Abstract
Temozolomide (TMZ) is the most commonly used alkylating agent in glioma chemotherapy. However, growing resistance to TMZ remains a major challenge for clinicians. Recent evidence emphasizes the key regulatory roles of non-coding RNAs (lncRNAs and miRNAs) in tumor biology, including the chemoresistance of cancers. However, little is known about the role and regulation mechanisms of lncRNA cancer X-inactive specific transcripts (XIST) in glioma tumorigenesis and chemotherapy resistance. In the present study, higher XIST expression was observed in glioma tissues and cell lines, which was related to poorer clinicopathologic features and shorter survival time. XIST knockdown alone was sufficient to inhibit glioma cell proliferation and to amplify TMZ-induced cell proliferation inhibition. Moreover, XIST knockdown can sensitize TMZ-resistant glioma cells to TMZ. XIST can inhibit miR-29c expression by directly targetting TMZ-resistant glioma cells. DNA repair protein O6-methylguanine-DNA methytransferase (MGMT) plays a key role in TMZ resistance; transcription factor specificity protein 1 (SP1), a regulator of DNA mismatch repair (MMR) key protein MSH6, has been reported to be up-regulated in TMZ-resistant glioma cell lines. In the present study, we show that XIST/miR-29c coregulates SP1 and MGMT expression in TMZ-resistant glioma cell lines. Our data suggest that XIST can amplify the chemoresistance of glioma cell lines to TMZ through directly targetting miR-29c via SP1 and MGMT. XIST/miR-29c may be a potential therapeutic target for glioma treatment.
Keywords: chemoresistance, glioma, lncRNA-XIST, miR-29c, Temozolomide (TMZ)
Introduction
As the most common brain cancer, glioma accounts for >60% of primary brain tumors in adults [1,2]. Surgery followed by radiotherapy, with temozolomide (TMZ) adjuvant chemotherapy, is the standard treatment for glioma; however, the prognosis remains poor due to the growing resistance to TMZ [1,3]. The pathogenesis of glioma is largely unknown, although it has been shown that genetic changes in many genes are associated with the development of glioma [4–6]. Therefore, investigating the pathogenesis of glioma and the mechanism of chemoresistance acquisition are critically important for the diagnosis and treatment of this fatal disease.
In recent years, emerging evidence has regarded non-coding RNAs, including lncRNAs and miRNAs as major regulators of normal development and diseases, including cancer [7–9]. Under different circumstances, lncRNAs and miRNAs can play a role in tumorigenesis, tumor inhibition, or both [10–12]. The lncRNA X inactive specific transcript (XIST) has been found to be up-regulated and acts as a carcinogen in glioblastoma [13], ovarian cancer [14], and non-small-cell lung cancer [15]. In addition, its role in cancer resistance to chemotherapy has also been reported [16,17]; however, little is known about its expression pattern, biological function and potential mechanism of glioma progression, and the chemoresistance of glioma to TMZ. In addition to XIST, the role of miR-29c in cancers has been extensively studied. Through inhibiting cancer cell proliferation, invasion, and/or migration, miR-29c acts as a tumor suppressor in gastric cancer [18], pancreatic cancer [19], colorectal cancer [20], and so on. More importantly, miR-29c has been reported to regulate the radioresistance of cancer cells in lung cancer [21].
It has been recently discovered that the interactions between lncRNAs and miRNAs affect post-transcriptional regulation by inhibiting the available miRNA activity. According to previous studies, lncRNA can act as a specific ‘sponge’ for miRNA to reduce their regulation of mRNA [22]. Whether XIST can interact with miR-29c to affect glioma cell proliferation and its chemoresistance to TMZ remain to be uncovered.
In the present study, the expression levels of XIST in glioma tissues and the peritumoral brain edema (PTBE) tissues, the relationship between XIST expression and the clinical features in patients with glioma, and the effects of XIST on glioma cell proliferation and chemoresistance to TMZ were evaluated. Further, we revealed that the interaction between XIST and miR-29c regulates the chemosensitivity to TMZ-based chemotherapy through specificity protein 1 (SP1) and O6-methylguanine-DNA methytransferase (MGMT). Our findings provide a novel understanding of the function of XIST/miR-29c/SP1/MGMT in the sensitivity of glioma to chemotherapy and the mechanism involved.
Materials and methods
Cell lines, tissues, and transfection
With the approval of the Ethical Committee of Xiangya Hospital, the Central South University (CSU), we collected 69 paired glioma tissues as well as the PTBE tissues. All the samples were obtained from patients who underwent surgical resection at Xiangya Hospital, CSU (Changsha, China). All the tissue samples were snap-frozen and stored at –80°C in liquid nitrogen. The clinical features of patients are listed in Table 1. Univariate and multivariate analyses of factors related to oversurvival using the COX proportional hazard model is listed in Table 2.
Table 1. Correlation of the expression of XIST with clinicopathologic features.
| Characteristics | n | Relative XIST expression | P-value | |
|---|---|---|---|---|
| High | Low | |||
| Age | 0.921 | |||
| <45 years | 28 | 14 | 14 | |
| ≥45 years | 41 | 21 | 20 | |
| Gender | 0.537 | |||
| Female | 31 | 17 | 14 | |
| Male | 38 | 18 | 20 | |
| Tumor size | 0.003 | |||
| <5 cm | 32 | 10 | 22 | |
| ≥5 cm | 37 | 25 | 12 | |
| PTBE | 0.116 | |||
| ≥1 cm | 36 | 15 | 21 | |
| <1 cm | 33 | 20 | 13 | |
| WHO stage | <0.001 | |||
| I + II | 33 | 6 | 27 | |
| III + IV | 36 | 29 | 7 | |
Table 2. Univariate and multivariate analyses for factors related to oversurvival using the COX proportional hazard model.
| Characteristics | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | ||
| Age | <45 compared with ≥45 | 0.747 (0.422–1.324) | 0.318 | N.A. | |
| Gender | female compared with male | 0.908 (0.521–1.583) | 0.735 | N.A. | |
| Tumor size | ≥5 cm compared with <5 cm | 0.825 (0.472–1.444) | 0.502 | N.A. | |
| PTBE | ≥1 cm compared with <1 cm | 1.030 (0.591–1.795) | 0.917 | N.A. | |
| WHO stage | I + II compared with III + IV | 0.428 (0.242–0.756) | 0.004 | 0.571 (0.306–1.067) | 0.079 |
| XIST expression | high compared with low | 2.560 (1.438–4.557) | 0.001 | 2.037 (1.083–3.831) | 0.027 |
N.A., not applicable.
