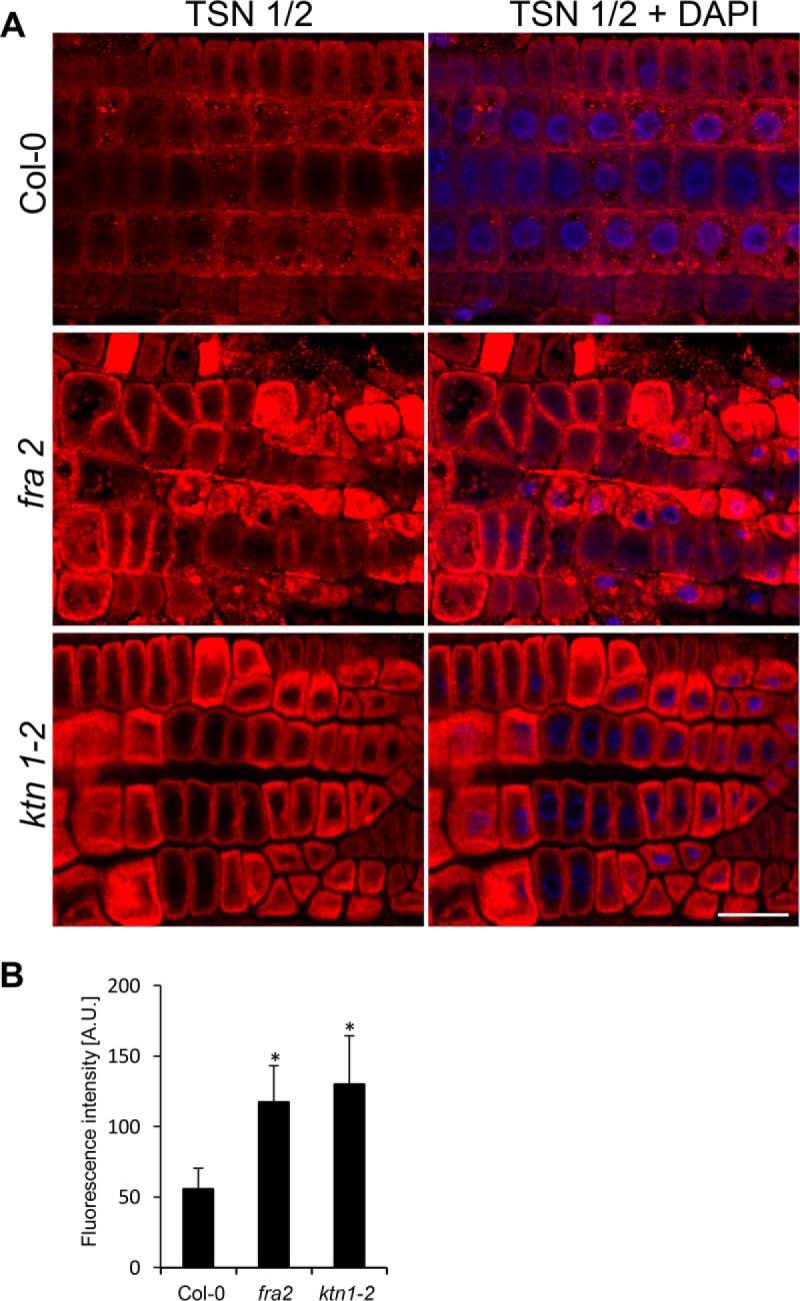
A, immunolocalization of TSN1/2 (TUDOR-staphylococcal nuclease protein 1/2) in root epidermal cells of Arabidopsis wild type Col-0 and KATANIN 1 mutants fra2 and ktn1-2. B, fluorescence intensity quantification of immunolabeled TSN1/2 in root epidermal cells of wild type Col-0 and fra2 and ktn1-2 mutants. Maximum intensity projections from z-stack images (15 μm thick) of root epidermal cells were used for measurements. At least five individual root tips were analyzed. Differences between both mutants and Col-0 were statistically significant (p ≤ 0.05) according to Student's t test. Bar, 20 μm.
Defective Microtubule Severing Altered Actin Regulatory Proteins and Actin Organization
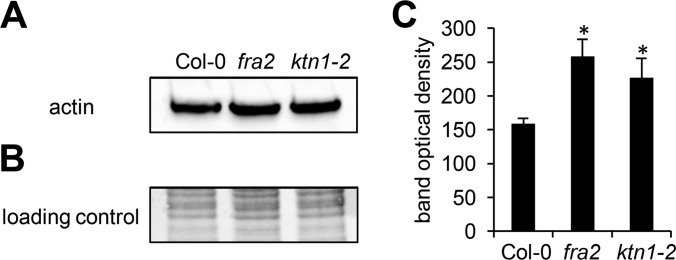
Notably, defective microtubule severing in the mutants affected abundances of actin and actin-binding proteins (ABPs). Thus we have found up-regulation of profilin 1 in roots of the ktn1-2 mutant and up-regulation of actin 7 in fra2 aerial parts (Table II). The abundance of actin-binding vacuole-type proton ATPase subunit B1 was also increased in aerial parts of fra2 (Table II). Verification of actin abundances in the mutants and Col-0 control using immunoblotting with an antibody recognizing denatured monomeric G-actin approved the proteomic data (Fig. 9). In addition, actin-depolymerizing factor 3 was negatively regulated in the ktn1-2 aerial parts (Table II). Importantly, visualization of F-actin by staining with fluorescently labeled phalloidin showed that proteomic changes of actin 7 isoform (up-regulation) and actin-depolymerizing factor 3 (ADF3, down-regulation) led to reorientation and disorganization of actin filaments in leaves of fra2 and ktn1-2 mutants. Thus, actin filaments in the leaf epidermal cells of both mutants were more distorted, less abundant, and also less bundled in these cells (Fig. 10). These results suggested a tight cross-talk between microtubules and organization of the actin cytoskeleton in KATANIN 1 mutants.
Fig. 9.
Immunoblotting analysis of actin in aerial parts of Arabidopsis wild type Col-0 and KATANIN 1 mutants fra2 and ktn1-2. A, immunoblots probed with anti-actin antibody recognizing both monomeric G-actin and filamentous F-actin. B, visualization of transferred proteins on nitrocellulose membranes. C, optical density quantification of bands in A. Asterisks indicate significant differences between mutants and wild type at p ≤ 0.05 according to the Student's t test. Error bars represent standard deviations.
Fig. 10.
Filamentous actin (F-actin) organization in leaf epidermal cells of Arabidopsis wild type Col-0 and KATANIN 1 mutants fra2 and ktn1-2. A, Alexa phalloidin was used for F-actin visualization. B, F-actin orientation and degree of isotropy analyzed by CytoSpectre software. Note reorientation of F-actin and higher anisotropy in both mutants in contrast to Col-0. Moreover, actin filaments are less prominent and look distorted in both mutants. Bar, 20 μm.
