Abstract
We previously examined Salmonella proteome within infected host cells and found differential expression of many proteins with defined functional roles such as metabolism or virulence. However, the precise roles of other altered proteins in Salmonella pathogenesis are largely unknown. A putative transcriptional regulator, YdcR, was highly induced intracellularly whereas barely expressed in vitro, implicating potential relevance to bacterial infection. To unveil its physiological functions, we exploited quantitative proteomics of intracellular Salmonella and found that genetic ablation of ydcR resulted in severe repression of SrfN, a known virulence factor. Immunoblotting, qRT-PCR, and β-galactosidase assays further demonstrate YdcR-dependent transcription and expression of srfN. Moreover, we found physical interaction of YdcR with the promoter region of srfN, suggesting direct activation of its transcription. Importantly, a Salmonella mutant lacking ydcR was markedly attenuated in a mouse model of infection. Our findings reveal that YdcR temporally regulates the virulence factor SrfN during infection, thus contributing to Salmonella pathogenesis. Our work also highlights the utility of combining quantitative proteomics and bacterial genetics for uncovering the functional roles of transcription factors and likely other uncharacterized proteins as well.
Salmonella enterica serovar Typhimurium (S. typhimurium) is one of the leading food-borne bacterial pathogens (1). Successful infection is highly dependent on the type III secretion systems (T3SSs) encoded by Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2)1 (2). T3SS functions as a molecular syringe that is able to translocate bacterial virulence factors (effector proteins) directly into host cytosol. These T3SS effector proteins, upon delivery, can modulate various host cellular processes to promote bacterial infection as well as survival within host cells (3). As an intracellular bacterial pathogen, S. typhimurium has the capability to invade nonphagocytic epithelial cells and proliferate with a membrane-bound compartment known as the Salmonella-containing vacuoles (SCVs) (4).
To understand how bacterial pathogens adapt to their intracellular niches, we previously studied the proteome of intracellular Salmonella within epithelial cells at distinct stages of infection (5, 6). At 6 h post-infection (hpi), Salmonella upregulates multiple pathways involved in acquisition of metal ions such as iron, suggesting a general shortage of these ions within the host (5). At a later stage of infection (e.g. 18 hpi), intracellular Salmonella undergoes massive metabolic reprogramming and favors glycolysis as well as mixed acid fermentation for its energy requirements, whereas the tricarboxylic acid (TCA) cycle and most respiration pathways are repressed (6). Overall, our quantitative proteomic measurements suggest extensive bacterial adaptations to infected host epithelial cells.
In addition, the large-scale proteomic data sets also reveal many other differentially regulated proteins and/or pathways during Salmonella infection. Of interest are transcription factors that were induced during the course of infection. We hypothesize that such regulators are likely to play important roles in bacterial pathogenesis. For instance, one of the MarR family transcription factors, SlyA, was 2-fold upregulated at 18 hpi. SlyA has been shown to contribute to Salmonella intracellular survival within macrophages and other aspects of bacterial physiology as well (7, 8, 9). Interestingly, we also identified a putative transcription factor YdcR that was highly induced throughout the infection process, though this protein was barely expressed (i.e. not detected in our proteomic measurements) in bacteria cultured in vitro. YdcR is homologous to the family of a MocR/GabR-type transcriptional regulator that falls into the GntR superfamily, one of the most widespread families of bacterial transcription factors that regulate diverse biological processes (10, 11), but the function of YdcR in Salmonella pathogenesis remains elusive.
Herein we quantitatively studied the protein expression of a Salmonella mutant lacking ydcR (ΔydcR) within host cells in comparison to that of its parental strain, which provides a powerful means to uncover potential YdcR-regulated proteins under physiological conditions. Notably, we identified a Salmonella virulence factor SrfN whose expression within infected host cells is strictly dependent on the presence of YdcR. Furthermore, we found evidence that YdcR directly control its expression on the transcriptional level by binding to the promoter region of the srfN gene. Importantly, the loss of YdcR resulted in attenuated bacterial virulence in a murine infection model. Our results underscore the power of combining quantitative proteomics and bacterial genetics to study the regulatory roles of diverse prokaryotic transcription factors.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Cell Lines, and Culture Conditions
The S. typhimurium SL1344 was used in this study, and all Salmonella strains were routinely grown on LB plates with 1.5% agar and 30 μg/ml streptomycin at 37 °C. A single colony picked from the plates was inoculated into LB broth with 30 μg/ml streptomycin, and then the overnight culture was diluted 1:20 into 3 ml of LB broth supplemented with 0.3 m NaCl (to induce the expression of Salmonella SPI-1 T3SS). The bacteria were harvested for infection assays when they grew to the mid-exponential phase (OD600 ≈ 0.9). HeLa cells (a human epithelial cell line) were grown in Dulbecco's Modified Eagle Medium (DMEM, Hyclone, Logan, UT) supplemented with 10% fetal bovine serum (FBS, Gibco, Life Technologies, Carlsbad, CA) under an atmosphere of 5% CO2 at 37 °C.
Molecular Cloning and Construction of Bacterial Mutants
The S. typhimurium ydcR deletion mutant (ΔydcR) was constructed using the standard homologous recombination method as previously described (5). The lambda red recombination system (12, 13) was used to construct the ydcR deletion mutant with kanamycin resistance and bacterial strains chromosomally expressing 3×FLAG-tagged YdcR and SrfN. Briefly, the sequences encoding the 3×FLAG epitope were inserted in-frame at the C termini of the genes of interest right before the stop codon. Successful deletion or tagging of the target genes was confirmed by both sequencing and PCR analyses. For constructing the complementation strain that harbors a plasmid-borne ydcR gene in the ΔydcR background (ΔydcR+pYdcR), the ydcR fragment was amplified, digested, and inserted into a plasmid with an arabinose-inducible promoter and a C-terminal 3×FLAG tag. For β-galactosidase assays, the upstream region (819 bp) of the srfN gene was cloned and inserted into the upstream of lacZ in the single copy plasmid pNN387 (14). All primers and strains that were used in this study are listed in the supplemental Table S1.
Experimental Design and Statistical Rationale
To uncover potential YdcR-regulated proteins, we performed proteomic analyses of the intracellular ΔydcR mutant at 18 hpi, whereas the wild-type strain was used as a control. Salmonella infection and subsequent isolation from host cells was performed as previously described, with minor modifications (6). Briefly, Salmonella invasion of HeLa cells was performed when cell monolayers reached 70–85% confluence, and the infection was carried out for 45 min in Hanks' balanced salt solution (HBSS) with a multiplicity of infection (MOI) of 30. Subsequently, cell monolayers were treated with 100 μg/ml gentamicin for 1 h to selectively eliminate extracellular S. typhimurium. Cells were then washed with HBSS and fresh DMEM containing 10 μg/ml gentamicin was added. At 18 hpi, cell monolayers were washed extensively with PBS, and lysed in a buffer containing 20 mm Tris-HCl (pH 7.6), 150 mm NaCl, and 0.1% Triton X-100. To recover intracellular bacteria, collected cell lysates were centrifuged at 600 × g for 5 min, and then the supernatant was centrifuged at 4000 × g for 20 min. The bacterial pellets were immediately washed with RIPA buffer to remove residual host proteins. The final pellets were resuspended in the SDS-PAGE sample buffer and heated at 95 °C for 5 min. Then bacterial protein samples were prefractionated by 10% SDS-PAGE, processed into 8 gel bands, and subjected to in-gel trypsin digestion as previously described (15). The resulting peptide samples were reconstituted in HPLC-grade water for LC-MS/MS analyses on a hybrid ion trap-Orbitrap mass spectrometer (LTQ Orbitrap Velos, Thermo Scientific) coupled with nanoflow reversed-phase liquid chromatography (EASY-nLC 1000, Thermo Scientific). The capillary column (75 μm × 150 mm) with a laser-pulled electrospray tip (Model P-2000, Sutter instruments) was home-packed with 4 μm, 100 Å Magic C18AQ silica-based particles (Michrom BioResources Inc., Auburn, CA). Eluted peptides from the capillary column were electrosprayed directly onto the mass spectrometer for MS and MS/MS analyses in a data-dependent acquisition mode. One full MS scan (m/z 350–1500) was acquired and then MS/MS analyses were performed on the 10 most intense ions. Dynamic exclusion was set with repeat duration of 24 s and exclusion duration of 12 s. In total, three independent biological replicates of intracellular Salmonella samples were analyzed in 48 LC-MS/MS experiments.
