Abstract
Objective: Development of the gastrointestinal tract and immune system can be modulated by the gut microbiota. Establishment of the intestinal microbiota, in its turn, is affected by host and environmental factors. As such, development of the gut microbiota is greatly impacted in preterm infants, who have an immature gut and are exposed to factors like hospitalization, caesarean section, antibiotics, and respiratory support.
Design: We analyzed fecal microbiota composition and activity of ten preterm infants (gestational age 25–30 weeks; birthweight 630–1750 g) during the first six postnatal weeks through metaproteomics (LC-MS/MS) and 16S-rRNA gene sequencing.
Results: A gestational-age-dependent microbial signature is observed, enabling microbiota-based differentiation between extremely preterm (25–27 weeks gestation) and very preterm (30 weeks gestation) infants. In very preterm infants, the intestinal microbiota developed toward a Bifidobacterium-dominated community and was associated with high abundance of proteins involved in carbohydrate and energy metabolism. Extremely preterm infants remained predominantly colonized by facultative anaerobes and were associated with proteins involved in membrane transport and translation. Delayed colonization by obligate anaerobes could be associated with antibiotic treatment and respiratory support.
Conclusion: We speculate that gestational age and its associated intensity of care (e.g. antibiotics and respiratory support) affects intestinal microbiota composition and activity in preterm infants. As the gut microbiota plays a major role in development of the neonate, gestational age and its associated factors could set the stage for early and later life health complications via interference with microbiota development.
During birth and rapidly thereafter, microbes colonize the human gastrointestinal (GI) tract and eventually form a stable, adult-like microbial population (1–3). It is generally believed that the first colonizers are facultative anaerobes, primarily Staphylococcus, Streptococcus, Enterococcus, and Enterobacter, who create an anaerobic environment to allow for colonization by obligate anaerobes like Bifidobacterium, Bacteroides, and Clostridium (4). In early life, the intestinal microbiota is dynamic, and its development is highly susceptible to host and environmental factors (5). Establishment of the intestinal microbiota has shown to be greatly impacted by preterm birth (6). An abnormal pattern of microbial colonization is characterized in preterm infants, with high levels of facultative anaerobes and delayed colonization with obligate anaerobes like Bifidobacterium (7–9). Furthermore, it has been shown that dominance of anaerobes occurs around postconceptional age 33–36 weeks, underlining the substantial influence of gestational age (GA) on microbiota development (10). In addition, mode of delivery, type of feeding, and the use of antibiotics are factors that have shown to independently affect microbiota composition in preterm infants (11). More frequent than term infants, preterm infants are exposed to caesarean section, hospitalization, antibiotic treatment, delayed introduction of enteral feeding, and formula feeding, contributing to a bacterial community rich in facultative anaerobes (11). Respiratory support is a potential influencer of microbiota development but is only occasionally mentioned in current studies. Administration of air or an air–oxygen mixture might interfere with microbiota development, particularly with colonization of anaerobic bacteria. Altogether, the preterm infant is very likely to develop an altered intestinal microbiota, which can be associated with adverse early and later life health outcomes. Host–microbe interactions influence GI-tract and immune system development (12), and disturbances in microbiota development have been related to development of disorders like necrotizing enterocolitis, infant colic, atopy, inflammatory bowel disease, and obesity (13–16).
Despite increasing knowledge about microbiota composition in preterm infants, knowledge about the functional signatures of the intestinal microbiota remains limited. A metaproteomics case study of one preterm infant revealed that bacterial activity transits toward more complex metabolic functions in time (17). The temporal increase in functional complexity has been confirmed by metabolomics in a bigger set of preterm infants, in which metabolic complexity was related to weaning (18). The same group also showed an increase in specific metabolites prior to necrotizing enterocolitis diagnosis in preterm infants (19). To our knowledge, no further data are available about microbiota activity during the first weeks of a preterm infant's life. In the present study, 16S-rRNA gene sequencing and metaproteomics are combined to study microbiota development during the first six postnatal weeks in preterm infants and to identify the factors associated with this development.
EXPERIMENTAL PROCEDURES
Subjects and Sample Collection
This study was part of an observational, nonintervention study involving (pre)term infants admitted to the neonatal intensive care unit or the pediatric ward of Isala in Zwolle, the Netherlands. The board from the Medical Ethical Committee of Isala Zwolle concluded that this study does not fall under the scope of the Medical Research Involving Human Subjects Act (WMO). Informed consent was obtained from both parents of all individual participants included in the study. Ten preterm infants were included for fecal microbiota characterization. Five infants (infants A–E) were born extremely preterm (EP, 25–27 weeks gestation) and five (infants F–J) were born very preterm (VP, 30 weeks gestation). Infant clinical characteristics are shown in Table I. Meconium and fecal samples were collected during the first six postnatal weeks. For metaproteomics analysis, meconium and fecal samples collected at week one, two, three, four, and six were used. For infant H, samples collected daily during the first two postnatal weeks were also included for metaproteomics analysis, resulting in a total of 64 samples for LC-MS/MS (Table S1). For 16S-rRNA sequencing, meconium and fecal samples collected daily during the first two postnatal weeks and collected at weeks three, four, and six were used, resulting in 116 samples (Table S1). Samples were stored temporally at −20 °C until transfer to −80 °C.
Table I. Infant clinical characteristics.
| Infant | gender | GA (week + day) | BW (gr) | Delivery mode | FI | #AB | AB1 | AB2 | Maternal AB | Days until full EF* | Human milk (% per week)** | Discharge*** | Respiratory support (days)**** |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Female | 25 + 2 | 680 | Vaginal | Yes | 2 | Amx/Ctz/Va | Ctz/Va | Yes | 10 | 35, 89, 100, 100, 100 | 82 | 76 |
| B | Male | 26 + 4 | 670 | C-section | Yes | 2 | Amx/Ctz/Va | Mem/Va/Erm | No | 17 | 13, 47, 40, 21, 15 | 72 | 74 |
| C | Male | 26 + 6 | 630 | C-section | Yes | 2 | Amx/Ctz/Va | Ctz/Va/Mtz | No | 14 | 15, 63, 100, 100, 100 | 85 | 66 |
| D | Male | 27 + 0 | 1095 | C-section | No | 2 | Amx/Ctz/Erm | Ctz/Va | Yes | 10 | 41, 93, 22, 49, 100 | 61 | 58 |
| E | Male | 27 + 5 | 925 | C-section | Yes | 2 | Amx/Ctz | Ctz/Va | No | 16 | 3, 32, 98, 100, 100 | 64 | 57 |
| F | Female | 30 + 4 | 1260 | C-section | Yes | 1 | Amx/Ctz | - | No | 12 | 7, 71, 100, 100, 93 | 7 | 6 |
| G | Female | 30 + 4 | 1600 | C-section | No | 0 | - | - | No | 10 | 2, 87, 100, 100, 66 | 4 | 4 |
| H | Female | 30 + 4 | 1220 | C-section | No | 1 | Ctz/Va | - | No | 12 | 15, 79, 100, 100, 100 | 10 | 0 |
| I | Male | 30 + 6 | 1750 | Vaginal | No | 1 | Amx/Ctz | - | No | 8 | 45, 100, 100, 100, 100 | 8 | 2 |
| J | Female | 30 + 1 | 1675 | C-section | No | 1 | Amx/Ctz | - | No | 9 | 10, 95, 56, 9, 0 | 13 | 5 |
BW: birth weight, FI: food intolerance, #AB: number of antibiotic treatments, AB1: first course antibiotics, AB2: second course antibiotics, EF: enteral feeding, Amx: amoxicillin, Ctz: ceftazidime, Erm: erythromycin, Mem: meropenem, Mtz: metronidazole, Va: vancomycin.
