Abstract
Breast cancer is the second leading cause of cancer-associated mortality in females in the USA. Hsa-miR-599 was demonstrated to function as a tumour suppressor during cancer progression. However, the function and mechanism of the hsa-miR-599 in human breast cancer remain elusive. Thus, the aim of the present study was to investigate the potential role of hsa-miR-599 in breast cancer biology. The expression levels of hsa-miR-599 in 40 pairs of surgical specimens and human breast cancer cell lines were detected using quantitative polymerase chain reaction analysis. The overexpression of hsa-miR-599 was established by transfecting mimics into the MCF-7 and MDA-MB-231 cell lines. Cell counting kit-8, colony formation and transwell assays were used to investigate the potential function of hsa-miR-599 in MCF-7 and MDA-MB-231 cell lines. Luciferase assays combined with western blot analysis was performed to validate the regulation of a putative target of hsa-miR-599. The results demonstrated that hsa-miR-599 was downregulated in the breast cancer tissues and breast cancer cell lines. Overexpression of hsa-miR-599 was revealed to inhibit the viability and proliferation of cell in vitro and tumour growth in vivo. The results of the luciferase assay indicate that bromodomain containing 4 (BRD4) is a direct target of hsa-miR-599. Furthermore, the xenograft mouse model demonstrated that overexpressed hsa-miR-599 reduced BRD4 expression. These results suggest that hsa-miR-599 serves as an oncosuppressive microRNA that impairs breast cancer tumorigenesis and progression by directly targeting BRD4. Furthermore, increased BRD4 expression partially reversed the suppressive effect of hsa-miR-599. In conclusion, the results of the present study demonstrated that hsa-miR-599 suppressed breast cancer progression by downregulating BRD4. The overexpression of hsa-miR-599 may be considered as a novel therapeutic target for the treatment of patients with breast cancer.
Keywords: hsa-miR-599, breast cancer, BRD4, migration and invasion, proliferation
Introduction
Breast cancer is the second leading cause of cancer-associated mortality in females in the USA, and it had an estimated annual diagnosis rate of 246,660 people and annual mortality rate of 40,450 cases in 2016 (1). Currently, the primary therapeutic strategy for breast cancer patients is surgery, while chemotherapy, radiotherapy and hormonal therapy are also used. Despite the impressive advances in breast cancer treatment, relapse and metastasis remain inevitable in breast cancer patients (2). Therefore, determining the molecular mechanisms underlying breast cancer migration and invasion may facilitate the identification of novel therapeutic targets and consequently lead to the improvement of prognosis in the future.
MicroRNAs (MiRNAs) are noncoding RNAs with 20–22 nucleotides. They post-transcriptionally modulate gene expression by binding to the 3′-untranslated region (3′-UTR) of the target mRNAs (3). As is well known, miRNAs are involved in tumour cell proliferation, migration and invasion (4). Moreover, altered miRNA expression is involved in breast cancer pathogenesis via the modulation of oncogenes and tumour suppressors that subsequently affect the downstream signalling pathway (5–7). For instance, miR-411, miR-490-3p and miR-145 were significantly down-regulated in breast cancer cases and played a critical role in the suppression of tumour metastasis (8–10). Although the pathogenesis of breast cancer metastasis was already investigated, the mechanism was not completely illustrated. Crucial miRNAs that might play significant roles in breast cancerogenesis must be identified. The newly discovered miRNA hsa-miR-599 plays an important role in the development of hepatocellular carcinoma (11). However, the mechanism underlying the development and progression of breast cancer remains unknown.
In this study, we firstly detected the expression level of hsa-miR-599 in breast cancer tissues and metastatic cell lines. Secondly, we investigated the role of hsa-miR-599 in breast cancer proliferation, migration and invasion in vitro and in vivo. Finally, bromodomain containing 4 (BRD4) was identified as a direct target of hsa-miR-599. The results revealed that hsa-miR-599 functioned as a tumour suppressor in breast cancer.