MaxQuant (http://maxquant.org/, Version 1.5.3.30) was used to generate peak lists from MS raw files, and Andromeda was used to search S. typhimurium LT2 protein database (strain LT2/SGSC1412/ATCC 700720, 11/7/2011, downloaded from UniProt with 5199 sequences) augmented with the reversed sequence of each entry in the database. The precursor mass tolerance was set at 20 ppm, and the fragment mass tolerance for CID MS/MS was set at 0.8 Da. Trypsin was selected as the digestive enzyme with a maximum of two missed cleavages. Oxidation (M) was set as a variable modification. There were no fixed modifications. The minimum ratio counts were set at 2. The false discovery rate (FDR) of peptides and proteins was controlled at < 1%. The MaxQuant software was used to calculate the label-free quantitation (LFQ) intensity for each protein. The obtained LFQ intensity values were further processed by using the Perseus software (version 1.5.4.1). Logarithmic values (Log2) of the LFQ intensity were used and the missing values were replaced with random numbers from a normal distribution (width = 0.3, shift = 1.8). The p value of each protein was obtained by using the two-tailed Student's t test. Altered proteins with average fold changes > 2 or < 0.5 and p values < 0.05 were considered significant.
Western Blotting Analyses
Salmonella strains expressing 3×FLAG-tagged proteins were used to infect HeLa cells in 6-well plates with an MOI of 30. At various time points (indicated) after infection, mammalian cells were lysed and crude fractions of intracellular bacteria were obtained by differential centrifugation. Isolated bacterial pellets were resuspended in the SDS-PAGE sample buffer and run by 10% SDS-PAGE. Then gel-separated bacterial proteins were further transferred to polyvinylidene difluoride (PVDF) membranes. Immunoblotting analyses were carried out with primary antibodies specific for Salmonella DnaK as a loading control (Enzo Life Sciences, Farmingdale, NY) (1:5,000) or FLAG (CWBIO, Beijing, China) (1:2,000) and horseradish peroxidase (HRP)-conjugated secondary antibody (CWBIO, China) (1:5,000). For all immunoblotting assays, at least three independent experiments were performed.
Quantitative Real-Time PCR
HeLa cells were infected by Salmonella wild-type and ΔydcR strains as described above. Intracellular bacteria were isolated at 18 hpi from host cells for RNA extraction. Total RNA was harvested by using an EasyPure RNA Kit (TransGen Biotech, Beijing, China) and then treated with DNase I. Reverse transcription of the RNA samples was carried out with TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech). Real-time RT-PCR analyses were performed on an Applied Biosystems ViiA™ 7 Real-Time PCR System by using UltraSYBR Mixture (Low ROX) (CWBIO). To quantitatively compare transcriptional levels of srfN, cysN, nupC, yfgC, pipB, STM2234, and frdB, the housekeeping 16S rRNA gene was used for normalization and mRNA levels were analyzed using the comparative threshold cycle number (2−ΔΔCt) method (16).
Expression and Purification of Recombinant Proteins
FLAG-tagged YdcR was expressed in the Salmonella ΔydcR+pYdcR strain with the addition of 0.1% arabinose to induce protein expression. The bacterial cells (from 400 ml culture) were lysed in 30 ml of ice cold PBS buffer via sonication and cell lysates were clarified by centrifugation at 5000 × g for 10 min three times. The resulting supernatants were used for YdcR purification by immunoprecipitating with anti-FLAG M2 agarose beads (50% slurry, Sigma). Briefly, the agarose beads were incubated overnight with the clarified lysates at 4°C under rotation, and collected by centrifugation at 1500 × g for 3 min. Then the beads were washed three times with a buffer containing 20 mm Tris-HCl (pH 7.5)/150 mm NaCl and proteins were eluted with 150 ng/μl 3×FLAG peptides/20 mm Tris-HCl (pH 7.5)/150 mm NaCl. Finally, the 3×FLAG peptides were removed from the purified proteins by ultrafiltration prior to use.
Electrophoretic Mobility Shift Assays (EMSA)
Putative DNA promoter regions were amplified by PCR, purified with a Gel Extraction Kit (TransGen Biotech) and dissolved in water. Then purified YdcR proteins were incubated with 40 nm DNA fragments in 20 μl of binding buffer (10 mm Tris-HCl (pH 7.5), 100 mm KCl, 1 mm EDTA, 0.1 mm DTT, 5% v/v glycerol, and 10 μg/ml BSA). Molar ratios between DNA fragment and YdcR were set at 1:0, 1:2.5, 1:5, and 1:10 (that is indicated as 0, 100, 200, 400 in Fig. 6B respectively). The reaction mixtures were incubated at room temperature for 30 min and then loaded onto 8% native polyacrylamide gels. Electrophoresis was performed using 0.5×TBE buffer (44.5 mm Tris, 44.5 mm boric acid, and 1 mm EDTA) in ice bath. The gel was stained with Gelstain (TransGen Biotech) and photographed by using a Tanon-1600 Gel Image System (Tanon, Shanghai, China).
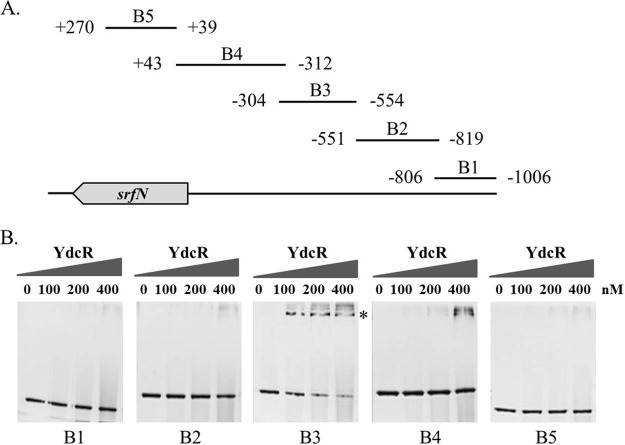
Fig. 6.
Determination of YdcR-binding sites in the promoter region of srfN by EMSA. A, Various DNA fragments covering different regions upstream of srfN were tested in the EMSA experiments. The numbers represent the positions of the fragments with respect to the translation start site. B, EMSAs were performed by incubating serial concentrations of purified YdcR (ranging from 0 to 400 nm) with the DNA fragments B1, B2, B3, B4, and B5 prior to electrophoretic separation.