* Days to reach full enteral feeding (>140 ml/kg/day).
** Percentage of total feeding (enteral + parenteral).
*** Days until discharge.
**** Respiratory support as mechanical ventilation and/or CPAP in days.
Protein Extraction
Proteins were extracted mechanically by repeated bead beating as described previously (20). In short, ∼0.125 g of meconium or feces were resuspended in 375 μl PBS, mixed by vortexing, and covered with gaseous nitrogen. Cells were lysed mechanically by five times bead beating with 0.1 mm zirconia/silica beads using the Precellys®24 instrument at 6.5 ms-1 for 45 s (Bertin Technologies, Montigny le Bretonneux, France). The mixture was centrifuged to remove beads (10,000 g; 4 °C; 5 min) and cell debris (14,000 g; 4 °C; 8 min). Proteins were quantified using the Qubit® Protein Assay Kit on a Qubit®2.0 fluorometer (Life Technologies, Carlsbad, CA).
In Gel-Digestion Procedures
Protein extracts were diluted in PBS to obtain a 3 μg/μl concentration. 40 μl of each sample were mixed with 20 μl loading buffer and subsequently 50 μl were loaded on precast 10% acrylamide gels (PreciseTM Protein Gels, Thermo Scientific, Rockford, IL) using the Mini-PROTEAN® Tetra Electrophoresis System (Bio-Rad Laboratories, Hercules, CA) according to manufacturer instructions. After short electrophoresis (20 mA; 10 min) to allow for the complete sample to enter, the gels were stained with Coomassie Brilliant Blue. Proteins were reduced by incubating the SDS gels in 50 mm dithiothreitol (60 min; 60 °C) while gently shaking. The gels were washed with water, followed by protein alkylation by incubation in 100 mm iodoacetamide (60 min; room temperature). The protein-containing fraction of the gel was cut out with a clean scalpel, placed on parafilm, and further processed into 1 mm2 pieces. In addition, a nonprotein-containing fraction was taken along as negative control. The gel pieces obtained were transferred to a 1.5 ml Eppendorf Protein LoBind tube and placed in 5 ng/μl trypsin solution to allow for in-gel digestion (overnight; room temperature). Protein digests were sonicated and centrifuged (14,000 rpm; 5 min). The pH of the obtained supernatant was adjusted to 2–4 with 10% trifluoroacetic acid. The peptide solutions were desalted and concentrated using in-house made C18 stage tip microcolumns as described previously (21). Sample volumes were reduced to 10 μl using a SpeedVac vacuum centrifuge at 35 °C, and increased to 50 μl with 1 ml/l formic acid in water. Samples were analyzed by nano-LC-LTQ-Orbitrap-MS as previously described (22).
Database Construction
The obtained MS/MS spectra were searched against the publicly available Human Microbiome Project reference genomes from the gastrointestinal tract, containing 457 bacterial genomes (2014, http://www.hmpdacc.org/HMRGD/). A smaller in-house database was constructed to be more representative of the study group and to decrease the chance of false matches. For this database, representative bacterial genera were selected based on the genera identified by using the Human Microbiome Project database or by 454 pyrosequencing. The proteomes of species within these genera were obtained from Uniprot (http://www.uniprot.org/proteomes/) and merged into one database together with the proteomes of human, cow, candida spp. and common contaminants (e.g. trypsin and keratin). This led to an in-house database containing 87 bacterial species and a total size of 438,537 sequences (Table S2).
LC-MS/MS Data Analysis
The mass spectrometry data have been deposited to the ProteomeXchange Consortium (23) via the PRIDE partner repository with dataset identifier PXD005574. Obtained MS/MS spectra were analyzed with MaxQuant 1.3.0.5 (24) using the “Specific Trypsin/P” digestion mode with maximally two missed cleavages, match between runs on in default settings, LFQ on in default settings, and default settings for the Andromeda search engine (first search 20 ppm peptide tolerance, main search 6 ppm tolerance, ion trap MS/MS fragment match tolerance of 0.5 Da, carbamidomethyl set as a fixed modification, while variable modifications were set for protein N-terminal acetylation and M oxidation that were completed by nondefault settings for deamidation of N and Q, the maximum number of modifications per peptide was five) (25). False discovery rates were set to 0.01 on peptide and protein level. Minimally, two peptides were necessary for protein identification, of which at least one is unique and at least one is unmodified. After filtering, 1641 protein groups could be identified, of which 953 were bacterial derived. MaxQuant creates protein groups containing one or more proteins. 1021 protein groups with >2 proteins were created, meaning that those proteins cannot be discriminated based on the measured peptides. In case of ambiguous protein assembly, the protein with the highest peptide count and highest number of unique peptides in its group was selected for further analysis. For each sample, intensity-based absolute quantification (iBAQ) intensities were used for the generation of taxonomic and functional profiles (26). For taxonomic classification, no further ranking than genus level was applied because of high protein sequence homology among species from the same genus. For functional classification, protein identifiers (IDs) were assigned to Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology (KO) identifiers and functionally annotated using the KEGG Brite database on hierarchy level B. When one protein could be classified into multiple functional categories, iBAQ intensity values were balanced between these categories. Sample proteome, KO identifier, taxonomic, and functional profiles and corresponding clinical data were imported in Canoco multivariate statistics software v5 for principal component analysis, redundancy analysis and principal response curve analysis. Here, a p value of less than 0.05 was used as threshold for statistical significance. Analyses were generally performed using Canoco's default settings. Specific settings are described in the figure captions.
