Abstract
Protein phosphorylation is known to regulate pathogenesis, mycelial growth, conidiation and stress response in Pyricularia oryzae. However, phosphorylation mediated regulatory networks in the fungal pathogen remain largely to be uncovered. In this study, we identified 1621 phosphorylation sites of 799 proteins in mycelia of P. oryzae, including 899 new p-sites of 536 proteins and 47 new p-sites of 31 pathogenicity-related proteins. From the sequences flanking the phosphorylation sites, 19 conserved phosphorylation motifs were identified. Notably, phosphorylation was detected in 7 proteins that function upstream of Pmk1, but not in Pmk1 and its downstream Mst12 and Sfl1 that have been known to regulate appressorium formation and infection hyphal growth of P. oryzae. Interestingly, phosphorylation was detected at the site Ser240 of Pmp1, which is a putative protein phosphatase highly conserved in filamentous fungi but not characterized. We thus generated Δpmp1 deletion mutants and dominant allele PMP1S240D mutants. Phenotyping analyses indicated that Pmp1 is required for virulence, conidiation and mycelial growth. Further, we observed that phosphorylation level of Pmk1 in mycelia was significantly increased in the Δpmp1 mutant, but decreased in the PMP1S240D mutant in comparison with the wild type, demonstrating that Pmp1 phosphorylated at Ser240 is important for regulating phosphorylation of Pmk1. To our surprise, phosphorylation of Mps1, another MAP kinase required for cell wall integrity and appressorium formation of P. oryzae, was also significantly enhanced in the Δpmp1 mutant, but decreased in the PMP1S240D mutant. In addition, we found that Pmp1 directly interacts with Mps1 and the region AA180–230 of Pmp1 is required for the interaction. In summary, this study sheds new lights on the protein phosphorylation mediated regulatory networks in P. oryzae.
Phosphorylation of proteins is coordinately controlled by protein kinases and phosphatases, and is a key posttranslational modification in living cells that dynamically regulates a wide range of important biological processes, including cell cycle, cell differentiation, proliferation and metabolic control (1, 2). Deregulated phosphorylation because of abnormal expression and/or modification of protein kinases and phosphatases is a hallmark of cancers and other diseases, and many disease-related kinases and phosphatases are thus becoming prominent drug targets (3).
Biological functions of protein phosphorylation have been extensively and intensively investigated by large-scale knockout or silence of individual protein kinases or phosphatases genes and phosphoproteome analyses in diverse organisms in the past decade. In Neurospora crassa, deletion mutants for 77 kinase genes (4) and 24 phosphatase genes (5) were generated, of which 57% and 91% exhibited at least one growth or developmental phenotype. RNAi-mediated silencing of 695 kinase or putative kinase genes and 146 phosphatase genes in human cells showed that 29 of them are candidate clock components (6). Phosphoproteome analyses can greatly accelerate cell signaling research by providing critical information about regulatory sites and dynamics. Over a dozen of extensive phosphoproteome studies have been reported in the budding yeast Saccharomyces cerevisiae (7, 8) and fission yeast Schizosaccharomyces pombe (9). Large-scale comparative phosphoproteomics identified conserved phosphorylation sites (p-sites) in rice and Arabidopsis (10). Systematic studies of the tissue-specific protein expression and phosphorylation in mouse (11) and rat (12) identified more than 30,000 p-sites and revealed the tissue specificity of phosphorylation.
Protein phosphorylation is also a key player during the life cycles of plant fungal pathogens. For the wheat scab fungal pathogen Fusarium graminearum, gene knockout mutants were generated for more than 90 distinct protein kinases (13). Biological assays indicated that these protein kinases are important for changes in 17 phenotypes, including mycelial growth, conidiation, pathogenesis, stress responses and sexual reproduction. Recently, large-scale phosphoproteomic analyses showed that, in plant fungal pathogens such as F. graminearum, Alternaria brassicicola and Botrytis cinerea, protein phosphorylation is involved in many biological processes, including mycelial growth, metabolic process, transport, DNA dependent transcription regulation, response to stimulus and signal transduction (14, 15). However, protein phosphorylation information of plant fungal pathogens is still limited and is expected to be extensively explored in more organisms in order to provide a global view on protein interaction networks and signaling pathways mediated by protein phosphorylation.
Pyricularia oryzae is a filamentous ascomycete that causes the rice blast disease, which is the most devastating disease of cultivated rice (16), and is also the first plant fungal pathogen to have its genome sequenced (17). Infection cycle by the fungal pathogen involves several key steps, including conidium production, attachment of conidium to the plant surface, appressorium formation from germ tube tips of conidium, penetration and invasive growth of infection hyphae (18). Several protein kinase cascades play important roles in the infection cycle of P. oryzae (19). Two p21-activated kinases (PAK)1, Mst20 and Chm1, are important for the conidium production and morphogenesis (20). The cyclic AMP-protein kinase A (cAMP-PKA) pathway plays a key regulatory role during the surface recognition for appressorium formation and turgor generation in appressorium (18, 21). Pmk1 is a mitogen-activated protein kinase (MAPK) homologous to the Fus3/Kss1 in Saccharomyces cerevisiae, and is essential to appressorium formation, penetration, and invasive growth of infection hyphae (22). The Pmk1 MAPK cascade involves three core protein kinases, Mst11, Mst7, and Pmk1 (23). Mst11 and Mst7 are MEKK and MEK, respectively. The Mck1-Mps1 pathway is another MAPK cascade that is important for cell wall integrity and the appressorium penetration and invasive growth of infection hyphae (24, 25). In addition, the Ssk1-Osm1 MAPK pathway regulates cellular turgor for response to hyperosmotic stress (26, 27). The SNF1/AMP-activated protein kinase (AMPK) pathway was reported to contribute to peroxisomal maintenance and lipid metabolism, as well as maintaining cell wall porosity and turgor pressure in appressorium (28). Identification and functional characterization of these protein kinases signify important roles of protein phosphorylation during mycelial growth, asexual development and pathogenesis of P. oryzae. Recently, a comparative phosphoproteome was conducted with mycelia, conidia, germinated conidia, and appressoria of a wild type P. oryzae and a ΔcpkA mutant. The study identified 2924 p-sites on 1514 proteins, and a phosphorregulated transcription factor was proved to be involved in regulating glycerol metabolism (29). However, the genome of P. oryzae contains more than 12,000 genes (17, 30), approximate 80 of them are predicted to encode protein kinases (17, 31). As described above, only a few of the protein kinases genes have been functionally characterized. It has been reported that 60% of yeast genes encode phosphoproteins (32), and p-sites identified in yeast are still under saturation after dozen of phosphoproteome studies (8). Thus, we believe that phosphoproteins and their p-sites in P. oryzae remain largely to be identified and characterized. Furthermore, hundreds of pathogenicity-related proteins have been studied, including diverse types of transcription factors (33), autophagy-associated proteins (34), septins (35), exocyst components (36), RhoGAP proteins (37), regulators of G-protein signaling proteins (38, 39), peroxidase (40), chitin synthase genes (41), light regulation related proteins (42, 43), and hypoxia-responsive proteins (44). However, links between phosphorylation and their functions are rarely disclosed. Therefore, more phosphoproteomic analyses are expected to shed light on the regulatory networks and functions of protein phosphorylation during mycelial growth, conidiation and pathogenesis of P. oryzae.
In this study, we identified 1621 p-sites of 799 proteins, including 899 new p- sites of 536 proteins and 47 new p-sites of 31 pathogenicity-related proteins, and 19 conserved phosphorylation motifs in the phosphoproteins. Importantly, we found that the protein phosphatase Pmp1 is phosphorylated at the site Ser240 in mycelia and is required to regulate phosphorylation of Pmk1 and Mps1, two MAPK kinases previously reported to be essential to pathogenicity of P. oryzae. Furthermore, we found that Pmp1 directly interacts with Mps1 and the region AA180–230 of Pmp1 is required for the interaction. In addition, we demonstrated that Pmp1 is important for pathogenicity, conidiation and mycelial growth. In general, our results provide new insights into phosphorylation mediated regulatory networks in P. oryzae.
EXPERIMENTAL PROCEDURES
Strains and Cultures
P. oryzae strain P131 (30) was maintained on Yeast Extract Glucose medium (YEG) plates or Oatmeal Tomato Agar (OTA) plates at 28 °C. For phosphoproteomic analysis, three independent duplicates were used. For each duplicate, mycelia harvested from YEG plates were fragmented in 1 ml of liquid YEG, and then transferred into 200 ml of liquid YEG, and cultured at 25 °C for 48 h with shaking at 180 rpm. Previously described protocols were followed for isolation of fungal DNAs and protoplasts, and for assaying the colony growth, conidiation and virulence (45, 46).
Sample Preparations
Mycelia were harvested with filtration from YEG culture were immediately frozen and grinded in liquid N2 for protein extraction. Total proteins were extracted from mycelia as described (47) but with protease inhibitor mixture (1:100 dilution) and phosphatase inhibitor mixture 2 (1:1000 dilution) (Sigma-Aldrich, St. Louis, MO). The extracts were centrifuged at 26,900 × g for 30 min and the resulted supernatants were collected as protein samples, in which protein concentrations were measured with the Bradford method (Bio-Rad, Hercules, CA). Protein samples (10 mg) diluted in the urea lysis buffer at the concentration of 5 mg/ml were digested with trypsin as described previously (47). After incubation at 37 °C for 16 h, the digestion reaction was stopped with trifluoroacetic acid (TFA) (0.4%) at pH < 2, and then the peptide mixtures were desalted with 500 mg Sep-Pak tC18 cartridges (Waters, Milford, MA) and dried prior to strong cation exchange (SCX) fractionation.