β-Galactosidase Assays
Salmonella cells were grown in LB with or without the addition of 0.2% arabinose at 37 °C to an OD600 of ∼1.0. Bacterial cells from 1.2 ml of culture were pelleted at 14,000 × g for 2 min. Bacterial pellets were then resuspended in 1.2 ml of Z buffer (60 mm Na2HPO4, 40 mm NaH2PO4, 10 mm KCl, and 1 mm MgSO4) plus 50 mm β-mercaptoethanol (freshly added). Then 30 μl of chloroform and 15 μl of 0.1% SDS were added and mixed upon vortexing. The assays were started by the addition of 240 μl of 4 mg/ml o-nitrophenyl-d-galactopyranoside (ONPG). Upon the observation of a faint yellow color, the reaction was stopped by the addition of 600 μl of 1 m Na2CO3 and the reaction time was noted. Finally, samples were centrifuged at 14,000 × g for 2 min, and the OD420 of the supernatant was recorded. Assay units were calculated as 1000 × OD420/(OD600) (total reaction time).
Growth Curve Experiments and HeLa Cell Infection Assays
To assess bacterial growth in LB broth, an overnight culture of either the wild-type or the ΔydcR mutant strain was diluted 1:300 into 3 ml of LB broth. The optical density of bacterial culture was monitored for 13 h. For Salmonella infection assays, HeLa cells in 6-well plates were infected with different strains at an MOI of 10. After 45 min, infected cells were washed with HBSS and incubated in DMEM supplemented with 100 μg/ml gentamicin for 1 h to kill extracellular bacteria. Cells were then lysed and viable intracellular bacteria were enumerated by colony-forming units (CFU) assays to determine the rate of bacterial invasion. Otherwise, cells were washed with HBSS and fresh DMEM containing 10 μg/ml gentamicin was added. At 18 hpi, cells were then lysed and viable intracellular bacteria were enumerated by CFU assays to determine the rate of intracellular bacterial replication.
Mouse Infection Experiments
Animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the Chinese Association for Laboratory Animal Sciences (CALAS) and approved by the ethics committee of Renji Hospital, School of Medicine, Shanghai Jiao Tong University (Shanghai, China). Wild-type S. typhimurium and the ydcR deletion mutant with kanamycin resistance were cultured in LB to the exponential growth phase (OD600 = 0.9) before competitive infection. Twenty female BALB/c mice of 6 weeks old were intraocularly injected with a mixed inoculum (∼1 × 105 CFU/mouse) containing an equivalent number of each test strain in 0.1 m HEPES buffer (pH 8.0) with 0.9% NaCl. Mice were sacrificed at 72 hpi. The spleen, liver, and cecum were harvested and homogenated for the determination of bacterial burdens by plating on LB plates with and without kanamycin (30 μg/ml) simultaneously. The competitive index (CI) was calculated from CFU assays as follows: (mutant/wild-type)output/(mutant/wild-type)input. The competitive infection data were analyzed using the two-tailed Student's t test.
RESULTS
Profound Induction of YdcR in Intracellular Salmonella During Infection of Host Epithelial Cells
Previously we studied intracellular Salmonella proteome within infected HeLa cells and found extensive reprogramming of bacterial metabolic pathways (5, 6). In addition, our large-scale proteomic datasets also reveal many other proteins with altered expression levels during infection, though most of them cannot be categorized into distinct biological processes. Furthermore, for some differentially regulated proteins very little information is available regarding their molecular functions or potential roles in bacterial pathogenesis. We reasoned that some of these proteins could be of vital interest in understanding the mechanisms underlying Salmonella infection. One of those altered proteins is YdcR, which is annotated as a putative GntR family regulatory protein, and it was profoundly induced at both 6 hpi and 18 hpi (Fig. 1A). In contrast, this protein was barely expressed for in vitro cultured Salmonella. Interestingly, such a pattern of protein expression is reminiscent of that of some SPI-2 encoded virulence factors that are essential for survival and replication of intracellular Salmonella. Indeed, the abundance of two SPI-2 effectors, PipB and PipB2, were both induced in a similar manner as YdcR (Fig. 1A). To further confirm the proteomic changes, next we constructed a Salmonella strain chromosomally expressing 3×FLAG-tagged YdcR and infected HeLa cells as described under Experimental Procedures. After isolation of intracellular bacteria at different time points post-infection, the bacterial lysates were probed with a FLAG-specific antibody to detect YdcR. Consistent with the proteomic data, immunoblotting analyses reveal that YdcR was highly induced upon Salmonella infection of HeLa cells whereas its expression levels were rather low for in vitro as well as internalized bacteria early during infection (i.e. at 1 hpi) (Fig. 1B). Collectively, these data suggest that YdcR is exclusively produced from intracellular Salmonella, reminiscent of those SPI-2 encoded virulence proteins.
Fig. 1.
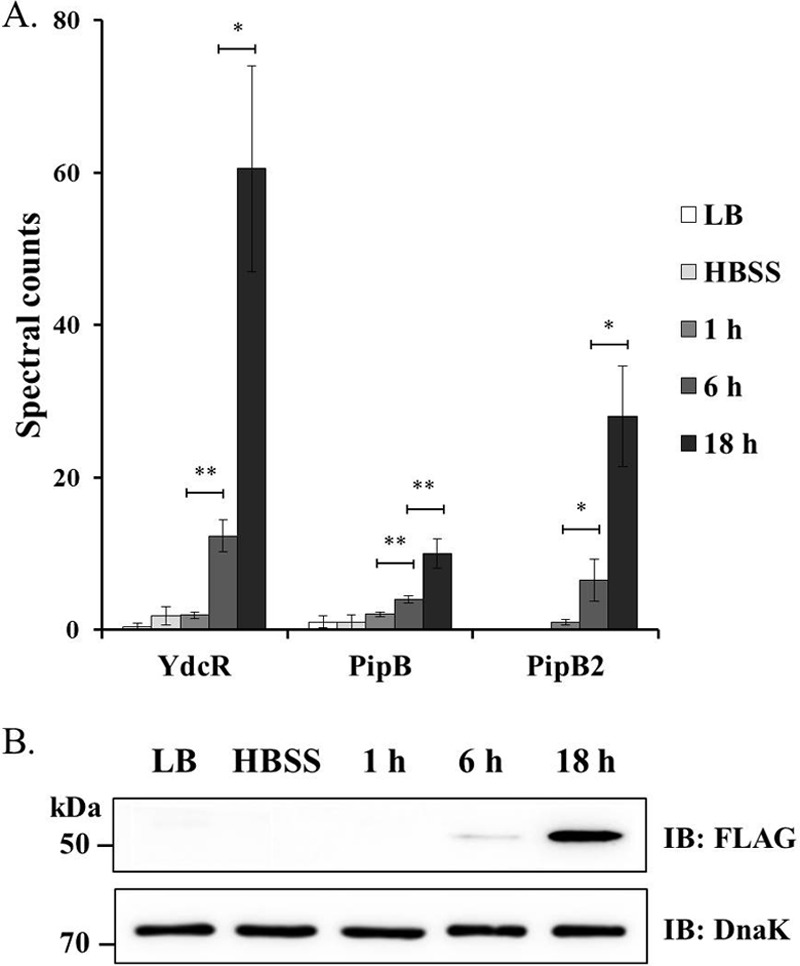
Upregulation of Salmonella YdcR at 18 hpi. A, The spectral counts of YdcR and two SPI-2 effectors (PipB, PipB2) derived from three independent LC-MS/MS measurements. Intracellular Salmonella was isolated from infected host epithelial cells at various time points (1 h, 6 h, and 18 h post-infection) and subjected to LC-MS/MS analyses together with bacteria cultured in LB media and those incubated further in HBSS prior to invasion (the extracellular population). These data were obtained from our previous studies (5, 6) and the original proteomic dataset can be found at http://iai.asm.org/content/83/7/2897/suppl/DCSupplemental and http://pubs.acs.org/doi/suppl/10.1021/acs.jproteome.6b00793. Asterisks indicate significant differences (*, p < 0.05; **, p < 0.01). B, Representative immunoblotting data of YdcR (and DnaK as a loading control) by using host cells infected by the Salmonella strain chromosomally expressing 3×FLAG-tagged YdcR.
