ABSTRACT
Across metazoans, innate immunity is vital in defending organisms against viral infection. In mammals, antiviral innate immunity is orchestrated by interferon signaling, activating the STAT transcription factors downstream of the JAK kinases to induce expression of antiviral effector genes. In the nematode Caenorhabditis elegans, which lacks the interferon system, the major antiviral response so far described is RNA interference (RNAi), but whether additional gene expression responses are employed is not known. Here we show that, despite the absence of both interferon and JAK, the C. elegans STAT homolog STA-1 orchestrates antiviral immunity. Intriguingly, mutants lacking STA-1 are less permissive to antiviral infection. Using gene expression analysis and chromatin immunoprecipitation, we show that, in contrast to the mammalian pathway, STA-1 acts mostly as a transcriptional repressor. Thus, STA-1 might act to suppress a constitutive antiviral response in the absence of infection. Additionally, using a reverse genetic screen, we identify the kinase SID-3 as a new component of the response to infection, which, along with STA-1, participates in the transcriptional regulatory network of the immune response. Our work uncovers novel physiological roles for two factors in viral infection: a SID protein acting independently of RNAi and a STAT protein acting in C. elegans antiviral immunity. Together, these results illustrate the complex evolutionary trajectory displayed by innate immune signaling pathways across metazoan organisms.
IMPORTANCE
Since innate immunity was discovered, a diversity of pathways has arisen as powerful first-line defense mechanisms to fight viral infection. RNA interference, reported mostly in invertebrates and plants, as well as the mammalian interferon response and JAK/STAT pathway are key in RNA virus innate immunity. We studied infection by the Orsay virus in Caenorhabditis elegans, where RNAi is known to be a potent antiviral defense. We show that, in addition to its RNAi pathway, C. elegans utilizes an alternative STAT pathway to control the levels of viral infection. We identify the transcription factor STA-1 and the kinase SID-3 as two components of this response. Our study defines C. elegans as a new example of the diversity of antiviral strategies.
INTRODUCTION
RNA viruses, a highly diverse family known to infect organisms of almost all kingdoms of life, also represent an important burden on human health. Indeed, their highly mutagenic and adaptive nature is an ever-growing challenge for diagnosis and treatment and the underlying understanding of host-pathogen interaction. The first and critical step in mounting successful antiviral defense is the conserved innate immune response; however, its complexity is yet to be fully apprehended. Several pathways have been reported for various organisms that differ in their presence or relative importance. To date, the main pathways can be divided in two categories: protein-based innate immunity and RNA-based innate immunity. Jawed vertebrates rely mostly on the powerful interferon (IFN) system, whereas most other eukaryotes take advantage of the RNA interference (RNAi) machinery (1, 2).
In mammals, the initiation of the interferon response in response to RNA viruses depends on the RNA helicase RIG-I (retinoic acid-inducible gene I product) and its paralogs. RIG-I senses double-stranded RNA (dsRNA) intermediates that are generated during the replication of single-stranded RNA (ssRNA) viruses and initiates production of the type 1 interferon and inflammatory cytokines via activation of the transcription factors interferon regulatory factor IRF3/7 and NF-κB (3, 4). Interferons can in turn activate the JAK/STAT signaling pathway to induce an antiviral state and mediate viral control. In essence, binding of interferon to the type 1 interferon receptor (IFNAR) leads to activation of the receptor-associated tyrosine kinases JAK1 and TYK2, and eventually to the phosphorylation of the STAT transcription factors. Upon dimerization, STAT transcription factors can undergo translocation to the nucleus where they activate the expression of antiviral genes and inflammatory response genes (5). On the other hand, antiviral RNAi relies on the processing by the endoribonuclease Dicer of long double-stranded viral RNA molecules occurring during the viral cycle. These dsRNA molecules appear particularly during replication of the RNA genome by the virally encoded RNA-dependent RNA polymerase. After processing by Dicer, the resulting small interfering RNAs (siRNA) are loaded in the RNA-induced silencing complex (RISC) through the binding of the siRNA to a protein of the Argonaute family. Recognition of the target viral RNA by the active RISC eventually leads to its degradation (1).
In principle, all the components required for RNAi are still present in higher vertebrates, but this might simply reflect other biological functions such as microRNA (miRNA)-based gene regulation. It has also been proposed that the IFN-based innate immunity and antiviral RNAi might be incompatible (6, 7) or that the evolution of Dicer in mammalian somatic cells renders it inactive for long dsRNA processing (8).
However, some evidence supports potential roles for RNAi-based antiviral immunity in mammals (9–11). Similarly, the prominence of the RNAi pathway in fighting viruses in invertebrates may obscure other mechanisms that the innate immune system may use to combat viruses in these organisms. One such example is the nematode Caenorhabditis elegans, where antiviral immunity has previously been shown to involve a potent RNAi response (12).
A single virus, the Orsay virus, has been so far described to infect C. elegans in the wild (12). This small bipartite single-stranded RNA virus is efficiently targeted by the nematode RNAi machinery to prevent its replication. Surprisingly, DRH-1, a conserved helicase related to RIG-I, is essential for the antiviral RNAi pathway in C. elegans instead of the classical interferon response triggered by its mammalian counterpart RIG-I in mammals (Fig. 1A) (13–15). Interestingly, a previous analysis of gene expression changes upon viral infection revealed evidence for the induction of antiviral response genes upon viral infection independent of the RNAi pathway (16). However, neither the upstream signaling pathway linking these gene expression changes to viral infection nor the extent to which gene expression alterations directly contribute to antiviral defense are known. We therefore set out to uncover new signaling pathways involved in sensing and transducing viral infection.
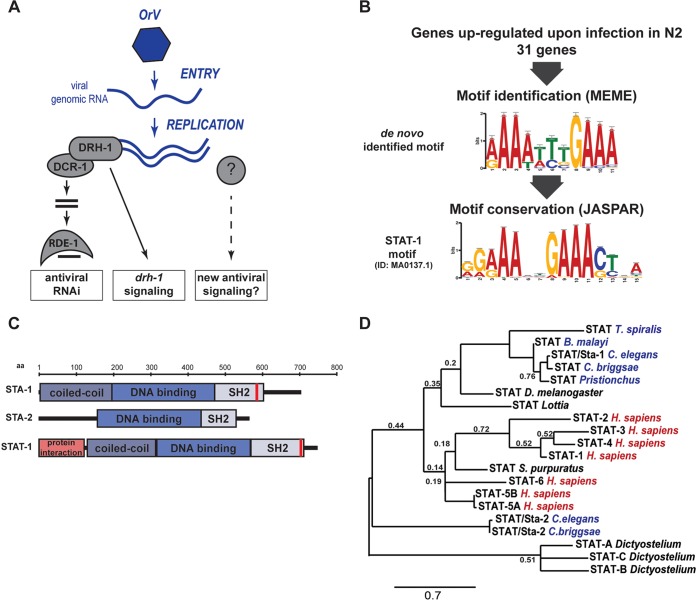
FIG 1 .
Identification of a STAT transcription factor signature in immune response against the Orsay virus (OrV) in C. elegans. (A) Schematic representation of known antiviral pathways in C. elegans. (B) MEME motif enrichment in genes regulated upon infection and conservation of the de novo-identified motif by Tom-tom. ID, identifier. (C) Schematic representation of conserved domains between the human STAT-1 and C. elegans STATs. The red bars depict the predicted phosphorylation sites (20). aa, amino acids. (D) Phylogenetic analysis of STAT transcription factors. Nematodes are indicated in blue, and vertebrates are indicated in red. Trichinella spiralis, Brugia malayi, Caenorhabditis briggsae, Drosophila melanogaster, Homo sapiens, and Strongylocentrotus purpuratus STATs are shown on the tree. The tree is based on full-length sequences. Alignment was performed with Muscle. Branch support values are from bootstrap using 1,000 iterations. Only values lower than 0.8 are depicted. The bar shows a branch length of 0.7 nucleotide substitutions per position.
RESULTS
Identification of a STAT transcription factor as a modulator of the infection response.