Abundances of Cytoskeletal Proteins and Transcript Levels of Corresponding Genes Are Not Correlated in the KATANIN1 Mutants
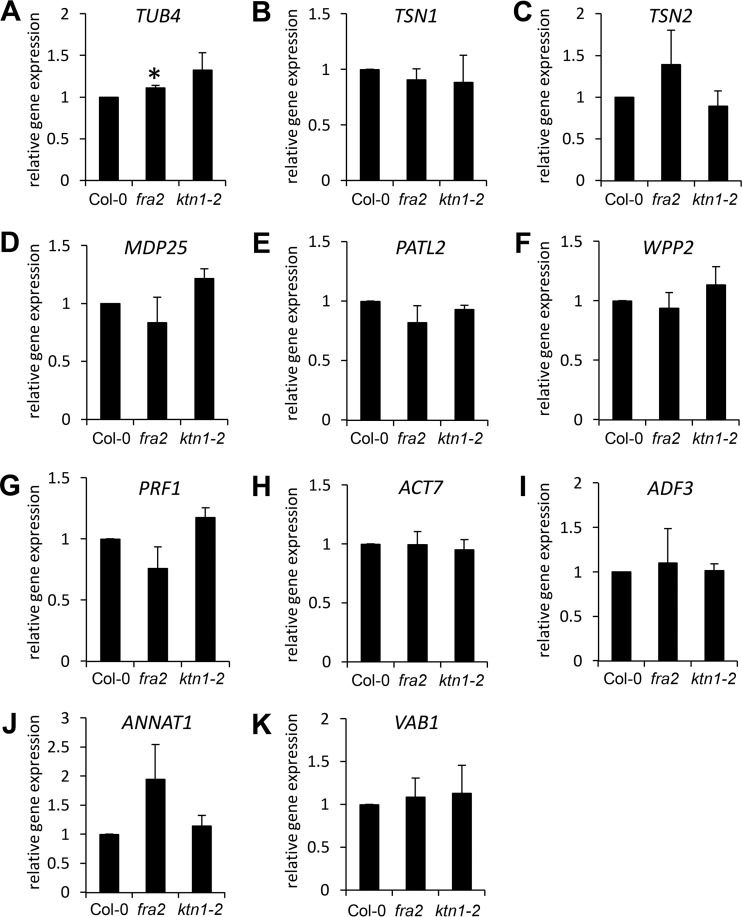
KATANIN 1 mutants exhibit severe developmental defects that may cause general transcriptional reprogramming. This may imply that the changes of cytoskeletal proteins in KATANIN 1 mutants resulted from altered transcriptional regulation and not from direct effects of impaired microtubule-severing activity. Therefore, we quantitatively examined expression levels of genes encoding the most-important 11 cytoskeletal and cytoskeleton-related proteins found by proteomic analysis. In all cases, except TUBULIN β-4 (TUB4) in fra2, we have found only minor insignificant changes in the mRNA levels of both KATANIN 1 mutants as compared with the control (Fig. 11). This indicates that changes in protein abundances were not caused by transcriptional alterations in the mutants.
Fig. 11.
Quantitative expression levels of TUBULIN β-4 (TUB4; A); TUDOR-STAPHYLOCOCCAL NUCLEASE (TSN1; B); TSN2 (C); MICROTUBULE-DESTABILIZING PROTEIN 25 (MDP25; D); PATELLIN 2 (PATL2; E); WPP DOMAIN-CONTAINING PROTEIN 2 (WPP2; F); and PROFILIN 1 (PRF1; G) in roots, as well as ACTIN 7 (ACT7; H); ACTIN-DEPOLYMERIZING FACTOR 3 (ADF3; I); ANNEXIN 1 (ANNAT1; J); and V-TYPE PROTON ATPASE SUBUNIT B1 (VAB1; K) in aerial parts of Col-0, fra2, and ktn1-2. Asterisk in A indicates significant difference between fra2 mutant and wild type at p ≤ 0.05 according to Student's t test. Error bars represent standard deviations.
Proteins Involved in Hormonal Homeostasis Showed Altered Abundances in KATANIN 1 Mutants
Generally, it is known that microtubule cytoskeleton is sensitive to hormones (40), whereas abundance of TUBULIN α2 (TUA2) is controlled by gibberellic acid in Arabidopsis (41). In addition, KATANIN 1 participates in the regulation of microtubule reorganization induced by gibberellic acid and ethylene (26). Interestingly, we have found several proteins involved in the regulation of auxin, ethylene (Table II), and gibberellic acid (Tables III and IV) homeostasis. Moreover, a screening of abscisic acid-responsive elements in the promoter sequences of genes encoding identified differentially regulated proteins indicated that proteins involved in ABA response might be also affected by impaired microtubule severing (Table V). Thus, our study also provides new protein candidates for the cross-talk between hormones and microtubules.
Table III. List of differentially abundant proteins in fra2 and ktn1-2 mutants (as compared with Col-0 wild type; p ≤ 0.05), which are responsive to gibberellic acid as reported in transcriptomic study (86).
| Accession | Protein name | Fold change fra2 vs. Col-0 | Fold change ktn1-2 vs. Col-0 | |
|---|---|---|---|---|
| gi18391066 | 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase 1 | 1.78 (root) | Stomatal movements, pollen development (85) | |
| gi334182565 | Salt tolerance-related protein | 0.57 (aerial parts) | Salt tolerance (87) | |
| gi79313261 | PYK10-binding protein 1 | 0.54 (root) | 0.55 (root) | Defense (88) |
| gi15235401 | Glutathione S-transferase F2 | 2.4 (root) | 3.27 (root) | Stress tolerance (89) |
| gi79326500 | Putative cinnamyl alcohol dehydrogenase 9 | 2.95 (aerial parts) | Lignification (90) |
Table IV. List of differentially abundant proteins in fra2 and ktn1-2 mutants (as compared with Col-0 wild type; p ≤ 0.05) containing GA-responsive element ACGTGTC (86) in their promoter sequence (1000 kb upstream of ATG).