SCX Chromatographic Fractionation and Immobilized Metal Affinity Chromatography (IMAC) Enrichment
The digested peptides were dissolved in 200 μl of SCX buffer A (30% acetonitrile (ACN) in 7 mm KH2PO4 solution, pH 2.65) and then fractionated on Agilent 1100 HPLC equipped with a SCX column (9.4 × 200 mm, PolyLC, Columbia, MD) as described (47). A total of 12 peptide fractions were separately collected from 1.5 to 49.5 min with a 4-min interval. The elution conditions were for the first 2 min with buffer A, for 2 to 30 min with linear gradient elution of 0–25% buffer B (30% ACN in 7 mm KH2PO4 and 350 mm KCl solution, pH 2.65), for 30 to 45 min with linear gradient of 25–100% buffer B, and for the last 5 min with 100% buffer B. The 12 fractions were then concentrated with an Eppendorf Vacufuge concentrator 5301 (Eppendorf AG, Hamburg, Germany) to two thirds of the original volume, and desalted with 50 mg-tC18 Sep-Pak cartridges (Waters) prior to phosphopeptide enrichment.
Phosphopeptide enrichment was performed with IMAC as described (47). The eluates from IMAC were acidified with 2.5% TFA to pH below 2.5, and desalted with Pierce graphite spin column (Thermo Scientific, Rockford, IL), and dried as described above for LC-MS/MS analysis.
NanoLC-MS/MS Analysis
The 12 phosphopeptide-enriched samples from IMAC were each resuspended in 20 μl of 5% ACN in 0.4% formic acid, and then analyzed with a Bruker nanoLC - amaZon speed ETD ion trap mass spectrometer system (Bruker Daltonics, Fremont, CA). A C18 analytical column (0.1 × 150 mm, 3 μ, 200 Å, Bruker) was used to separate the peptides with gradient elution from 2–32% ACN in 0.1% formic acid for 90 min at the flow rate of 500 nL/min. Each sample was analyzed twice.
For data acquisition, the MS/MS were acquired in ion trap using CID, followed by neutral loss-triggered ETD and MS3-CID. The neutral loss masses were set as 32.67, 49.00, 38.67, and 58.00. When the neutral loss was observed among the top 5 most intense ions in a MS/MS spectrum, the ion(s) showing neutral loss would undergo MS3 -CID and the precursor ion would be fragmented with ETD. The enhanced resolution mode (8100 m/z/s) and ultra-scan mode (32500 m/z/s) were selected for MS scan and MS/MS scan, respectively. The maximum accumulation time was set at 50 ms and the accumulation target was set at 400000. The MS scan range was set between 400 and 1400 m/z. The averages and rolling averages were set as 5 and 2, respectively. The absolute and relative thresholds of precursor were set at 20,000 and 0.2%, respectively. Exclusion was active after 2 spectra and the excluded spectra were released after 0.5 min. However, when the intensity of a current precursor was 5 times greater than that of the previous precursor, the current precursor would be run for MS/MS.
Data Analysis
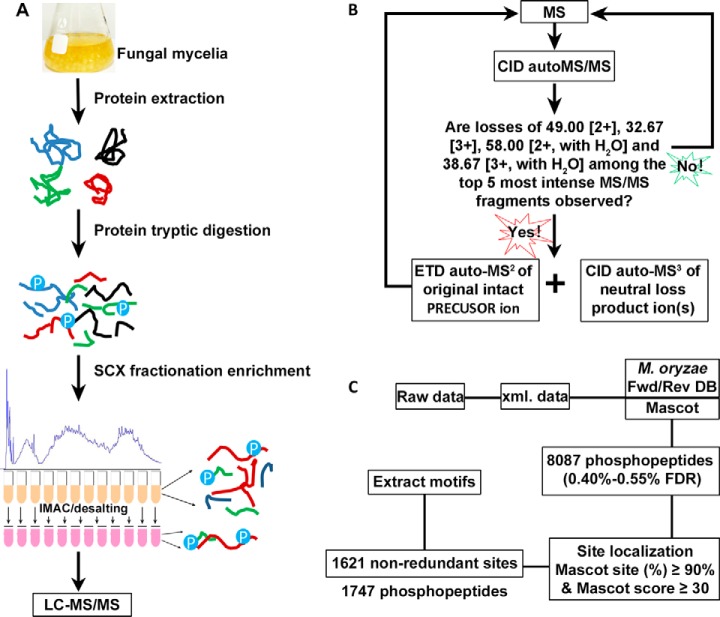
Raw data were converted into xml files via Bruker DataAnalysis 4.1 using the default Ion Trap.m with several changes: (1) 5 min to 95 min for spectral range selection, (2) 100,000 for intensity threshold of positive ion, (3) 30,000 for maximum number of compounds, (4) 0 min for retention time window, and (5) 5, 6, and 3 for maximum charge of MS full scan, MS MaxRes scan and MS(n), respectively. Xml files from the same biological duplicate were combined together with Bruker ProteinScape 3.1 and then searched against a concatenated database containing the 12,991 predicted protein entries of M. oryzae 70–15 (MG8) (http://www.broadinstitute.org) and the reversed (decoy) sequence database with Mascot 2.2.07 (Matrix Science, London, UK). Search parameters included a fixed modification of carboxyamidomethylation on Cys; variable modification of oxidation on Met and phosphorylation on Ser, Thr, and Tyr; trypsin digestion; two missed cleavages allowed; 0.6 Da for MS tolerance and 0.6 Da for MS/MS tolerance; significance value at p < 0.05. Significant hits were assessed with mascot score at 39.0, 39.0, and 39.2 for the three biological duplicates. For each peptide, the top hit compound only was accepted; and in case MS/MS spectra matching peptides from more than one identified proteins, the highest scoring peptide (rank 1 peptide) was accepted. The false discovery rate (FDR) at the peptide level of each biological duplicate was calculated with a Perl script fdr_table.pl downloaded from Matrix Science. P-site assignments with site (%) ≥ 90% and mascot peptide score (i.e. Meta score) ≥ 30 were accepted for further analysis.
Motif Analysis
Phosphopeptides were pre-aligned against the M. oryzae_70–15 (MG8) protein database with peptidextender (http://schwartzlab.uconn.edu/pepextend). Prealigned peptides were 15 amino acids in lengths with the p-sites at the center. Phosphopeptides with the p-site adjacent to the N- or C terminus within 7 amino acid range were not included in this analysis. Motif analysis was performed with the motif-x (48) under the following parameters: 15 amino acids width, occurrence threshold at 20, significance threshold at 1 × 10−6 and the M. oryzae_70–15 (MG8) protein database as the background file.
Phosphorylation motifs were classified as acidic, basic, and proline-directed as described (49). Motifs were assigned to typical kinases through online search of human protein reference database (http://www.hprd.org/serine motifs/) (50) and PHOSIDA posttranslational modification database (http://phosida.de/) (51).
Gene Ontology (GO) Annotation
Phosphoproteins were functionally annotated with Blast2GO under the parameters: E-Value-Hit-Filter of 1 × 10−6, annotation Cutoff of 55, and GO Weight of 5 (52, 53). BlastP was performed against the NCBInr database with an expect value of 1 × 10−6. The M. oryzae_70–15 (MG8) protein database was used as the reference database for GO enrichment analysis. Enriched terms met a 0.05 FDR threshold, calculated by Blast2GO using Fisher's exact test.
PMP1 Gene Replacement Vectors and Mutants
To generate the PMP1 gene replacement construct pKOPMP1, the 1.3-kb upstream and 1.5-kb downstream fragments of the PMP1 gene were independently amplified and cloned into the PstI-EcoRI and HindIII-ClaI sites of pKOV21 (46). After linearization with NotI, pKOPMP1 was transformed into the wild-type strain P131 to generate PMP1 deletion mutants, which were first identified by PCR and further confirmed by Southern blot hybridization. Genomic DNAs of putative Pmp1 mutants and wild-type strain P131 were digested by SalI, and Southern blot hybridization was performed as described (46). To generate the PMP1 gene complementary construct pHBPMP1, the PMP1 gene with 1.7-kb upstream sequences was amplified and cloned into the EcoRI-BamHI site of pGTN that contained the eGFP-TrpC terminator sequences with pEASY-Uni Seamless Cloning and Assembly Kit (TransGen Biotech, Beijing, China). The PMP1S240D allele, pHBPMP1S240D, was generated from pHBPMP1 by PCR-based site-directed mutagenesis (54). After linearization with NotI, pHBPMP1 and pHBPMP1S240D were independently transformed into a Δpmp1 mutant.
Protoplasts were isolated and transformed as described (46). Media were supplemented with 250 μg/ml hygromycin B (Roche, Branchburg, NJ) or 400 μg/ml neomycin (Ameresco, Solon, OH) to select hygromycin- or neomycin-resistant transformants.