In order to identify key factors regulating the immune state of C. elegans after infection by Orsay virus (OrV), we set out to identify conserved regulatory motifs for the genes that participate in the transcriptional response to infection. Previously we identified a set of such putative antiviral response genes that were upregulated upon infection with the Orsay virus in the laboratory reference strain N2 (16). Interestingly, some of these genes, also upregulated upon infection of the antiviral RNAi-deficient rde-1 mutant, were not upregulated in another domesticated strain of C. elegans, JU1580, which is also hypersensitive to the Orsay virus (13, 16). The JU1580 strain lacks the RIG-I homolog DRH-1 (Fig. 1A). This suggested that DRH-1 might be required for a transcriptional response to infection, independently of RNAi. However, as strains JU1580 and N2 differ at many loci besides drh-1, including some infection response genes (12, 13), we decided to test this further by performing microarray analysis to compare infection-induced genes in strain N2, a drh-1 knockout in the N2 strain, JU1580, and JU1580 carrying a transgene containing the N2 drh-1 locus (JU1580 drh-1 rescue strain). drh-1 mutants infected with Orsay virus showed changes in gene expression similar to those of infected JU1580 animals (see Fig. S1 in the supplemental material), in particular, the lack of upregulation of a set of genes induced in wild-type animals. Importantly, this shows that there must be gene expression changes that are induced independently of the level of viral replication, as both JU1580 and drh-1 have much higher virus titers than strain N2 or the JU1580 drh-1 rescue strain. This indicates that there may be signaling pathways governing antiviral gene expression in C. elegans (Fig. S1).
Orsay virus (OrV) infection induces gene expression changes that depend on the genotype of the host. The heatmap shows changes in gene expression, as measured by Affymetrix array, occurring in each of the indicated strains after infection by the Orsay virus. Download FIG S1, TIF file, 1 MB (1MB, tif) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To gain insights into the nature of the signaling events following viral infection, we extracted the promoters from the set of genes upregulated in strain N2 after OrV infection (Data Set S1) and searched for associated motifs (E value of <0.1 against a background model drawn from the nucleotide composition of the input sequences) using the motif prediction software MEME (17). We then compared these motifs to known transcription factor DNA binding motifs in the JASPAR core database using Tom-tom (18, 19). Remarkably, we identified an enriched motif with similarity to the one of STAT transcription factors (Fig. 1B), well-known to have a conserved role in antiviral defense in mammals. There are two STAT homologs, STA-1 and STA-2, in C. elegans (Fig. 1C). STA-1 has all the classical functional STAT domains: a coiled-coil domain for protein-protein interaction, a DNA binding domain, an SH2 domain, and a putative tyrosine phosphorylation motif (20). STA-2 is similar but lacks the coiled-coil domain as well as the tyrosine phosphorylation motif (21). Consistent with this, a phylogenetic tree with C. elegans STA-1 and STA-2 and representative STATs from other organisms suggests that STA-1 is closely related to mammalian STATs but that STA-2 is more divergent (Fig. 1D). Interestingly, STA-2 has previously been implicated in an antifungal response (21), whereas STA-1 has been linked to developmental signaling in C. elegans (22). We therefore speculated that STA-1 and/or STA-2 might have a previously unappreciated role in antiviral defense.
Upstream sequences of genes upregulated upon infection Download DATA SET S1, TXT file, 0.03 MB (31.6KB, txt) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
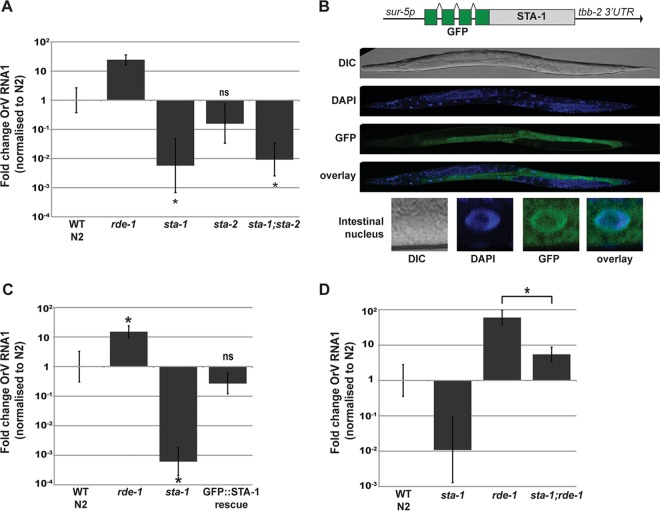
To evaluate a potential role for STAT signaling in antiviral defense in C. elegans, we used reverse transcription-quantitative PCR (RT-qPCR) to quantify the viral loads of sta-1 and sta-2 mutants following infection by the Orsay virus. For controls, we used wild-type N2 animals and rde-1 mutants, which are hypersensitive to infection due to the lack of the Argonaute protein RDE-1, essential for the initiation of antiviral RNAi (Fig. 1A) (12, 23). Surprisingly, sta-1 mutants were 100-fold less permissive to infection than wild-type animals, whereas sta-2 mutants showed wild-type sensitivity (Fig. 2A). Double mutants lacking both STAT homologs were no more permissive than sta-1 single mutants were. We conclude that STA-1 but not STA-2 acts in regulating antiviral defense. Consistent with these observations of viral load, induction of a viral response reporter gene, comprising gfp driven by the promoter of the viral response gene sdz-6 (16), was reduced in sta-1 mutants compared to strain N2 (P = 0.0061) and increased in rde-1 mutants (P = 0.0034) (Fig. S2). Next we generated a single-copy, intrachromosomal sur-5::gfp::sta-1 transgene, which drives expression of a green fluorescent protein (GFP)–STA-1 fusion protein in the intestine and other somatic tissues (Fig. 2B). GFP–STA-1 accumulated on chromatin in the nuclei of intestinal cells (Fig. 2B). Importantly, the sur-5::gfp::sta-1 transgene restored wild-type sensitivity to the Orsay virus in a strain lacking endogenous sta-1 (Fig. 2C). Finally, we asked whether STA-1 acts independently of the antiviral RNAi pathway using epistasis analysis (Fig. 2D). In rde-1 mutants lacking the antiviral RNAi response (Fig. 1A), the Orsay virus accumulates to much higher levels than in the wild-type N2 strain. However, sta-1; rde-1 double loss-of-function mutants show significantly reduced viral loads compared to rde-1 single mutants, suggesting that sta-1 acts in parallel or downstream of rde-1 (Fig. 2D). Together, these results identify the C. elegans STAT transcription factor STA-1 as acting in addition to the antiviral RNAi pathway in the immune response to viral infection in C. elegans.
FIG 2 .
STA-1 is a key transcription factor in the immune response upon Orsay virus (OrV) infection. (A, C, and D) Viral loads in different strains measured by RT-qPCR on the Orsay virus RNA1 genome after 3 days of infection. The wild-type (WT) control N2 strain, rde-1(ne219) strain, and strains with sta-1(ok587) and sta-2(ok1860) mutations were infected. Values that were significantly different (P < 0.01) from the value for the wild-type control strain (unless otherwise indicated) by a two-tailed Mann-Whitney U test are indicated with an asterisk. Values that were not significantly different (ns) are indicated. n = 6 for panels A and C, and n = 4 for panel D. (B) Expression of a single-copy transgene coding for a GFP::STA-1 fusion protein under the control of a sur-5 promoter in adult animals. DIC, differential interference contrast.
sta-1 reduces activation of an infection sensor. Activation of the sdz-6 viral sensor. The promoter of the sdz-6 gene drives the expression of the GFP in the infected intestine. Animals were scored for GFP signal after 3 days of infection in three biological replicates with an average of 125 animals scored for each condition. A t test was performed to compare the average fraction of positive animals (on and dim) after infection in wild-type (WT) and rde-1 animals and in WT and sta-1 animals. Download FIG S2, TIF file, 0.8 MB (849.1KB, tif) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
STA-1 is a repressor of infection response genes.