| Accession no. | Protein name | Position (upstream of ATG) | Fold change |
Function | |
|---|---|---|---|---|---|
| fra2 vs. Col-0 | ktn1-2 vs. Col-0 | ||||
| gi334183935 | Dehydrin ERD14 | 843−850 | 0.22 (aerial parts) | 0.52 (aerial parts) | Stress response (91) |
| gi15233111 | Cysteine synthase C1 | 472−479 | 3.55 (root) | 2.61 (root) | Defense (92) |
| gi79326500 | Putative cinnamyl alcohol dehydrogenase 9 | 428−435 | 2.95 (aerial parts) | Lignification (90) | |
| gi18406229 | TRAF-like protein | 131−138 | 1.45 (roots) | Unknown | |
| gi15227259 | Cyclophilin ROC3 | 128−135 | 0.43 (roots) | Defense (93) | |
| gi15234637 | Photosystem II subunit Q-2 | 149−156 | 1.45 (aerial parts) | Photosystem II assembly (94) | |
| gi15238217 | Sulfite reductase | 89−96 | 0.43 (roots) | Sulfate assimilation, growth and development (95) | |
Table V. List of differentially abundant proteins in fra2 and ktn1-2 mutants (as compared with Col-0 wild type; p ≤ 0.05) containing abscisic acid-responsive element (ABRE) in their promoter sequence (1000 kb upstream of ATG).
| ABRE | Accession no. | Protein name | Position (upstream of ATG) | Fold change |
Function | |
|---|---|---|---|---|---|---|
| fra2 vs Col-0 | ktn1-2 vs Col-0 | |||||
| ACACGTGTC | gi334183935 | Dehydrin ERD14 | 571−562 | 0.22 (aerial parts) | 0.52 (aerial parts) | Stress response (91) |
| ACACGTGGC | gi15220216 | Annexin 1 | 49−40 | 1.67 (aerial parts) | 0.55 (aerial parts) | Actin-binding, calcium signaling (96, 97) |
| ACACGTGTA | gi15220854 | Alkenal/one oxidoreductase | 339−330 | 0.52 (aerial parts) | Removal of reactive carbonyls (98) | |
| gi15228498 | UDP-glucose pyrophosphorylase 1 | 74−65 | 1.43 (roots) | Cellulose and callose formation, growth, development (99, 100) | ||
| gi30687411 | Dihydrolipoamide succinyltransferase | 56−47 | fra2 unique (aerial parts) | Unknown | ||
| ACACGTGTT | gi15233272 | Triose-phosphate isomerase | 928−919 | 1.49 (roots) | Glycolysis, gluconeogenesis (101) | |
| CCACGTGGC | gi15242459 | Mitochondrial HSO70 2 | 79−70 | 1.66 (roots) | ||
| CCACGTGTT | gi79326500 | Putative cinnamyl alcohol dehydrogenase 9 | 959−950 | 2.95 (aerial parts) | Lignification (90) | |
| CCACGTGTC | gi15227259 | Cyclophilin ROC3 | 75−66 | 0.43 (roots) | Defense (93) | |
| gi15234637 | Photosystem II subunit Q-2 | 78−69 | 1.45 (aerial parts) | Photosystem II organization (94) | ||
| CTACGTGTC | gi18406229 | TRAF-like protein | 155−146 | 1.45 (roots) | Unknown | |
| GCACGTGTC | gi15238217 | Sulfite reductase | 91−82 | 0.43 (roots) | Sulfate assimilation, growth and development (95) | |
| CTACGTGTT | gi15242451 | AIG2-like protein | 62−53 | 2.72 (roots) | Unknown | |
| CCACGTGTG | gi18417239 | Photosystem II reaction center PSB28 protein | 164−155 | 2.79 (aerial parts) | Photosystem II assembly (102) | |
DISCUSSION
Although proteomics are effective in identification of proteins regulating cytoskeletal organization, it has been only rarely exploited for this aim in current plant-oriented research. Targeted proteomic approaches were used to identify microtubule-binding proteins (42), whereas proteomic analysis of detergent-resistant and -sensitive membranous fractions combined with cytoskeletal inhibitor treatments revealed the cytoskeleton-dependent distribution of plasma membrane proteins (43). Recently, changes in the abundance of proteins involved in the vesicular transport, RNA nuclear export, and ABA response were reported in response to the actin-depolymerizing drug latrunculin B (44). In mammalian cells, where microtubule severing is more complex as in plants, a proteomic approach was adopted to define the protein interaction module consisting of katanin, katanin-like protein isoforms, and various microtubule-associated proteins (28). However in plants, neither altered cytoskeletal protein profiles nor cross-talk to the actin cytoskeleton was reported in the mutants defective in microtubule-associated protein so far. This study combining genetic (mutants), proteomic, biochemical, and cell biological approaches provides new evidence that defects in microtubule severing by KATANIN 1 caused significant changes in abundances of tubulins, MAP, actin, and ABPs concomitant to global reorganization of microtubules and actin cytoskeleton. In this way, this study revealed a feedback mechanism in the regulation of microtubule organization and uncovered a novel cross-talk mechanism between microtubule and actin cytoskeleton in Arabidopsis.
Defective Microtubules and Actin Cytoskeleton in KATANIN 1 Mutants
It is generally accepted that defective microtubule severing impairs cell elongation and promotes isotropic growth leading to profound phenotypic manifestations in KATANIN 1 mutants (20, 21, 26). KATANIN 1 is activated by Rho GTPase ROP6 via binding to the activator RIC1 (19). Microtubule severing by KATANIN 1 is also regulated by the microtubule-associated protein SPIRAL2 defining where the severing occurs (18). Our proteomic analysis revealed that roots of fra2 mutant exerted a decreased abundance of TUB4 and increased abundance of TSN1, TSN2, as well as microtubule-destabilizing protein MDP25.
TSN proteins are components of cytoplasmic messenger ribonucleoprotein (mRNP) complexes called stress granules. Such stress granules are sites of post-transcriptional gene silencing, and TSN proteins are important for stress-induced mRNA decapping thus modulating the abiotic stress responses. Importantly, stress granules assemble under stress conditions in a KATANIN 1-dependent manner, and TSN proteins co-localize and move along cortical microtubules (32). This might suggest their potential role in the regulation of microtubule organization. Our data indicate that abundances and subcellular accumulation of TSN proteins are dependent on microtubule severing. MDP25 is a plasma membrane-associated protein, which dissociates from the membrane in a calcium-dependent manner exerting inhibition of microtubule polymerization (37). Interestingly, MDP25 overexpression leads to reduced cell elongation and cortical microtubule reorientation (37), similarly to KATANIN 1 mutants. Along with reorganization of microtubules in both KATANIN 1 mutants, it suggests a feedback microtubule control and possible link between MDP25 and microtubule severing in Arabidopsis. Moreover, MDP25 is also capable of calcium-dependent binding to F-actin and its severing (38). Another link to the actin cytoskeleton is provided by co-expression of TSN2 with ACTIN7 (Fig. 4). Our proteomic analysis revealed alterations in actin and important ABPs (PRF1 and ADF3) in KATANIN 1 mutants, very likely contributing to disturbances in the actin organization. This was consistent with reoriented and distorted actin filaments, which we observed in leaf epidermal cells of fra2 and ktn1-2 mutants. Moreover, we found up-regulation of annexin 1 in aerial parts of both mutants. Interestingly, phosphorylation of annexin A2 (closely related to annexin 1) was found to be essential for actin cytoskeleton dynamics (45). Cross-talk between microtubule and the actin cytoskeleton was widely documented (46–48). Quantitative live cell imaging showed that reorganization and reassembly of actin microfilaments is dependent on microtubules following drug-induced depolymerization (49). Altered actin organization related to microtubule severing was not described yet. Here, we show that disturbed microtubule severing in the ktn1-2 mutant might also have direct impact on the actin organization, which is mediated by increased abundances of MDP25 and profilin 1 but decreased abundance of ADF3. In fact, the equilibrium of profilins and ADFs directs the actin filaments to polymerization or depolymerization (50). MDP25 is a promising candidate potentially linking KATANIN 1 with actin microfilaments. Further targeted analyses would be beneficial to experimentally study interactions between katanin and ABPs.