Human glioma cell lines: U251, U373, LN229, U118, and LN229, as well as a normal cell line, normal human astrocytes (NHA) were obtained from the American Type Culture Collection (ATCC, U.S.A.), cultured in 10% FBS (Gibco, U.S.A.) supplemented RPMI-1640 medium (Invitrogen, U.S.A.) at 37 °C with 5% v/v CO2.
LN229 and U251 glioma cells, normally sensitive to TMZ, were cultured in incremental concentrations of TMZ up to 400 µM over several weeks in our laboratory with stepwise selection and the subculture of resistant clones as described by Zhang et al. [23].
MiR-29c mimic or miR-29c inhibitor (GenePharma, China) was transfected into the indicated target cells to achieve miR-29c overexpression or miR-29c inhibition by using Lipofectamine 2000 (Invitrogen). SiRNA-XIST was used to achieve knockdown of XIST (GeneCopoeia, China).
Real-time PCR
TRIzol reagent (Invitrogen) was used for total RNA extraction following the manufacturer’s instructions. By using miRNA-specific primer, total RNA was reverse transcribed and the miScript Reverse Transcription Kit (Qiagen, Germany) was used for miR-29c qRT-PCR. The SYBR Green PCR Master Mix (Qiagen) was used following the manufacturer’s instructions. The Ct method was used to evaluate the relative expression and normalized to U6 expression. The First-strand cDNA Synthesis Kit (Promega, U.S.A.) was used to perform the reverse transcription to determine XIST expression following the manufacturer’s instructions. The expression of GAPDH mRNA was regarded as an internal control.
Western blotting
RIPA buffer (Cell Signaling Technology, U.S.A.) was used to homogenize the cells. The expression of SP1 and MGMT in glioma cells was detected by performing immunoblotting. Cells were lysed, cultured, or transfected in 1% PMSF supplemented RIPA buffer. Protein was loaded on to SDS/PAGE minigel, and then transferred on to PVDF membrane. The blots were probed with the following antibodies: anti-SP1 (Cat# EPR6662 (B), Abcam, U.S.A.), anti-MGMT (Cat# EPR4397, Abcam, U.S.A.), and anti-GAPDH (Cat# 6C5, Abcam, U.S.A.) at 4°C overnight, and incubated with HRP–conjugated secondary antibody (1:5000). Signals were visualized using ECL Substrates (Millipore, U.S.A.). The protein expression was normalized to endogenous GAPDH.
Luciferase activity
LN229 cells were cultured overnight after being seeded into a 24-well plate, cotransfected with the wt-XIST or mut-XIST reporter gene plasmid containing a 5-bp mutation in the predicted binding site of miR-29c and miR-29c mimics or miR-29c inhibitor. Forty-eight hours after transfection, Dual Luciferase Reporter Assay System (Promega, U.S.A.) was used to perform the luciferase assays.
RNA immunoprecipitation
LN229/TMZ and U251/TMZ cell lysates were used for RNA immunoprecipitation (RIP). The Imprint RNA Immunoprecipitation Kit (Sigma, U.S.A.) was used in RIP with the AGO2 antibody (ab32381, Abcam, U.S.A.), which is a key component of the miRNA-containing RNA-induced silencing complex (RISC). AGO2 was used as positive controls and IgG as the negative controls. The levels of XIST and miR-29c in the precipitates were determined using real-time PCR.
MTT assay
Twenty four hours after seeding into 96-well plates (5000 cells per well), cells were transfected with siRNA-XIST. Twenty four hours post-transfection, cells were exposed to TMZ (0, 7.5, 15, 30, 60, 120, 240, and 480 μM) for another 24 h. Then, 20 μl MTT (at a concentration of 5 mg/ml; Sigma–Aldrich) was added, and the cells were incubated for an additional 4 h in a humidified incubator. DMSO (200 μl) was added after the supernatant discarded to dissolve the formazan. OD490 nm value was measured. The viability of the untreated cells (control) was defined as 100%, and the viability of cells from all other groups was calculated separately from that of the control group.
BrdU incorporation assay
By measuring 5-Bromo-2-deoxyuridine (BrdU) incorporation, the DNA synthesis in proliferating cells was determined. BrdU assays were conducted at 24 and 48 h after glioma cells were transfected with siRNA-XIST. Cells were seeded in 96-well culture plates at a density of 2 × 103 cells/well, cultured for 24 or 48 h, then incubated with a final concentration of 10 μM BrdU (BD Pharmingen, San Diego, CA, U.S.A.) for 2 h. When the incubation period ended, the medium was removed, the cells were fixed for 30 min at RT, incubated with peroxidase-coupled anti-BrdU antibody (Sigma–Aldrich) for 60 min at RT, washed three times with PBS, incubated with peroxidase substrate (tetramethylbenzidine) for 30 min, and the 450-nm absorbance values were measured for each well. Background BrdU immunofluorescence was determined in cells not exposed to BrdU but stained with the BrdU antibody.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Statistics analysis
Data from three independent experiments were presented as mean ± S.D., processed using SPSS 17.0 statistical software (SPSS, U.S.A.). Paired Student’s t test was used to compare the expression of miR-29c and XIST in glioma tissues and normal tissues. P-values of <0.05 were considered statistically significant.