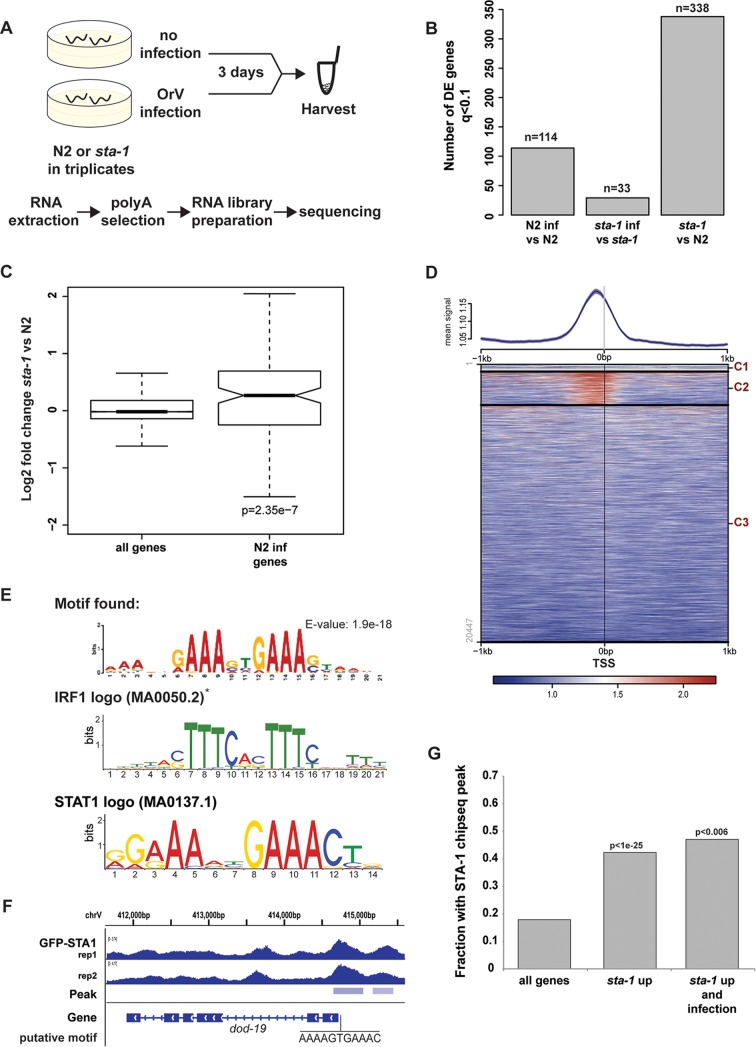
In order to test whether the low permissivity of sta-1 mutants to viral infection reflects a role for STA-1 in the regulation of antiviral response genes, we performed transcriptome sequencing (RNA-seq) analysis of wild-type strain N2 and sta-1 mutant animals with or without OrV infection (Fig. 3A). We then selected transcripts altered significantly after OrV infection (DESeq q value [false-discovery rate] of <0.1; Data Set S2). We observed a robust response to infection in N2 animals, whereas fewer transcripts were significantly altered in sta-1 mutants upon infection (Fig. 3B). However, sta-1 mutants displayed a constitutive deregulation of gene expression compared to N2 animals in the absence of OrV infection (Fig. 3B). Furthermore, genes that changed expression significantly upon infection in N2 showed a strong trend to be constitutively upregulated in sta-1 mutants, while this was not the case when all genes were considered (Fig. 3C). These data suggest that STA-1 largely acts as a transcriptional repressor of an antiviral gene expression program. However, this antiviral gene expression program includes genes that are up- and downregulated upon infection, either directly or indirectly (Fig. 3B). We therefore propose that the lower permissivity of sta-1 mutants to viral infection is caused by either a constitutive deregulation of the antiviral defense gene expression program or control by STA-1 of OrV essential host genes.
FIG 3 .
STA-1 acts at the promoter of virus response genes and represses their expression. (A) Schematic representation of the RNA-seq experiment performed with strain N2 and sta-1(ok587) animals. (B) Representation of the number of genes showing differential expression (DE) upon infection in strain N2 (N2 inf), infection in sta-1 mutant animals, or infection in sta-1 mutants compared to N2, as measured by RNA-seq. (C) Gene expression measured by RNA-seq in a sta-1 mutant animals normalized to the N2 control. All genes or a subset of genes that show upregulation upon infection in N2 animals are depicted. The box shows the interquartile range, and the whiskers extend to the greatest value ≤1.5 times the interquartile range from the box. The significance of the enrichment is represented as the P value of the Wilcoxon unpaired test. (D) ChIP-sequencing after immunoprecipitation of the GFP::STA-1 transgene. (Top) The ChIP signal is plotted 1 kb upstream and 1 kb downstream of all genes anchored at TSS as in references 52 and 53. The heatmap was generated using k-means clustering (three clusters, C1 [cluster 1], C2, and C3]) of all transcription start sites (TSS) using the GFP::STA-1 signal for clustering. (E) Motif identification and conservation by Centrimo (54) for the 1-kb sequence centered around the summits of the peaks in cluster 2 (C2) in panel D. The asterisk indicates that the IRF1 motif identified by Tom-tom matches the reverse complement of the motif found. (F) Example of a upregulated sta-1 gene showing binding of STA-1 by ChIP-seq in two independent biological replicates (rep1 [replicate 1 ] and rep2). The putative STA-1 binding motif is indicated. (G) ChIP peak enrichment in different sets of genes. The three sets of genes were genome-wide genes (all genes), genes upregulated in sta-1 versus strain N2 (sta-1 up), and genes differentially expressed upon infection and upregulated in sta-1 (sta-1 up and infection).
Differential gene expression measured by RNA-seq. Filtered data set for genes showing differential expression (P < 0.1) in at least one sample. Download DATA SET S2, XLSX file, 0.2 MB (184.4KB, xlsx) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next we asked whether STA-1, similarly to STATs in mammals, might bind to DNA in a sequence-specific manner to regulate infection response genes. We therefore performed chromatin immunoprecipitation-DNA sequencing (ChIP-seq) analysis of GFP::STA-1 to determine its genomic binding pattern in noninfected animals expressing the sur-5::gfp::sta-1 transgene but lacking endogenous sta-1. Through GFP::STA-1 ChIP-seq, we identified 2,133 STA-1 peaks across the genome. STA-1 was enriched close to transcription start sites (TSS) of about 20% of all genes, with an enriched binding peak located ~200 bp upstream of their TSS (Fig. 3D and Data Set S3). These data are in agreement with STA-1 acting as a specific DNA-binding transcription factor to regulate gene expression. To test further the association of STA-1 binding with the infection gene expression response, we used the sequences surrounding the STA-1 peaks in order to search for enriched motifs. Importantly, we recovered an enriched motif that was nearly identical to the motif predicted from our gene expression analysis (Fig. 1B) and matching the mammalian IRF and STAT motifs in the JASPAR core database and the consensus core interferon-sensitive response element (ISRE) TTCNNTTT (Fig. 3E and F) (24). Intriguingly, we additionally identified a separate highly enriched motif with strong similarity to the consensus sequence for GATA-like transcription factors (Fig. S3). The predicted GATA and STAT motifs were not found at the same set of genes, suggesting that GATA-like transcription factors may be able to recruit STA-1 to DNA independently of STA-1 DNA binding, consistent with previous studies of mammalian cells (25). This may serve to expand the number of sites bound by STA-1 beyond those containing a core STAT binding element.
Discovery of a GATA motif enriched in the STA-1-bound genes. (A) Enrichment of the IRF motif (as depicted in Fig. 3) across 1,375 peaks (out of 3,211 total peaks). (B) Enrichment of a GATA motif across 503 peaks (out of a total of 3,211 peaks) and logo representation. Download FIG S3, TIF file, 0.9 MB (892.2KB, tif) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Association between ChIP peak calling and gene names Download DATA SET S3, XLSX file, 0.5 MB (504.1KB, xlsx) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We next tested the association between STA-1 DNA binding and gene expression. STA-1 binding was strongly enriched at genes with increased expression in sta-1 mutant animals (P < 1e−25 [Fig. 3G]) (Fig. S4) compared to all genes. However, it is worth noting that STA-1 binding was also enriched at the promoters of the smaller number of genes downregulated in the mutant (11/46 downregulated genes versus 122/379 upregulated genes). Thus, although STA-1 largely acts as a constitutive repressor of gene expression, it may have a more complex role at the promoters of some genes. The intersection of STA-1 binding with all N2 infection response genes, including those not upregulated in sta-1 mutants was not significant, which may indicate that other signaling pathways are involved in gene expression responses to infection or technical limitations of identifying STA-1 binding sites from ectopically expressed GFP–STA-1; however, STA-1 binding was strongly enriched at N2 response genes that were also upregulated in sta-1 mutants, although there were few of these genes (Fig. 3G).
STA-1 is bound to genes upregulated in sta-1 mutants and infection genes. The table shows the intersection for gene sets and STA-1 ChIP-seq peaks as depicted in Fig. 3G. Download FIG S4, TIF file, 0.6 MB (680.8KB, tif) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
STA-1 is required for normal life span.