Katanin Mutants Exerts Aberrant Nuclear Shape Likely Caused by Deregulation of WPP2 and HSC70–1
Roots of fra2 mutant possess substantially decreased levels of WPP domain-containing protein 2 (WPP2). WPP2 is localized to the nuclear envelope in interphase cells and to immature cell plates during cytokinesis (39). This was consistent with changed nuclear shapes in both KATANIN 1 mutants as revealed by DAPI staining. WPP proteins bind to WPP domain-interacting tail-anchored protein (WITs) and facilitate their nuclear envelope targeting (51). The same binding and targeting activity were assigned to heat shock cognate protein 70-1 (HSP70-1), which also interacts with WPPs (51). HSP70-1, unlike WPP2, showed an increased abundance in the fra2 roots, suggesting an altered equilibrium in these two mechanisms of WIT nuclear targeting. WITs are constituents of plant Klarsicht/ANC-1/Syne-1 homology (KASH)–Sad1/UNC-84 (SUN) complex controlling nuclear morphology and movement (52). Recently, it was shown that WIT2 proteins interact and recruit myosin XI-i to the nuclear envelope and link KASH–SUN complexes with actin cytoskeleton (53), which is reorganized in the KATANIN 1 mutants. Our results suggest that actin-dependent nuclear shape control in plants through WPP2 and WIT might be linked to microtubule severing by KATANIN 1.
Proteins Associated with Cell Plate Alterations in KATANIN 1 Mutants
In dividing cells, WPP2 might contribute to the emergence of obliquely oriented and misaligned cell plates in KATANIN 1 mutants (this study and Ref. 21), which was tested here by co-immunolocalization of KNOLLE, a bona fide cell plate marker (34), and microtubules in the phragmoplast. Additionally, patellin 2 (down-regulated in the ktn1-2 mutant roots) is a cell plate associated protein involved in the maturation of the cell plate (54). Recently, it was identified as a phosphorylation target of mitogen-activated protein kinase 4 (MPK4), whereas patellin phosphorylation by MPK4 altered its binding to phosphoinositides (55). The role of MPK4 in cell division is more complex, because it is also involved in phragmoplast formation through phosphorylation of phragmoplast-localized microtubule-associated protein MAP65-1 (9, 56–58). Together, WPP2 and patellin 2 may represent protein candidates co-regulated with KATANIN 1 during cytokinesis. In conclusion, phenotypes of fra2 and ktn1-2 mutants are likely determined by wider deregulation of developmentally important cytoskeletal and cell plate proteins.
Proteins Associated with Hormone Homeostasis in KATANIN 1 Mutants
It is well known that microtubule organization is responsive to plant hormones (40). KATANIN 1 was proposed as a mediator of GA- and ethylene-induced microtubule rearrangements in Arabidopsis (26). Mutant lue1 exhibits increased AtGA20ox1 expression levels, a key oxidase enzyme in the gibberellin biosynthesis. Hormonal responses of lue1 to ethylene and gibberellins caused inappropriate cortical microtubule reorientation during cell growth (22). Other proteins regulating GA biosynthesis in Arabidopsis, like TSN1 and TSN2 and glycine-rich RNA-binding protein 7 (GRP7) (59, 60), were differentially abundant in KATANIN 1 mutants. Although TSN1 expression positively correlates with AtGA20ox1 in Arabidopsis (59), GRP7 appears to have a negative role (60).
In fra2 we encountered also changed abundances of proteins involved in ethylene biosynthesis. Thus, 1-aminocyclopropane-1-carboxylate oxidase 2, catalyzing oxygen-dependent conversion of 1-aminocyclopropane-1-carboxylic acid to ethylene (61), was up-regulated. Moreover, 14-3-3-like protein GF14ω, found in Col-0 but not in the KATANIN 1 mutants, was reported to control ethylene synthesis through down-regulation of ubiquitin ligases targeting 1-aminocyclopropane-1-carboxylate synthase for degradation (62). In contrast, enzymes controlling synthesis of ethylene precursor methionine, such as methionine synthase, S-adenosylmethionine synthetase 4, as well as 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase (cobalamin-independent methionine synthase) (63), were down-regulated. Methionine is a precursor of S-adenosyl-l-methionine being a major methyl group donor for trans-methylation reactions (63). Adenosine kinase maintaining general S-adenosyl-l-methionine-dependent methylation activities (64, 65) was up-regulated in the fra2 mutant.
Our results point also to altered auxin homeostasis in KATANIN 1 mutants. S-Alkyl-thiohydroximate lyase SUR1 (also known as SUPERROOT1), a protein involved in auxin biosynthesis, was detected only in wild-type roots suggesting down-regulation in the KATANIN 1 mutants. Suppression of SUR1 results in heavy accumulation of auxin in Arabidopsis (66), and thus lower abundance in KATANIN 1 mutants may indicate some defects in auxin homeostasis. This might be also influenced by pyruvate dehydrogenase E1 component subunit α-2, also called IAA-ALANINE-RESISTANT 4 (IAR4), which was strongly decreased in abundance in both KATANIN 1 mutants, and it is implicated in IAA homeostasis (67, 68). Altogether, we show that, except for ethylene and GA, microtubule severing is likely linked to homeostasis of abscisic acid and auxin. The mechanism of hormonal regulation by KATANIN 1 is not known. One possible explanation is suggested by changes in enzymes responsible for methylation in the KATANIN 1 mutants, because they have been shown to control hormone homeostasis in plants (69).
Because of the substantial impact on agriculturally important crop traits, cytoskeleton and cytoskeleton-associated proteins serve as perspective subjects of genetic engineering for biotechnological applications (70). Our study strengthens this view and provides proteome framework for developmental defects related to KATANIN 1 function. Thus, genetic modification of KATANIN 1 may be considered as a tool to modify plant growth and development.