Salmonella YdcR is a Putative Transcriptional Regulator and is Highly Conserved Among a Number of Bacterial Species
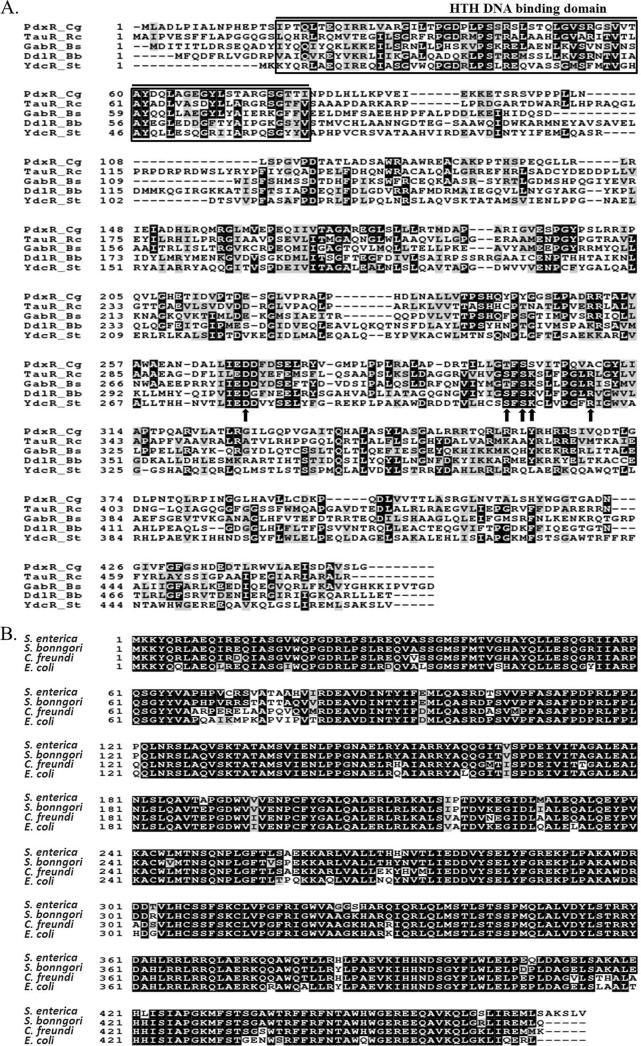
YdcR was initially annotated as a putative GntR family regulatory protein. Further sequence alignment with known regulators of this family reveals that it exhibits similarity to regulators PdxR, TauR, GabR, and DdlR, which belong to the MocR/GabR subfamily (Fig. 2A). In addition to an N-terminal helix-turn-helix (HTH) DNA-binding domain, the members of this subfamily (including YdcR) consist of a very large C-terminal domain that shows sequence similarity to class I aminotransferases. These enzymes often require pyridoxal 5′-phosphate (PLP) as a cofactor for exerting their functions. We found that YdcR also carries the conserved amino acid residues responsible for the interaction with PLP (17) (arrowed in Fig. 2A). In addition, a BLAST search against reference sequences in the NCBI database reveals homologues of YdcR in Salmonella bongori. Moreover, YdcR is found to be highly conserved among other bacterial genera including Citrobacter spp., Escherichia spp., Klebsiella spp., and Shigella spp. with >85% sequence similarity (some representative YdcR homologues are shown in Fig. 2B), although it has not been characterized in those bacteria. Therefore, it appears that YdcR homologs are at least not confined to nonpathogenic bacteria.
Fig. 2.
Multiple sequence alignment of YdcR with other transcription factors or its homologs from different bacteria. A, Amino acid sequence alignment shows YdcR is a putative MocR/GabR-type bacterial transcription factor. PdxR_Cg: PdxR of C. glutamicum ATCC 13032 (accession number Q8NS92), TauR_Rc: TauR of R. capsulatus ATCC BAA-309 (accession number D5AKX9), GabR_Bs: GabR of B. subtilis (accession number P94426), DdlR_Bb, DdlR of B. brevis (accession number C0ZDG2), YdcR_St: YdcR of S. typhimurium SL1344 (accession number Q8ZPC8). Arrows indicate those residues that are functional sites involved in PLP binding. B, Amino acid sequence alignment shows YdcR is highly conserved among several bacterial species. YdcR: S. enterica (accession number Q8ZPC8); S. bonngori (accession number S5N885); C. freundi (accession number A0A0D7); E. coli (accession number P77730). The protein sequences were aligned by using the CLUSTALW program (http://www.genome.jp/tools/clustalw/) and the figure was prepared with the BOXSHADE program (http://www.ch.embnet.org/software/BOX_form.html).
Comparative Proteomics Reveal Seven Candidates of YdcR-Regulated Proteins
To shed light on the functional roles of YdcR, if there is any, in Salmonella pathogenesis, next we sought to determine the downstream proteins under its regulation in the exact context where its expression was highly induced. To uncover YdcR-regulated proteins, we exploited quantitative proteomics to compare intracellular Salmonella protein expression between the wild-type and the isogenic ΔydcR strains. As Salmonella YdcR was most produced at 18 hpi, we isolated bacteria from infected host cells at this point. In total, we identified 1563 bacterial proteins from three biological replicates with at least two unique peptides. Fig. 3A shows the protein-level volcano plot of detected proteins from intracellular Salmonella strains. By using peptide intensity-based LFQ, our proteomic datasets reveal fairly high similarity of Salmonella proteomes between the wild-type and ΔydcR strains. In fact, with our significance criteria (fold ratios > 2 and p value < 0.05), only eight bacterial proteins differed in the Salmonella strain lacking the ydcR gene (Table I). Among these, four proteins were upregulated (YfgC, PipB, STM2234, and FrdB) and the other four were downregulated (YdcR, SrfN, CysN, and NupC). A complete list of all identified proteins is provided as supplemental Table S2. Notably, the positive control for our quantitative proteomic experiments, YdcR, was the most depressed protein in the entire dataset (Fig. 3A, in the left upper corner). Interestingly, in the volcano plot the only protein near YdcR is SrfN (STM0082), indicating its marked repression in the ΔydcR strain. Furthermore, like YdcR, SrfN was not detected in the ydcR-deficient strain, though it was abundantly expressed in wild-type Salmonella within infected HeLa cells (Table I). Therefore, our proteomic analyses identified SrfN together with other six proteins as candidate proteins under YdcR regulation either in a direct or indirect manner.
Fig. 3.