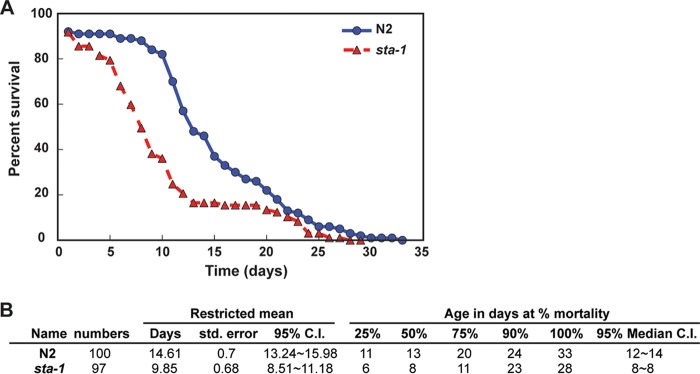
The lower permissivity of sta-1 mutants to infection raises the question of whether there are negative fitness consequences associated with STA-1 deficiency that might act as trade-offs between resistance to infection and optimal growth. No obvious defects in development or fecundity were observed in sta-1 mutants, consistent with published data (20). However, we observed a significant decrease in the median life span of sta-1 mutants (Fig. 4). This may be due to the constitutive activation of pathogen response genes in sta-1 mutants imposing a cost on animal development and/or physiology as has been shown in other systems, e.g., insects (26–28).
FIG 4 .
STA-1 is required for normal life span. (A) Survival plot showing the relationship between the survival and age of the wild-type N2 and sta-1 mutant animals. (B) Descriptive statistics of the life span experimental data in panel A. The associated log rank test Bonferroni P value is 2.7e−6. std. error, standard error; 95% C.I., 95% confidence interval.
The SID-3 kinase acts upstream of STA-1 in regulation of the accumulation of the Orsay virus.
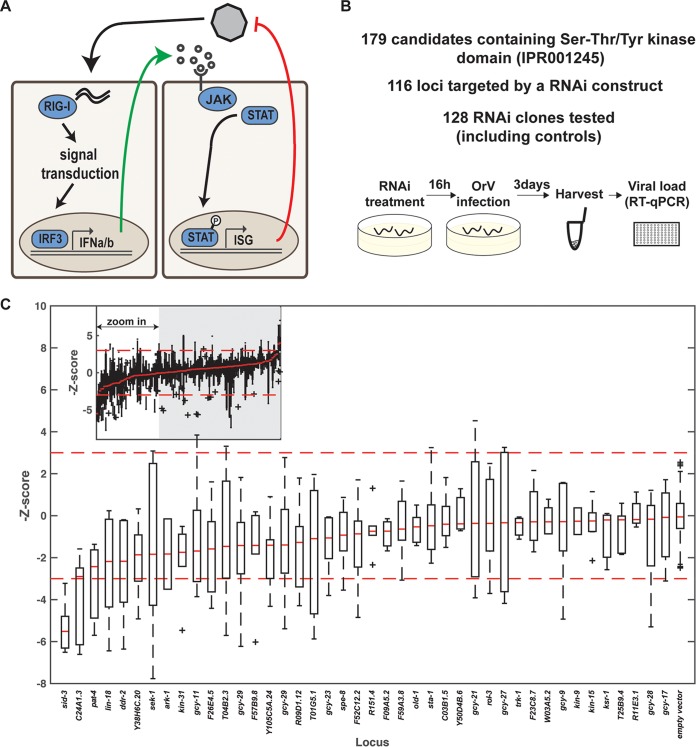
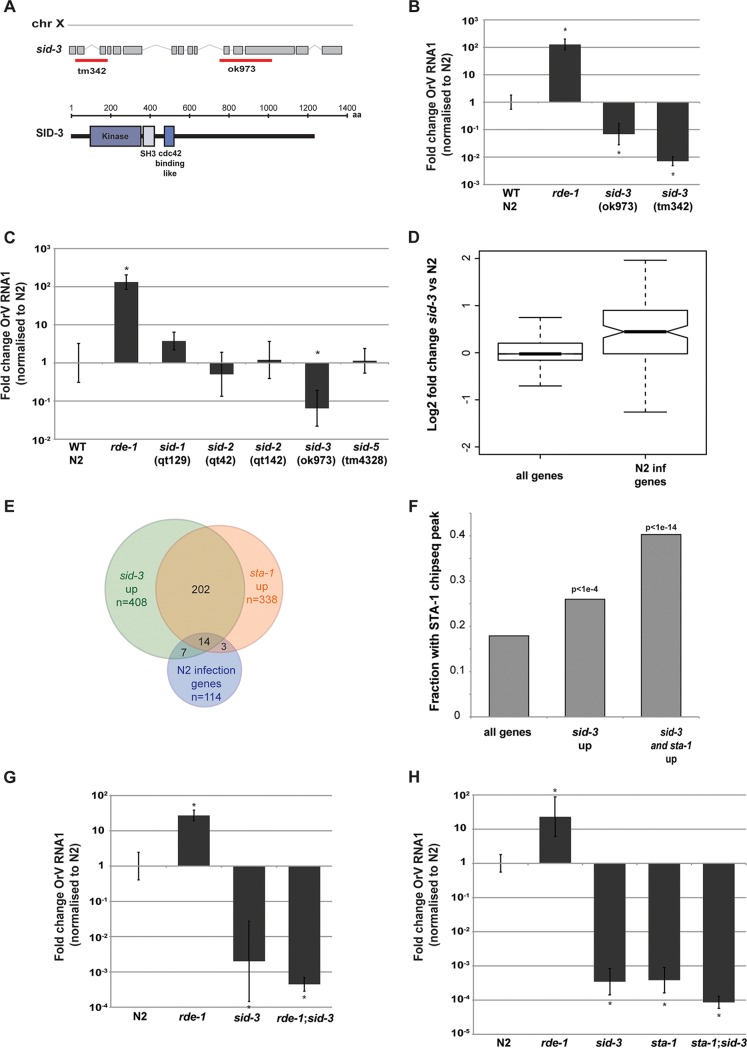
Having demonstrated that STA-1 acts as a constitutive repressor of antiviral response genes, we wondered how STA-1 activity was inhibited in response to viral infection. In mammals, STAT proteins are regulated by JAK tyrosine kinase phosphorylation (Fig. 5A). There is no conserved homolog of the JAK kinases in C. elegans; however, the canonical tyrosine phosphorylation site on STA-1 is conserved. Thus, other kinases may regulate STA-1 activity (20). There is also a growing body of evidence of JAK-independent phosphorylation of STAT transcription factors, including serine phosphorylation (29, 30). Additionally, tyrosine and serine/threonine kinases play an essential role in antibacterial and antifungal defense in C. elegans (31, 32). To identify potential kinases upstream of STA-1, we performed an RNAi screen, testing genes with the serine/threonine/tyrosine-protein kinase catalytic domain IPR001245 (33). The C. elegans genome encodes 176 proteins with this domain, 116 of which were available as clones as part of C. elegans genome-wide RNAi libraries (34, 35). Following RNAi by feeding, we infected animals with the Orsay virus and then quantified the viral load for each of the 116 candidates and additional controls (Fig. 5B and C and Data Set S4). Using a stringent cutoff (|Z-score| > 3), we identified only a single regulator of viral load, sid-3. RNAi of sid-3 resulted in ~100-fold reduction of viral RNA accumulation compared to the control (Fig. 5C). We confirmed that sid-3 is required for permissivity to viral infection by testing two independent deletion mutants of sid-3 (Fig. 6A and B). SID-3 is a tyrosine kinase, implicated in systemic RNAi, and is presumed to assist in the import of dsRNA into the cell during experimental RNAi (36). However, this role in RNAi is not likely to be linked to its role in antiviral defense, because other genes required for systemic RNAi, such as sid-1, sid-2, and sid-5, do not show a significant difference in antiviral sensitivity from wild-type strain N2 (37) (Fig. 6C).
FIG 5 .
The tyrosine kinase SID-3 enables efficient viral replication. (A) Activation of the JAK/STAT signaling pathway in RNA virus infection in humans. Upon recognition of the viral RNA by a sensor like RIG-I, type 1 interferon (alpha/beta interferon) is released in the environment, recognized by the IFN receptor, and activates the JAK tyrosine kinase to trigger nuclear translocation of activated STAT transcription factors. IFNa/b, alpha/beta interferon; ISG, interferon-stimulated genes; P, phosphate. (B) Overview of the RNAi screen. (C) RNAi treatment leading to increased resistance to OrV infection. The viral RNA accumulation after RNAi treatment is represented by the Z-score of the ΔΔCT values (relative to empty vector [eV]). The red broken lines represent the 99% confidence interval of the control RNAi (eV), calculated as plus or minus 2.7 standard deviations. The red bars represent the median of the biological replicates for the genes tested or for controls. The plus signs represent outliers. The inset at the top of the graph depicts all tested clones.