Results
Expression of XIST in glioma tissues and its relationship with the clinical features in patients with glioma
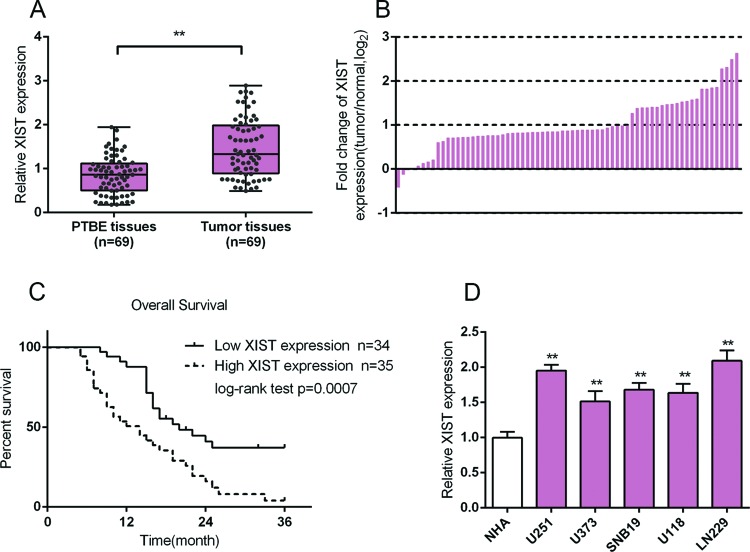
In order to investigate the function of XIST in glioma, we first evaluated the expression of XIST in 69 paired glioma tissues and PTBE tissues using real-time PCR assays. The results showed that XIST expression was up-regulated in glioma tissues, compared with that of the PTBE tissues (Figure 1A). In 33.33% (23/69) glioma tissues, XIST had a fold-change of more than 2 compared with the PTBE tissues (Figure 1B). We then divided these samples into two groups according to XIST expression: a high XIST expression group (above the median XIST expression, n=35) and a low XIST expression group (below the median XIST expression, n=34) (Table 1). As shown in Table 1, lower XIST expression was observed more frequently in patients with advanced WHO stage (III + IV) (P<0.001) and larger tumor size (≥5, P=0.003). The survival and pathologic features of 69 patients were analyzed using the COX risk proportional regression model. Univariate analysis showed that WHO stage and XIST expression caused significant differences in survival time; multivariate analysis showed that high XIST expression was of high risk (HR =2.037, 95% CI =1.083–3.831) (Table 2). The survival time of patients with glioma with high expression of XIST was shorter than that in patients with low expression of XIST (P=0.0007, Figure 1C). We also monitored the expression levels of XIST in all five glioma cell lines, and the results showed that XIST expression was up-regulated in all the five glioma cell lines, U251, U373, LN229, U118, and LN229. The expression of XIST was higher in U251 and LN229 cells than in the other three cell lines (Figure 1D); therefore, U251 and LN229 cell lines were selected as further cell models.
Figure 1. Expression of XIST in glioma tissues and its relationship with the clinical features in patients with glioma.
(A,B) Expression of XIST in 69 paired glioma tissues and PTBE tissues were determined using real-time PCR assays. Fold changes of XIST expression was exhibited as log2 (tumor/normal). (C) Kaplan–Meier overall survival curves for 69 patients with glioma classified according to relative XIST expression level. (D) The expression levels of XIST in five glioma cell lines, U251, U373, LN229, U118, LV229, and a normal cell line, NHA, were determined using real-time PCR. The data are presented as mean ± S.D. of three independent experiments; **P<0.01.
Effects of XIST on glioma cell proliferation and chemoresistance to TMZ
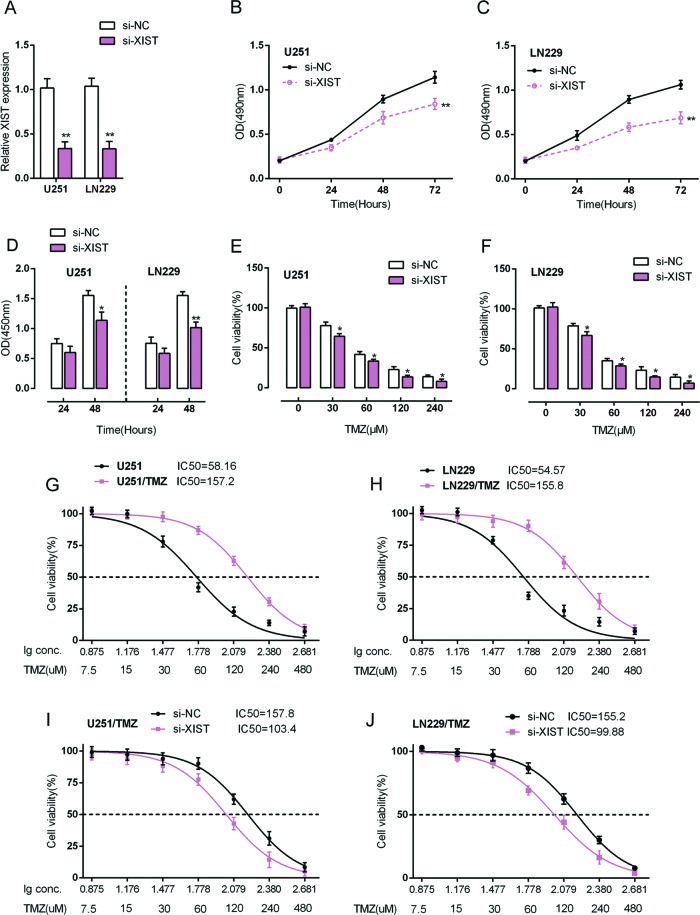
U251 and LN229 cells were transfected with si-XIST to achieve XIST knockdown, as verified using real-time PCR assays (Figure 2A). Cell viability of si-XIST-transfected U251 and LN229 cells was determined using MTT assays. The results showed that XIST knockdown significantly suppressed the cell viability in U251 and LN229 cells (Figure 2B,C). Further, the DNA synthesis capability was also evaluated using BrdU assays. Similar to MTT assays, the DNA synthesis capability was significantly suppressed by XIST knockdown (Figure 2D). These data further demonstrate the role of XIST in promoting glioma cell proliferation. We then investigated whether XIST affects the chemoresistance of glioma cells to TMZ.
Figure 2. Effects of XIST on glioma cell proliferation and chemoresistance to TMZ.