Western Blot Assay
Total proteins were isolated from mycelia and separated on a 12% SDS-PAGE gel and transferred to PVDF Membrane (Merck Millipore, Shanghai, China). Phosphorylation of the Pmk1 and Mps1 MAP kinases was detected with the PhophoPlus p44/42 MAP kinase antibody (Cell Signaling Technology, Beverly, MA) as described previously (55, 56). The anti-actin (Abmart, Shanghai, China) was used as a loading control.
Yeast Two-Hybrid Assays
Protein-protein interactions were assayed with the Matchmaker yeast two-hybrid system (Clontech, Mountain View, CA). ORFs of the PMK1 and MPS1 were amplified and cloned into pGBKT7 (Clontech) as bait constructs. PMP1 ORF and its fragments were amplified and cloned into pGADT7 as prey constructs. The resulting bait and prey constructs were co-transformed in pairs into yeast strain AH109. The Leu+/Trp+ transformants were isolated and assayed for growth on S.D.-Trp-Leu-His medium and β-galactosidase activities. Empty prey construct with Mps1 bait construct or with empty bait construct were used as negative controls. Primers used in this study were listed in supplemental Table S6.
RESULTS
Phosphoproteome in Mycelia of P. oryzae
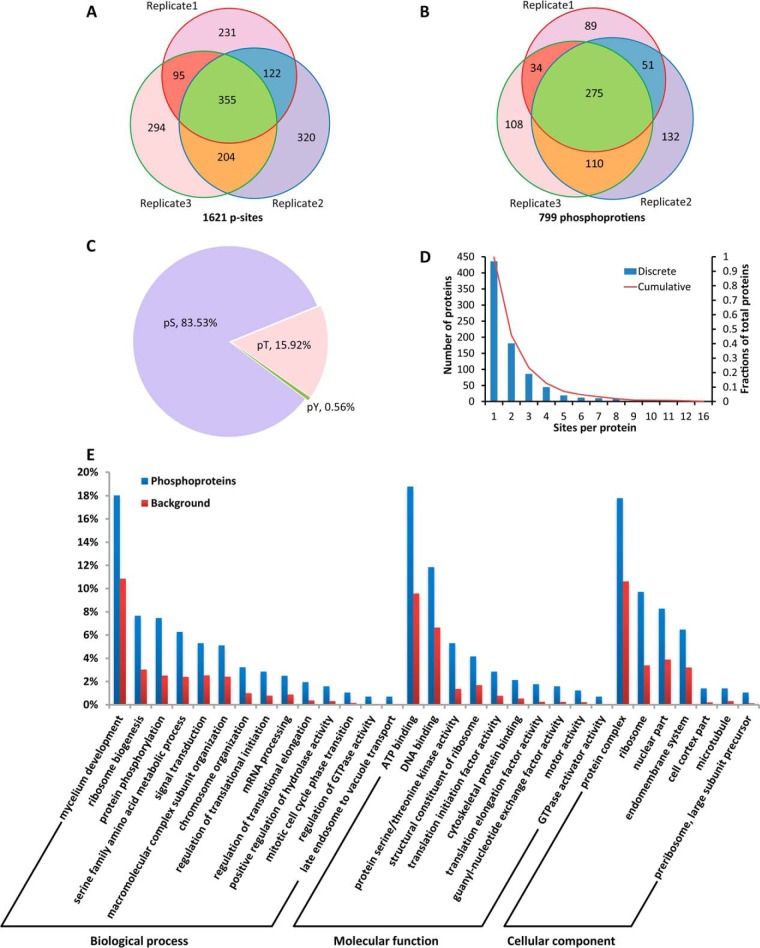
Hyphal growth is an essential phase in the life cycle of P. oryzae. To uncover the phosphorylation events in this phase, we profiled the phosphoproteome in mycelia of P. oryzae as outlined in Fig. 1. Phosphopeptides from mycelia of three duplicates were enriched with SCX/IMAC approach (Fig. 1A) and detected with neutral loss triggered ETD strategy on amaZon Speed ETD mass spectrometer (Fig. 1B). A total of 8087 phosphopeptides from 1187 proteins were identified with p < 0.05 and protein score ≥ 39.0, and the FDRs at the peptide level were 0.40%, 0.50%, and 0.55% for the three biological duplicates calculated with the target and decoy strategy (Fig. 1C, supplemental Table S1). Confident phosphopeptides were collected with a threshold of mascot site (%) ≥ 90% and mascot score ≥ 30 generated from Mascot search engine. In total, 1621 non-redundant p-sites from 1747 phosphopeptides of 799 proteins were identified in three independently duplicated experiments (supplemental Table S2A). Among the 1621 confident p-sites, 355 p-sites were overlapped in all three duplicates, accounting for 21.9% of total p-sites, and 47%–59% of p-sites were shared between two duplicates (Fig. 2A). Among the 799 phosphoproteins, 275 proteins were overlapped in all three duplicates, accounting for 34% of total phosphoproteins, and 57.5%–73.1% of phosphoproteins were shared between two duplicates (Fig. 2B). An early study identified 1393 p-sites of 831 proteins in mycelia of the P. oryzae strain 70–15 (29). Compared with that study, 480 p-sites of 332 proteins were overlapped, accounting for 29.6%–34.4% of the p-sites and ∼40% of proteins between the two studies. To our surprise, our study included 641 p-sites that were detected in conidia and germinated conidia by the previous study (29). In addition, our study included 899 p-sites that were not described in the early study (supplemental Fig. S1, supplemental Table S2B–S2E).
Fig. 1.
Schematic for identifying phosphoproteins and phosphopeptides via liquid chromatography tandem mass spectrometry. A, Workflow of sample preparation. Mycelial proteins were extracted and trypsin-digested. Total proteins (10 mg) were used for trypsin digestion and the tryptic peptides were subjected to SCX fractionation and IMAC phosphopeptide enrichment. B, A neutral loss triggered ETD strategy for data acquisition using an amaZon speed ETD mass spectrometer. C, Process of data analysis.
Fig. 2.
Phosphorylation events in mycelia of P. oryzae. A, A Venn diagram depicting numbers of p-sites identified in the three biological duplicates and p-site reproducibility. B, A Venn diagram depicting numbers of phosphoproteins identified in the three biological duplicates and phosphoprotein reproducibility. C, Distribution of phosphorylated Ser, Thr and Tyr sites. D, A histogram depicting numbers of p-sites observed per protein. E, A histogram indicating enriched GO terms of phosphoproteins.
The 1621 p-sites of 799 proteins identified in this study were consisted of 1354 pSer, 258 pThr and 9 pTyr sites in a relative abundance of 83.53%, 15.92%, and 0.56%, respectively (Fig. 2C). The average number of p-sites per protein was 2.03. Approximately 45.8%, 12.9%, and 0.9% of the phosphoproteins contained ≥ 2, ≥ 4, and ≥ 10 p-sites, respectively. The most highly phosphorylated protein was MGG_02655T0, in which 16 p-sites were identified (Fig. 2D). Its orthologs are widely conserved in fungi, but none of them have been functionally characterized.
The specific recognition of substrates by a typical protein kinase is based on the presence of a consensus phosphorylation site in substrates (57). To uncover the consensus, we conducted motif-x analysis on the 1621 p-sites of 799 phosphoproteins identified in mycelia of P. oryzae. In total, 19 distinct types of consensus sequences around the p-sites were extracted, which could be classified into 6 acidic, 7 basic and 6 proline-directed motifs. The peptides containing acidic, basic, and proline-directed motifs accounted for 17.1%, 38.5%, and 44.4%, respectively. The 19 phosphorylation motifs consisted of 16 pSer motifs and 3 pThr motifs, but not pTyr motif. As compared with a previous phosphoproteome study (29), 8 of 19 motifs were newly identified for the fungal pathogen. Notably, there were three proline-directed motifs for which typical kinases have not been determined according to previous studies (50, 51) (supplemental Fig. S3, supplemental Table S5).
Gene Ontology and Enrichment Analysis of Phosphoproteins Identified in Mycelia
Gene ontology (GO) annotation and enrichment analysis were conducted on the 799 identified phosphoproteins for comprehensive understanding the phosphorylation events occurred in mycelia of P. oryzae. A total of 576 proteins could be functionally annotated with the gene ontology by Blast2GO (52, 53), distributing in 364 biological process terms, 140 cellular component terms, and 265 molecular function terms (supplemental Table S3). Top 15 GO terms of biological process, molecular function and cellular component were displayed by pie charts (supplemental Fig. S2). Unsurprisingly, the mycelium development process was the biggest piece (88 proteins) in biological process pie chart. The second and third biggest pieces were the metabolic process (46 proteins) and protein phosphorylation process (31 proteins), respectively. The top 3 molecular function terms were ATP binding (91 phosphoproteins), zinc ion binding (52 phosphoproteins), and DNA binding (48 phosphoproteins). Nucleus (66 proteins), ribosome (42 proteins), and integral components of membrane (41 proteins) were the three most frequent cellular components.