DNA Extraction
DNA was extracted from feces by the repeated bead beating plus phenol/chloroform method as described previously (27). DNA was quantified using a NanoDrop ND-2000 spectrophotometer (Thermo Scientific, Wilmington, DE) and by using a Qubit®2.0 fluorometer (Life Technologies, Carlsbad, CA).
454 Pyrosequencing
Amplification of the V3-V5 regions of the 16S-rRNA gene was performed using the Bifidobacterium optimized 357F and 926Rb primers as described previously (28). For each sample, the reverse primer included a unique barcode sequence to allow for demultiplexing. PCR and 454 pyrosequencing (GS Junior, Roche) were performed by LifeSequencing S.L. (Valencia, Spain) as described previously (28). Sequencing data are available in the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under study accession PRJEB18915.
Sequencing Data Analysis
Pyrosequencing data were analyzed using the Quantitative Insights Into Microbial Ecology (QIIME) software package (v1.8) (29). Fasta data were demultiplexed and filtered using default settings. Sequences were denoised using Acacia (30), followed by chimera removal using the Usearch algorithm (31). UCLUST software (32) was used to pick de novo operational taxonomic units (OTUs) with 97% sequence similarity. A representative sequence from each OTU was picked and taxonomy assigned using the SILVA 111 reference database (33) clustered at 97% similarity and complying with the six taxonomic levels of a Ribosomal Database Project classifier. The obtained OTU table was filtered for OTUs with a total observation count of less than two and for OTUs that were present in less than two samples. This resulted in the identification of 2789 OTUs and the remaining of 975,238 sequences, representing 7332 ± 3022 reads per sample (mean ± S.D.).
To compare the fecal microbial communities between and within infants, weighted unifrac distances were determined. The core microbiota was identified using the QIIME compute_core_microbiome.py script. OTUs present in at least 70% of the samples were considered to be part of the core microbiota. To study (dis)similarities in microbiota composition and relate changes in microbiota composition to clinical data, principal component analysis and redundancy analysis were performed using the Canoco multivariate statistics software v5. Specific settings are described in the figure captions.
16S-rRNA Gene Sequence Similarity
All 16S-rRNA gene sequences from members of the Enterobacter (2515) and Klebsiella (1783) genus were downloaded from the SILVA small subunit r126 RefNR database (http://www.arb-silva.de/). Enterobacter sequences were blasted against Klebsiella sequences and vice versa, and the average similarity of the hits was determined.
Experimental Design and Statistical Rationale
This study included ten preterm infants from which fecal samples were collected during the first six postnatal weeks. 64 and 116 samples were included for metaproteomics and 16S-rRNA gene sequencing, respectively. Samples from different infants were collected at similar time points and, in that way, served as biological replicates. In this case, five infants were born EP, and five infants were born VP. Data processing and statistical analysis are described in detail in sections LC-MS/MS Data Analysis and Sequencing Data Analysis.
RESULTS
Microbiota Development Is Highly Variable during the First Two Postnatal Weeks
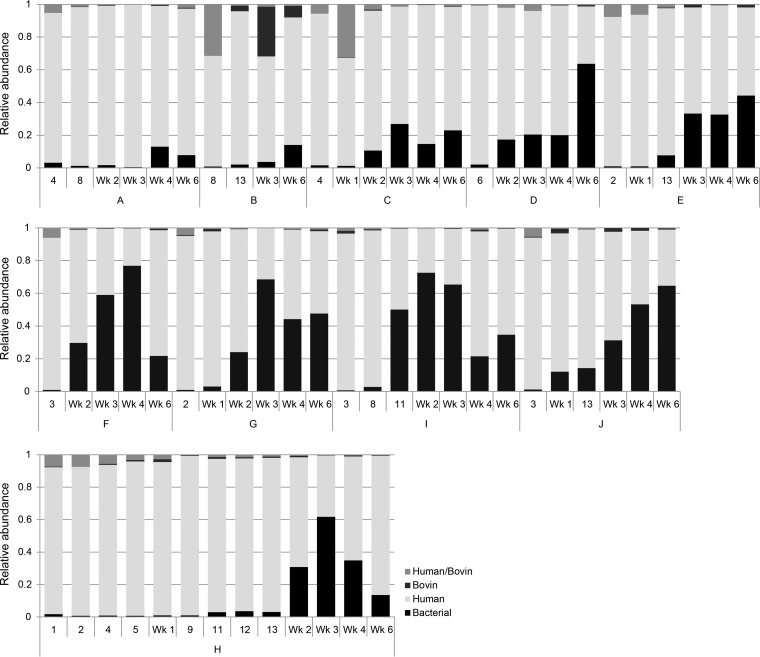
Analyses of the fecal metaproteome revealed that the proportion of bacterial proteins was low (0.7–12.1%) till the second postnatal week in all preterm infants (Fig. 1). Microbiota composition, as determined by 16S-rRNA sequencing, showed high inter-and intraindividual variation during these first two postnatal weeks (Fig. S1A and Fig. S1B). In all infants, the core microbiota consisted of Enterococcus and Staphylococcus, present in 73 and 97% of the samples, respectively (Table II). Other genera, including Propionibacterium and Enterobacter, were identified as highly abundant during the first two postnatal weeks, but their abundances were more sample specific (Fig. S2).
Fig. 1.
Distribution of bacterial-, human-, and bovin-derived fecal proteins. Relative abundances were calculated using iBAQ intensities. Human- and bovin-derived homologous proteins are indicated as Human/Bovin. Mec: meconium, A–J: individual infants.
Table II. Core fraction and relative abundance of the bacterial genera identified in fecal samples during the first two postnatal weeks. Core microbiota and relative abundances of genera were identified using fecal samples from all preterm infants collected during the first two postnatal weeks.
| Phylum | Genus | Core* | Rel. abundance** |
|---|---|---|---|
| Actinobacteria | Bifidobacterium | - | 0.040 ± 0.137 |
| Propionibacterium | - | 0.059 ± 0.128 | |
| Firmicutes | Staphylococcus | 0.97 | 0.338 ± 0.330 |
| Enterococcus | 0.73 | 0.146 ± 0.253 | |
| Streptococcus | - | 0.011 ± 0.036 | |
| Veillonella | - | 0.015 ± 0.046 | |
| Clostridium | - | 0.021 ± 0.084 | |
| Lactobacillales; Other | - | 0.005 ± 0.013 | |
| Proteobacteria | Enterobacter | - | 0.058 ± 0.203 |
| Escherichia-Shigella | - | 0.001 ± 0.003 | |
| Enterobacteriaceae; Other | - | 0.001 ± 0.002 |
* Fraction of samples in which the genus is identified. Genera were considered to belong to the core microbiota when identified in at least 70% of the samples.