(A) siRNA-NC/siRNA-XIST was transfected into LN229 and U251 cells. The expression of XIST was verified using real-time PCR assays. (B,C) After transfection with the indicated vectors, the cell viability of LN229 and U251 cells was determined using MTT assays. (D) After transfection with the indicated vectors, the cell proliferation of LN229 and U251 cells was determined using BrdU assays. (E,F) LN229 and U251 cells were transfected with the indicated vectors. Twenty four hours post-transfection, cells were treated with a series dose of TMZ (0, 30, 60, 120, 240 μM) for 24 h. The cell viability was determined using MTT assays, and the data were presented as a percentage normalized to the viability of cells with 0 μM TMZ treatment. The data are presented as mean ± S.D. of three independent experiments; *P<0.05, **P<0.01. (G,H) LN229, TMZ-resistant LN229 (LN229/TMZ), U251, TMZ-resistant U251 (U251/TMZ) cells were treated with a series dose of TMZ (7.5, 15, 30, 60, 120, 240, and 480 μM) for 24 h, and the cell viability of the indicated cells was determined using MTT assays. Data were displayed as a percentage normalized to the viability of cells with no TMZ treatment. The abscissa was the logarithm of TMZ concentration (log-conc.). LC50 represented the concentration of TMZ when cell viability was reduced to 50%. (I,J) LN229/TMZ and U251/TMZ cells were transfected with siRNA-NC/siRNA-XIST. Twenty four hours after transfection, cells were treated with a series dose of TMZ (7.5, 15, 30, 60, 120, 240, and 480 μM) for another 24 h; the cell viability was then determined using MTT assays.
Si-XIST-transfected U251 and LN229 cells were treated with a series of concentrations of TMZ (0, 30, 60, 120, 240 μM); and then cell viability was evaluated using MTT assays. The results showed that glioma cell viability was repressed by TMZ treatment in a dose-dependent manner; TMZ-induced repression of glioma cell viability could be amplified by XIST knockdown (Figure 2E,F). These data suggest that XIST is involved in regulation of glioma cell proliferation either in the presence or absence of TMZ; in addition, XIST may affect the chemoresistance of glioma cells to TMZ.
We validated that XIST knockdown can enhance the repressive effect of TMZ on glioma cell proliferation; we then examined the effect of XIST on regulating the chemosensitivity of glioma cells to TMZ. LN229, LN229/TMZ, U251, and U251/TMZ cells were treated with a series of doses of TMZ (7.5, 15, 30, 60, 120, 240, and 480 μM) for 24 h and then monitored for cell viability. The cell viability of untreated cells was defined as 100%. The results showed that for U251 cells, the TMZ concentration to reduce cell viability to 50% was approximately 58.16 μM (lC50 =58.16); for U251/TMZ cells this value was 157.2 μM (lC50 =157.2) (Figure 2G). Similar results were observed for LN229 cells, the TMZ concentration to reduce LN229 cell viability to 50% was approximately 54.57 μM (lC50 =54.57), for LN229/TMZ cells 155.8 μM (lC50 =155.8) (Figure 2H). We transfected LN229/TMZ and U251/TMZ cells with siRNA-NC/siRNA-XIST, and then repeated the above assays to validate the effect of XIST on glioma cells’ chemosensitivity. Results showed that XIST knockdown amplified TMZ-induced repression of glioma cells viability and reduced the lC50 values to 103.4 (LN229/TMZ) and 99.88 (U251/TMZ) (Figure 2I,J). These data suggested that XIST may exacerbate the chemoresistance of glioma cells to TMZ-based chemotherapy. However, the mechanism by which XIST regulates the chemoresistance of glioma cells to TMZ remains to be investigated.
XIST regulated miR-29c by directly targetting in TMZ-resistant glioma cells
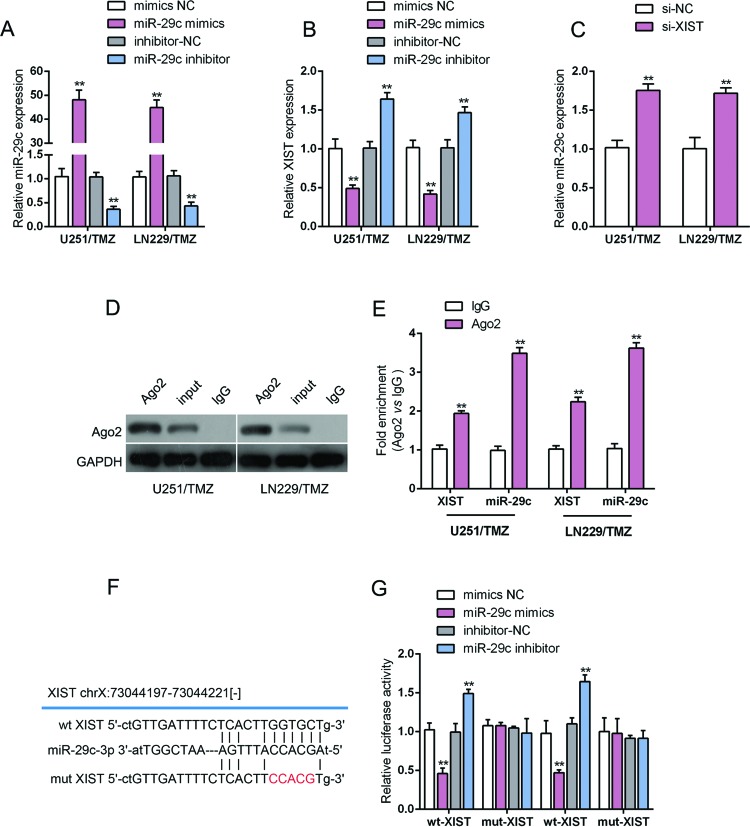
In view of the key role of miR-29c in cancers, in particular the chemosensitivity of cancer cells [16,17]; we further investigated whether XIST regulates the chemoresistance of glioma cell to TMZ through miR-29c. Mimics NC/miR-29c mimics or inhibitor NC/miR-29c inhibitor was transfected into LN229/TMZ and U251/TMZ cells to achieve miR-29c overexpression or inhibition, and the transfection efficiency was verified using real-time PCR (Figure 3A). The expression of XIST in these cells was monitored using real-time PCR. The results showed that XIST expression was up-regulated by miR-29c inhibition while down-regulated by ectopic miR-29c (Figure 3B). Then the expression of miR-29c in response to XIST knockdown was monitored in LN229/TMZ and U251/TMZ cells. The results showed that miR-29c expression was also negatively regulated by XIST (Figure 3C). To investigate the mechanism by which XIST and miR-29c regulates each other, we further performed RIP assay to verify the direct binding of miR-29c and XIST. RIP results showed that miR-29c and XIST were associated with the AGO2 in LN229/TMZ and U25/TMZ cells (Figure 3D). In RNA extracted from precipitated AGO2 protein, miR-29c levels were 3.5 times higher than IgG, and XIST levels were more than two times higher than IgG (Figure 3E). To confirm the interaction between XIST and miR-29c, a wt-XIST luciferase reporter gene vector, as well as a mut-XIST luciferase reporter gene vector containing a 5-bp mutation at the putative binding site of miR-29c was constructed (Figure 3F). The indicated vectors were cotransfected into TMZ-resistant glioma cell line U251/TMZ with mimics NC/miR-29c mimics or inhibitor NC/miR-29c inhibitor, and the luciferase activity was then monitored using dual luciferase assays. Results showed that the luciferase activity of the wt-XIST luciferase reporter vector was notably suppressed in response to miR-29c mimics transfection while amplified in response to miR-29c inhibitor transfection, compared with control groups (Figure 3G). Additionally, the effect of miR-29c mimics or inhibitor on luciferase activity was offset by mutations in XIST (Figure 3G). These data suggest that XIST directly binds to miR-29c to inhibit its expression, thereby affecting the chemoresistance of glioma cells to TMZ.