Enrichment analysis was furtherly conducted to reveal in which biological process, molecular function and cellular component terms that phosphorylation events were enhanced. In general, GO-annotated phosphoproteins were significantly enriched in 84 biological processes, 41 molecular functions and 30 cellular components (supplemental Table S4). After reducing these GO terms into the most specific ones, 14 biological processes, 10 molecular functions, and 7 cellular components terms were generated and plotted as histograms (Fig. 2E). Phosphorylation was enriched in proteins that are involved in hyphal development (GO:0043581, 1.7-fold), signal transduction (GO:0007165, 2.1-fold), RNA processing (GO:0006396, 2.8-fold), ribosome biogenesis processes (GO:0042254, 2.5-fold), chromosome organization (GO:0051276, 3.2-fold), regulation of translational initiation (GO:0006446, 3.6-fold) and elongation (GO:0006448, 5.2-fold), and mitotic cell cycle phase transition (GO:0044772, 6.7-fold). Notably, 80% proteins annotated for regulation of GTPase activity (GO:0043087) and late endosome to vacuole transport (GO:0045324) were phosphorylated. Phosphorylation of the proteins regulating translation initiation factor activity, translation elongation factor activity, ribosomal structural constituents, protein serine/threonine kinase activity, and GTPase activator activity were enriched 3.7-, 7.1-, 2.5-, 3.9-, and 44.8-fold, respectively. In terms of cellular component, phosphorylation was enriched in ribosome, nuclear part, endomembrane system, cell cortex part, microtubule, and pre-ribosome.
Phosphorylation of Pathogenesis-related Proteins
The phosphorylation and dephosphorylation are dynamic regulations of protein activities and signal transduction. According to literature, more than 450 protein-encoding genes have been characterized in P. oryzae, but only a few studies focused on phosphorylation regulation of their activities. Thus, we checked which of phosphoproteins identified in this study are related with pathogenesis, and found that at least 64 characterized proteins were phosphorylated on 110 p-sites, including 82 p-sites of 46 pathogenicity-related proteins. In comparison with a previous study (29), 47 p-sites of 31 pathogenicity-related proteins were newly identified (Table I). Phosphorylation was observed on 7 proteins of the Pmk1 pathway proteins (Mst7 and Mst11, Rga1, Cdc24, Mst20, Chm1, and Rgs1) (20, 23, 37, 39), 15 transcription factors (33, 58, 59), 6 RhoGAP proteins (37), 3 autophagy proteins (34), 3 light regulation related proteins (42, 43), 2 core septins (35), and 1 a spatial regulator of septation Sep1(60).
Table I. Identified phosphorylation of pathogenicity-related proteins and other reported proteins. As compared with the previous study (29), the newly identified p-sites in the present study were shown in boldface.
| Accession | Protein | P-sites | Function | Pathogenicity |
|---|---|---|---|---|
| MGG_00454 | MoATG13 | S356, S736 | initiation of autophagy | defect |
| MGG_03139 | MoATG18 | S341 | recycling | defect |
| MGG_03459 | MoATG26 | S27, S32, S270, S593, T793 | selective autophagy | No defects observed |
| MGG_06320 | CHM1 | S77 | PAK kinase on MAPK pathway | defect |
| MGG_12821 | MST20 | S273, S288, S612 | PAK kinase on MAPK pathway | No defects observed |
| MGG_14847 | MST11 | S557 | MEK on Pmk1 MAPK pathway | defect |
| MGG_00800 | MST7 | S358 | MEKK on Pmk1 MAPK pathway | defect |
| MGG_06482 | Mkk1 | S127 | MEKK on Mps1 MAPK pathway | unknown |
| MGG_02962 | CNF1 | S410 | Zn2Cys6 transcription factor (TF) | defect |
| MGG_00096 | FZC63 | S258 | Zn2Cys6 transcription factor (TF) | No defects observed |
| MGG_05153 | FZC73 | S44 | Zn2Cys6 transcription factor (TF) | No defects observed |
| MGG_15139 | FZC85 | S237 | Zn2Cys6 transcription factor (TF) | No defects observed |
| MGG_17841 | GPF1 | S540, S599, S643 | Zn2Cys6 transcription factor (TF) | defect |
| MGG_04108 | TAS1 | S672 | Zn2Cys6 transcription factor (TF) | No defects observed |
| MGG_06848 | CONx8 | S125, S615 | Cys2-His2 (C2H2) zinc finger TFs | No defects observed |
| MGG_02474 | GPF2 | S188 | Cys2-His2 (C2H2) zinc finger TFs | defect |
| MGG_03451 | GCF6 | S192, S544 | Cys2-His2 (C2H2) zinc finger TFs | defect |
| MGG_00504 | MoNSDC | S82, S247 | Cys2-His2 (C2H2) zinc finger TFs | No defects observed |
| MGG_02505 | MoREI1 | S196, T219, S490 | Cys2-His2 (C2H2) zinc finger TFs, RNA-binding proteins | defect |
| MGG_05709 | MGG_05709 | S291 | bHLH protein, putative TF | defect |
| MGG_09560 | EXP5 | T1259 | karyopherin, orthologue of exportin-5 | defect |
| MGG_04478 | Fim1 | S94,T112 | actin binding protein | unknown |
| MGG_02696 | HEX1 | S300 | hexagonal peroxisome, Woronin body | defect |
| MGG_06070 | Fis1 | S51, T55, S215, S528 | mitochondrial fission protein | defect |
| MGG_04377 | LRG1 | S648 | RhoGAP protein | defect |
| MGG_04186 | MoRga1 | S210, T315, S528, S574, S577, S590, S592 | RhoGAP protein | defect |
| MGG_06390 | MoRga2 | S363, S366, S699, S727, S805 | RhoGAP protein | No defects observed |
| MGG_03048 | MoRga3 | S53, S542 | RhoGAP protein | defect |
| MGG_09531 | MoRga4 | S878 | RhoGAP protein | defect |
| MGG_09303 | MoRga5 | T304 | RhoGAP protein | defect |
| MGG_13013 | MoChs6 | S634 | chitin synthases | defect |
| MGG_13014 | MoChs5 | S1688 | chitin synthases | No defects observed |
| MGG_09639 | MoAgs1 | S1801, S1804, S1887, S1900 | a-1,3-glucan synthase | defect |
| MGG_02961 | MoAnd1 | T1323, S1871 | cell cortex protein | defect |
| MGG_04009 | MobZIP08 | S201 | a basic leucine zipper TF | No defects observed |
| MGG_07705 | MoCps1 | S59 | acyl-CoA ligase-like protein | defect |
| MGG_05133 | MoCrz1 | S305 | calcineurin-dependent TF | defect |
| MGG_01342 | MoHik2 | S1084 | histidine kinase | defect |
| MGG_06696 | MoHik6 | S579 | histidine kinase | defect |
| MGG_06564 | MoPac2 | S358, S360, S402, S406 | Gti1/Pac2 family domain protein | defect |
| MGG_05332 | MoPlc2 | T254 | phospholipase C | defect |
| MGG_15018 | MoPlc5 | S201 | phospholipase C | unknown |
| MGG_07003 | MoSak1 | S898 | Snf1-activating kinase | defect |
| MGG_06930 | MoSip2 | S42, S346 | beta-subunit of Snf1 kinase | defect |
| MGG_06726 | MoSep4 | S318 | Septin | defect |
| MGG_03087 | MoSep5 | S368 | Septin | defect |
| MGG_04100 | SEP1 | S479, S1369 | Septation initialization, Cdc7/SepH homolog | defect |
| MGG_11534 | MoSre1 | S215 | sterol regulatory element-binding protein | defect |
| MGG_08556 | MoVea | T259 | velvet gene | defect |
| MGG_06399 | MoYAK1 | S326, Y580, S924 | a dual-specificity tyrosine-regulated protein kinase | defect |
| MGG_05280 | NOXR | S321 | NADPH oxidase | defect |
| MGG_02773 | MoMcm1 | S41 | MADS-box protein | defect |
| MGG_09471 | NTH1 | S12, S45, T361 | neutral trehalase | defect |
| MGG_00529 | PEX6 | S490 | PEROXIN | defect |
| MGG_14517 | Rgs1 | S159, S399, S401, S510 | regulator of G-protein signaling | defect |
| MGG_05857 | RPD3 | S443 | the histone deacetylase complex | unknown |
| MGG_13498 | Sin3 | S1369 | histone deacetylase complex | unknown |
| MGG_03538 | WC1 | S1019 | blue light receptor | unknown |
| MGG_08084 | MoSsd1 | S196, T1280, S1282 | orthologue of a cell wall assembly regulator ScSsd1 | defect |
| MGG_00501 | TDG2 | S411 | Tra1-dependent gene TF | defect |
| MGG_05247 | MGD1 | S24 | Tra1-dependent NAD-specific glutamate dehydrogenase | defect |
| MGG_02378 | GLD2 | S9 | Tra1-dependent glutamate decarboxylase | unknown |
| MGG_06439 | Tea4 | S11 | cell end marker protein | defect |
| MGG_07503 | TPX1 | T41, T59 | thioredoxin peroxidase | defect |
The Pmk1 MAPK pathway is well known to regulate infection-related morphogenesis, particularly in differentiation of mature appressoria and invasive hyphae in P. oryzae (22, 23). More than 16 components of the Pmk1 pathway have been identified (18, 19). This study showed nine of them were phosphorylated in the mycelia (Table I) (20, 23, 37, 39), including Mst50 and Ras1 that were phosphorylated at ambiguous site(s) (supplemental Table S1). Notably, 7, 4, 3, and 3 p-sites were detected for Rga1, Rgs1, Cdc24 and Mst20, respectively, indicating that they are hyper-phosphorylated. To our surprise, all these detected phosphoproteins function upstream of Pmk1, whereas phosphorylation was not detected for Pmk1 (23) and its downstream targets (Mst12 and Sfl1) (61, 62). Moreover, Mkk1, Mst11, and Mst7 were detected to be phosphorylated at Ser127, Ser557, and Ser358 in mycelia, respectively, which differed from those sites that have been previously reported to be important for activation of the downstream proteins (23, 55, 63).