** Mean ± S.D.
Gestational Age Is Predictive for Microbial Signatures during Early Microbiota Development
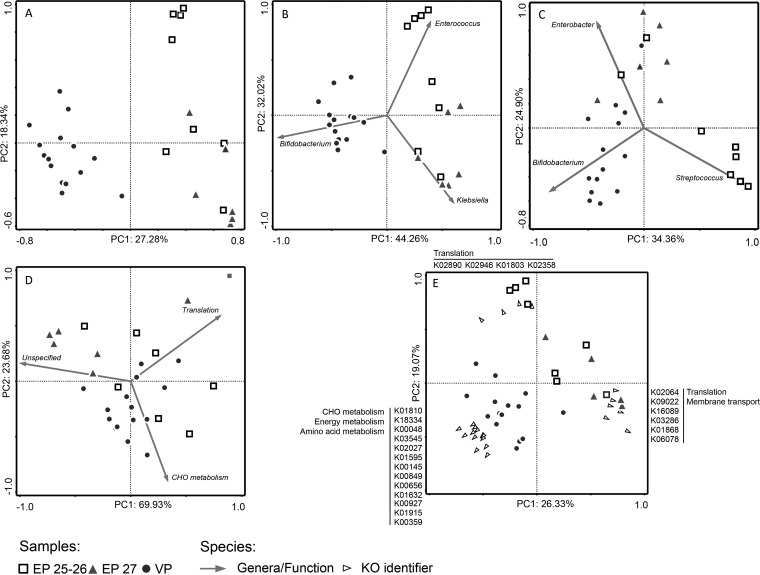
From the third postnatal week onward, the proportion of bacterial-derived proteins rapidly increased (Fig. 1). However, this process was delayed in EP infants (infants A–E). Strikingly, the delay was most obvious in EP infants born at 25–26 weeks gestation (infants A–C). In addition, ordination analysis of the fecal bacterial proteome revealed a clear separation between samples from EP and VP infants but also between samples from EP infants born at 25–26 or 27 weeks gestation (Fig 2A). For further analysis, the EP infants were therefore stratified as born EP at 25–26 or 27 weeks gestation (EP25-26; EP27).
Fig. 2.
Principal component analysis of the fecal bacterial community using (A) proteome profiles, (B) protein-based taxonomic profiles, (C) 16S-rRNA gene-based microbiota profiles, (D) protein-based KEGG Brite functional profiles, and (E) protein-based KEGG orthology identifier profiles. A–D: Taxa and KEGG Brite level B categories fitting into the ordination space from 75–100% are shown. E: KEGG orthology identifiers fitting into the ordination space from 50–100% are shown.
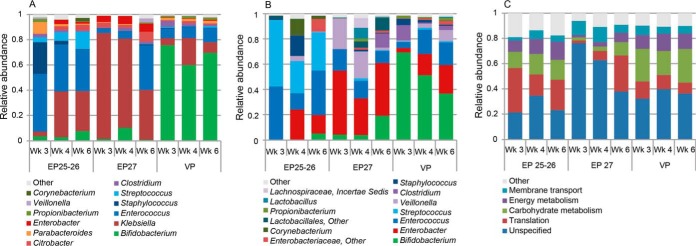
The GA-related separation of the fecal bacterial proteomes by ordination analysis could be explained by taxonomic differences (Fig 2B). VP infants were associated with increased abundance of Bifidobacterium-derived proteins, while EP25-26 and EP27 infants were associated with increased abundance of Enterococcus- and Klebsiella-derived proteins, respectively. These differences remained throughout postnatal weeks 3–6 (Fig. S3A). Such separation of samples could also be observed based on 16S-rRNA sequencing data, associated with the abundance of Streptococcus, Enterobacter, and Bifidobacterium (Fig 2C). However, microbiota composition became more similar over time between EP25-26, EP27, and VP infants (Fig. S3B). In general, EP infants were colonized with a higher proportion of aerobic and facultative anaerobic bacteria compared with VP infants (Fig. S4). Bacterial-protein-based taxonomic classification revealed that the biggest proportion (66–90%) of identified proteins derived from Klebsiella, Bifidobacterium, and Enterococcus (Fig 3A). Based on 16S-rRNA gene sequencing data, Bifidobacterium, Enterobacter, and Enterococcus comprised the most abundant genera (42–87%) (Fig 3B). Blasting revealed that all SILVA-derived 16S-rRNA gene sequences from Enterobacter hit Klebsiella and vice versa, with an average 16S-rRNA gene similarity of 97.7 ± 1.7% and 98.1 ± 1.5% (mean ± S.D.), respectively. This means that these genera cannot be distinguished based on their 16S-rRNA gene sequence, which could lead to misclassification and contributes to dissimilar findings when comparing sequencing data with metaproteomics data. The abundance of Bifidobacterium, Enterobacter/Klebsiella, Enterococcus, Streptococcus, and Clostridium correlated significantly (Spearman correlation p < 0.01) between the protein- and 16S-based approach (Table S3).
Fig. 3.
Protein-based (A) and 16S-based (B) taxonomic profiles and protein-based functional profiles (C) in EP25-26, EP27 and VP infants during postnatal weeks 3–6. Per time point, average relative abundances for each GA group are shown. Relative abundances were calculated using iBAQ intensities and read counts for protein- and 16S-based profiles, respectively. Mec: meconium. Profiles per sample can be found in Fig. S6.
Divergence between Bacterial Activity in Preterm Infants of Varying GA
Fecal bacterial proteins were matched to their corresponding KO ID and could be classified into 21 KEGG Brite functional categories. Proteins involved in translation, carbohydrate (CHO)1 metabolism, energy metabolism, membrane transport, and unspecified processes were most abundant (64–93%) (Fig 3C).
Ordination analysis using the KEGG Brite functional profiles revealed no clear functional differences related to GA (Fig 2D). Similar functional processes were covered by different bacterial genera (Fig. S5). In VP infants, Bifidobacterium was the main genus involved in each functional process. In EP infants, metabolic processes CHO and energy metabolism were predominantly covered by Enterococcus. In EP27 infants, membrane transport proteins were mostly derived from Klebsiella, while in EP25-26 infants these derived from both Klebsiella and Enterococcus. Thus, different bacteria cover similar functional processes, leading to comparable KEGG Brite functional profiles.