Figure 3. XIST regulated miR-29c by directly targetting in TMZ-resistant glioma cells.
(A) LN229/TMZ and U251/TMZ cells were transfected with mimics NC/miR-29c mimics or inhibitor NC/miR-29c inhibitor. Expression of miR-29c was verified using real-time PCR assays. (B) LN229/TMZ and U251/TMZ cells were transfected with mimics NC/miR-29c mimics or inhibitor NC/miR-29c inhibitor. Expression of XIST in response to miR-29c overexpression or inhibition was determined using real-time PCR assays. (C) LN229/TMZ and U251/TMZ cells were transfected with siRNA-NC/siRNA-XIST. Expression of miR-29c in response to XIST knockdown was determined using real-time PCR assays. (D,E) Association of miR-29c and XIST with AGO2 in LN229/TMZ and U251/TMZ cells. Detection of AGO2 and IgG using Western blotting (D). Detection of miR-29c and XIST using real-time PCR (E). (F) wt-XIST and mut-XIST luciferase reporter gene vectors were constructed by mutating the putative binding site of miR-29c in XIST. (G) The indicated vectors were cotransfected into the TMZ-resistant glioma cells (U251/TMZ) with miR-29c mimics or miR-29c inhibitor. The luciferase activity in each group was then determined using dual luciferase assays. The data are presented as mean ± S.D. of three independent experiments; **P<0.01.
XIST/miR-29c axis regulated glioma cell chemoresistance to TMZ through DNA mismatch repair pathway
After confirming that XIST directly binds to miR-29c to regulate its expression, next, we assessed the combined effect of XIST and miR-29c on glioma cell proliferation and chemoresistance to TMZ. TMZ-resistant U251/TMZ and LN229/TMZ cells were cotransfected with si-XIST and miR-29c inhibitor. The cell proliferation of U251/TMZ and LN229/TMZ cells was significantly suppressed by XIST knockdown, whereas promoted by miR-29c inhibition; the suppressive effect of XIST knockdown on glioma cell proliferation coule be partially reversed by miR-29c inhibition (Figure 4A,B). Furthermore, 24 h after transfection, cells were treated with a series dose of TMZ (7.5, 15, 30, 60, 120, 240, and 480 μM) for 24 h; the cell viability was then determined using MTT assays to assess the combined effect of XIST and miR-29c on glioma cell chemoresistance to TMZ. Data were displayed as described; the TMZ concentration to reduce cell viability to 50% (lC50) for U251/TMZ and LN229/TMZ was significantly reduced by XIST knockdown, whereas increased by miR-29c, indicating that XIST knockdown reduced the chemoresistance of glioma cell to TMZ whereas miR-29c inhibition played an opposite role. Moreover, the effect of XIST knockdown on glioma cell chemoresistance could be partially reversed by miR-29c inhibition (Figure 4C,D).
Figure 4. XIST/miR-29c axis regulated glioma cell chemoresistance to TMZ through DNA mismatch repair (MMR) pathway.
U251/TMZ and LV229/TMZ cells were cotransfected with si-XIST and miR-29c inhibitor. (A,B) The cell viability and DNA synthesis capability were determined using MTT and BrdU assays. (C,D) Twenty four hours after transfection, cells were treated with a series dose of TMZ (7.5, 15, 30, 60, 120, 240, and 480 μM) for 24 h; the cell viability was then determined using MTT assays. Data were displayed as described. (E) The protein levels of MSH6 were determined using Western blot assays. (F) The protein levels of SP1 and MGMT were determined using Western blot assays. The data are presented as mean ± S.D. of three independent experiments; *P<0.05, **P<0.01. #P<0.05, ##P<0.01, compared to si-XIST + inhibitor-NC group.
TMZ is one of the most widely used alkylating agents, which modify DNA in several positions, one of which is O6-methylguanine MeG (O6MeG). This modified guanine preferentially pairs with thymine during DNA replication, triggering a DNA mismatch repair (MMR) pathway that ultimately causes DNA double-strand breaks and induces apoptosis [24–26]. Methylation damage can be reversed by MGMT [26]. In addition, SP1 has been reported to regulate one of the key MMR proteins, MSH6 [27]. Here, we investigated whether the MMR pathway were involved in XIST/miR-29c regulation of glioma cell chemoresistance to TMZ by measuring the protein levels of MSH6, MGMT, and SP1. MSH6, SP1, and MGMT protein levels were significantly reduced by XIST knockdown, whereas increased by miR-29c inhibition; the suppressive effects of XIST knockdown on the indicated proteins could be significantly reversed by miR-29c inhibition (Figure 4E,F). These data indicate that the MMR pathway is involved in XIST/miR-29c regulation of glioma cell chemoresistance to TMZ.