Pmp1 is a Novel Virulence Factor and Important for Dephosphorylation of MAP Kinase Pmk1 and Mps1 in Mycelia
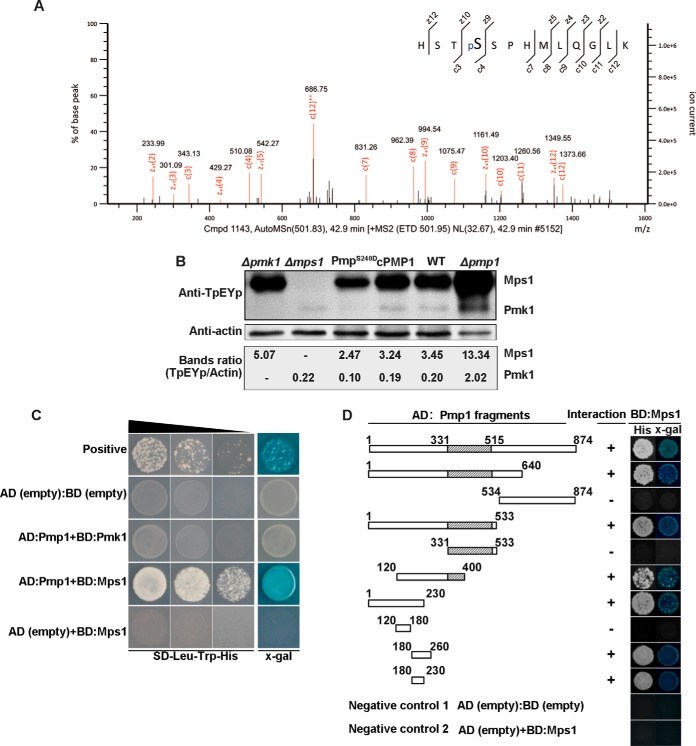
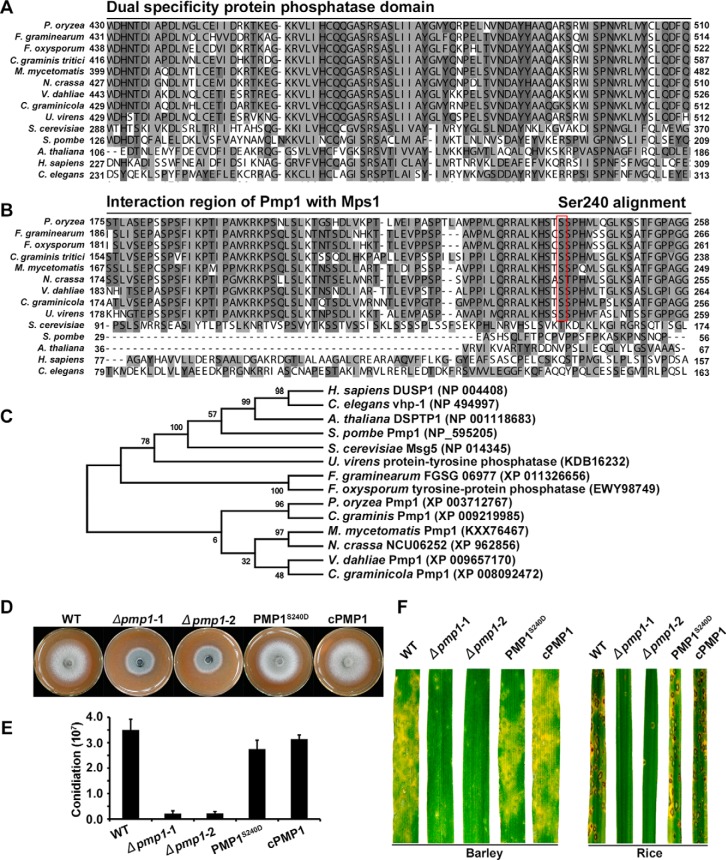
We noted that Pmp1 (MGG_15140) was phosphorylated at the site Ser240 in the mycelia (Fig. 3A). Pmp1 is a putative protein phosphatase that contained dual specificity protein phosphatase domain (Fig. 4A). In S. cerevisiae, the dual specificity protein phosphatase Msg5p negatively regulates the MAPK kinase Fus3p (64, 65), which is the ortholog of Pmk1 in P. oryzae (22). Because Pmk1 was dephosphorylated in mycelia, we reasoned that Pmp1 with phosphorylation at Ser240 site may function as an ortholog of Msg5p and is possibly involved in regulating phosphorylation levels of Pmk1. To verify this speculation, we generated two PMP1 knockout mutants by replacing the gene with hygromycin phosphotransferase coded gene HPH (supplemental Fig. S4). Western blot assay with anti-TpEYp antibody showed that the phosphorylation level of Pmk1 was significantly increased in the Δpmp1 mutants as compared with the wild type P131 and the complemented transformant (Fig. 3B). We also constructed the site-directed dominant PMP1S240D allele and transformed it into a Δpmp1 mutant. Further Western blot assay showed that the phosphorylation level of Pmk1 was significantly decreased in the PMP1S240D transformants as compared with the wild type P131 and the complemented strain (Fig. 3B). To our surprise, phosphorylation of Mps1, which is another MAPK kinase essential to pathogenicity, was also significantly increased in the Δpmp1 mutant and decreased in PMP1S240D mutant (Fig. 3B). We further did yeast two-hybrid assays to verify whether Pmp1 directly interacts with Pmk1 and Mps1, and obtained results showing that Pmp1 could directly interact with Mps1 (Fig. 3C) by the region AA180–230 (Fig. 3D). However, Pmp1 was unable to physically interact with Pmk1 (Fig. 3C), suggesting that Pmp1-mediated dephosphorylation of Pmk1 is indirect or may require a mediator. Blast search and sequence alignment indicated that Pmp1 orthologous proteins are widely distributed but are very divergent in eukaryotes (Fig. 4A–4C). Notably, the region interacting with Mps1 and the site pSer240 in Pmp1 are conserved only in filamentous fungi but not in the orthologous proteins from yeasts, plants and animals (Fig. 4B). A phylogenetic tree which was constructed with the full sequence comparison of dual specific protein phosphatase from different organisms showed that the Pmp1 in P. oryzae is distant those in the yeasts, plants and animals but highly similar to those from ascomycete fungi (Fig. 4C).
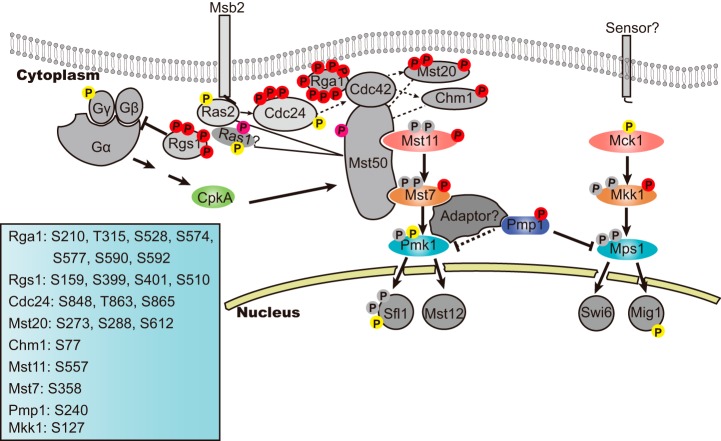
Fig. 3.
Phosphatase Pmp1 regulates phosphorylation of Pmk1 and Mps1 in mycelia of P. oryzae. A, A MS/MS spectrum depicting phosphorylation of Pmp1 at Ser240. B, Western blot showing phosphorylation of Mps1 and Pmk1 in mycelia of the wild-type stain P131, Δpmk1, Δmps1, Δpmp1, Δpmp1 complemented strain cPmp1 and a Pmp1S240D allele transformant of Δpmp1. Total proteins were extracted from mycelia of the strains and phosphorylation of Mps1 (46 kDa) and Pmk1 (42 kDa) were detected with anti-TpEYp antibody as previously described (55, 56). C, Yeast two-hybrid assay showing Pmp1 interacts Mps1 but not Pmk1. Yeast cells expressing the prey construct of Pmp1 and bait constructs of Pmk1 or Mps1 were assayed for growth on SD-Leu-Trp-His plates and for β-galactosidase activities (x-gal), respectively. Yeast cells expressing the empty prey construct pGADT7 with empty bait construct pGBKT7 or with Mps1 bait construct were used as two negative controls, and pGADT7-T prey construct with pGBKT7–53 bait construct were used as a positive control. D, Pmp1 fragment prey constructs for determining Pmp1 regions that interacts with Mps1(the left) and the yeast two-hybrid assays (right). The same negative controls were used as described in Fig. 3C.
Fig. 4.