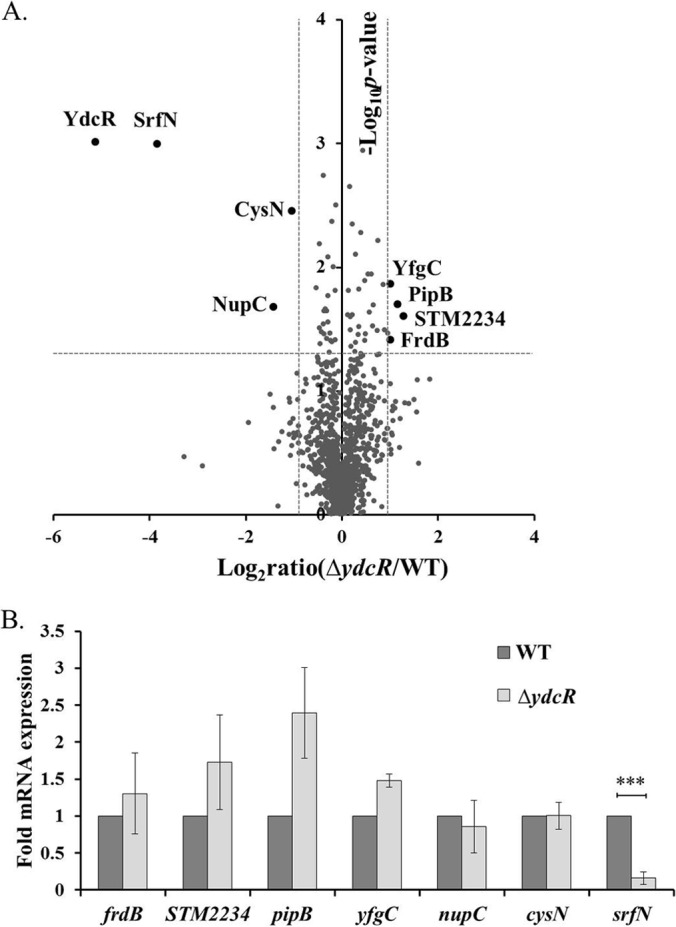
YdcR positively regulates the expression of SrfN. A, A volcano plot of intracellular Salmonella proteins detected by LC-MS/MS experiments. The fold changes were calculated by dividing LFQ intensity values from the ΔydcR mutant by those from the wild-type strain at 18 hpi. The logarithmic values of the average fold changes are reported on the x axis. The y axis plots negative logarithmic p values determined from the t test on three biological replicates. Dotted lines denote 2-fold (vertical) and p < 0.05 cutoff (horizontal). B, qRT-PCR analyses of mRNA samples extracted from intracellular Salmonella at 18 hpi (n = 3). Asterisks indicate significant differences (***, p < 0.001).
Table I. Differentially expressed Salmonella proteins in the ΔydcR strain at 18 hpi.
| Gene | Protein description | Abundancea |
Foldd | p valuee | |
|---|---|---|---|---|---|
| WTb | ΔydcRc | ||||
| ydcR | Putative GntR family regulatory protein | 1.9E7 | n.df | −34.8 | 0.001 |
| STM0082 | Putative secreted protein | 9.6E6 | n.d | −14.2 | 0.001 |
| nupC | NUP family nucleoside transport protein | 3.1E6 | 1.0E6 | −3.1 | 0.021 |
| cysN | Sulfate adenylyltransferase subunit 1 | 1.3E6 | 6.3E5 | −2.1 | 0.004 |
| STM2234 | Putative tail fiber assembly protein | 1.1E6 | 2.6E6 | 2.4 | 0.025 |
| pipB | Pathogenicity island encoded protein: SPI5 | 2.6E6 | 5.9E6 | 2.2 | 0.020 |
| yfgC | TPR repeat-containing protein YfgC | 2.5E6 | 5.1E6 | 2.0 | 0.014 |
| frdB | Fumarate reductase | 8.7E5 | 1.7E6 | 2.0 | 0.039 |
a Averaged LFQ intensity from three biological replicates.
b Wild-type bacteria isolated from host cells at 18 hpi.
c The ΔydcR strain isolated from host cells at 18 hpi.
d Fold change in protein abundance (missing values were replaced by random numbers and negative values indicate lower levels in the ΔydcR samples).
e p values were calculated by using the paired Student's t-test.
f n.d, not detected in LC-MS/MS experiments.
YdcR Transcriptionally Regulates the Expression of Salmonella SrfN
Next, we sought to investigate if YdcR indeed regulates these altered proteins on the transcriptional level. We measured the mRNA levels of these seven genes (frdB, STM2234, pipB, yfgC, nupC, cysN, and srfN) by qRT-PCR analyses of extracted RNA samples from intracellular bacteria (Fig. 3B). At 18 hpi, only the transcript level of srfN was markedly altered (6.3-fold lower in the ΔydcR mutant) with our significance criteria (fold > 2 and p < 0.05). The mRNA level of pipB increased by 2.4-fold, yet this change was not statistically significant (p = 0.08). These results demonstrate that only the transcription of srfN is highly dependent on the presence of a functional ydcR gene.
Futhermore, β-galactosidase assays were used to establish the transcriptional control of the srfN gene by YdcR. To do this, we introduced a plasmid containing a lacZ transcriptional fusion to the downstream region of the srfN promoter and assayed the activities of β-galactosidase in vitro. Like the ydcR deletion mutant, the wild-type Salmonella showed similar β-galactosidase activities (Fig. 4A), suggesting that the wild-type strain failed to increase the level of lacZ gene transcription under this condition. In fact, such observation is consistent with our findings indicating that YdcR is barely expressed under in vitro culturing conditions (Fig. 1). To circumvent this issue and facilitate the examination of YdcR-regulated srfN transcription in vitro, we further introduced a YdcR-expressing plasmid into the ΔydcR strain, whose expression is under the control of arabinose. Using this strain, we detected a significant increase of β-galactosidase activities, indicating YdcR-dependent transcriptional activation of the srfN-lacZ fusion. In line with our hypothesis that YdcR regulates the expression of SrfN at the transcriptional level, we detected the markedly increased β-galactosidase activity only when the inducible ydcR-carrying plasmid and the inducer arabinose were present (Fig. 4B). Taken together, these results indicate that YdcR controls the expression of the srfN gene at the transcriptional level.
Fig. 4.
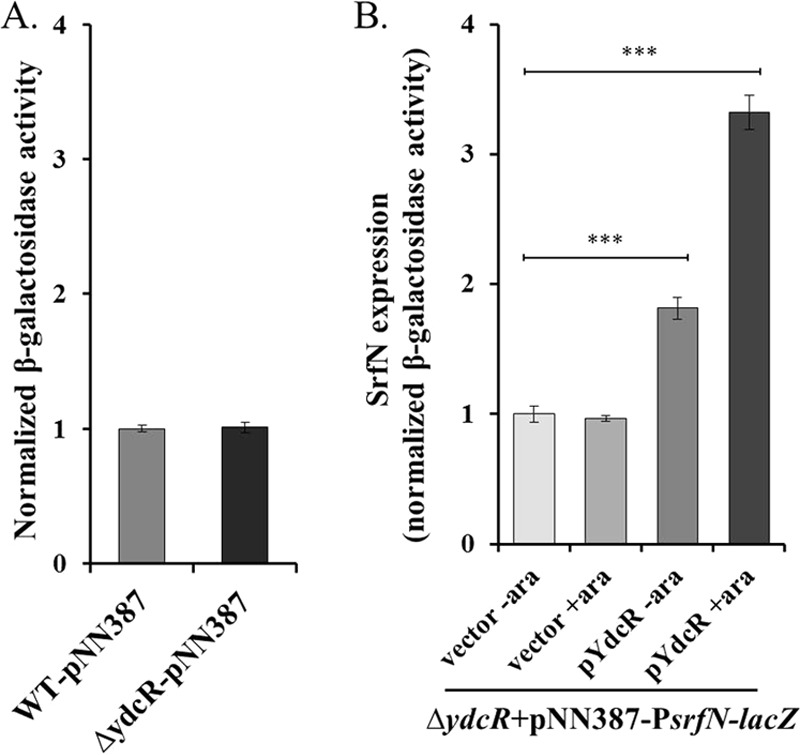
YdcR activates the expression of srfN at the transcriptional level. A, β-Galactosidase activity in the wild-type and ΔydcR mutant strains harboring a plasmid encoding lacZ fusion with the promoter region of srfN. β-Galactosidase activities from three independent measurements are shown with values normalized to that of the wild-type strain. B, β-Galactosidase activity assays were performed with the ΔydcR mutant strain harboring a plasmid encoding a lacZ fusion with the promoter region of srfN and complemented with a plasmid-encoded copy of ydcR or empty vector, under noninducing (no arabinose) or inducing (with 0.2% arabinose) conditions as indicated. β-Galactosidase activities from three independent measurements are shown with values normalized to that of the ΔydcR strain complemented with an empty plasmid under noninducing conditions. Asterisks indicate significant differences (***, p < 0.001).