Functional (dis)similarities between infants born at varying GA were further explored at the protein level using KO-ID-based profiles. Ordination analysis revealed a clear separation between EP and VP infants (Fig 2E). EP infants were associated with a higher abundance of KO identifiers within functional categories translation and membrane transport. VP infants were associated with increased abundance of KO identifiers within CHO and energy metabolism. This could also be observed from the ten most abundant KO identifiers per GA group (Table III). The ten most abundant proteins accounted for 63.5%, 77.6%, and 58.8% of total proteins in EP25-26, EP27, and VP infants, respectively. In VP infants, the top ten KO identifiers mainly represented proteins involved in CHO and energy metabolism, while in EP infants they represented proteins involved in translation and membrane transport. Murein lipoprotein was particularly abundant in EP27 infants but decreased over time, resulting in profiles more similar to EP25-26 and VP infants (Fig. S3C).
Table III. Top ten KEGG orthology identifiers per gestational age group during postnatal weeks 3–6. Per gestational age group, average relative abundances for each KEGG orthology identifier were calculated based on iBAQ intensities.
| GA group | KO ID | Protein | KEGG Brite functional category | Rel. abundance* |
|---|---|---|---|---|
| EP 25–26 | K01803 | Triosephosphate isomerase | CHO metabolism, Energy metabolism | 0.213 |
| K06078 | Murein lipoprotein | x | 0.162 | |
| K02994 | Small subunit ribosomal protein S8 | Translation | 0.063 | |
| K02950 | Small subunit ribosomal protein S12 | Translation | 0.053 | |
| K02358 | Elongation factor Tu | Translation, Transport and catabolism | 0.034 | |
| K00100 | NADH-dependent butanol dehydrogenase A | CHO metabolism, Lipid metabolism, Xenobiotics biodegr. and metabolism | 0.031 | |
| K00134 | Glyceraldehyde 3-phosphate dehydrogenase | CHO metabolism, Energy metabolism, Signal transduction | 0.023 | |
| K02040 | Phosphate transport system substrate-binding protein | Membrane transport, Signal transduction | 0.019 | |
| K05878 | Dihydroxyacetone kinase | Lipid metabolism | 0.019 | |
| K03286 | OmpA-OmpF porin, OOP family | Membrane transport | 0.018 | |
| EP 27 | K06078 | Murein lipoprotein | x | 0.537 |
| K01803 | Triosephosphate isomerase | CHO metabolism, Energy metabolism | 0.062 | |
| K03286 | OmpA-OmpF porin, OOP family | Membrane transport | 0.053 | |
| K03647 | Nrd∣ family protein | x | 0.032 | |
| K09475 | Outer membrane pore protein C | Signal transduction | 0.021 | |
| K02986 | Small subunit ribosomal protein S4 | Translation | 0.02 | |
| K00134 | Glyceraldehyde 3-phosphate dehydrogenase | CHO metabolism, Energy metabolism, Signal transduction | 0.014 | |
| K02950 | Small subunit ribosomal protein S12 | Translation | 0.014 | |
| K04744 | LPS-assembly protein | Membrane transport | 0.012 | |
| K02871 | Large subunit ribosomal protein L13 | Translation | 0.011 | |
| VP | K06078 | Murein lipoprotein | x | 0.132 |
| K00656 | Formate C-acetyltransferase | CHO metabolism | 0.128 | |
| K01632 | Xylulose-5-p/fructose-6-p phosphoketolase | Energy metabolism | 0.072 | |
| K01803 | Triosephosphate isomerase | CHO metabolism, Energy metabolism | 0.071 | |
| K01810 | Glucose-6-phosphate isomerase | CHO metabolism | 0.042 | |
| K00927 | Phosphoglycerate kinase | CHO metabolism, Energy metabolism | 0.038 | |
| K00134 | Glyceraldehyde 3-phosphate dehydrogenase | CHO metabolism, Energy metabolism, Signal transduction | 0.031 | |
| K01915 | Glutamine synthetase | AA metabolism, CHO metabolism, Energy metabolism, Signal transduction | 0.026 | |
| K00359 | Pyridine nucleotide-disulfide oxidoreductase | x | 0.025 | |
| K02906 | Large subunit ribosomal protein L3 | Translation | 0.022 |
GA: gestational age, KO: KEGG orthology, ID: identifier, CHO: carbohydrate, EP: extremely preterm, VP: very preterm.
A Bifidobacterium-Dominated Community Is Associated with Active Metabolism toward Human Milk Oligosaccharide Degradation
VP infants showed to have a Bifidobacterium-dominated community, active in CHO and energy metabolism. To metabolize complex CHO structures such as human milk oligosaccharides (HMOs), ABC transporters and glycolytic enzymes are required, including galactosidases, fucosidases, and sialidases. The glycolytic enzymes identified in our dataset were β-galactosidases derived from Bifidobacterium, Enterobacter, Streptococcus, and Clostridium. Bifidobacterium-derived β-galactosidases were identified in all preterm infants but were more abundant in VP than in EP infants (p = 0.026) (Table S4). Enterobacter-, Streptococcus-, and Clostridium-derived β-galactosidases could only be identified in very low abundance and just in a few samples. Similar accounted for ABC transporters for oligosaccharides, Bifidobacterium-derived ABC transporters were more abundant in VP than in EP infants (p = 8.1E–06), while those Klebsiella and Eubacterium derived were rarely identified. ABC transporters for oligopeptides could only be identified in a few samples from both EP and VP infants.
Respiratory Support and Antibiotic Treatment Influences Microbiota Succession
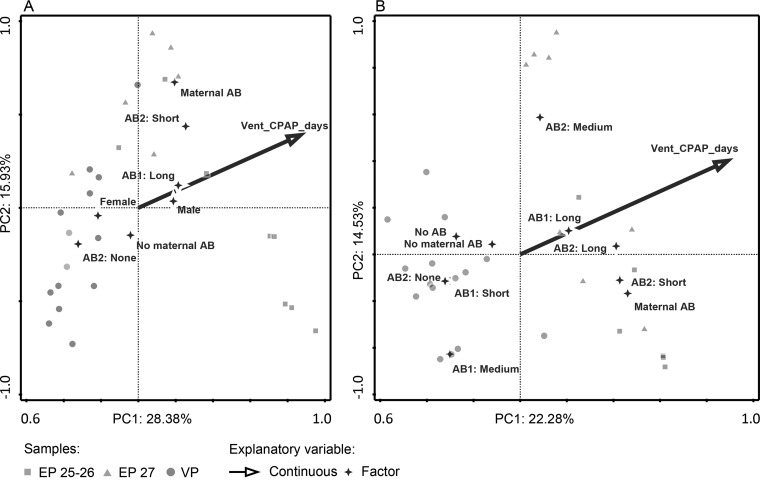
The effect of clinical characteristics in association with microbiota composition and function were analyzed by redundancy analysis. The GA-based separation of fecal microbiota composition and protein KO ID profiles were mainly driven by the duration of respiratory support and antibiotic-treatment-related factors (Fig. 4). Respiratory support explained 24.1% and 4.9% of the variation in microbiota composition based on 16S- and protein KO ID data, respectively. Antibiotic-treatment-related factors explained 25.6% and 44.6% of the variation, respectively, and comprised the number and duration of treatment and the administration of maternal antibiotics (Fig. 4). Other factors, including mode of delivery, birth weight, feeding intolerance, proportion of human milk feeding, and days until discharge did not have a significant influence. To provide more support for the association between microbiota composition and clinical factors, this analysis was repeated in all additional EP25-26 (n = 14), EP27 (n = 17), and VP30 (n = 6) infants from the complete cohort, albeit solely based on 16S-rRNA gene sequencing (Illumina MiSeq, to be published elsewhere). Ventilation remained the main factor associated with microbiota composition (4.7%, p = 0.002). In addition, maternal antibiotics and duration of the first and second course of antibiotic treatment, respectively, explained 2%, 2.6%, and 1.8% of the observed variation in microbiota composition (p < 0.05).