FIG 6 .
SID-3 and STA-1 regulate a common set of genes. (A) Schematic representation of the different sid-3 alleles available. The sid-3 ok973 allele induces a deletion of 1,330 bp and insertion of 12 nucleotides, deleting exons 11 and 12 and part of exon 13, after the tyrosine kinase domain. The sid-3 tm342 allele is a null allele, where an early 821-bp-long deletion (exon 1 to exon 3) and insertion of 7 nucleotides lead to a premature stop codon. chr X, chromosome X. (B and C) Viral load after 3 days of infection, measured by RT-qPCR on the Orsay virus RNA1 genome. *, P < 0.01 by two-tailed Mann-Whitney test. n = 6 for each strain. (D) RNA-seq analysis showing an enrichment for infection genes in sid-3(ok973) upregulated genes. (E) Overlap between genes showing differential expression by RNA-seq upon infection or upregulated in sta-1 or upregulated in sid-3. (F) Overlap between STA-1-bound genes, as shown by ChIP-seq and sid-3 upregulated genes. (G and H) Viral load in the indicated strains, measured by RT-qPCR on the Orsay virus RNA1 genome after 3 days of infection. The sid-3 allele used is sid-3(ok973). In panel G, *, P < 0.01 by two-tailed Mann-Whitney test and n = 6 for each strain. In panel H, *, P < 0.05 by two-tailed Mann-Whitney test and n = 3 for each strain except N2 and n = 4 for strain N2.
RNAi screen data Download DATA SET S4, XLSX file, 0.1 MB (101.7KB, xlsx) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We therefore wondered whether SID-3 might act in the same pathway as STA-1. We quantified gene expression in sid-3 mutants using RNA-seq. Similar to what we observed for sta-1 mutants, genes that changed expression significantly upon infection in strain N2 showed a strong trend to be constitutively upregulated in sid-3 mutants, while this was not the case when all genes were considered (Fig. 6D and Data Set S2). Furthermore, the genes upregulated in sid-3 mutant animals showed a striking overlap with the genes upregulated in sta-1 mutant animals (P = 1.2e−15). Additionally, sid-3 upregulated genes were enriched for antiviral response genes (P = 5.55e−10) (Fig. 6E and Fig. S5). Interestingly, among the viral infection response genes that showed constitutive upregulation in both sta-1 and sid-3 mutants (Data Set S2), three genes have been previously implicated in the control of adult life span (C32H11.9, dod-21, and dod-23), of which two are tandem paralogs of human EPXH1, an epoxide hydrolase gene, suggesting a putative role in detoxification. We also noticed the presence of dct-17, involved in the innate immune response and localized to the membrane raft, an important compartment for RNA virus replication (38, 39). Moreover, genes upregulated in sid-3 mutants, including those shared with sta-1, were enriched for STA-1 binding by ChIP-seq, suggesting that sid-3 acts upstream of sta-1 in the antiviral gene expression response (Fig. 6F). We conclude that SID-3 and STA-1 act in the same process to regulate an innate antiviral immunity program.
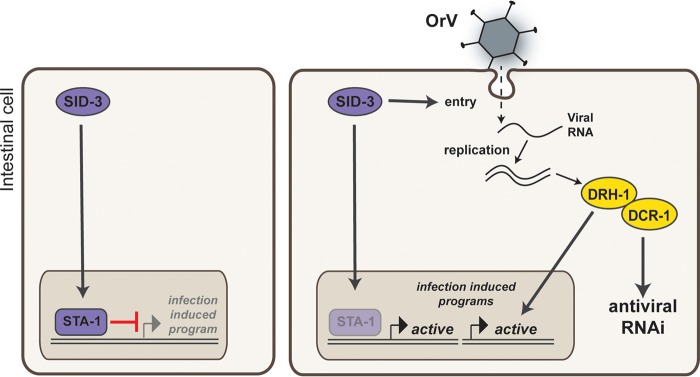
Finally, we addressed the relationship between SID-3 and the antiviral RNAi pathway using epistasis analysis. Interestingly, rde-1;sid-3 double loss-of-function mutants showed the same permissivity to OrV infection as sid-3 mutants (Fig. 6G). This is in contrast to what we observed in sta-1;rde-1 mutants (Fig. 2D). Additionally, sta-1;sid-3 double mutants also showed a very low permissivity to OrV infection (Fig. 6H). One likely interpretation of these data is that SID-3 acts both upstream of antiviral RNAi and upstream of a STA-1-dependent antiviral gene expression program to regulate the response to OrV infection. We propose a model (Fig. 7) whereby in uninfected animals, SID-3 is required to maintain repression of viral infection response genes by STA-1. Upon infection, this signaling is curtailed, inducing an antiviral response program.
FIG 7 .
A STA-1 pathway controls the response to viral infection in C. elegans, a model.
DISCUSSION
Here, we uncover a hitherto unappreciated role for the C. elegans homolog of the mammalian STAT family of transcription factors. We demonstrate that the tyrosine kinase SID-3 and the STAT transcription factor STA-1 are two new key factors in the antiviral response in C. elegans. Additionally, we uncover a novel potential signaling pathway linking the kinase SID-3, previously reported only for systemic RNA interference, to STAT signaling. Our results have intriguing implications both for innate immunity in C. elegans and for the evolution of eukaryotic signaling pathways.
Regulation of antiviral gene expression in C. elegans.
Previously there has been some debate over the extent to which gene expression responses to infection in C. elegans represent specific pathogen response pathways or more-general responses to stress. Our findings that a STAT family transcription factor is responsible for constitutively repressing infection response genes and that this is relieved upon infection suggest that at least some part of the transcriptional response to infection is specific. Moreover, constitutive upregulation of these genes, as demonstrated in mutants lacking sta-1, leads to low permissivity to viral infection, although further examination of the functions of these genes will be needed to understand exactly how their expression is involved in viral infectivity.
How is the constitutive repression of antiviral response genes by sta-1 relieved upon viral infection? The fact that sid-3 mutants show loss of sta-1 repression suggests that phosphorylation of STA-1 is required for it to exert its repressive function. Whether the phosphorylation is directly mediated by a signaling cascade downstream of the kinase activity of SID-3 or conversely is caused by disruption of the SID-3 complex upon infection (as would potentially be mimicked by the sid-3 mutation) is still a point of discussion. Viral infection may also lead to loss of SID-3 activity and concomitant loss of STA-1 phosphorylation. The differences in gene expression observed between sta-1 mutant animals and infected animals could then be explained by the difference between the set of transcription factors active in an infected cell compared to the noninfected sta-1 mutant.
An interesting clue to the potential role of SID-3 in viral infection comes from its proposed role in endocytosis (36). Nonenveloped viruses require the endosomal pathway for entry into cells; it is therefore plausible that viral entry into endosomes may lead to titration of SID-3 away from its role in signaling to STA-1, leading to relief of repression. Such an early role for sid-3 in infection is consistent with the ability of sid-3 mutation to suppress the sensitivity of RNAi pathway mutants. Indeed, Dave Wang and colleagues demonstrate in this issue that sid-3 is required for viral entry (40). Thus, we hypothesize that the dependence of the virus on sid-3 for its entry has been exploited by C. elegans as a mechanism to regulate antiviral gene expression. Further work will be required to refine this signaling pathway.
Evolution of STAT signaling.
It is remarkable that the STAT family of transcription factors has a role in antiviral gene regulation in both mammals and C. elegans, despite the fact that both upstream and downstream genes in the pathway are not conserved in mammals and C. elegans. Even more remarkably, the mechanism whereby STAT transcription factors contribute to gene expression regulation are quite distinct in nematodes from that in mammals, as while STA-1 is constitutively bound to DNA and acts as a transcriptional repressor in C. elegans, STATs are activated in response to infection. Despite this, there appear to be intriguing parallels between STA-1 activity and the mammalian STAT pathway, in particular the association that we uncovered between STA-1 binding and GATA transcription and the conserved role of STATs in viral gene expression responses in C. elegans and mammals.
Our results highlight the remarkable plasticity of signaling pathways through evolution, whereby a central module may retain the same function over millions of years while acquiring distinct regulatory partners. This ability to both retain and diversify functionality simultaneously is clearly particularly important in the immune system, where the specific nature of the threats involved changes rapidly while their general modes of action may not. It will be intriguing to explore whether STATs in other metazoans have similarly diversified while retaining antiviral activity—it is possible that the C. elegans mode of STAT action that we uncover here is in fact the original conserved role of STAT transcription factors in viral responses. If so, vestiges of this function may be retained in mammalian cells, particularly those without expression of the JAK pathway.