In conclusion, genetic disruption of microtubule severing in fra2 and ktn1-2 mutants shows a strong impact on the abundance of tubulins, MAP, actin, and ABPs and on the organization of both microtubules and actin filaments. Thus, our study opens a door to investigate these new aspects of feedback microtubule control and cross-talk between microtubules and actin cytoskeleton in plants involving KATANIN 1.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (71) partner repository with the dataset identifier PXD005917 (http://www.ebi.ac.uk/pride/archive/). In addition to “.raw” data files, the “.msf” results files areavailable to download. They can be viewed free of charge using Proteome Discoverer demo/viewer (https://portal.thermo-brims.com/).
Supplementary Material
Acknowledgments
We thank George Komis and Miroslav Ovečka for critical reading of the manuscript and George Komis for help with CytoSpectre software. Tony Arick created computer script to sum ion intensities for multiple samples. We thank Dr. Masayoshi Nakamura for kindly providing ktn1-2 seeds and Drs. Ioannis Adamakis and Emanuel Panteris for providing fra2 seeds. We also thank Dr. Panagiotis Moschou and Prof. Gerd Juergens for kindly providing anti-TSN1/2 and anti-KNOLLE primary antibodies. The mass spectrometry proteomics analysis was performed at the Institute for Genomics, Biocomputing and Biotechnology, Mississippi State University, with partial support from Mississippi Agriculture and Forestry Experimental Station.
Footnotes
Author contributions: J. S. designed research; T. T., O. S., T. P., and I. L. performed research; T. T., O. S., and T. P. analyzed data; T. T., O. S., and J. S. wrote the paper. All authors reviewed the manuscript.
* This work was supported by Grant 15-19284S from the Czech Science Foundation GAČR and National Institutes of Health-INBRE Award 4P20GM103476-15. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- MAP
- microtubule-associated protein
- ABA
- abscisic acid
- ABP
- actin-binding protein
- ADF
- actin-depolymerizing factor
- GA
- giberellic acid
- IAA
- indole-3-acetic acid
- MDP25
- microtubule-destabilizing protein 25
- TUB4
- TUBULIN β-4
- TSN
- TUDOR-staphylococcal nuclease protein
- WPP2
- WPP domain-containing protein 2
- FDR
- false discovery rate
- BSE
- m-maleimidobenzoyl-N-hydroxysuccinimide ester
- ANOVA
- analysis of variance
- PII
- precursor ion intensity
- M-MLV
- Moloney murine leukemia virus
- GO
- gene ontology.
REFERENCES
- 1. Cyr R. J. (1994) Microtubules in plant morphogenesis: role of the cortical array. Annu. Rev. Cell Biol. 10, 153–180 [DOI] [PubMed] [Google Scholar]
- 2. Kost B., Mathur J., and Chua N.-H. (1999) Cytoskeleton in plant development. Curr. Opin. Plant Biol. 2, 462–470 [DOI] [PubMed] [Google Scholar]
- 3. Goddard R. H., Wick S. M., Silflow C. D., and Snustad D. P. (1994) Microtubule components of the plant cell cytoskeleton. Plant Physiol. 104, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wasteneys G. O., and Ambrose J. C. (2009) Spatial organization of plant cortical microtubules: close encounters of the 2D kind. Trends Cell Biol. 19, 62–71 [DOI] [PubMed] [Google Scholar]
- 5. Sedbrook J. C., and Kaloriti D. (2008) Microtubules, MAPs and plant directional cell expansion. Trends Plant Sci. 13, 303–310 [DOI] [PubMed] [Google Scholar]
- 6. Hamada T. (2007) Microtubule-associated proteins in higher plants. J. Plant Res. 120, 79–98 [DOI] [PubMed] [Google Scholar]
- 7. Gardiner J. (2013) The evolution and diversification of plant microtubule-associated proteins. Plant J. 75, 219–229 [DOI] [PubMed] [Google Scholar]
- 8. Šamajová O., Komis G., and Šamaj J. (2013) Emerging topics in the cell biology of mitogen-activated protein kinases. Trends Plant Sci. 18, 140–148 [DOI] [PubMed] [Google Scholar]
- 9. Beck M., Komis G., Müller J., Menzel D., and Samaj J. (2010) Arabidopsis homologs of nucleus- and phragmoplast-localized kinase 2 and 3 and mitogen-activated protein kinase 4 are essential for microtubule organization. Plant Cell 22, 755–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Müller J., Beck M., Mettbach U., Komis G., Hause G., Menzel D., and Samaj J. (2010) Arabidopsis MPK6 is involved in cell division plane control during early root development, and localizes to the pre-prophase band, phragmoplast, trans-Golgi network and plasma membrane. Plant J. 61, 234–248 [DOI] [PubMed] [Google Scholar]
- 11. Komatsu S., Yang G., Khan M., Onodera H., Toki S., and Yamaguchi M. (2007) Over-expression of calcium-dependent protein kinase 13 and calreticulin interacting protein 1 confers cold tolerance on rice plants. Mol. Genet. Genomics 277, 713–723 [DOI] [PubMed] [Google Scholar]
- 12. Yalovsky S., Bloch D., Sorek N., and Kost B. (2008) Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol. 147, 1527–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Q., Lin F., Mao T., Nie J., Yan M., Yuan M., and Zhang W. (2012) Phosphatidic acid regulates microtubule organization by interacting with MAP65–1 in response to salt stress in Arabidopsis. Plant Cell 24, 4555–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hartman J. J., Mahr J., McNally K., Okawa K., Iwamatsu A., Thomas S., Cheesman S., Heuser J., Vale R. D., and McNally F. J. (1998) Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell 93, 277–287 [DOI] [PubMed] [Google Scholar]
- 15. Stoppin-Mellet V., Gaillard J., Timmers T., Neumann E., Conway J., and Vantard M. (2007) Arabidopsis katanin binds microtubules using a multimeric microtubule-binding domain. Plant Physiol. Biochem. 45, 867–877 [DOI] [PubMed] [Google Scholar]
- 16. Nakamura M. (2015) Microtubule nucleating and severing enzymes for modifying microtubule array organization and cell morphogenesis in response to environmental cues. New Phytol. 205, 1022–1027 [DOI] [PubMed] [Google Scholar]
- 17. Stoppin-Mellet V., Gaillard J., and Vantard M. (2006) Katanin's severing activity favors bundling of cortical microtubules in plants. Plant J. 46, 1009–1017 [DOI] [PubMed] [Google Scholar]
- 18. Wightman R., Chomicki G., Kumar M., Carr P., and Turner S. R. (2013) SPIRAL2 determines plant microtubule organization by modulating microtubule severing. Curr. Biol. 23, 1902–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin D., Cao L., Zhou Z., Zhu L., Ehrhardt D., Yang Z., and Fu Y. (2013) Rho GTPase signaling activates microtubule severing to promote microtubule ordering in Arabidopsis. Curr. Biol. 23, 290–297 [DOI] [PubMed] [Google Scholar]
- 20. Bichet A., Desnos T., Turner S., Grandjean O., and Höfte H. (2001) BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25, 137–148 [DOI] [PubMed] [Google Scholar]