Fig. 4.
Redundancy analysis of the fecal bacterial community using (A) 16S-rRNA gene-based microbiota profiles and (B) protein-based KEGG orthology identifier profiles. Clinical factors that significantly explain the variation are shown. AB: antibiotic, AB1: duration of first course of antibiotics, AB2: duration of second course of antibiotics, Vent_CPAP_days: duration of ventilation and/or continuous positive airway pressure in days.
DISCUSSION
The current study implements metaproteomics and 16S-rRNA gene sequencing in a cohort of preterm infants to get insight into the establishment and activity of their intestinal microbiota. Metatranscriptomics and proteomics have only recently been applied to study GI and microbiota function in preterm infants (17, 34, 35). As such, a previous study showed that metaproteomics data are consistent with metagenomics and 16S-rRNA gene analysis and that bacterial activity transits toward more complex metabolic functions in time (17). However, those findings were based on analysis of the fecal metaproteome of one preterm infant (28 weeks GA) during the first three postnatal weeks. The current study adds up to these data by using a combination of metaproteomics and 16S-rRNA gene sequencing to study microbiota development during the first six postnatal weeks in ten preterm infants of varying GA.
Analysis of the fecal proteome indicated low bacterial load till the second postnatal week in all preterm infants. Low bacterial load might have contributed to the observed variation in microbiota composition during the first two postnatal weeks. From the third postnatal week onward, microbiota composition and function could be distinguished between infants of varying GA. Bacterial colonization was delayed in EP25-26 and EP27 infants compared with VP infants, emphasizing that microbial colonization pattern is related to GA.
Facultative anaerobes remained dominant throughout the first six postnatal weeks in EP infants. Colonization with obligate anaerobes was delayed, which is consistent with previous findings (7–9). In VP infants, obligate anaerobe and beneficial early-life colonizer Bifidobacterium became predominant from the third postnatal week. This is in concordance with a previous study showing that dominance of obligate anaerobes occurs around postconceptional age 33–36 weeks in preterm infants (10). The development toward a Bifidobacterium-dominant microbiota as observed in VP infants, but not in EP infants, is more representative of microbiota development in term, vaginally born, breast-fed infants, which is considered most beneficial during early-life development.
The observed differences between proteome- and 16S-based taxonomy in EP infants could indicate that microbiota composition is not representative for metabolic activity of the microbiota. However, it should be noted that misclassification of bacterial genera could occur in the case of high 16S-rRNA gene similarity, which is the case for members of Enterobacter and Klebsiella. Applying two complementary approaches, like metaproteomics and 16S-rRNA gene sequencing, is therefore of great added value for data interpretation. In general, 16S-rRNA gene sequencing confirmed the findings obtained by metaproteomics, showing clear taxonomic differences between infants born at varying GA, with high abundance of Bifidobacterium in VP infants.
In addition to taxonomic differences, our data show clear differences in bacterial activity between GA groups. VP infants were associated with a high abundance of proteins involved in CHO and energy metabolism, while proteins involved in membrane transport and translation were highly abundant in EP infants. Proteins related to HMO degradation were particularly abundant in VP infants, coinciding with the high abundance of Bifidobacterium. Bifidobacterium species are considered beneficial early-life colonizers predominantly colonizing the infant gut due to their ability to metabolize complex CHO structures, including HMOs. These findings indicate a well-established and metabolically active microbiota in VP infants, whereas in EP infants, the protein profile indicates a more active role on the generation and maintenance of biomass.
Our findings indicate that GA is associated with microbiota establishment and its activity in preterm infants. However, one should be aware of the factors that are associated with preterm birth and keep in mind that these factors can be greatly related to GA, including the extent and duration of intensive care. In this study, the observed differences in microbiota development between EP and VP infants were mainly driven by respiratory support and antibiotic treatment, which extent is negatively correlated to GA. One of the respiratory support strategies is continuous positive airway pressure in which pressure is combined with air/oxygen administration, thereby allowing air to reach the GI-tract (36). This might create an oxygen-rich environment in the GI-tract, which could impede the passage and survival of obligate anaerobes and hence lead to establishment of an intestinal microbiota dominated by aerobes and facultative anaerobes. Indeed, our findings show dominance of aerobic and facultative anaerobic bacteria in EP infants. These infants all received respiratory support throughout the six weeks of the study period. These findings are supported by a previous study, showing that duration of respiratory support in preterm infants is associated with predominance of fecal aerobes/facultative anaerobes and with the onset of Staphylococcal late-onset sepsis (37). In addition to longer respiratory support, the duration and number of antibiotic treatment showed to be significant drivers of the observed GA-dependent microbiota development. Ceftazidime and amoxicillin are broad-spectrum β-lactam antibiotics, targeting Gram-negative and -positive bacteria. It has been shown that Bifidobacterium is sensitive to β-lactam antibiotics (38, 39) and that treatment with amoxicillin can greatly influence the composition of Bifidobacterium species in infant intestinal microbiota (40). Vancomycin particularly targets Gram-positive bacteria and has been shown to affect Bifidobacterium (38, 39). Prolonged and multiple antibiotic treatment in EP infants could therefore delay or prohibit establishment of a Bifidobacterium-dominated microbiota. Previous studies indeed show that antibiotic treatment greatly affects microbiota development in preterm infants (41–43). Although we were able to replicate our findings considering the role of respiratory support and antibiotic treatment on microbiota development in the complete cohort, the effect appeared to be less apparent. This variation could be due to differences in the distribution of GA in the groups, with 50% (5/10) of infants being born at 30 weeks gestation in the current study and only 16% (6/37) in the complete cohort. As factors such as respiratory support and antibiotic treatment strategies are associated with GA, further studies are needed to unravel the true contribution of these factors to microbiota development.