YdcR Positively Controls the Expression of Salmonella Virulence Factor SrfN
Salmonella SrfN was first identified by Osborne et al. as an SsrB-regulated virulence factor that contributes to bacterial fitness in a murine model of infection (18). In our previous studies of intracellular Salmonella proteome, we also demonstrated a steady increase of the SrfN expression during infection (data sampled at 1, 6, and 18 hpi) (5, 6). The SrfN induction profile is like that of YdcR, also supporting our hypothesis. To further verify the proteomic observations, next we constructed Salmonella strains chromosomally expressing 3×FLAG-tagged SrfN in the wild-type and ydcR deletion backgrounds. Then these bacterial strains were used to infect host epithelial cells and at 18 hpi intracellular Salmonella was harvested and probed for the expression of SrfN. Consistent with the previous LC-MS/MS measurements, immunoblotting analyses using specific antibodies show abundant SrfN expression in the wild-type bacteria whereas it was barely detected in the ΔydcR strain (Fig. 5A). Next, we complemented the deletion mutant with a plasmid-encoded copy of ydcR. When the expression of YdcR was induced by the addition of 0.02% arabinose into the cell culture medium, the expression of SrfN was readily detected by immnunoblotting analysis. With the addition of more arabinose (0.2%), furthermore, we observed higher levels of both YdcR and SrfN (Fig. 5A). Similar results were obtained with the wild-type bacteria complemented with a YdcR-expressing plasmid. In contrast, the expression level of PipB did not differ significantly for Salmonella strains chromosomally expressing 3×FLAG-tagged PipB in the wild-type and ydcR deletion backgrounds (Fig. 5B), indicating the specific regulation of SrfN by YdcR. Taken together, these data demonstrate that YdcR positively regulates the expression of Salmonella virulence factor SrfN.
Fig. 5.
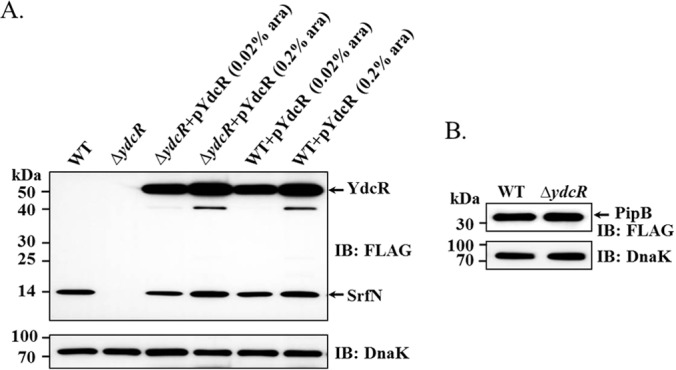
Western blotting analyses confirmed YdcR-dependent expression of Salmonella SrfN during infection. A, HeLa cells were infected by Salmonella strains chromosomally expressing 3×FLAG-tagged SrfN in various genetic backgrounds (wild-type, ΔydcR, and those complemented with plasmid-encoded YdcR) with an MOI of 30. Then intracellular bacteria were isolated at 18 hpi and probed for the expression levels of SrfN and YdcR. Representative immunoblots of 3×FLAG-tagged YdcR and SrfN are shown with DnaK as a loading control. As indicated, 0.02% and 0.2% arabinose were added to the cell culture medium during infection to induce the expression of YdcR. B, HeLa cells were infected by Salmonella strains chromosomally expressing 3×FLAG-tagged PipB in the wild-type and ΔydcR mutant strains with an MOI of 30. Then intracellular bacteria were isolated at 18 hpi and probed for the expression levels of PipB.
YdcR Directly Binds to the Promoter Region of the srfN Gene
Like other transcriptional regulators of the GntR family, YdcR also has an N-terminal helix-turn-helix (HTH) DNA-binding domain. Thus, we next explored whether YdcR can directly interact with the promoter region of srfN by conducting electrophoretic mobility shift assays (EMSA). Purified YdcR proteins were incubated with DNA fragments spanning from −1006 bp to +270 bp with respect to the translation start site of the srfN operon (Fig. 6A). Among the five DNA fragments (B1-B5) under investigation, the addition of YdcR to the reaction mixture caused a specific shift in the mobility of DNA fragment B3, extending from position −554 bp to −304 bp. In contrast, no obvious changes were seen for the other DNA fragments (Fig. 6B). Therefore, these results indicate that the 250 bp of DNA fragment B3 harbors the essential elements for mediating direct binding to YdcR.
Salmonella YdcR Contributed to Bacterial Fitness in a Mouse Model of Infection
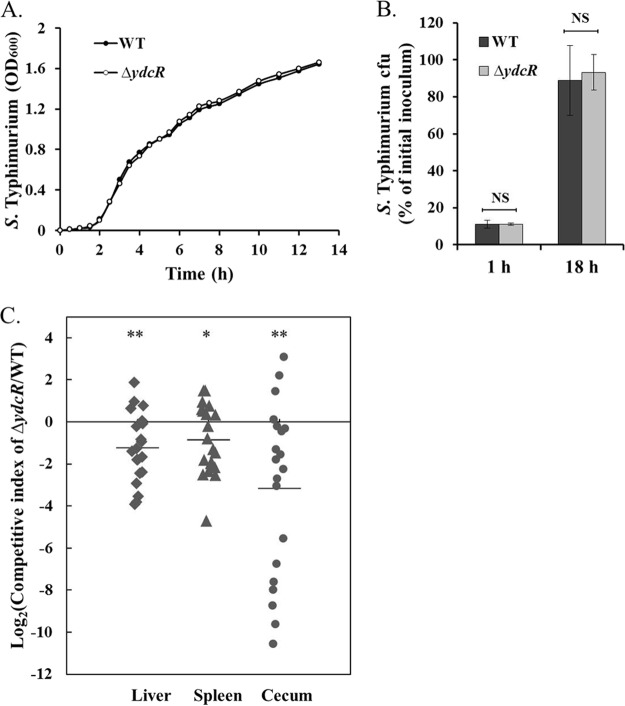
As we have established that YdcR directly controls the expression of a known virulence factor, next we set out to characterize the phenotype of a Salmonella strain lacking ydcR (ΔydcR) in the context of bacterial infection. In vitro growth studies reveal that proliferation of the ΔydcR mutant in LB broth was indistinguishable from that of the wild-type Salmonella (Fig. 7A). Next we sought to investigate whether deleting ydcR has any influence on bacterial invasion of host cells. As demonstrated in Fig. 7B, the Salmonella strain lacking ydcR invaded HeLa cells as efficiently as the wild-type bacteria. At 18 hpi, by enumerating viable intracellular bacteria we also found that upon internalization the ΔydcR mutant replicated equally well compared with its parental strain (Fig. 7B). Therefore, these data collectively suggest that genetic ablation of ydcR did not compromise Salmonella growth in vitro, bacterial invasion or intracellular multiplication during infection of host epithelial cells.