MATERIALS AND METHODS
Nematode culture and strains.
We grew C. elegans under standard conditions at 20°C. The wild-type strain was var. Bristol N2. The food source used was Escherichia coli strain HB101 (Caenorhabditis Genetics Center, University of Minnesota, Twin Cities, MN, USA). Detailed information about all strains generated and used in this study can be found in Table S1 in the supplemental material.
Strains used in this study Download TABLE S1, PDF file, 0.03 MB (33.2KB, pdf) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Viral filtrate.
Stably infected populations of sensitive animals (JU1580) were transferred to 2-liter liquid cultures with E. coli HB101 food and grown for 7 days. The supernatant of the culture was harvested on ice and filtered with a 0.22-µm filter. The resulting viral filtrate was aliquoted and stored at −80°C.
Viral infection.
Two wild-type or three mutant larval stage 4 (L4) animals were added to seeded 50-mm plates. Sixteen hours later, 20 µl of Orsay virus filtrate was added to the edge of the bacterial lawn. Animals were collected 3 days after infection. The animals were washed off the plates with M9 buffer. The animal pellets were washed another three times by pelleting the animals either by gravity on ice or by centrifugation at 800 × g for 2 min in a swinging bucket centrifuge, snap-frozen in liquid nitrogen, and stored at −80°C.
Life span assay.
One hundred animals of each strain were selected and transferred to ten 50-mm nematode growth medium (NGM) plates. Adult animals were transferred every 2 days to fresh plates for the duration of egg laying. Their survival was measured by movement of the head. Animals showing no head movement were gently touched twice with a worm pick and observed for 30 s following each touch. Animals that did not move were considered dead and removed from the plate. The survival curves were plotted and analyzed using OASIS (Online Application for the Survival Analysis) (41).
The experiment was reproduced three times with one representative example illustrated.
RT-qPCR.
Lysis of the worm pellets was performed with 5 µl of worm pellet and 45 µl of the lysis solution from the Power SYBR green Cells-to-CT kit (Ambion). Ten freeze-thaw cycles and 30 min shaking at room temperature were performed before the lysis incubation step. The reverse transcription-quantitative PCR (RT-qPCR) was performed according to the manufacturer’s instructions. The qPCR was run on a Step One Plus real-time PCR system (Applied Biosystems). The analysis was done using the ΔΔCT method. The threshold cycle (CT) values of four to six biological replicates per experiment were pooled to generate the average ΔCT for each strain and an associated standard error (SD). The ΔΔCT value was calculated with ΔCT in strain N2 as the calibrator. The 2−ΔΔCT was plotted as the fold change, and the interval of confidence is represented by error bars as 2−ΔΔCT + SD(ΔCT) and 2−ΔΔCT − SD(ΔCT). The significance of the differences observed was tested by a two-tailed Mann-Whitney U test at a significance level of P < 0.01. The experiments were repeated independently at least twice, and one representative example is shown in the figures.
DNA constructs.
The viral sensor construct and the gfp::sta-1 construct were generated by Gateway cloning using the MultiSite Gateway three-fragment vector construction kit (Life Technologies). Gateway entry clones containing each of the following were generated by standard techniques: sur-5 promoter, sdz-6 promoter, sta-1 coding sequence, eGFP(F64L/S65T) (eGFP stands for enhanced green fluorescent protein), and tbb-2 3′ untranslated region (3′UTR). The single-copy transgene was generated by transposase-mediated integration (MosSCI), as described previously (42, 43), at insertion site ttTi5605 on chromosome II. Injection mixes contained 20 ng/µl of vector, 20 ng/µl of Mos1 transposase (only for MosSCI), and 5 ng/μl of a pharynx marker. Integration of the extrachromosomal array was performed by ethyl methanesulfonate (EMS) treatment (50 mM EMS for 4 h).
Sensor scoring.
Scoring of the GFP was made by eye under a Leica MZ16F fluorescence stereomicroscope. Three biological replicates of the infection were analyzed, with at least 110 animals per replicate scored by eye, as follows: on for strong GFP signal, dim for weak/medium GFP signal, and off for no GFP.
Imaging.
Adult animals were harvested from a plate; they were washed off in M9 buffer. After three additional washes in M9 buffer, the pelleted animals were fixed in 0.5 ml of precooled methanol at −20°C for 10 min. The pellet was washed three times in Tris-buffered saline containing 0.1% Tween 20 (TBST) and then incubated for 10 min in a 4′,6′-diamidino-2-phenylindole (DAPI) solution (0.5 mg/ml in TBST). The pellet of animals was washed three times in TBST. Five microliters of the pelleted animals was pipetted directly onto a Cel-line diagnostic microscope slide (Thermo Scientific) and imaged using a Leica SP8 upright microscope.
Microarray.
To obtain a putative set of signaling response genes triggered by viral infection but not responding to the level of viral RNA, the array was first normalized to the expression of cul-6, as we and others have shown this to be closely linked to viral RNA levels (16, 44). Microarray data were processed using rma, and annotations were obtained through the Bioconductor pipeline in the R programming environment. Differentially regulated genes in any condition were identified by using t test, P < 0.01, and a twofold difference upon infection. Hierarchical clustering on the drh-1 array was carried out on the set of differentially regulated genes using the hclust function in R and Ward’s method.
Motif analysis.
To specifically identify potential viral response genes, we normalized the array by cul-6, as this removed variation in the final level of infection. Qualitatively, this identified trends similar to those previously published (16), such as putative antibacterial response genes upregulated specifically in strain JU1580 and drh-1 upon infection. We identified genes that were >40% induced relative to cul-6 in strain N2 upon infection and used BiomaRt to download the upstream sequences (Data Set S1). These sequences were used as input for MEME, attempting to find 0 or 1 site per gene. We screened these manually to avoid spurious hits and then compared potential motifs identified to the JASPAR core database. To test enrichment of STAT motifs in STA-1 chromatin immunoprecipitation-DNA sequencing (ChIP-seq) peaks, we used FIMO to search for the STAT-1 core motif within the 500-bp sequences upstream of genes containing a STA-1 binding site expressed at an increased level (DESeq q < 0.1) in sta-1 mutants relative to strain N2, applying a false-discovery rate cutoff of 0.2. We compared this to a random set of genes chosen from the transcriptome sequencing (RNA-seq) data (see below).
RNA-seq.
Animals were grown on 50-mm NGM plates, infected or not, and harvested as described in “Viral infection” above. Total RNA was extracted using TRIsure (Bioline, UK). RNA library preparation was performed with the NEBNext Ultra RNA Library Prep kit for Illumina with total RNA purified with the NEBNext poly(A) mRNA magnetic isolation module, according to the manufacturer’s instructions (NEB, USA).
RNA-seq analysis.
RNA-seq data were aligned to the C. elegans transcriptome WS190 using bowtie2. Counts were obtained from resulting BAM files using BEDTools (45), and these were used to generate normalized data tables using DESeq (46) (Data Set S2). Significance between intersecting data sets was calculated by a Fisher’s exact test (Fig. S4 and Fig. S5). A cutoff of mean 25 normalized reads (normalized according to DESeq’s negative binomial distribution) for at least one condition was used, and significantly altered genes were selected (DESeq Benjamini-Hochberg multiple-test correction q < 0.1).
sta-1, sid-3, and infection by the Orsay virus lead to the regulation of a shared set of genes. (A) Table showing the percentage of intersection for gene sets as depicted in Fig. 6. (B) Table showing the intersection for gene sets and STA-1 ChIP-seq peaks as depicted in Fig. 6. The data used in Fig. 3 are shaded in the tables. Download FIG S5, TIF file, 0.9 MB (975.6KB, tif) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ChIP sequencing.