Overall, our findings indicate that GA is positively associated with abundance of Bifidobacterium and negatively associated with abundance of facultative anaerobic bacteria. Development of the intestinal microbiota most likely suffers from exposure to respiratory support and antibiotic treatment. A high extent of exposure to these factors is common in EP infants, pressuring the bacterial community to become rich in facultative anaerobes and particularly active in translation and membrane transport. VP infants are to a lesser extent exposed to respiratory support and antibiotic treatment, allowing for development toward a more stable, metabolically active, Bifidobacterium-dominated microbiota. A microbial signature characterized by low abundance of Bifidobacterium and high abundance of facultative anaerobes has been associated with several negative health outcomes in early life, including necrotizing enterocolitis and late-onset sepsis (44, 45). In addition, disturbances in microbiota development have been related to development of disorders in later life. In light of this, our data indicate that GA and its associated intensity of care could greatly influence early and later life health of preterm infants by interfering with microbiota development.
DATA AVAILABILITY
The mass spectrometry data have been deposited to the ProteomeXchange Consortium (22) via the PRIDE partner repository with dataset identifier PXD005574. 16S-rRNA gene sequencing data are available in the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under study accession PRJEB18915. Detailed information regarding protein identifications and taxonomic and functional classifications can also be found in Supplemental Table S5.
Supplementary Material
Footnotes
Author contributions: R.D.Z., L.J.G.J., and S.B. performed research; R.D.Z., S.B., and C.B. analyzed data; D.v.Z.-G., R.M., R.A.v.L., I.B.R., C.B., and J.K. designed research; D.v.Z.-G., R.A.v.L., I.B.R., R.M.v.E., C.B., and J.K. provided scientific input; R.M., L.J.G.J., and S.B. reviewed the manuscript, and R.D.Z. wrote the paper.
* Authors RM, RMvE, IBR, and JK are employees of Nutricia Research BV, The Netherlands. Authors RDZ and CB are financially supported by Nutricia Research BV, The Netherlands.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CHO
- carbohydrate
- EP
- extremely preterm
- GA
- gestational age
- GI
- gastrointestinal
- HMO
- human milk oligosaccharide
- iBAQ
- intensity-based absolute quantification
- KO
- KEGG orthology
- OTU
- operational taxonomic unit
- rRNA
- ribosomal RNA
- VP
- very preterm.
REFERENCES
- 1. Palmer C., Bik E. M., DiGiulio D. B., Relman D. A., and Brown P. O. (2007) Development of the human infant intestinal microbiota. PLoS Biol. 5, e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koenig J. E., Spor A., Scalfone N., Fricker A. D., Stombaugh J., Knight R., Angenent L. T., and Ley R. E. (2011) Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 108, 4578–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yatsunenko T., Rey F. E., Manary M. J., Trehan I., Dominguez-Bello M. G., Contreras M., Magris M., Hidalgo G., Baldassano R. N., Anokhin A. P., Heath A. C., Warner B., Reeder J., Kuczynski J., Caporaso J. G., Lozupone C. A., Lauber C., Clemente J. C., Knights D., Knight R., and Gordon J. I. (2012) Human gut microbiome viewed across age and geography. Nature 486, 222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Penders J., Thijs C., Vink C., Stelma F. F., Snijders B., Kummeling I., van den Brandt P. A., and Stobberingh E. E. (2006) Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521 [DOI] [PubMed] [Google Scholar]
- 5. Rodriguez C., Taminiau B., Brévers B., Avesani V., Van Broeck J., Leroux A., Gallot M., Bruwier A., Amory H., Delmée M., and Daube G. (2015) Faecal microbiota characterisation of horses using 16 rDNA barcoded pyrosequencing, and carriage rate of Clostridium difficile at hospital admission. BMC Microbiol. 15, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Westerbeek E. A., van den Berg A., Lafeber H. N., Knol J., Fetter W. P., and van Elburg R. M. (2006) The intestinal bacterial colonisation in preterm infants: A review of the literature. Clinical Nutrition 25, 361–368 [DOI] [PubMed] [Google Scholar]
- 7. Arboleya S., Binetti A., Salazar N., Fernández N., Solís G. Hernández-Barranco A., Margolles A., de Los Reyes-Gavilán C. G., and Gueimonde M. (2012) Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiology Ecology 79, 763–772 [DOI] [PubMed] [Google Scholar]
- 8. Jacquot A., Neveu D., Aujoulat F., Mercier G., Marchandin H., Jumas-Bilak E., and Picaud J.-C. (2011) Dynamics and clinical evolution of bacterial gut microflora in extremely premature patients. J. Pediatrics 158, 390–396 [DOI] [PubMed] [Google Scholar]
- 9. Westerbeek E. A., Slump R. A., Lafeber H. N., Knol J., Gorgi G., Fetter W. P., and van Elburg R. M. (2013) The effect of enteral supplementation of specific neutral and acidic oligosaccharides on the faecal microbiota and intestinal microenvironment in preterm infants. Eur. J. Clin. Microbiol. Infect. Dis. 32, 269–276 [DOI] [PubMed] [Google Scholar]
- 10. La Rosa P. S., Warner B. B., Zhou Y., Weinstock G. M., Sodergren E., Hall-Moore C. M., Stevens H. J., Bennett W. E. Jr., Shaikh N., Linneman L. A., Hoffmann J. A., Hamvas A., Deych E., Shands B. A., Shannon W. D., and Tarr P. I. (2014) Patterned progression of bacterial populations in the premature infant gut. Proc. Natl. Acad. Sci. U.S.A. 111, 12522–12527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collado M. C., Cernada M., Neu J., Pérez-Martínez G., Gormaz M., and Vento M. (2015) Factors influencing gastrointestinal tract and microbiota immune interaction in preterm infants. Pediatric Research 77, 726–731 [DOI] [PubMed] [Google Scholar]
- 12. Martin R., Nauta A. J., Ben Amor K., Knippels L. M., Knol J., and Garssen J. (2010) Early life: Gut microbiota and immune development in infancy. Benef Microbes 1, 367–382 [DOI] [PubMed] [Google Scholar]
- 13. Nylund L., Satokari R., Salminen S., and de Vos W. M. (2014) Intestinal microbiota during early life—Impact on health and disease. Proc. Nutr. Soc. 73, 457–469 [DOI] [PubMed] [Google Scholar]
- 14. Wopereis H., Oozeer R., Knipping K., Belzer C., and Knol J. (2014) The first thousand days—Intestinal microbiology of early life: Establishing a symbiosis. Pediatric Allergy Immunology 25, 428–438 [DOI] [PubMed] [Google Scholar]