The expression levels and correlations of miR-29c, SP1, and MGMT in tumor and PTBE tissues
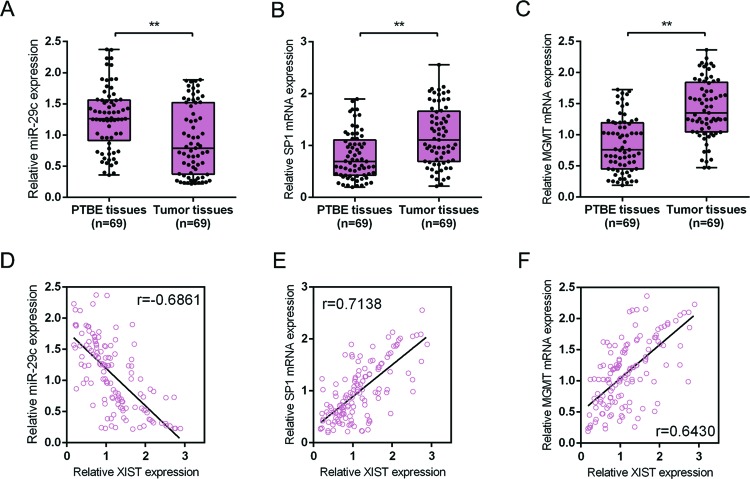
Finally, to further confirm the above findings, the expression of miR-29c, SP1, and MGMT in glioma tissues and the PTBE tissues was monitored using real-time PCR assays. The results showed that in tumor tissues, miR-29c expression was down-regulated while SP1 and MGMT mRNA expression was up-regulated as compared with in PTBE tissues (Figure 5A-C). Spearman’s rank correlation coefficient was performed to analyze the correlation between XIST and miR-29c, between XIST and PS1, between XIST and MGMT. The results showed that XIST was inversely correlated with miR-29c, positively correlated with PS1, positively related with MGMT (Figure 5D-F). Taken together, these data suggest that XIST can inhibit miR-29c expression by directly binding to miR-29c and subsequently up-regulate the expression of SP1 and MGMT to promote the chemoresistance of glioma cells to TMZ.
Figure 5. The expression levels and correlations of miR-29c, SP1, and MGMT in tumor and PTBE tissues.
(A–C) Expression of miR-29c, SP1, and MGMT mRNA in 69 paired glioma tissues and PTBE tissues was determined using real-time PCR assays. The data are presented as mean ± S.D. of three independent experiments; **P<0.01. (D–F) The correlation between XIST and miR-29c, SP1, and MGMT mRNA expression, respectively, was analyzed using Spearman’s rank correlation analysis.
Discussion
TMZ-based chemotherapy is the most commonly used treatment for glioma; however, due to the acquisition of chemoresistance of glioma cells to TMZ, the efficacy is very limited [1,3,28]. Although changes in a number of genetic factors, such as PTEN, VEGF, and EGFR, are associated with the pathogenesis of glioma and the chemoresistance of glioma cells to TMZ [3,29,30], the molecular biology of chemoresistance of glioma cells to TMZ is still largely unknown.
Studies have demonstrated that approximately 18% of these ncRNAs are associated with human tumors, compared with only 9% of human protein-coding genes [31], suggesting that lncRNA can act as a major contributor to carcinogenesis and cancer progression. Moreover, the role of dysregulated lncRNAs in the chemoresistance of many cancers have garnered increased scientific interest in recent years. Accumulating evidence confirms that lncRNAs can affect the sensitivity of cancer cells to chemotherapy. For example, a well-characterized lncRNA, HOTAIR, contributes to the chemoresistance of lung adenocarcinoma and glioma via inhibiting p21 expression [32,33]. Another lncRNA, UCA1, enhances 5-fluorouracil resistance of colorectal cancer by inhibiting miR-204-5p [34]. Low XIST expression predicts drug response in PDXs associated with a significant reduction in the breast cancer stem cells population [17]. In the present study, we first evaluated the expression of XIST and its relationship with the clinical features in patient with glioma. XIST expression was significantly up-regulated in glioma tissues and cell lines, compared with PTBE tissues and NHA cell line, respectively; further, a higher XIST expression was correlated with a poorer prognosis in patients with glioma, including larger tumor size, advanced WHO stages, and shorter OS. XIST knockdown significantly suppressed glioma cell proliferation in the presence or absence of TMZ treatment. Upon TMZ treatment, XIST acted on glioma cell proliferation in a dose-dependent manner, indicating the potential role of XIST in regulating the chemoresistance of glioma cell to TMZ.
We then conducted a series of doses of TMZ treatment on non-TMZ-resistant and TMZ-resistant glioma cell lines (U251, U251/TMZ, LN229, and LN229/TMZ). The cell viability of non-TMZ-resistant glioma cells was significantly suppressed by TMZ treatment in a dose-dependent manner; however, the suppressive effects of TMZ on TMZ-resistant glioma cells were attenuated. Then, XIST knockdown was conducted to investigate its role in glioma chemoresistance to TMZ. XIST knockdown reduced the lC50 values of both LN229/TMZ and U251/TMZ cells, indicating that XIST aggregated the chemoresistance of glioma cell to TMZ. However, the mechanism by which XIST affects the chemoresistance of glioma cells remains to be investigated.