Animals were grown on NGM plates seeded with thick E. coli HB101 food and harvested as a mixed-stage population. Frozen animals were ground to a fine powder, fixed in phosphate-buffered saline (PBS) containing 1% formaldehyde for 10 min, quenched with 0.125 µM glycine, and then washed three times in PBS with protease inhibitors. The pellet was resuspended in 1 ml of FA buffer (50 mM HEPES–KOH [pH 7.5], 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 150 mM NaCl with protease inhibitors) per 4 ml of ground worm powder. Extract was sonicated to a size range of 200 to 1,000 bp using a Diagenode Bioruptor Pico with a setting of 18 pulses, each pulse lasting 30 s followed by a 30-s pause. The extract was spun for 10 min at 16,000 × g at 4°C, and the soluble fraction was flash frozen in liquid nitrogen and stored at −80°C until use. Each ChIP was prepared in 500 µl of FA buffer containing 1% Sarkosyl. Fifteen micrograms of an anti-GFP antibody (ab290) was incubated with 3 mg of extract. In addition, 10% of extract was saved as a reference. After overnight rotation at 4°C, 40 µl of blocked and washed magnetic protein A Dynabeads (Invitrogen) was added, and the incubation continued for 2 additional hours. The beads were washed at room temperature twice for 5 min in FA buffer, once in FA buffer with 500 mM NaCl for 10 min, once in FA buffer with 1 M NaCl for 5 min, once in TEL buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.0]) for 10 min, and twice in Tris-EDTA (TE) (pH 8.0) for 5 min. DNA was eluted twice with 57-μl elution buffer (1% SDS in TE with 250 mM NaCl) at 65°C for 15 min each time. Eluted DNA was incubated with 20 μg of RNase for 30 min at 37°C and then with 20 μg of proteinase K for 1 h at 55°C. Input DNA was also diluted in 114-µl elution buffer and treated with ChIP samples. Cross-links were reversed overnight at 65°C. DNA was purified on PureLink PCR purification columns (Invitrogen). The libraries were prepared using a modified TruSeq ChIP Library preparation kit protocol (https://ethanomics.files.wordpress.com/2012/09/chip_truseq.pdf). Size selection was performed using Agencourt AMPure XP beads (Beckman Coulter).
ChIP-seq analysis. (i) Alignment to reference genome.
ChIP-seq and RNA-seq libraries were sequenced using Illumina HiSeq. Reads were aligned to the WS220/ce10 assembly of the C. elegans genome using BWA v. 0.7.7 (47) with default settings (BWA-backtrack algorithm). The SAMtools v. 0.1.19 “view” utility was used to convert the alignments to BAM format. Normalized ChIP-seq coverage tracks were generated using the R implementation of BEADS algorithm (48, 49).
(ii) Summed ChIP-seq input.
We generated summed input BAM files by combining good-quality ChIP-seq input experiments from different extracts (eight experiments). The same summed inputs were used for BEADS normalization and peak calls.
(iii) Peak calls.
Initial ChIP-seq peaks were called using MACS v. 2.1.1 (50) with permissive 0.7 q-value cutoff and fragment size of 150 bp against summed ChIP-seq input. To generate combined peak calls, we used the modified irreproducible discovery rate (IDR) procedure (https://www.encodeproject.org/software/idr/) with an IDR threshold of 0.05 to combine replicates (Data Set S3). The pipeline for generating IDR peaks is available here: https://github.com/Przemol/biokludge/blob/master/macs2_idr/macs2_idr.ipy.
(iv) Mean signal distribution plots and heatmaps.
The summarized signal profile and heatmap for GFP::STA-1 were generated using SeqPlots exploratory analyses and plotting software (51).
RNAi screen.
RNAi clones from the Ahringer library (34, 35) were isolated on agar plates containing carbenicillin (50 μg/ml). Single colonies were picked in 2 ml LB plus ampicillin (50 μg/ml) and grown for 9 h with shaking at 37°C. Bacteria were then seeded onto 50-mm NGM plates containing isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM), carbenicillin (25 μg/ml), and amphotericin B (Fungizone) (2.5 μg/ml). Two days after seeding, two larval stage 4 (L4) animals were added and then grown at 20°C. After 16 h, plates were inoculated with 20 μl Orsay virus filtrate. Animals were collected after 3 days, and viral relative genome copy number was measured by reverse transcription-quantitative PCR (RT-qPCR). For each round of the screen, we used the following internal controls: N2 strain grown on empty vector (E. coli L4440 strain) as the normalization control, drh-1 mutant fed on GFP RNAi as a positive control for infection, and N2 strain fed on drh-1 RNAi clone as a positive control for RNA interference (RNAi). The full list of the RNAi clones and number of replicates used in this study is available in Data Set S4. Each replicate of the screen was performed with three biological replicates per RNAi clone treatment, and the screen was repeated at least twice. The plates with accidental fungal contamination and the plates where the RNAi treatment led to embryonic lethality were removed from the analysis. The resulting exact numbers of biological replicates are indicated in Data Set S4. Empty vector treatment was included on each qPCR plate analyzed and used as the calibrator. The ΔΔCT values were transformed into Z-scores, calculated as follows: Z-scorei = (ΔΔCTi − µΔΔCTempty vector, n = 161)/standard deviation(ΔΔCTempty vector), where µ indicates mean and i is the gene. Boxplots of the Z-score for each treatment are represented in Fig. 5. Individual values used for the analysis are available in Data Set S4.
Data availability.
High-throughput sequencing data sets are accessible through the GEO repository. All Illumina sequencing files are available from the GEO database (GSE95230 and GSE99586).
ACKNOWLEDGMENTS
We thank David Jordan for scientific discussions and help with data visualization, Jérémie Le Pen for scientific discussions and providing reagents, Alyson Ashe for providing reagents, Alex Appert for technical support with ChIP sequencing experiment, Kay Harnish and Sylviane Moss for high-throughput sequencing. We thank the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440) for providing some strains used in this study.
This work was supported by Cancer Research UK grant C13474/A18583, Wellcome Trust grant 104640/Z/14/Z, and European Research Council (ERC) grant 260688 to E. A. Miska, M. Tanguy, and L. Véron, by Wellcome Trust grant 101863 to J. Ahringer and P. Stempor, and Medical Research Council grant MC-A652-5PZ80 to Peter Sarkies.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/mBio.00940-17.
Citation Tanguy M, Véron L, Stempor P, Ahringer J, Sarkies P, Miska EA. 2017. An alternative STAT signaling pathway acts in viral immunity in Caenorhabditis elegans. mBio 8:e00924-17. https://doi.org/10.1128/mBio.00924-17.
REFERENCES
- 1.Ding S-W, Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Platanias LC. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 3.Kell AM, Gale M Jr.. 2015. RIG-I in RNA virus recognition. Virology 479-480:110–121. doi: 10.1016/j.virol.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 5.Ivashkiv LB, Donlin LT. 2014. Regulation of type I interferon responses. Nat Rev Immunol 14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Girardi E, Lefèvre M, Chane-Woon-Ming B, Paro S, Claydon B, Imler J-L, Meignin C, Pfeffer S. 2015. Cross-species comparative analysis of Dicer proteins during Sindbis virus infection. Sci Rep 5:10693. doi: 10.1038/srep10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seo GJ, Kincaid RP, Phanaksri T, Burke JM, Pare JM, Cox JE, Hsiang TY, Krug RM, Sullivan CS. 2013. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 14:435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P. 2013. A retrotransposon-driven Dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 155:807–816. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. 2013. Antiviral RNA interference in mammalian cells. Science 342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benitez AA, Spanko LA, Bouhaddou M, Sachs D, tenOever BR. 2015. Engineered mammalian RNAi can elicit antiviral protection that negates the requirement for the interferon response. Cell Rep 13:1456–1466. doi: 10.1016/j.celrep.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Lu J, Han Y, Fan X, Ding S-W. 2013. RNA interference functions as an antiviral immunity mechanism in mammals. Science 342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Félix M-A, Ashe A, Piffaretti J, Wu G, Nuez I, Bélicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, Sanroman M, Miska EA, Wang D. 2011. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol 9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashe A, Bélicard T, Le Pen J, Sarkies P, Frézal L, Lehrbach NJ, Félix M-A, Miska EA. 2013. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. Elife 2:e00994. doi: 10.7554/eLife.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R, Yigit E, Li W-X, Ding S-W. 2009. An RIG-I-like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog 5:e1000286. doi: 10.1371/journal.ppat.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coffman SR, Lu J, Guo X, Zhong J, Jiang H, Broitman-Maduro G, Li W-X, Lu R, Maduro M, Ding S-W. 2017. Caenorhabditis elegans RIG-I homolog mediates antiviral RNA interference downstream of Dicer-dependent biogenesis of viral small interfering RNAs. mBio 8:e00264-17. doi: 10.1128/mBio.00264-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarkies P, Ashe A, Le Pen J, McKie MA, Miska EA. 2013. Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome Res 23:1258–1270. doi: 10.1101/gr.153296.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 18.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. 2007. Quantifying similarity between motifs. Genome Biol 8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathelier A, Fornes O, Arenillas DJ, Chen C-Y, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, Zhang AW, Parcy F, Lenhard B, Sandelin A, Wasserman WW. 2016. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44:D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Levy DE. 2006. C. elegans STAT: evolution of a regulatory switch. FASEB J 20:1641–1652. doi: 10.1096/fj.06-6051com. [DOI] [PubMed] [Google Scholar]