Fig. 7.
Characterization of the ydcR-deficient Salmonella strain. A, Growth curves of the wild-type and ΔydcR mutant in the LB medium. B, The rate of bacterial invasion and intracellular growth determined by CFU assays (n = 3). NS, not significant. C, Deletion of ydcR led to decreased Salmonella fitness in a murine infection model. Asterisks indicate significant differences (*, p < 0.05; **, p < 0.01).
We further tested the growth fitness of the ΔydcR mutant in a mouse model of infection. BALB/c mice were intraocularly coinfected with an equivalent mixture of wild-type and ΔydcR mutant strains. At 72 hpi, viable bacteria from the spleen, liver, and cecum were enumerated and a competitive index was calculated. The geometric mean Log2(competitive index) for the ΔydcR mutant was −1.2, −0.9, and −3.2 in the liver, spleen, and cecum, respectively (Fig. 7C). Therefore, the ΔydcR mutant was significantly out-competed by the wild-type strain during systemic infection.
DISCUSSION
YdcR is a MocR/GabR-type transcriptional regulator with a predicted DNA-binding HTH motif. Our proteome data indicate highly induced levels of YdcR in intracellular Salmonella during infection of HeLa cells, though its functions and/or physiological roles in Salmonella infection biology has not been reported thus far. In fact, numerous MocR/GabR-type proteins were found in bacterial genomes. Nevertheless, only four regulator proteins of this family (GabR, PdxR, TauR, and DdlR) have been characterized to some extent (19, 20, 21, 22, 23). For instance, in the presence of PLP and γ-aminobutyrate (GABA), GabR activates the transcription of gabT and gabD, which encode GABA aminotransferase and succinate semialdehyde dehydrogenase, respectively (19, 20). PdxR, on the other hand, activates the transcription of pdxST genes encoding the PLP synthase. In vivo analyses indicate that PLP and PL can lower the transcription of pdxS and pdxT (21). It appears that the four characterized regulators are all involved in regulation of a narrow range of downstream genes (i.e. specific metabolic pathways). In addition, YdcR homologs are found to be present in many bacterial species with high sequence similarity (>85%) such as Citrobacter, Escherichia, Enterobacter, Klebsiella, and Shigella. Clearly, there is still a knowledge gap in understanding the regulatory mechanisms of many other MocR/GabR-type transcription factors and perhaps more importantly the downstream proteins directly under their control.
The goal of our current study is to uncover the functional roles of YdcR in the context of Salmonella infection of host epithelial cells. We took advantage of quantitative proteomics and examined specifically the Salmonella proteome within infected cells in that YdcR was only abundantly expressed during bacterial intracellular replication. Comparative analyses of Salmonella protein expression in the wild-type and ydcR-lacking strains permitted us to identify proteins whose expression levels was YdcR-dependent. By using this quantitative approach, we found that expression of Salmonella virulence factor SrfN was strictly dependent on YdcR during infection, suggesting a potentially important role of this MocR/GabR-type transcriptional regulator in bacterial pathogenesis. Next, we provided multiple lines of evidence to support that YdcR, a Salmonella transcription factor highly induced within infected host cells, positively regulates the expression of the bacterial virulence factor SrfN. First, we used qRT-PCR to measure the transcript levels of srfN and found the regulation of YdcR on the expression of SrfN indeed occurs on the transcriptional level. Further β-galactosidase assays of Salmonella strains harboring a lacZ fusion to the promoter region of the srfN gene also confirmed that YdcR can mediate transcriptional regulation of srfN in bacteria cultured in vitro. Additionally, immunoblotting assays indicated YdcR-mediated expression of SrfN in intracellular Salmonella during infection, consistent with our proteomic findings. Lastly, we carried out EMSA experiments by incubating purified YdcR with DNA fragments encompassing various regions upstream of srfN and found specific interaction of YdcR with a fragment spanning from position −554 bp to −304 bp, indicating its direct regulatory role in Salmonella virulence factor SrfN. Importantly, competitive infection reveals that the loss of YdcR led to reduced bacterial fitness in a murine infection model, thereby suggesting its physiological role in Salmonella pathogenesis.
srfN was initially discovered by Osborne et al. as a gene regulated by SsrB, a master regulator of Salmonella SPI-2 T3SS (18, 24). The mRNA level of srfN was reduced 8-fold in ΔssrB cells compared with the wild-type bacteria under SPI-2-inducing conditions and SsrB controls srfN directly through binding to the srfN cis-regulatory element. In line with this finding, we observed concurrent induction of SrfN and some SPI-2 encoded T3SS effectors at 18 hpi (i.e. PipB and PipB2) within infected epithelial cells. Osborne et al. also demonstrated that SrfN was a fitness factor and contributed to bacterial proliferation during systemic infection, though direct translocation of SrfN into host cells was not observed (18). Later Yoon et al. confirmed the requirement of srfN to attain full mouse virulence and demonstrated the translocation of SrfN into macrophage cells by using both cAMP and β-lactamase-based translocation assays (25). Thus, previous inability to observe the translocation of SrfN during infection might be attributed to the relatively lower sensitivity of immunoblotting assays. Intriguingly, SrfN was found to be translocated in a T3SS-independent manner. Yoon and colleagues also showed that inactivation of SPI-2 T3SS even led to increased protein expression and translocation levels of SrfN during Salmonella infection of macrophage cells (25), arguing against its positive regulation by SsrB.
Our current study uncovered that the expression of SrfN is positively regulated by YdcR in S. typhimurium. Interestingly, Salmonella bongori encodes both YdcR and SrfN as well, despite its lack of SsrB and SPI-2 T3SS (18). Thus, it is tempting to speculate that in S. bongori SrfN is likely to be regulated by YdcR as well. Yet unlike YdcR homologs being widely distributed in Citrobacter spp. and Klebsiella spp. (Fig. 2B), SrfN homologs are only present in specific bacterial species of these genera such as C. rodentium and K. pneumoniae (with >60% sequence identity). In light of SrfN being a virulence factor, therefore, its transcriptional regulation by YdcR may be a trait evolutionarily confined to pathogenic bacteria. In support of this evolutionary confinement among Salmonella, Navarre et al. suggested potential negative regulation of srfN by the histone-like nucleoid structuring protein H-NS that has an important role in regulating Salmonella virulence genes (26). Navarre et al. found H-NS can selectively silence horizontally acquired genes and their microarray data revealed 6.7-fold repression of srfN expression by H-NS (26). It is interesting to speculate that srfN is initially repressed by H-NS in vitro, whereas during infection of host cells YdcR (and SsrB) may come into play and activate the expression of srfN. In light of the different promoter regions that YdcR and SsrB bind to, it is likely that the expression of srfN requires both regulators. Apparently, further studies would be needed to elucidate the precise regulatory mechanisms of srfN. For instance, it would be interesting to determine whether there is any crosstalk between YdcR and SsrB in regulating srfN or other proteins.
In conclusion, we have utilized comparative proteomics to uncover the potential substrates of a transcriptional regulator. By profiling the proteomes of the ydcR deletion mutant together with wild-type S. typhimurium, we unveiled a direct regulatory role of YdcR in the expression of SrfN. By encoding a transcriptional regulator, therefore, Salmonella has evolved a mechanism that specifically turns on the production of the virulence factor SrfN and hence contributes to bacterial virulence. Furthermore, our approach may be broadly applied to aid the characterization of other bacterial mutants lacking observable phenotypes in conventional assays.