Functional characterization of Pmp1. Alignments of the dual specificity domain (A) and the interaction region of Pmp1 (B) from P. oryzae with orthologs from other organisms were performed using the ClustalX program. C, A phylogenetic tree of Pmp1 orthologs, generated by the MEGA7 program, with 500 bootstraps. The sequences of dual specificity protein phosphatase families were from organisms as follows: Pyricularia oryzae Pmp1 (XP_003712767), Madurella mycetomatis Pmp1 (KXX76467), Neurospora crassa NCU06252 (XP_962856), Gaeumannomyces graminis Pmp1 (XP_009219985), Fusarium graminearum FGSG_06977 (XP_011326656), Verticillium dahlia Pmp1 (XP_009657170), Colletotrichum graminicola dual specificity phosphatase (XP_008092472), Fusarium oxysporum tyrosine-protein phosphatase (EWY98749), Ustilaginoidea virens protein-tyrosine phosphatase (KDB16232), Homo sapiens DUSP1 (NP_004408), Schizosaccharomyces pombe Pmp1 (NP_595205), Saccharomyces cerevisiae MSG5 (NP_014345), Arabidopsis thaliana DSPTP1 (NP_001118683), Caenorhabditis elegans vhp-1 (NP_494997). D, Colony growth of wild-type strain P131 (WT), pmp1 deletion mutants (Δpmp1-1 and Δpmp1-2), PmpS240D allele transformant and complemented strain cPmp1 on OTA medium for 5 days. E, Conidiation of the same set of strains. F, Lesions formed by the same set of strains on barley (Left) and rice (right) leaves. Barley and rice seedlings were sprayed with conidial suspensions, and cultured for 5 days and 7 days, respectively.
In addition, we characterized phenotype changes in the Δpmp1 mutants and observed that the mutants were significantly reduced in mycelial growth and in conidiation as compared with the wild type strain P131 and the complementary transformants (Fig. 4D and 4E). Infection assays showed that the Δpmp1 mutants were also dramatically reduced in virulence to both rice and barley (Fig. 4F). However, PmpS240D strains were undistinguished from the wild-type P131 in mycelial growth, conidiation and virulence (Fig. 4D–4F). These results indicate that PMP1 is an important gene for mycelial growth, conidiation, and pathogenicity.
DISCUSSION
General Characteristics of the Phosphoproteome in Mycelia of P. oryzae
In the present study, we identified a total of 1621 confident p-sites of 799 proteins in mycelia of the P. oryzae p131 strain, including 355 p-sites that shared in all three duplicates (Fig. 2A, supplemental Table S2). In addition, 47%–59% of the p-sites and 57.4%–73.1% of the phosphoproteins were shared between two duplicates in this study (Fig. 2B). As compared with an early study in which 1393 p-sites of 831 proteins were identified in mycelia of the 70–15 strain (29), 480 p-sites of 332 proteins were overlapped, accounting for 29.6%–34.4% of the p-sites and ∼40% of the proteins identified in mycelia between the two studies (supplemental Fig. S1). These overlap percentages are much higher than those reported in S. cerevisiae, in which, about 12% of p-sites and 28% of phosphoproteins were shared between any two experiments (8). However, this study included 899 new p-sites in 536 proteins that were not described in the previous study (supplemental Table S2) (29). For the differences in p-sites and phosphoproteins identified between the two studies, there may be following reasons: (1) Different culture conditions were applied. In our study, mycelia were cultured in the yeast extract glucose medium for 48 h and then harvested for the protein analysis, whereas in the previous study, mycelia were cultured in the minimal medium for 5 days (2). Different strains were used. In this study, a field isolate P131 was used, whereas in the previous study, a laboratory strain 70–15 was used. These two strains also differ at the genome level (30) (3). Different protocols were used for sample preparation and detection. In the previous study, phosphopeptides were enriched by IMAC and TiO2 combination approach and analyzed with Q-Exactive hybrid mass spectrometer, followed with MaxQuant data search (29). In our study, SCX/IMAC enrichment approach was implemented for phosphopeptide enrichment, and neutral loss triggered ETD strategy was used for data acquisition by ion trap amaZon speed ETD, followed by Mascot search. So far, only a few studies focused on phosphoproteome of P. oryzae, which are undoubtedly insufficient for covering all the phosphorylation events. Thus, more phosphoproteome studies of P. oryzae are encouraged to enhance the understanding of phosphorylation in P. oryzae.
It is believed that fungi do not have any tyrosine kinases (TKs). However, the present study identified 0.56% pTyr sites in mycelia of P. oryzae, which was quite like the previous study (29), but was slight lower than those (1.4%-2.8%) detected in phosphoproteins from yeasts, mammals and higher plants (8–12). Some previous studies reported that a fungal-specific lineage of protein kinases (FslKs) may function as TK functional analogs. In addition, some serine or threonine kinases as dual specificity kinases can phosphorylate Tyr sites (66–69). Thus, further studies will be required to identify protein kinases that are responsible for the phosphorylation of Tyr sites in P. oryzae.
Phosphorylation of Pathogenicity-Related Proteins
In the present study, we detected phosphorylation of 46 pathogenicity-related proteins at 82 p-sites in mycelia of P. oryzae (Table I). The p-sites identified in this study will be invaluable for uncovering how phosphorylation regulates functions of the proteins.
Sep1 is essential for determining the position and frequency of cell division sites in P. oryzae (60). In this study, we detected pSer479 of Sep1 in addition to Ser1369, which has been indicated as a candidate target of PKA kinase because the p-site could be detected in the wild type, but not in the ΔcpkA mutant (29). pSer479 occurred in the acid motif pSD, which can be recognized by CK2 (50). Therefore, CK2 together with PKA may mediate the phosphorylation of Sep1. To verify this, further investigation will be required.
We also detected pSer557 of Mst11 and pSer358 of Mst7, which are different from the pSer453 and pSer458 of Mst11, and the pSer212 and pThr216 of Mst7 that have been determined as important p-sites for activating the Pmk1 cascade and for appressorium formation (23, 55). In addition, pSer127 was detected in the MEK Mkk1, which functions upstream of Mps1 and has pThr369 and pThr375 that are important for phosphorylation and activation of Mps1 (63). It would be interesting to investigate whether and how the pathogenicity-associated functions of Mst11, Mst7, and Mkk1 are affected by these newly identified phosphorylation sites.
Rho GAP proteins play important roles in mycelial growth and infection processes of P. oryzae (37). The present study showed that 6 Rho GAP proteins were phosphorylated in mycelia, including LRG1, and MoRga1. All 6 Rho GAP proteins specifically interacted with some Rho GTPases, and their encoding gene deletion mutants displayed one or more defects in mycelial growth, conidiation, and appressorium formation. In particular, LRG1 and MoRga1 are important for pathogenicity (37). However, functional regulation of these proteins by phosphorylation has not been reported and should be investigated.
It should be also noted that mycelia cultured in rich medium were used in this study. Phosphorylation of proteins at many p-sites identified in this study is likely a response to nutrient signals or stress, which are encountered by the fungus during cultivation. Thus, phosphorylation at these p-sites may be not closely related with function regulation of the proteins during infection.
Phosphorylation Site Consensuses
In this study, we extracted 19 conserved phosphorylation motifs from 1621 p-sites identified in mycelia of P. oryzae, including 3 proline-directed motif that have not been assigned to typical protein kinases (supplemental Fig. S3, supplemental Table S5). For identifying protein kinases that specifically recognize the three motifs, the deletion mutants of protein kinase-encoding genes in F. graminearum, N. crassa and Aspergillus nidulans (4, 13, 70) will be invaluable because P. oryzae is a close relative to these fungi (17), although a similar set of protein kinase gene deletion mutants has not been generated.
Pmp1 Regulates Phosphorylation of Pmk1 and Mps1 MAP Cascades and is Important for Virulence in P. oryzae
Previous studies have reported that functional Pmk1 for appressorium formation requires phosphorylated at TEY residue (23) and the phosphorylation is required to phosphorylate Mst12 (61) and Sfl1 (62). Consistently, the present study showed that Pmk1 with its targets Mst12 and Sfl1 was not phosphorylated in mycelia of P. oryzae. Interestingly, we found that Pmp1 was phosphorylated at the site Ser240 in mycelia (Fig. 3A) and proved that the phosphorylation of Pmk1 was significantly increased in the Δpmp1 mutant, but decreased in the site-directed dominant PMP1S240D allele mutant (Fig. 3B) indicating that dephosphorylation of Pmk1 in mycelia of P. oryzae is regulated by Pmp1 and that phosphorylation at the site Ser240 of Pmp1 is important for dephosphorylating Pmk1. Similarly, the dual specificity protein phosphatase Msg5p dephosphorylates MAP kinase Fus3p in S. cerevisiae (64), which is the ortholog of Pmk1 in P. oryzae. To our surprise, phosphorylation of Mps1, another MAP kinase required for pathogenicity (24), was also significantly increased in the Δpmp1 mutant, and decreased in the site-directed dominant PMP1S240D allele mutant (Fig. 3B). Further yeast two-hybrid assays indicated that Pmp1 can directly interact with Mps1. These results clearly showed that Pmp1 regulates dephosphorylation of Mps1.
Although orthologous proteins of Mps1 and Pmp1 are widely distributed, dephosphorylation of Mps1 by Pmp1 has not been reported in any other fungi. Sequence alignment and phylogenetic tree indicated Pmp1 orthologous proteins are highly diversified in eukaryotes (Fig. 4A and 4C). Notably, the region interacting with Mps1 and the pSer240 site in Pmp1 are conserved only in the orthologous proteins from filamentous fungi but not in those from yeasts, plants and animals (Fig. 4B). It would be interesting to investigate whether similar mechanisms of interaction and p-sites exist in filamentous fungi, and to identify and compare the interaction regions and p-sites of Pmp1 orthologs important for dephosphorylating targets in yeasts, plants and animals.