- 15. Patel R. M., and Denning P. W. (2015) Intestinal microbiota and its relationship with necrotizing enterocolitis. Pediatric Research 78, 232–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Weerth C., Fuentes S., and de Vos W. M. (2013) Crying in infants: On the possible role of intestinal microbiota in the development of colic. Gut Microbes 4, 416–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Young J. C., Pan C., Adams R. M., Brooks B., Banfield J. F., Morowitz M. J., and Hettich R. L. (2015) Metaproteomics reveals functional shifts in microbial and human proteins during a preterm infant gut colonization case. Proteomics 15, 3463–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stewart C. J., Skeath T., Nelson A., Fernstad S. J., Marrs E. C., Perry J. D., Cummings S. P., and Berrington J. E., and Embleton N. D. (2015) Preterm gut microbiota and metabolome following discharge from intensive care. Sci. Rep. 5, 17141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stewart C. J., Embleton N. D., Marrs E. C., Smith D. P., Nelson A., Abdulkadir B., Skeath T., Petrosino J. F., Perry J. D., Berrington J. E., and Cummings S. P. (2016) Temporal bacterial and metabolic development of the preterm gut reveals specific signatures in health and disease. Microbiome 4, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kolmeder C. A., de Been M., Nikkilä J., Ritamo I., Mättö J., Valmu L., Salojärvi J., Palva A., Salonen A., and de Vos W. M. (2012) Comparative metaproteomics and diversity analysis of human intestinal microbiota testifies for its temporal stability and expression of core functions. PLoS ONE 7, e29913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature Protocols 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 22. Lu J., Boeren S., de Vries S. C., van Valenberg H. J., Vervoort J., and Hettinga K. (2011) Filter-aided sample preparation with dimethyl labeling to identify and quantify milk fat globule membrane proteins. J. Proteomics 75, 34–43 [DOI] [PubMed] [Google Scholar]
- 23. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavida I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.-W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 25. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 26. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 27. Moles L., Gomez M., Heilig H., Bustos G., Fuentes S., de Vos W., Fernández L., Rodríquez J. M., and Jiménez E. (2013) Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS ONE 8, e66986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sim K., Cox M. J., Wopereis H., Martin R., Knol J., Li M. S., Cookson W. O., Moffatt M. F., and Kroll J. S. (2012) Improved detection of bifidobacteria with optimised 16S rRNA-gene based pyrosequencing. PLoS ONE 7, e32543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E. Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., and Knight R. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bragg L., Stone G., Imelfort M., Hugenholtz P., and Tyson G. W. (2012) Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat. Methods 9, 425–426 [DOI] [PubMed] [Google Scholar]
- 31. Edgar R. C., Haas B. J., Clemente J. C., Quince C., and Knight R. (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edgar R. C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 [DOI] [PubMed] [Google Scholar]
- 33. Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza B., Peplies J., Glöckner F. O. (2013) The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang P., Smith B., Qvist N., Nielsen C., Wan J. M., Sit W. H., Jensen T. K., Wang H., and Sangild P. T. (2013) Intestinal proteome changes during infant necrotizing enterocolitis. Pediatric Research 73, 268–276 [DOI] [PubMed] [Google Scholar]
- 35. Knight J. M., Davidson L. A., Herman D., Martin C. R., Goldsby J. S., Ivanov I. V., Donavan S. M., and Chapkin R. S. (2014) Non-invasive analysis of intestinal development in preterm and term infants using RNA-sequencing. Sci. Rep. 4, 5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jaile J. C., Levin T., Wung J. T., Abramson S. J., Ruzal-Shapiro C., and Berdon W. E. (1992) Benign gaseous distension of the bowel in premature infants treated with nasal continuous airway pressure: A study of contributing factors. AJR Am. J. Roentgenol. 158, 125–127 [DOI] [PubMed] [Google Scholar]
- 37. Shaw A. G., Sim K., Randell P., Cox M. J., McClue Z. E., Li M.-S., Donaldson H., Langford P. R., Cookson W. O. C. M., Moffatt M. F., and Kroll J. S. (2015). Late-onset bloodstream infection and perturbed maturation of the gastrointestinal microbiota in premature infants. PLoS ONE 10, e0132923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Delgado S., Flórez A. B., and Mayo B. (2005) Antibiotic susceptibility of Lactobacillus and Bifidobacterium species from the human gastrointestinal tract. Curr. Microbiol. 50, 202–207 [DOI] [PubMed] [Google Scholar]
- 39. Moubareck C., Gavini F., Vaugien L., Butel M. J., and Doucet-Populaire F. (2005) Antimicrobial susceptibility of bifidobacteria. J. Antimicrob. Chemother. 55, 38–44 [DOI] [PubMed] [Google Scholar]
- 40. Mangin I., Suau A., Gotteland M., Brunser O., and Pochart P. (2010) Amoxicillin treatment modifies the composition of Bifidobacterium species in infant intestinal microbiota. Anaerobe 16, 433–438 [DOI] [PubMed] [Google Scholar]
- 41. Arboleya S., Sanchez B., Milani C., Duranti S., Solís G., Fernández N., de los Reyes-Gavilán C. G., Ventura M., Margolles A., and Gueimonde M. (2015) Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J. Pediatr. 166, 538–544 [DOI] [PubMed] [Google Scholar]
- 42. Fricke W. F. (2014) The more the merrier? Reduced fecal microbiota diversity in preterm infants treated with antibiotics. J. Pediatr. 165, 8–10 [DOI] [PubMed] [Google Scholar]
- 43. Greenwood C., Morrow A. L., Lagomarcino A. J., Altaye M., Taft D. H., Yu Z., Newburg D. D., Ward D. V., and Schibler K. R. (2014) Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of Enterobacter. J. Pediatr. 165, 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Groer M. W., Luciano A. A., Dishaw L. J., Asmeade T. L., Miller E., and Gilbert J. A. (2014) Development of the preterm infant gut microbiome: A research priority. Microbiome 2, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vangay P., Ward T., Gerber J. S., and Knights D. (2015) Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe 17, 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited to the ProteomeXchange Consortium (22) via the PRIDE partner repository with dataset identifier PXD005574. 16S-rRNA gene sequencing data are available in the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under study accession PRJEB18915. Detailed information regarding protein identifications and taxonomic and functional classifications can also be found in Supplemental Table S5.