Materials and methods
Cell culture
Breast cancer cell lines MCF-7 (TCHu74), MDA-MB-231 (TCHu104) were purchased from the Shanghai Cell Bank, Chinese Academy of Sciences. BT474 (HTB-20™), and T-47D (HTB-133) and normal mammary epithelial cell line MCF 10A (CRL-10317) were obtained from the American Type Culture Collection (Manassas, VA, USA). The cell identity was confirmed by STR analysis. The MCF-7, BT474, MDA-MB-231 and T-47D cells were cultured in Dulbecco's modified Eagle's medium (DMEM; HyClone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) (HyClone; GE Healthcare Life Sciences). The MCF-10A cells were cultured in DMEM/F12 medium (HyClone; GE Healthcare Life Sciences) containing 5% horse serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 1% penicillin/streptomycin (HyClone; GE Healthcare Life Sciences), 20 ng/ml of EGF (Gibco; Thermo Fisher Scientific, Inc.), 0.5 µg/ml of hydrocortisone (Sigma, St. Louis, MO, USA). All cells were incubated in a humidified atmosphere with 5% CO2 and humidified sphere of 95% at 37°C.
Cell transfection
An hsa-miR-599 mimic (sense 5′-CUGUCCACAGUGUGUUUGAUAAG-3′) were chemically synthesised by Shanghai GenePharma Co., Ltd. (Shanghai, China). RNA with no homology to any human genomic sequence was used as negative control (NC) (sense 5′-ACUACUGAGUGACAGUAGA-3′). For convenience, the hsa-miR-599 mimic and the NC were designated as miR-599 and NC, respectively. A BRD4 overexpression plasmid (sc-425356-ACT) and control vector (sc-437275) were obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). During cell transfection, the cells were seeded in six-well plates and then cultured until 50 to 70% confluency was reached in 1 day. Transfection was performed using a Lipofectamine® 2000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocols. The transfection mixture was replaced in a medium containing 10% FBS after 6 to 8 h.
Cell viability assay
Cell viability was measured using a Cell Counting Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology, Haimen, China). The cells were transfected with miR-599 and control and then cultured overnight. Subsequently, the cells were trypsinised and seeded at 3,000 cells/well in a 96-well plate. After culturing at indicated time periods (0, 24, 48 and 72 h), 10 µl of the CCK-8 solution was added into each well. The resulting mixtures were incubated at 37°C. After 3 h, the absorbance of each well was measured using a Multiskan MK3 spectrophotometer set at a wavelength of 450 nm.
Colony formation assay
The cells transfected with the miR-599 mimic and control were seeded at 200 cells/well in the 6-well plates. After 1 week of culture, the colonies were stained with 0.5% crystal violet (Beyotime Institute of Biotechnology), and the images of the stained colonies were captured using a CKX41 light microscope. The number of the colonies was counted using the images.
Cell migration and invasion assay
Cell migration and invasion were assessed by performing a Boyden chamber assay. For the invasion assay, the upper sides of the filters were coated with 50 µl of Matrigel (BD Biosciences, Bedford, MA, USA). Cells were harvested at 48 h post-transfection. Approximately 5×104 cells with 200 µl of serum-free medium were seeded in the upper chamber. The lower chamber was filled with medium supplemented with 5% FBS. After incubation at 37°C with 5% CO2 for 8 h (migration) or 12 h (invasion), the cells on the lower filter were fixed with methanol and stained with crystal violet. The stained cells were then counted under a light microscope (CKX41; Olympus Corporation, Tokyo, Japan).
Luciferase assay
BRD4 3′-UTR-luciferase reporter vectors were created by ligating the BRD4 3′-UTR polymerase chain reaction (PCR) products into the XhoI and NotI restriction sites of the psiCHECK-2™ Vector (Promega Corporation, Madison, WI, USA). The mutant 3′-UTR regions were chemically synthesised and ligated into the psiCHECK-2™ vector. The cells were cultured in 24-well plates, and each well was transfected with 250 ng of vectors with 50 nM Hsa-miR-599 mimic or control. After 48 h of co-transfection, the luciferase activity was measured using a dual-luciferase reporter assay system (Promega Corporation) according to the manufacturer's instructions.