DATA AVAILABILITY
The proteomics data reported in this paper have been deposited to the iProx database (URL: http://www.iprox.org/) and are available under the accession number IPX0000899001.
Supplementary Material
Acknowledgments
We thank the members of the Liu laboratory for careful review of this manuscript. We also thank Dr. Guifang Jia for the use of the Applied Biosystems ViiA™ 7 Real-Time PCR System, Dr. Aixin Yan for the kind gift of the plasmid pNN387, and Dr. Yicheng Sun and Gaixian Ren for providing the λ Red recombination system.
Footnotes
Author contributions: Y.L., Q.L., M.L., and X.L. designed research; Y.L., Q.L., L.Q., and T.D. performed research; L.Q., Z.W., J.F., and M.H. contributed new reagents or analytic tools; Y.L., J.S., and X.L. analyzed data; Y.L., J.S., and X.L. wrote the paper.
* This work was supported by grants from the National Natural Science Foundation of China (21475005 and 21622501) and the Thousand Young Talents program of the Chinese government.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- SPI-1
- Salmonella pathogenicity island 1
- SPI-2
- Salmonella pathogenicity island 2
- HBSS
- Hanks balanced salt solution
- MOI
- multiplicity of infection
- FDR
- false discovery rate
- LFQ
- label-free quantitation
- CFU
- colony forming units
- SCVs
- Salmonella-containing vacuoles
- TCA
- tricarboxylic acid
- DMEM
- Dulbecco's Modified Eagle Medium
- FBS
- fetal bovine serum
- PVDF
- polyvinylidene difluoride
- EMSA
- electrophoretic mobility shift assays
- CI
- competitive index
- HTH
- helix-turn-helix
- PLP
- pyridoxal 5′-phosphate.
REFERENCES
- 1. Ohl M. E., and Miller S. I. (2001) Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52, 259–274 [DOI] [PubMed] [Google Scholar]
- 2. Galán J. E. (2001) Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17, 53–86 [DOI] [PubMed] [Google Scholar]
- 3. van der Heijden J., and Finlay B. B. (2012) Type III effector-mediated processes in Salmonella infection. Future Microbiol. 7, 685–703 [DOI] [PubMed] [Google Scholar]
- 4. Bakowski M. A., Braun V., and Brumell J. H. (2008) Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic 9, 2022–2031 [DOI] [PubMed] [Google Scholar]
- 5. Liu Y., Zhang Q., Hu M., Yu K., Fu J., Zhou F., and Liu X. (2015) Proteomic analyses of intracellular Salmonella enterica serovar Typhimurium reveal extensive bacterial adaptations to infected host epithelial cells. Infect. Immun. 83, 2897–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Y., Yu K., Zhou F., Ding T., Yang Y., Hu M., and Liu X. (2017) Quantitative proteomics charts the landscape of Salmonella carbon metabolism within host epithelial cells. J. Proteome Res. 16, 788–797 [DOI] [PubMed] [Google Scholar]
- 7. Libby S. J., Goebel W., Ludwig A., Buchmeier N., Bowe F., Fang F. C., Guiney D. G., Songer J. G., and Heffron F. (1994) A cytolysin encoded by Salmonella is required for survival within macrophages. Proc. Natl. Acad. Sci. U.S.A. 91, 489–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Linehan S. A., Rytkönen A., Yu X. J., Liu M., and Holden D. W. (2005) SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infect. Immun. 73, 4354–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Navarre W. W., Halsey T. A., Walthers D., Frye J., McClelland M., Potter J. L., Kenney L. J., Gunn J. S., Fang F. C., and Libby S. J. (2005) Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol. Microbiol. 56, 492–508 [DOI] [PubMed] [Google Scholar]
- 10. Haydon D. J., and Guest J. R. (1991) A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 63, 291–295 [DOI] [PubMed] [Google Scholar]
- 11. Jain D. (2015) Allosteric control of transcription in GntR family of transcription regulators: A structural overview. IUBMB Life 67, 556–563 [DOI] [PubMed] [Google Scholar]
- 12. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uzzau S., Figueroa-Bossi N., Rubino S., and Bossi L. (2001) Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. U.S.A. 98, 15264–15269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elledge S. J., and Davis R. W. (1989) Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev. 3, 185–197 [DOI] [PubMed] [Google Scholar]
- 15. Hu M., Liu Y., Yu K., and Liu X. (2014) Decreasing the amount of trypsin in in-gel digestion leads to diminished chemical noise and improved protein identifications. J. Proteomics 109, 16–25 [DOI] [PubMed] [Google Scholar]
- 16. Livak K. J., and Schmitgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 17. Bramucci E., Milano T., and Pascarella S. (2011) Genomic distribution and heterogeneity of MocR-like transcriptional factors containing a domain belonging to the superfamily of the pyridoxal-5′-phosphate dependent enzymes of fold type I. Biochem. Biophys. Res. Commun. 415, 88–93 [DOI] [PubMed] [Google Scholar]
- 18. Osborne S. E., Walthers D., Tomljenovic A. M., Mulder D. T., Silphaduang U., Duong N., Lowden M. J., Wickham M. E., Waller R. F., Kenney L. J., and Coombes B. K. (2009) Pathogenic adaptation of intracellular bacteria by rewiring a cis-regulatory input function. Proc. Natl. Acad. Sci. U.S.A. 106, 3982–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Belitsky B. R., and Sonenshein A. L. (2002) GabR, a member of a novel protein family, regulates the utilization of gamma-aminobutyrate in Bacillus subtilis. Mol. Microbiol. 45, 569–583 [DOI] [PubMed] [Google Scholar]
- 20. Okuda K., Kato S., Ito T., Shiraki S., Kawase Y., Goto M., Kawashima S., Hemmi H., Fukada H., and Yoshimura T. (2015) Role of the aminotransferase domain in Bacillus subtilis GabR, a pyridoxal 5′-phosphate-dependent transcriptional regulator. Mol. Microbiol. 95, 245–257 [DOI] [PubMed] [Google Scholar]
- 21. Jochmann N., Götker S., and Tauch A. (2011) Positive transcriptional control of the pyridoxal phosphate biosynthesis genes pdxST by the MocR-type regulator PdxR of Corynebacterium glutamicum ATCC 13032. Microbiology 157, 77–88 [DOI] [PubMed] [Google Scholar]
- 22. Wiethaus J., Schubert B., Pfänder Y., Narberhaus F., and Masepohl B. (2008) The GntR-like regulator TauR activates expression of taurine utilization genes in Rhodobacter capsulatus. J. Bacteriol. 190, 487–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takenaka T., Ito T., Miyahara I., Hemmi H., and Yoshimura T. (2015) A new member of MocR/GabR-type PLP-binding regulator of D-alanyl-D-alanine ligase in Brevibacillus brevis. FEBS J. 282, 4201–4217 [DOI] [PubMed] [Google Scholar]
- 24. Tomljenovic-Berube A. M., Mulder D. T., Whiteside M. D., Brinkman F. S., and Coombes B. K. (2010) Identification of the regulatory logic controlling Salmonella pathoadaptation by the SsrA-SsrB two-component system. PLoS Genet. 6, e1000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoon H., Ansong C., McDermott J. E., Gritsenko M., Smith R. D., Heffron F., and Adkins J. N. (2011) Systems analysis of multiple regulator perturbations allows discovery of virulence factors in Salmonella. BMC Syst. Biol. 5, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Navarre W. W., Porwollik S., Wang Y., McClelland M., Rosen H., Libby S. J., and Fang F. C. (2006) Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics data reported in this paper have been deposited to the iProx database (URL: http://www.iprox.org/) and are available under the accession number IPX0000899001.