- 21. Burk D. H., Liu B., Zhong R., Morrison W. H., and Ye Z. H. (2001) A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827 [PMC free article] [PubMed] [Google Scholar]
- 22. Meier C., Bouquin T., Nielsen M. E., Raventos D., Mattsson O., Rocher A., Schomburg F., Amasino R. M., and Mundy J. (2001) Gibberellin response mutants identified by luciferase imaging. Plant J. 25, 509–519 [DOI] [PubMed] [Google Scholar]
- 23. Luptovčiak I., Samakovli D., Komis G., and Šamaj J. (2017) KATANIN 1 is essential for embryogenesis and seed formation in Arabidopsis. Front. Plant Sci. 8, 728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Panteris E., Adamakis I.-D., Voulgari G., and Papadopoulou G. (2011) A role for katanin in plant cell division: microtubule organization in dividing root cells of fra2 and lue1 Arabidopsis thaliana mutants. Cytoskeleton 68, 401–413 [DOI] [PubMed] [Google Scholar]
- 25. Komis G., Luptovčiak I., Ovečka M., Samakovli D., Šamajová O and Šamaj J. (2017) Katanin effects on dynamics of cortical microtubules and mitotic arrays in Arabidopsis thaliana revealed by advanced live-cell imaging. Front. Plant Sci. 8, 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bouquin T., Mattsson O., Naested H., Foster R., and Mundy J. (2003) The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J. Cell Sci. 116, 791–801 [DOI] [PubMed] [Google Scholar]
- 27. Vasconcelos E. J. R., Pacheco A. C. L., Gouveia J. J. S., Araujo F. F., Diniz M. C., Kamimura M. T., Costa M. P., Maggioni R., Araujo-Filho R., Costa R. B., and de Oliveira D. M. (2007) Profilins, Formins and Katanins as Flagellar Proteins of Leishmania spp.: A Genome-based, Multi-step Bioinformatics-driven Description. October 14–17, 2007, Boston, MA 2007 IEEE 7th International Symposium on BioInformatics and BioEngineering, pp. 880–887, IEEE, New York [Google Scholar]
- 28. Cheung K., Senese S., Kuang J., Bui N., Ongpipattanakul C., Gholkar A., Cohn W., Capri J., Whitelegge J. P., and Torres J. Z. (2016) Proteomic analysis of the mammalian Katanin Family of microtubule-severing enzymes defines Katanin p80 subunit B-like 1 (KATNBL1) as a regulator of mammalian Katanin microtubule-severing. Mol. Cell. Proteomics 15, 1658–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hajduch M., Ganapathy A., Stein J. W., and Thelen J. J. (2005) A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol. 137, 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Conesa A., and Götz S. (2008) Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gutierrez-Beltran E., Moschou P. N., Smertenko A. P., and Bozhkov P. V. (2015) Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 27, 926–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smékalová V., Luptovčiak I., Komis G., Šamajová O., Ovečka M., Doskočilová A., Takáč T., Vadovič P., Novák O., Pechan T., Ziemann A., Košútová P., and Šamaj J. (2014) Involvement of YODA and mitogen activated protein kinase 6 in Arabidopsis post-embryogenic root development through auxin up-regulation and cell division plane orientation. New Phytol. 203, 1175–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lauber M. H., Waizenegger I., Steinmann T., Schwarz H., Mayer U., Hwang I., Lukowitz W., and Jürgens G. (1997) The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139, 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kartasalo K., Pölönen R.-P., Ojala M., Rasku J., Lekkala J., Aalto-Setälä K., and Kallio P. (2015) CytoSpectre: a tool for spectral analysis of oriented structures on cellular and subcellular levels. BMC Bioinformatics 16, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Panteris E., Apostolakos P., and Galatis B. (2006) Cytoskeletal asymmetry in Zea mays subsidiary cell mother cells: a monopolar prophase microtubule half-spindle anchors the nucleus to its polar position. Cell Motil. Cytoskeleton 63, 696–709 [DOI] [PubMed] [Google Scholar]
- 37. Li J., Wang X., Qin T., Zhang Y., Liu X., Sun J., Zhou Y., Zhu L., Zhang Z., Yuan M., and Mao T. (2011) MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis. Plant Cell 23, 4411–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qin T., Liu X., Li J., Sun J., Song L., and Mao T. (2014) Arabidopsis microtubule-destabilizing protein 25 functions in pollen tube growth by severing actin filaments. Plant Cell 26, 325–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patel S., Rose A., Meulia T., Dixit R., Cyr R. J., and Meier I. (2004) Arabidopsis WPP-domain proteins are developmentally associated with the nuclear envelope and promote cell division. Plant Cell 16, 3260–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shibaoka H. (1994) Plant hormone-induced changes in the orientation of cortical microtubules–alterations in the cross-linking between microtubules and the plasma-membrane. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45, 527–544 [Google Scholar]
- 41. Gallardo K., Job C., Groot S. P., Puype M., Demol H., Vandekerckhove J., and Job D. (2002) Proteomics of Arabidopsis seed germination. A comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol. 129, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hamada T., Nagasaki-Takeuchi N., Kato T., Fujiwara M., Sonobe S., Fukao Y., and Hashimoto T. (2013) Purification and characterization of novel microtubule-associated proteins from Arabidopsis cell suspension cultures. Plant Physiol. 163, 1804–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szymanski W. G., Zauber H., Erban A., Gorka M., Wu X. N., and Schulze W. X. (2015) Cytoskeletal components define protein location to membrane microdomains. Mol. Cell. Proteomics 14, 2493–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Takáč T., Bekešová S., and Šamaj J. (2017) Actin depolymerization-induced changes in proteome of Arabidopsis roots. J. Proteomics 153, 89–99 [DOI] [PubMed] [Google Scholar]
- 45. de Graauw M., Tijdens I., Smeets M. B., Hensbergen P. J., Deelder A. M., and van de Water B. (2008) Annexin A2 phosphorylation mediates cell scattering and branching morphogenesis via cofilin activation. Mol. Cell. Biol. 28, 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blancaflor E. B. (2000) Cortical actin filaments potentially interact with cortical microtubules in regulating polarity of cell expansion in primary roots of maize (Zea mays L.). J. Plant Growth Regul. 19, 406–414 [DOI] [PubMed] [Google Scholar]
- 47. Collings D. A. (2008) in Plant Microtubules–Development and Flexibility (Nick P., ed) pp. 47–82, Springer-Verlag, Berlin [Google Scholar]