It has been recently discovered that the interaction between lncRNAs and miRNAs affects post-transcriptional regulation by inhibiting the available miRNA activity. According to previous studies, lncRNA can act as a specific ‘sponge’ for miRNAs to attenuate their regulatory effect on mRNAs [22]. In view of the important role of miR-29c in regulating cancer cell growth and invasion [18–20], as well as glioma chemoresistance to TMZ [35], we validated whether XIST affects the chemoresistance of glioma cells to TMZ through miR-29c. As shown by real-time PCR, XIST can reduce the detectable amount of miR-29c, while up-regulating miR-29c expression could reduce XIST expression, indicating that XIST exerted similar regulatory behaviors as miR-29c inhibitor, so we speculated that XIST might regulate miR-29c expression via the RNAi pathway at the post-transcriptional level. This would suggest that both miR-29c and XIST exist in the RISC. Since AGO2 is a key component of the RISC, we therefore performed RIP with the AGO2 antibody. In RNA extracted from precipitated AGO2 protein, we could detect both miR-29c and XIST with a more than 2–3.5-fold enrichment compared with IgG. These indicate that miR-29c and XIST are associated with each other in glioma cells. To further confirm the interaction between XIST and miR-29c, luciferase assays were performed. Consistent with RIP results, XIST interacted with miR-29c through direct binding to miR-29c, thereby degrading miR-29c most possibly through the RISC it carried. Furthermore, we also assessed the combined effect of XIST and miR-29c on glioma cell proliferation and chemoresistance to TMZ. XIST knockdown caused significant suppression of glioma cell proliferation and chemoresistance, whereas miR-29c inhibition promoted glioma cell proliferation and chemoresistance; the effect of XIST knockdown on the indicated TMZ-resistant glioma cells could be partially reversed by miR-29c inhibition. These data suggest that XIST can directly bind to miR-29c to inhibit its expression, thereby affecting glioma cell proliferation and chemoresistance to TMZ.
TMZ is the most widely used alkylating agent that modifies DNA at several positions, one of which is O6MeG. This modified guanine preferentially pairs with thymine during DNA replication which initiates the MMR pathway, and then ultimately causes DNA double-strand break and induces apoptosis [24–26]. The methylation damage can be reversed by MGMT [26]. Although various mechanisms that mediate the intrinsic or acquired resistance of TMZ have been recognized, MGMT is now considered to play a major role in mediating the resistance of TMZ and other alkylating agents [36]. The intracellular levels of the alkylating enzyme MGMT interfere with TMZ response in patients with glioblastoma multiform [37,38]. In addition to MGMT, SP1 has been reported to regulate a key MMR protein MSH6 [27] by affecting the promoter activity of MSH6. In the present study, we monitored the protein levels of MSH6, MGMT, and SP1 in response to cotransfecting XIST and miR-29c in TMZ-resistant glioma cell lines. XIST knockdown significantly reduced MSH6, SP1, and MGMT proteins, whereas miR-29c inhibition significantly increased MSH6, SP1, and MGMT proteins; the suppressive effect of XIST knockdown on these proteins could be partially reversed by miR-29c inhibition. These data suggest that XIST/miR-29c may modulate the chemoresistance of glioma cells to TMZ by modulating the MMR pathway.
In the glioma tissues, miR-29c expression was down-regulated, whereas SP1 and MGMT mRNA expression was up-regulated; moreover, XIST was inversely correlated with miR-29c, whereas positively correlated with SP1 and MGMT expression in glioma tissues, indicating that targetting XIST to rescue miR-29c expression, thereby inhibiting the chemoresistance of glioma cells to TMZ may be a promising strategy for improving the efficiency of TMZ-based chemotherapy.
Abbreviations
- AGO2
argonaute 2
- BrdU
5-bromo-2-deoxyuridine
- CI
confidence interval
- COX
Cox's proportional hazards regression model
- CSU
Central South University
- Ct
comparative cycle threshold
- EGFR
epidermal growth factor receptor
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HOTAIR
HOX transcriptional antisense RNA
- HR
hazard ratio
- HRP
horseradish peroxidase
- lncRNA
long non-coding RNA
- MGMT
O6-methylguanine-DNA methytransferase
- MMR
mismatch repair
- MSH6
mutS homolog 6
- mut
mutant-type
- ncRNA
non-coding RNA
- NHA
normal human astrocyte
- OD490 nm
optical density at a wavelength of 490 nm
- O6MeG
O6-methylguanine MeG
- OS
overall survival
- PDX
patient-derived xenograft
- PTBE
peritumoral brain edema
- PTEN
phosphatase and tensin homolog deleted on chromosome ten
- qRT-PCR
quantitative real-time polymerase chain reaction
- RIP
RNA immunoprecipitation
- RISC
RNA-induced silencing complex
- RIPA
radio-immunoprecipitation assay
- RPMI-1640
Roswell Park Memorial Institute 1640
- RT
room temperature
- si-XIST
siRNA for XIST
- SP1
specificity protein 1
- TMZ
temozolomide
- VEGF
vascular endothelial growth factor
- wt
wild-type
- XIST
X inactive specific transcript
Funding
This work was supported by the Hunan Provincial Science and Technology Foundation [grant number 2015JC3017]; and the Post-doctoral Research Funding.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Author contribution
Peng Du: wrote the manuscript, conceived and designed the experiments. Haiting Zhao: cellular experiment operation. Renjun Peng: molecular experiment operation. Qing Liu: data processing and statistical analysis. Gang Peng: figure preparation. Jian Yuan: figure preparation. Yiwei Liao: experimental guidance and data verification.