Western blot analysis
Equal amounts of proteins (30 µg) from the lysates of the cells were subjected to electrophoresis through a 10% SDS-PAGE (Beyotime Institute of Biotechnology) at 80 V for 30 min and at 100 V for 1.5 h. The proteins were then transferred onto polyvinylidene difluoride membranes. After blocking in 5% skimmed milk, the membranes were then incubated with the following diluted primary antibodies: Rabbit polyclonal BRD4 (Abcam, Cambridge, MA, USA), mouse monoclonal β-actin (Beyotime Institute of Biotechnology) overnight at 4°C and horseradish peroxidase-conjugated goat anti-rabbit antibody (Santa Cruz Biotechnology, Inc.) at room temperature for 2 h. Specific bands were visualised on an autoradiographic film using an enhanced chemiluminescence reagent (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China).
Quantitative PCR (qPCR)
The total RNA from cells was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and then reverse-transcribed using a PrimeScript RT Reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's instructions. qPCR was performed using a SYBR Premix Ex Taq. The specificity of the amplification was verified using a melting curve and electrophoresis in agarose gel. The following PCR conditions were used for the detection of the mRNAs: 95°C for 30 sec, followed by 40 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec. The β-actin and U6 small nuclear RNA were used as internal controls for the detection. The relative expression levels of miR-599 and BRD4 were calculated as the inverse log of ΔΔCq and normalised to the reference. The primers used for amplification were the following: miR-599 forward 5′-GUUGUGUCAGUUUAUCAAAC-3′ and reverse 5′-GUUGUGUCAGUUUAUCAAAC-3′. U6 forward 5′-TGCGGGTGCTCGCTTCGCAGC-3′ and reverse 5′-CCAGTGCAGGGTCCGAGGT-3′. BRD4 forward 5′-CATGGACATGAGCACAATCA-3′ and reverse 5′-TCATGGTCAGGAGGGTTGTA-3′. β-actin forward 5′-GATCATTGCTCCTCCTGAGC-3′ and reverse 5′-ACTCCTGCTTGCTGATCCAC-3′.
Clinical sample collection
Forty cases of surgically resected breast cancer samples (tumour and adjacent nontumour tissue) were obtained from Weifang People's Hospital. These samples were frozen in liquid nitrogen immediately and stored at −80°C. None of the patients received chemotherapy or radiotherapy before the surgery. All the patients gave their written informed consents, and the study was approved by the Ethics Committee of Weifang People's Hospital.
Animal studies
All animal studies were approved by the Animal Care and Welfare Committee of WeiFang Medical University and conducted in strict accordance with the guidelines of the National Animal Welfare Law of China. Four-week-old female BALB⁄c nude mice were purchased from the Laboratory Animal Center of Yangzhou University (Yangzhou, China) and maintained in a specific pathogen-free environment. Subcutaneous tumour xenografting was performed by subcutaneously injecting the mice with 200 µl PBS containing 1×106 MCF-7 cells transfected with miR-599 mimic or miR-NC. Tumour volume (mm3) was calculated every 5 days using the formula: V=0.5 × length × width2. After 25 days, the mice were euthanised, and the tumours were isolated, weighed, photographed and processed for immunohistochemistry.
Statistical analysis
All experiments were performed in triplicate. Unless otherwise indicated, the experimental values were expressed as mean ± SEM. Statistical significance was determined by unpaired Student's t-test using SPSS 13.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Hsa-miR-599 was downregulated in breast cancer tissues and cell lines
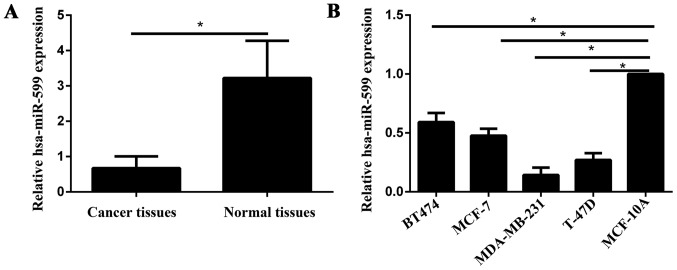
The expression level of hsa-miR-599 was measured in breast cancer specimens, and the adjacent normal tissues by qPCR. As shown in Fig. 1A, the hsa-miR-599 expression levels in the breast cancer tissues was significantly downregulated compared with those of the adjacent non-neoplastic tissues (P<0.05). In the breast cancer cell lines, hsa-miR-599 expression levels were downregulated in the low-invasive cell lines (MCF-7 and BT474) and high-invasive cell lines (MDA-MB-231 and T-47D) compared with those in the normal mammary epithelial cell line MCF-10A (Fig. 1B). Furthermore, in the low-invasive and high-invasive cell lines, the expression of hsa-miR-599 was low in the MCF-7 and MDA-MB-231, respectively. Therefore, MCF-7 and MDA-MB-231 were selected for further studies.