- 48. Havelková L., Nanda G., Martinek J., Bellinvia E., Sikorová L., Šlajcherová K., Seifertová D., Fischer L., Fišerová J., Petrášek J., and Schwarzerová K. (2015) Arp2/3 complex subunit ARPC2 binds to microtubules. Plant Sci. 241, 96–108 [DOI] [PubMed] [Google Scholar]
- 49. Sampathkumar A., Lindeboom J. J., Debolt S., Gutierrez R., Ehrhardt D. W., Ketelaar T., and Persson S. (2011) Live cell imaging reveals structural associations between the actin and microtubule cytoskeleton in Arabidopsis. Plant Cell 23, 2302–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li J., Blanchoin L., and Staiger C. J. (2015) Signaling to actin stochastic dynamics. Annu. Rev. Plant Biol. 66, 415–440 [DOI] [PubMed] [Google Scholar]
- 51. Brkljacic J., Zhao Q., and Meier I. (2009) WPP-domain proteins mimic the activity of the HSC70–1 chaperone in preventing mistargeting of RanGAP1-anchoring protein WIT1. Plant Physiol. 151, 142–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou X., Graumann K., Evans D. E., and Meier I. (2012) Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J. Cell Biol. 196, 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou X., Groves N. R., and Meier I. (2015) Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1. Nucleus 6, 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peterman T. K., Ohol Y. M., McReynolds L. J., and Luna E. J. (2004) Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol. 136, 3080–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suzuki T., Matsushima C., Nishimura S., Higashiyama T., Sasabe M., and Machida Y. (2016) Identification of phosphoinositide-binding protein PATELLIN2 as a substrate of Arabidopsis MPK4 MAP kinase during septum formation in cytokinesis. Plant Cell Physiol. 57, 1744–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smertenko A. P., Chang H.-Y., Sonobe S., Fenyk S. I., Weingartner M., Bögre L., and Hussey P. J. (2006) Control of the AtMAP65–1 interaction with microtubules through the cell cycle. J. Cell Sci. 119, 3227–3237 [DOI] [PubMed] [Google Scholar]
- 57. Kosetsu K., Matsunaga S., Nakagami H., Colcombet J., Sasabe M., Soyano T., Takahashi Y., Hirt H., and Machida Y. (2010) The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell 22, 3778–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Beck M., Komis G., Ziemann A., Menzel D., and Samaj J. (2011) Mitogen-activated protein kinase 4 is involved in the regulation of mitotic and cytokinetic microtubule transitions in Arabidopsis thaliana. New Phytol. 189, 1069–1083 [DOI] [PubMed] [Google Scholar]
- 59. Yan C., Yan Z., Wang Y., Yan X., and Han Y. (2014) Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J. Exp. Bot. 65, 5933–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Löhr B., Streitner C., Steffen A., Lange T., and Staiger D. (2014) A glycine-rich RNA-binding protein affects gibberellin biosynthesis in Arabidopsis. Mol. Biol. Rep. 41, 439–445 [DOI] [PubMed] [Google Scholar]
- 61. Ruduś I., Sasiak M., and Kępczyński J. (2013) Regulation of ethylene biosynthesis at the level of 1-aminocyclopropane-1-carboxylate oxidase (ACO) gene. Acta Physiol. Plant. 35, 295–307 [Google Scholar]
- 62. Yoon G. M., and Kieber J. J. (2013) 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell 25, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ravanel S., Gakière B., Job D., and Douce R. (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. U.S.A. 95, 7805–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moffatt B. A., Stevens Y. Y., Allen M. S., Snider J. D., Pereira L. A., Todorova M. I., Summers P. S., Weretilnyk E. A., Martin-McCaffrey L., and Wagner C. (2002) Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol. 128, 812–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Weretilnyk E. A., Alexander K. J., Drebenstedt M., Snider J. D., Summers P. S., and Moffatt B. A. (2001) Maintaining methylation activities during salt stress. The involvement of adenosine kinase. Plant Physiol. 125, 856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Boerjan W., Cervera M. T., Delarue M., Beeckman T., Dewitte W., Bellini C., Caboche M., Van Onckelen H., Van Montagu M., and Inzé D. (1995) Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7, 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. LeClere S., Rampey R. A., and Bartel B. (2004) IAR4, a gene required for auxin conjugate sensitivity in Arabidopsis, encodes a pyruvate dehydrogenase E1α homolog. Plant Physiol. 135, 989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Quint M., Barkawi L. S., Fan K.-T., Cohen J. D., and Gray W. M. (2009) Arabidopsis IAR4 modulates auxin response by regulating auxin homeostasis. Plant Physiol. 150, 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Westfall C. S., Muehler A. M., and Jez J. M. (2013) Enzyme action in the regulation of plant hormone responses. J. Biol. Chem. 288, 19304–19311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Komis G., Luptovciak I., Doskocilova A., and Samaj J. (2015) Biotechnological aspects of cytoskeletal regulation in plants. Biotechnol. Adv. 33, 1043–1062 [DOI] [PubMed] [Google Scholar]
- 71. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.-W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dalal J., Lewis D. R., Tietz O., Brown E. M., Brown C. S., Palme K., Muday G. K., and Sederoff H. W. (2016) ROSY1, a novel regulator of gravitropic response is a stigmasterol binding protein. J. Plant Physiol. 196, 28–40 [DOI] [PubMed] [Google Scholar]
- 73. Nishimura N., Sarkeshik A., Nito K., Park S.-Y., Wang A., Carvalho P. C., Lee S., Caddell D. F., Cutler S. R., Chory J., Yates J. R., and Schroeder J. I. (2010) PYR/PYL/RCAR family members are major in vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61, 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sjögren L. L., MacDonald T. M., Sutinen S., and Clarke A. K. (2004) Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 136, 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Weijers D., Franke-van Dijk M., Vencken R. J., Quint A., Hooykaas P., and Offringa R. (2001) An Arabidopsis minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128, 4289–4299 [DOI] [PubMed] [Google Scholar]
- 76. Romani I., Tadini L., Rossi F., Masiero S., Pribil M., Jahns P., Kater M., Leister D., and Pesaresi P. (2012) Versatile roles of Arabidopsis plastid ribosomal proteins in plant growth and development. Plant J. 72, 922–934 [DOI] [PubMed] [Google Scholar]