Integration of our results with previous results prompted an update of the MAP Pmk1 and the Mps1 cascades with Pmp1 as their new component (Fig. 5) (18, 19, 23, 29). In mycelia, Pmk1 and Mps1 are dephosphorylated, thus unable to phosphorylate downstream transcription factors. Pmp1 serving as a MAPK phosphatase regulates dephosphorylation of Pmk1 and Mps1, and the regulation is enhanced via the phosphorylation of Pmp1 on Ser240. An adaptor may exist to mediate the dephosphorylation of Pmk1 by Pmp1 because they are unable to directly interact with each other, whereas the Pmp1 directly interact with Mps1 for the dephosphorylation.
Fig. 5.
An updated Pmk1 and Mps1 MAPK signaling pathways involving Pmp1 and other components with phosphorylation sites. Phosphorylation sites identified in this study are labeled in red, and the positions of confident p-sites in individual proteins detected in this study are listed on bottom left corner; In pink are protein phosphorylation at ambiguous site observed in this study; In yellow, are distinct p-sites detected in mycelia in the previous phosphoproteome study (29); In gray, are phosphorylation sites reported by other previous studies (23, 55, 62, 63) but did not detected in mycelia by the two phosphoproteome studies.
This study showed that the Δpmp1 mutants are defective in mycelial growth, conidiation and virulence (Fig. 4D–4F) and differs from Δpmk1 and Δmps1 mutants, both of which are normal in mycelial growth (22, 24). It can be thus inferred that involvement of Pmp1 in mycelial growth is independent on the Pmp1-mediated dephosphorylation of Pmk1 and Mps1. Pmp1 may dephosphorylate some other proteins that function as negative regulators for mycelial growth. However, Pmp1, Mps1 and Pmk1 are important for conidiation and virulence. Further studies will be also required to reveal how the involvement of Pmp1 in virulence of P. oryzae is related with the phosphorylation regulation of Pmk1 and Mps1.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset) via the PRIDE partner repository with the dataset identifier PXD002364.
Supplementary Material
Footnotes
Author contributions: R.W., Q.X.L., and Y.P. designed research; R.W. and J.P. performed research; R.W., Q.X.L., and Y.P. analyzed data; Y.P., R.W., and Q.X.L. wrote the paper.
* This study was supported in part by the program for Changjiang Scholars and Innovative Research Team Project (IRT1042) and the 111 Project (B13006) from the Ministry of Education, China, and by the US National Institute on Minority Health and Health Disparities grant 8G12 MD007601. RW was a recipient of the Chinese Scholarship Council scholarship. We thank Dr. Mahdi Belcaid at the University of Hawaii, and Changfa Yin in China Agricultural University for assistance with data analysis. Phosphoprotein identification was done in the University of Hawaii.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PAK
- p21-activated kinases
- OTA
- Oatmeal Tomato Agar
- YEG
- Yeast Extract Glucose medium
- SCX
- strong cation exchange
- ETD
- Electron-Transfer Dissociation
- FDR
- false discovery rate
- GO
- gene ontology
- cAMP
- cyclic adenosine monophosphate
- MAPK
- mitogen-activated protein kinase
- MEK
- mitogen-activated protein kinase kinase
- MEKK
- mitogen-activated protein kinase kinase kinase
- AMPK
- SNF1/AMP-activated protein kinase
- CK2
- casein kinase 2
- DUSP
- dual-specific phosphatase.
REFERENCES
- 1. Hunter T. (2000) Signaling–2000 and beyond. Cell 100, 113–127 [DOI] [PubMed] [Google Scholar]
- 2. Pawson T., and Scott J. D. (2005) Protein phosphorylation in signaling–50 years and counting. Trends Biochem. Sci. 30, 286–290 [DOI] [PubMed] [Google Scholar]
- 3. Macek B., Mann M., and Olsen J. V. (2009) Global and site-specific quantitative phosphoproteomics: principles and applications. Annu. Rev. Pharmacol. Toxicol. 49, 199–221 [DOI] [PubMed] [Google Scholar]
- 4. Park G., Servin J. A., Turner G. E., Altamirano L., Colot H. V., Collopy P., Litvinkova L., Li L., Jones C. A., Diala F. G., Dunlap J. C., and Borkovich K. A. (2011) Global analysis of serine-threonine protein kinase genes in Neurospora crassa. Eukaryotic Cell 10, 1553–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ghosh A., Servin J. A., Park G., and Borkovich K. A. (2014) Global analysis of serine/threonine and tyrosine protein phosphatase catalytic subunit genes in Neurospora crassa reveals interplay between phosphatases and the p38 mitogen-activated protein kinase. G3 4, 349–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maier B., Wendt S., Vanselow J. T., Wallach T., Reischl S., Oehmke S., Schlosser A., and Kramer A. (2009) A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 23, 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., and Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 8. Amoutzias G. D., He Y., Lilley K. S., Van de Peer Y., and Oliver S. G. (2012) Evaluation and properties of the budding yeast phosphoproteome. Mol. Cell. Proteomics 11, M111 009555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilson-Grady J. T., Villen J., and Gygi S. P. (2008) Phosphoproteome analysis of fission yeast. J. Proteome Res. 7, 1088–1097 [DOI] [PubMed] [Google Scholar]
- 10. Nakagami H., Sugiyama N., Mochida K., Daudi A., Yoshida Y., Toyoda T., Tomita M., Ishihama Y., and Shirasu K. (2010) Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153, 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villen J., Haas W., Sowa M. E., and Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lundby A., Secher A., Lage K., Nordsborg N. B., Dmytriyev A., Lundby C., and Olsen J. V. (2012) Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat. Commun. 3, 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang C., Zhang S., Hou R., Zhao Z., Zheng Q., Xu Q., Zheng D., Wang G., Liu H., Gao X., Ma J. W., Kistler H. C., Kang Z., and Xu J. R. (2011) Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum. PLoS Pathog. 7, e1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rampitsch C., Tinker N. A., Subramaniam R., Barkow-Oesterreicher S., and Laczko E. (2012) Phosphoproteome profile of Fusarium graminearum grown in vitro under nonlimiting conditions. Proteomics 12, 1002–1005 [DOI] [PubMed] [Google Scholar]
- 15. Davanture M., Dumur J., Bataille-Simoneau N., Campion C., Valot B., Zivy M., Simoneau P., and Fillinger S. (2014) Phosphoproteome profiles of the phytopathogenic fungi Alternaria brassicicola and Botrytis cinerea during exponential growth in axenic cultures. Proteomics 14, 1639–1645 [DOI] [PubMed] [Google Scholar]
- 16. Dean R., Van Kan J. A., Pretorius Z. A., Hammond-Kosack K. E., Di Pietro A., Spanu P. D., Rudd J. J., Dickman M., Kahmann R., Ellis J., and Foster G. D. (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dean R. A., Talbot N. J., Ebbole D. J., Farman M. L., Mitchell T. K., Orbach M. J., Thon M., Kulkarni R., Xu J. R., Pan H., Read N. D., Lee Y. H., Carbone I., Brown D., Oh Y. Y., Donofrio N., Jeong J. S., Soanes D. M., Djonovic S., Kolomiets E., Rehmeyer C., Li W., Harding M., Kim S., Lebrun M. H., Bohnert H., Coughlan S., Butler J., Calvo S., Ma L. J., Nicol R., Purcell S., Nusbaum C., Galagan J. E., and Birren B. W. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986 [DOI] [PubMed] [Google Scholar]
- 18. Wilson R. A., and Talbot N. J. (2009) Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7, 185–195 [DOI] [PubMed] [Google Scholar]
- 19. Li G., Zhou X., and Xu J. R. (2012) Genetic control of infection-related development in Magnaporthe oryzae. Curr. Opin. Microbiol. 15, 678–684 [DOI] [PubMed] [Google Scholar]
- 20. Li L., Xue C., Bruno K., Nishimura M., and Xu J. R. (2004) Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol. Plant-Microbe Interact. 17, 547–556 [DOI] [PubMed] [Google Scholar]
- 21. Adachi K., and Hamer J. E. (1998) Divergent cAMP signaling pathways regulate growth and pathogenesis in the rice blast fungus Magnaporthe grisea. Plant Cell 10, 1361–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu J. R., and Hamer J. E. (1996) MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10, 2696–2706 [DOI] [PubMed] [Google Scholar]
- 23. Zhao X., Kim Y., Park G., and Xu J. R. (2005) A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell 17, 1317–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu J. R., Staiger C. J., and Hamer J. E. (1998) Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. U.S.A. 95, 12713–12718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jeon J., Goh J., Yoo S., Chi M. H., Choi J., Rho H. S., Park J., Han S. S., Kim B. R., Park S. Y., Kim S., and Lee Y. H. (2008) A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol. Plant-Microbe Interact. 21, 525–534 [DOI] [PubMed] [Google Scholar]
- 26. Dixon K. P., Xu J. R., Smirnoff N., and Talbot N. J. (1999) Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 11, 2045–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Motoyama T., Ochiai N., Morita M., Iida Y., Usami R., and Kudo T. (2008) Involvement of putative response regulator genes of the rice blast fungus Magnaporthe oryzae in osmotic stress response, fungicide action, and pathogenicity. Curr. Genet. 54, 185–195 [DOI] [PubMed] [Google Scholar]