References
- 1.Stupp R., et al. (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10, 459–466 [DOI] [PubMed] [Google Scholar]
- 2.Jemal A., et al. (2010) Cancer statistics, 2010. CA Cancer J. Clin. 60, 277–300 [DOI] [PubMed] [Google Scholar]
- 3.Minniti G., et al. (2009) Chemotherapy for glioblastoma: current treatment and future perspectives for cytotoxic and targeted agents. Anticancer Res. 29, 5171–5184 [PubMed] [Google Scholar]
- 4.Reifenberger G., et al. (2016) Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat. Rev. Clin. Oncol. 14, 434–452 [DOI] [PubMed] [Google Scholar]
- 5.Hoang-Xuan K. and Idbaih A. (2011) Advances in molecular genetics and treatment of gliomas. Bull. Acad. Natl. Med. 195, 11–20 [PubMed] [Google Scholar]
- 6.Nakada M., et al. (2011) Aberrant signaling pathways in glioma. Cancers (Basel) 3, 3242–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponting C.P., Oliver P.L. and Reik W. (2009) Evolution and functions of long noncoding RNAs. Cell 136, 629–641 [DOI] [PubMed] [Google Scholar]
- 8.Bach D.H., et al. (2017) The role of exosomes and miRNAs in drug-resistance of cancer cells. Int. J. Cancer 141, 220–230 [DOI] [PubMed] [Google Scholar]
- 9.Farazi T.A., et al. (2011) miRNAs in human cancer. J. Pathol. 223, 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou S., Wang J. and Zhang Z. (2014) An emerging understanding of long noncoding RNAs in kidney cancer. J. Cancer Res. Clin. Oncol. 140, 1989–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heneghan H.M., Miller N. and Kerin M.J. (2010) MiRNAs as biomarkers and therapeutic targets in cancer. Curr. Opin. Pharmacol. 10, 543–550 [DOI] [PubMed] [Google Scholar]
- 12.Ferracin M., Veronese A. and Negrini M. (2010) Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev. Mol. Diagn. 10, 297–308 [DOI] [PubMed] [Google Scholar]
- 13.Yao Y., et al. (2015) Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 359, 75–86 [DOI] [PubMed] [Google Scholar]
- 14.Ren C., et al. (2015) Functions and mechanisms of long noncoding RNAs in ovarian cancer. Int. J. Gynecol. Cancer 25, 566–569 [DOI] [PubMed] [Google Scholar]
- 15.Fang J., Sun C.C. and Gong C. (2016) Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem. Biophys. Res. Commun. 478, 811–817 [DOI] [PubMed] [Google Scholar]
- 16.Schouten P.C., et al. (2016) High XIST and low 53BP1 expression predict poor outcome after high-dose alkylating chemotherapy in patients with a BRCA1-like breast cancer. Mol. Cancer Ther. 15, 190–198 [DOI] [PubMed] [Google Scholar]
- 17.Salvador M.A., et al. (2013) The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin. Cancer Res. 19, 6520–6531 [DOI] [PubMed] [Google Scholar]
- 18.Yu B., et al. (2017) microRNA-29c inhibits cell proliferation by targeting NASP in human gastric cancer. BMC Cancer 17, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y., et al. (2016) MiR-29c inhibits cell growth, invasion, and migration of pancreatic cancer by targeting ITGB1. Onco. Targets Ther. 9, 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cristobal I., et al. (2015) M iR-29c downregulation contributes to metastatic progression in colorectal cancer. Ann. Oncol. 26, 2199–2200 [DOI] [PubMed] [Google Scholar]
- 21.Arechaga-Ocampo E., et al. (2017) Tumor suppressor miR-29c regulates radioresistance in lung cancer cells. Tumour Biol. 39, 1010428317695010. [DOI] [PubMed] [Google Scholar]
- 22.Paraskevopoulou M.D. and Hatzigeorgiou A.G. (2016) Analyzing miRNA-LncRNA interactions. Methods Mol. Biol. 1402, 271–286 [DOI] [PubMed] [Google Scholar]
- 23.Zhang J., et al. (2010) Acquired resistance to temozolomide in glioma cell lines: molecular mechanisms and potential translational applications. Oncology 78, 103–114 [DOI] [PubMed] [Google Scholar]
- 24.He J., et al. (2011) Expression of glioma stem cell marker CD133 and O6-methylguanine-DNA methyltransferase is associated with resistance to radiotherapy in gliomas. Oncol. Rep. 26, 1305–1313 [DOI] [PubMed] [Google Scholar]
- 25.Knizhnik A.V., et al. (2013) Survival and death strategies in glioma cells: autophagy, senescence and apoptosis triggered by a single type of temozolomide-induced DNA damage. PLoS ONE 8, e55665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bobola M.S., et al. (2007) Human glioma cell sensitivity to the sequence-specific alkylating agent methyl-lexitropsin. Clin. Cancer Res. 13, 612–620 [DOI] [PubMed] [Google Scholar]
- 27.Gazzoli I. and Kolodner R.D. (2003) Regulation of the human MSH6 gene by the Sp1 transcription factor and alteration of promoter activity and expression by polymorphisms. Mol. Cell Biol. 23, 7992–8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan T.Y., et al. (2014) Inhibition of EZH2 reverses chemotherapeutic drug TMZ chemosensitivity in glioblastoma. Int. J. Clin. Exp. Pathol. 7, 6662–6670 [PMC free article] [PubMed] [Google Scholar]
- 29.Nagane M., Huang H.J. and Cavenee W.K. (1997) Advances in the molecular genetics of gliomas. Curr. Opin. Oncol. 9, 215–222 [DOI] [PubMed] [Google Scholar]
- 30.Melin B. (2011) Genetic causes of glioma: new leads in the labyrinth. Curr. Opin. Oncol. 23, 643–647 [DOI] [PubMed] [Google Scholar]
- 31.Khachane A.N. and Harrison P.M. (2010) Mining mammalian transcript data for functional long non-coding RNAs. PLoS ONE 5, e10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z., et al. (2013) The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS ONE 8, e77293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jing L., et al. (2015) HOTAIR enhanced aggressive biological behaviors and induced radio-resistance via inhibiting p21 in cervical cancer. Tumour Biol. 36, 3611–3619 [DOI] [PubMed] [Google Scholar]
- 34.Bian Z., et al. (2016) LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci. Rep. 6, 23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao S., et al. (2016) miR-29c contribute to glioma cells temozolomide sensitivity by targeting O6-methylguanine-DNA methyltransferases indirectely. Oncotarget 7, 50229–50238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo J., et al. (2013) Research on DNA methylation of human osteosarcoma cell MGMT and its relationship with cell resistance to alkylating agents. Biochem. Cell Biol. 91, 209–213 [DOI] [PubMed] [Google Scholar]
- 37.Hegi M.E., et al. (2008) Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J. Clin. Oncol. 26, 4189–4199 [DOI] [PubMed] [Google Scholar]
- 38.Gerson S.L. (2004) MGMT: its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer 4, 296–307 [DOI] [PubMed] [Google Scholar]