- 77. De Castro R. D., Zheng X., Bergervoet J., De Vos C., and Bino R. J. (1995) β-Tubulin accumulation and DNA replication in imbibing tomato seeds. Plant Physiol. 109, 499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sundström J. F., Vaculova A., Smertenko A. P., Savenkov E. I., Golovko A., Minina E., Tiwari B. S., Rodriguez-Nieto S., Zamyatnin A. A. Jr., Välineva T., Saarikettu J., Frilander M. J., Suarez M. F., Zavialov A., Ståhl U., et al. (2009) Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat. Cell Biol. 11, 1347–1354 [DOI] [PubMed] [Google Scholar]
- 79. Ramachandran S., Christensen H. E., Ishimaru Y., Dong C. H., Chao-Ming W., Cleary A. L., and Chua N. H. (2000) Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol. 124, 1637–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sheoran I. S., Olson D. J., Ross A. R., and Sawhney V. K. (2005) Proteome analysis of embryo and endosperm from germinating tomato seeds. Proteomics 5, 3752–3764 [DOI] [PubMed] [Google Scholar]
- 81. Augustine R. C., Vidali L., Kleinman K. P., and Bezanilla M. (2008) Actin-depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J. 54, 863–875 [DOI] [PubMed] [Google Scholar]
- 82. Gilliland L. U., Pawloski L. C., Kandasamy M. K., and Meagher R. B. (2003) Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J. 33, 319–328 [DOI] [PubMed] [Google Scholar]
- 83. Gallardo K., Job C., Groot S. P., Puype M., Demol H., Vandekerckhove J., and Job D. (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol. 126, 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ma B., Qian D., Nan Q., Tan C., An L., and Xiang Y. (2012) Arabidopsis vacuolar H+-ATPase (V-ATPase) B subunits are involved in actin cytoskeleton remodeling via binding to, bundling, and stabilizing F-actin. J. Biol. Chem. 287, 19008–19017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhao Z., and Assmann S. M. (2011) The glycolytic enzyme, phosphoglycerate mutase, has critical roles in stomatal movement, vegetative growth, and pollen production in Arabidopsis thaliana. J. Exp. Bot. 62, 5179–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., and Yamaguchi S. (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15, 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Du J., Huang Y.-P., Xi J., Cao M.-J., Ni W.-S., Chen X., Zhu J.-K., Oliver D. J., and Xiang C.-B. (2008) Functional gene-mining for salt-tolerance genes with the power of Arabidopsis. Plant J. 56, 653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Matsushima R., Fukao Y., Nishimura M., and Hara-Nishimura I. (2004) NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. Plant Cell 16, 1536–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lee S.-H., Li C.-W., Koh K. W., Chuang H.-Y., Chen Y.-R., Lin C.-S., and Chan M.-T. (2014) MSRB7 reverses oxidation of GSTF2/3 to confer tolerance of Arabidopsis thaliana to oxidative stress. J. Exp. Bot. 65, 5049–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Eudes A., Pollet B., Sibout R., Do C.-T., Séguin A., Lapierre C., and Jouanin L. (2006) Evidence for a role of AtCAD 1 in lignification of elongating stems of Arabidopsis thaliana. Planta 225, 23–39 [DOI] [PubMed] [Google Scholar]
- 91. Kovacs D., Kalmar E., Torok Z., and Tompa P. (2008) Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 147, 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. García I., Rosas T., Bejarano E. R., Gotor C., and Romero L. C. (2013) Transient transcriptional regulation of the CYS-C1 gene and cyanide accumulation upon pathogen infection in the plant immune response. Plant Physiol. 162, 2015–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pogorelko G. V., Mokryakova M., Fursova O. V., Abdeeva I., Piruzian E. S., and Bruskin S. A. (2014) Characterization of three Arabidopsis thaliana immunophilin genes involved in the plant defense response against Pseudomonas syringae. Gene 538, 12–22 [DOI] [PubMed] [Google Scholar]
- 94. Allahverdiyeva Y., Suorsa M., Rossi F., Pavesi A., Kater M. M., Antonacci A., Tadini L., Pribil M., Schneider A., Wanner G., Leister D., Aro E.-M., Barbato R., and Pesaresi P. (2013) Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J. 75, 671–684 [DOI] [PubMed] [Google Scholar]
- 95. Khan M. S., Haas F. H., Samami A. A., Gholami A. M., Bauer A., Fellenberg K., Reichelt M., Hänsch R., Mendel R. R., Meyer A. J., Wirtz M., and Hell R. (2010) Sulfite reductase defines a newly discovered bottleneck for assimilatory sulfate reduction and is essential for growth and development in Arabidopsis thaliana. Plant Cell 22, 1216–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hu S., Brady S. R., Kovar D. R., Staiger C. J., Clark G. B., Roux S. J., and Muday G. K. (2000) Technical advance: identification of plant actin-binding proteins by F-actin affinity chromatography. Plant J. 24, 127–137 [DOI] [PubMed] [Google Scholar]
- 97. Davies J. M. (2014) Annexin-mediated calcium signalling in plants. Plants 3, 128–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Takagi D., Ifuku K., Ikeda K., Inoue K. I., Park P., Tamoi M., Inoue H., Sakamoto K., Saito R., and Miyake C. (2016) Suppression of chloroplastic alkenal/one oxidoreductase represses the carbon catabolic pathway in Arabidopsis leaves during night. Plant Physiol. 170, 2024–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Meng M., Geisler M., Johansson H., Harholt J., Scheller H. V., Mellerowicz E. J., and Kleczkowski L. A. (2009) UDP-glucose pyrophosphorylase is not rate limiting, but is essential in Arabidopsis. Plant Cell Physiol. 50, 998–1011 [DOI] [PubMed] [Google Scholar]
- 100. Park J.-I., Ishimizu T., Suwabe K., Sudo K., Masuko H., Hakozaki H., Nou I.-S., Suzuki G., and Watanabe M. (2010) UDP-glucose pyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol. 51, 981–996 [DOI] [PubMed] [Google Scholar]
- 101. López-Castillo L. M., Jiménez-Sandoval P., Baruch-Torres N., Trasviña-Arenas C. H., Díaz-Quezada C., Lara-González S., Winkler R., and Brieba L. G. (2016) Structural basis for redox regulation of cytoplasmic and chloroplastic triosephosphate isomerases from Arabidopsis thaliana. Front. Plant Sci. 7, 1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Mabbitt P. D., Wilbanks S. M., and Eaton-Rye J. J. (2014) Structure and function of the hydrophilic Photosystem II assembly proteins: Psb27, Psb28 and Ycf48. Plant Physiol. Biochem. 81, 96–107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (71) partner repository with the dataset identifier PXD005917 (http://www.ebi.ac.uk/pride/archive/). In addition to “.raw” data files, the “.msf” results files areavailable to download. They can be viewed free of charge using Proteome Discoverer demo/viewer (https://portal.thermo-brims.com/).