- 28. Zeng X. Q., Chen G. Q., Liu X. H., Dong B., Shi H. B., Lu J. P., and Lin F. (2014) Crosstalk between SNF1 pathway and the peroxisome-mediated lipid metabolism in Magnaporthe oryzae. PLoS ONE 9:e103124, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Franck W. L., Gokce E., Randall S. M., Oh Y., Eyre A., Muddiman D. C., and Dean R. A. (2015) Phosphoproteome analysis links protein phosphorylation to cellular remodeling and metabolic adaptation during Magnaporthe oryzae appressorium development. J. Proteome Res. 14, 2408–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xue M., Yang J., Li Z., Hu S., Yao N., Dean R. A., Zhao W., Shen M., Zhang H., Li C., Liu L., Cao L., Xu X., Xing Y., Hsiang T., Zhang Z., Xu J. R., and Peng Y. L. (2012) Comparative analysis of the genomes of two field isolates of the rice blast fungus Magnaporthe oryzae. PLoS Genet. 8, e1002869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kosti I., Mandel-Gutfreund Y., Glaser F., and Horwitz B. A. (2010) Comparative analysis of fungal protein kinases and associated domains. BMC Genomics 11, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yachie N., Saito R., Sugiyama N., Tomita M., and Ishihama Y. (2011) Integrative features of the yeast phosphoproteome and protein-protein interaction map. PLoS Comput. Biol. 7, e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu J., Cao H., Zhang L., Huang P., and Lin F. (2014) Systematic analysis of Zn2Cys6 transcription factors required for development and pathogenicity by high-throughput gene knockout in the rice blast fungus. PLoS Pathog. 10, e1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kershaw M. J., and Talbot N. J. (2009) Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc. Natl. Acad. Sci. U.S.A. 106, 15967–15972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dagdas Y. F., Yoshino K., Dagdas G., Ryder L. S., Bielska E., Steinberg G., and Talbot N. J. (2012) Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 336, 1590–1595 [DOI] [PubMed] [Google Scholar]
- 36. Gupta Y. K., Dagdas Y. F., Martinez-Rocha A. L., Kershaw M. J., Littlejohn G. R., Ryder L. S., Sklenar J., Menke F., and Talbot N. J. (2015) Septin-dependent assembly of the exocyst is essential for plant infection by Magnaporthe oryzae. Plant Cell 27, 3277–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ye W., Chen X., Zhong Z., Chen M., Shi L., Zheng H., Lin Y., Zhang D., Lu G., Li G., Chen J., and Wang Z. (2014) Putative RhoGAP proteins orchestrate vegetative growth, conidiogenesis and pathogenicity of the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 67, 37–50 [DOI] [PubMed] [Google Scholar]
- 38. Zhang H., Tang W., Liu K., Huang Q., Zhang X., Yan X., Chen Y., Wang J., Qi Z., Wang Z., Zheng X., Wang P., and Zhang Z. (2011) Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog. 7, e1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu H., Suresh A., Willard F. S., Siderovski D. P., Lu S., and Naqvi N. I. (2007) Rgs1 regulates multiple galpha subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. EMBO J. 26, 690–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mir A. A., Park S. Y., Abu Sadat M., Kim S., Choi J., Jeon J., and Lee Y. H. (2015) Systematic characterization of the peroxidase gene family provides new insights into fungal pathogenicity in Magnaporthe oryzae. Sci. Rep. 5, 11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kong L. A., Yang J., Li G. T., Qi L. L., Zhang Y. J., Wang C. F., Zhao W. S., Xu J. R., and Peng Y. L. (2012) Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 8, e1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deng Y. Z., Qu Z., and Naqvi N. I. (2015) Twilight, a novel circadian-regulated gene, integrates phototropism with nutrient and redox homeostasis during fungal development. PLoS Pathog. 11, e1004972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee K., Singh P., Chung W. C., Ash J., Kim T. S., Hang L., and Park S. (2006) Light regulation of asexual development in the rice blast fungus, Magnaporthe oryzae. Fungal Genet. Biol. 43, 694–706 [DOI] [PubMed] [Google Scholar]
- 44. Choi J., Chung H., Lee G. W., Koh S. K., Chae S. K., and Lee Y. H. (2015) Genome-wide analysis of hypoxia-responsive genes in the rice blast fungus, Magnaporthe oryzae. PLoS One 10, e0134939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen X. L., Yang J., and Peng Y. L. (2011) Large-scale insertional mutagenesis in Magnaporthe oryzae by agrobacterium tumefaciens-mediated transformation. Methods Mol. Biol. 722, 213–224 [DOI] [PubMed] [Google Scholar]
- 46. Yang J., Zhao X., Sun J., Kang Z., Ding S., Xu J. R., and Peng Y. L. (2010) A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae. Mol. Plant-Microbe Interact. 23, 112–123 [DOI] [PubMed] [Google Scholar]
- 47. Villen J., and Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chou M. F., and Schwartz D. (2011) Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinformatics 21, 3674–3676 [DOI] [PubMed] [Google Scholar]
- 49. Villen J., Beausoleil S. A., Gerber S. A., and Gygi S. P. (2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S. G., and Pandey A. (2007) A curated compendium of phosphorylation motifs. Nat. Biotechnol. 25, 285–286 [DOI] [PubMed] [Google Scholar]
- 51. Gnad F., Gunawardena J., and Mann M. (2011) PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 39, D253–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., and Robles M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 [DOI] [PubMed] [Google Scholar]
- 53. Gotz S., Garcia-Gomez J. M., Terol J., Williams T. D., Nagaraj S. H., Nueda M. J., Robles M., Talon M., Dopazo J., and Conesa A. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heckman K. L., and Pease L. R. (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2, 924–932 [DOI] [PubMed] [Google Scholar]
- 55. Qi L., Kim Y., Jiang C., Li Y., Peng Y., and Xu J. R. (2015) Activation of Mst11 and feedback inhibition of germ tube growth in Magnaporthe oryzae. Mol. Plant-Microbe Interact. 28, 881–891 [DOI] [PubMed] [Google Scholar]
- 56. Zhao X., and Xu J. R. (2007) A highly conserved MAPK-docking site in Mst7 is essential for Pmk1 activation in Magnaporthe grisea. Mol. Microbiol. 63, 881–894 [DOI] [PubMed] [Google Scholar]
- 57. Ubersax J. A., and Ferrell J. E. Jr. (2007) Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541 [DOI] [PubMed] [Google Scholar]
- 58. Cao H., Huang P., Zhang L., Shi Y., Sun D., Yan Y., Liu X., Dong B., Chen G., Snyder J. H., Lin F., and Lu J. (2016) Characterization of 47 Cys2-His2 zinc finger proteins required for the development and pathogenicity of the rice blast fungus Magnaporthe oryzae. New Phytol. 211, 1035–1051 [DOI] [PubMed] [Google Scholar]
- 59. Kong S., Park S. Y., and Lee Y. H. (2015) Systematic characterization of the bZIP transcription factor gene family in the rice blast fungus, Magnaporthe oryzae. Environ. Microbiol. 17, 1425–1443 [DOI] [PubMed] [Google Scholar]
- 60. Saunders D. G., Dagdas Y. F., and Talbot N. J. (2010) Spatial uncoupling of mitosis and cytokinesis during appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae. Plant Cell 22, 2417–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Park G., Bruno K. S., Staiger C. J., Talbot N. J., and Xu J. R. (2004) Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol. Microbiol. 53, 1695–1707 [DOI] [PubMed] [Google Scholar]
- 62. Li G., Zhou X., Kong L., Wang Y., Zhang H., Zhu H., Mitchell T. K., Dean R. A., and Xu J. R. (2011) MoSfl1 is important for virulence and heat tolerance in Magnaporthe oryzae. PLoS One 6, e19951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fujikawa T., Kuga Y., Yano S., Yoshimi A., Tachiki T., Abe K., and Nishimura M. (2009) Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 73, 553–570 [DOI] [PubMed] [Google Scholar]
- 64. Doi K., Gartner A., Ammerer G., Errede B., Shinkawa H., Sugimoto K., and Matsumoto K. (1994) MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 13:61–70, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhan X. L., Deschenes R. J., and Guan K. L. (1997) Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 11, 1690–1702 [DOI] [PubMed] [Google Scholar]
- 66. Zhao Z., Jin Q., Xu J. R., and Liu H. (2014) Identification of a fungi-specific lineage of protein kinases closely related to tyrosine kinases. PLoS One 9, e89813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hanks S. K., and Hunter T. (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9, 576–596 [PubMed] [Google Scholar]
- 68. Rudrabhatla P., Reddy M. M., and Rajasekharan R. (2006) Genome-wide analysis and experimentation of plant serine/ threonine/tyrosine-specific protein kinases. Plant Mol. Biol. 60, 293–319 [DOI] [PubMed] [Google Scholar]
- 69. Dhanasekaran N., and Premkumar Reddy E. (1998) Signaling by dual specificity kinases. Oncogene 17, 1447–1455 [DOI] [PubMed] [Google Scholar]
- 70. De Souza C. P., Hashmi S. B., Osmani A. H., Andrews P., Ringelberg C. S., Dunlap J. C., and Osmani S. A. (2013) Functional analysis of the Aspergillus nidulans kinome. PLoS One 8, e58008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset) via the PRIDE partner repository with the dataset identifier PXD002364.