- 21.Dierking K, Polanowska J, Omi S, Engelmann I, Gut M, Lembo F, Ewbank JJ, Pujol N. 2011. Unusual regulation of a STAT protein by an SLC6 family transporter in C. elegans epidermal innate immunity. Cell Host Microbe 9:425–435. doi: 10.1016/j.chom.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Levy DE. 2006. C. elegans STAT cooperates with DAF-7/TGF-β signaling to repress Dauer formation. Curr Biol 16:89–94. doi: 10.1016/j.cub.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 23.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123–132. doi: 10.1016/S0092-8674(00)81644-X. [DOI] [PubMed] [Google Scholar]
- 24.Darnell JE Jr, Kerr IM, Stark GR. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Paradis P, Aries A, Komati H, Lefebvre C, Wang H, Nemer M. 2005. Convergence of protein kinase C and JAK-STAT signaling on transcription factor GATA-4. Mol Cell Biol 25:9829–9844. doi: 10.1128/MCB.25.22.9829-9844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Libert S, Chao Y, Chu X, Pletcher SD. 2006. Trade-offs between longevity and pathogen resistance in Drosophila melanogaster are mediated by NFkappaB signaling. Aging Cell 5:533–543. doi: 10.1111/j.1474-9726.2006.00251.x. [DOI] [PubMed] [Google Scholar]
- 27.Jacot A, Scheuber H, Brinkhof MWG. 2004. Costs of an induced immune response on sexual display and longevity in field crickets. Evolution 58:2280–2286. doi: 10.1111/j.0014-3820.2004.tb01603.x. [DOI] [PubMed] [Google Scholar]
- 28.Armitage SAO, Thompson JJW, Rolff J, Siva-Jothy MT. 2003. Examining costs of induced and constitutive immune investment in Tenebrio molitor. J Evol Biol 16:1038–1044. doi: 10.1046/j.1420-9101.2003.00551.x. [DOI] [PubMed] [Google Scholar]
- 29.Villarino AV, Kanno Y, Ferdinand JR, O’Shea JJ. 2015. Mechanisms of Jak/STAT signaling in immunity and disease. J Immunol 194:21–27. doi: 10.4049/jimmunol.1401867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovarik P, Stoiber D, Eyers PA, Menghini R, Neininger A, Gaestel M, Cohen P, Decker T. 1999. Stress-induced phosphorylation of STAT1 at Ser727 requires p38 mitogen-activated protein kinase whereas IFN-gamma uses a different signaling pathway. Proc Natl Acad Sci U S A 96:13956–13961. doi: 10.1073/pnas.96.24.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan M-W, Ausubel FM. 2002. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 32.Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, Jin Y, Chisholm AD, Ewbank JJ. 2008. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol 18:481–489. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang H-Y, Dosztányi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Xenarios I, Yeh L-S, Young S-Y, Mitchell AL. 2017. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res 45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 35.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 36.Jose AM, Kim YA, Leal-Ekman S, Hunter CP. 2012. Conserved tyrosine kinase promotes the import of silencing RNA into Caenorhabditis elegans cells. Proc Natl Acad Sci U S A 109:14520–14525. doi: 10.1073/pnas.1201153109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashe A, Sarkies P, Le Pen J, Tanguy M, Miska EA. 2015. Antiviral RNA interference against Orsay virus is neither systemic nor transgenerational in Caenorhabditis elegans. J Virol 89:12035–12046. doi: 10.1128/JVI.03664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao L, Aizaki H, He J-W, Lai MMC. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol 78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim JY, Wang L, Lee J, Ou J-HJ. 26 July 2017 HCV induces the localization of lipid rafts to autophagosomes for its RNA replication. J Virol doi: 10.1128/JVI.00541-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang H, Chen D, Sandoval LE, Leung C, Wang D. 2017. An evolutionarily conserved pathway essential for Orsay virus infection of Caenorhabditis elegans. mBio 8:e00940-17. doi: 10.1128/mBio.00940-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J-S, Nam H-J, Seo M, Han SK, Choi Y, Nam HG, Lee S-J, Kim S. 2011. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One 6:e23525. doi: 10.1371/journal.pone.0023525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frøkjær-Jensen C, Davis MW, Ailion M, Jorgensen EM. 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods 9:117–118. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bakowski MA, Desjardins CA, Smelkinson MG, Dunbar TL, Dunbar TA, Lopez-Moyado IF, Rifkin SA, Cuomo CA, Troemel ER. 2014. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog 10:e1004200. doi: 10.1371/journal.ppat.1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheung M-S, Down TA, Latorre I, Ahringer J. 2011. Systematic bias in high-throughput sequencing data and its correction by BEADS. Nucleic Acids Res 39:e103. doi: 10.1093/nar/gkr425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stempor P. 2014. rBEADS - the R implementation of bias elimination algorithm for deep sequencing. Zenodo. [Google Scholar]
- 50.Feng J, Liu T, Zhang Y. 2011. Using MACS to identify peaks from ChIP-Seq data. Curr Protoc Bioinformatics Chapter 2:Unit 2.14. doi: 10.1002/0471250953.bi0214s34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stempor P, Ahringer J. 2016. SeqPlots - interactive software for exploratory data analyses, pattern discovery and visualization in genomics. Wellcome Open Res 1:14. doi: 10.12688/wellcomeopenres.10004.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen RA-J, Down TA, Stempor P, Chen QB, Egelhofer TA, Hillier LW, Jeffers TE, Ahringer J. 2013. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Res 23:1339–1347. doi: 10.1101/gr.153668.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. 2013. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife 2:e00808. doi: 10.7554/eLife.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bailey TL, Machanick P. 2012. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res 40:e128. doi: 10.1093/nar/gks433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Orsay virus (OrV) infection induces gene expression changes that depend on the genotype of the host. The heatmap shows changes in gene expression, as measured by Affymetrix array, occurring in each of the indicated strains after infection by the Orsay virus. Download FIG S1, TIF file, 1 MB (1MB, tif) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Upstream sequences of genes upregulated upon infection Download DATA SET S1, TXT file, 0.03 MB (31.6KB, txt) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
sta-1 reduces activation of an infection sensor. Activation of the sdz-6 viral sensor. The promoter of the sdz-6 gene drives the expression of the GFP in the infected intestine. Animals were scored for GFP signal after 3 days of infection in three biological replicates with an average of 125 animals scored for each condition. A t test was performed to compare the average fraction of positive animals (on and dim) after infection in wild-type (WT) and rde-1 animals and in WT and sta-1 animals. Download FIG S2, TIF file, 0.8 MB (849.1KB, tif) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential gene expression measured by RNA-seq. Filtered data set for genes showing differential expression (P < 0.1) in at least one sample. Download DATA SET S2, XLSX file, 0.2 MB (184.4KB, xlsx) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Discovery of a GATA motif enriched in the STA-1-bound genes. (A) Enrichment of the IRF motif (as depicted in Fig. 3) across 1,375 peaks (out of 3,211 total peaks). (B) Enrichment of a GATA motif across 503 peaks (out of a total of 3,211 peaks) and logo representation. Download FIG S3, TIF file, 0.9 MB (892.2KB, tif) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Association between ChIP peak calling and gene names Download DATA SET S3, XLSX file, 0.5 MB (504.1KB, xlsx) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
STA-1 is bound to genes upregulated in sta-1 mutants and infection genes. The table shows the intersection for gene sets and STA-1 ChIP-seq peaks as depicted in Fig. 3G. Download FIG S4, TIF file, 0.6 MB (680.8KB, tif) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNAi screen data Download DATA SET S4, XLSX file, 0.1 MB (101.7KB, xlsx) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains used in this study Download TABLE S1, PDF file, 0.03 MB (33.2KB, pdf) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
sta-1, sid-3, and infection by the Orsay virus lead to the regulation of a shared set of genes. (A) Table showing the percentage of intersection for gene sets as depicted in Fig. 6. (B) Table showing the intersection for gene sets and STA-1 ChIP-seq peaks as depicted in Fig. 6. The data used in Fig. 3 are shaded in the tables. Download FIG S5, TIF file, 0.9 MB (975.6KB, tif) .
Copyright © 2017 Tanguy et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
High-throughput sequencing data sets are accessible through the GEO repository. All Illumina sequencing files are available from the GEO database (GSE95230 and GSE99586).