Figure 1.
Expression levels of hsa-miR-599 in breast cancer tissues and cell lines. (A) Levels of hsa-miR-599 in the breast cancer tissues were significantly lower than in their corresponding adjacent non-neoplastic tissues (*P<0.05). (B) Levels of hsa-miR-599 were also lower in the MCF-7, BT474, MDA-MB-231 and T-47D cell lines, compared with the MCF-10A cell line (*P<0.05). Expression levels of hsa-miR-599 were determined by qPCR and normalized to U6.
Hsa-miR-599 suppressed the viability and proliferation of breast cancer cells in vitro
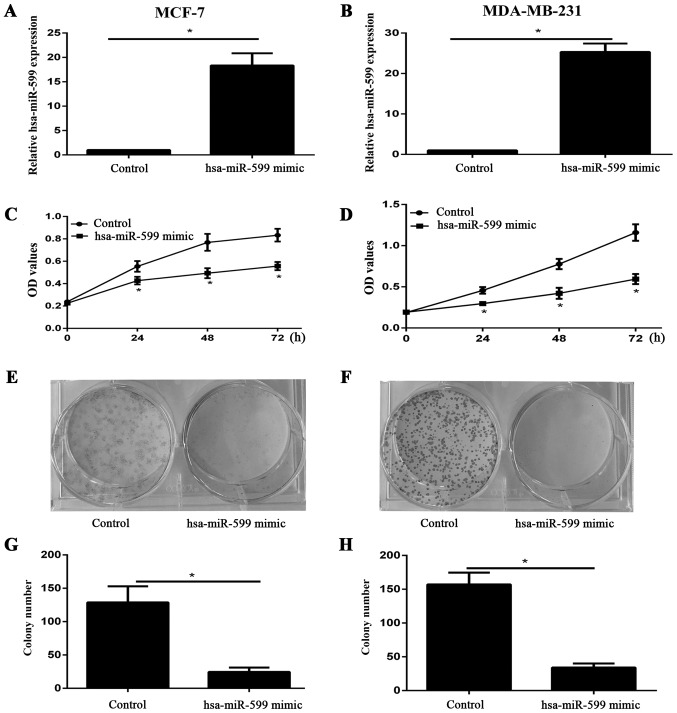
We examined the expression level of hsa-miR-599 in cells after the transfection of hsa-miR-599 mimics or control into MCF-7 and MDA-MB-231 through qPCR. The expression of hsa-miR-599 in the mimic group was higher than that in the control group (Fig. 2A and≈B). The viability of MCF-7 and MDA-MB-231 cells was investigated using CCK-8 assay after 24, 48, and 72 h of incubation. Hsa-miR-599 mimic evidently inhibited the viability of both cell lines (Fig. 2C and D). Furthermore, the results of the colony formation assay showed that the proliferation of hsa-miR-599 mimic cells was severely inhibited (Fig. 2E-H). These results suggested that hsa-miR-599 inhibited the growth of breast cancer cells.
Figure 2.
Overexpression of hsa-miR-599 inhibited the growth of breast cancer cells. qPCR analysis demonstrating the efficiency of hsa-miR-599 overexpression with hsa-miR-599 mimics in the (A) MCF-7 and (B) MDA-MB-231 cell line. Effect of hsa-miR-599 mimics induction on (C) MCF-7 and (D) MDA-MB-231 cell viability over 72 h using the Cell Counting Kit-8 assay (*P<0.05). Overexpression of hsa-miR-599 inhibited cell colony formation ability of (E) MCF-7 and (F) MDA-MB-231 cell line. Graphical representation of the clone number for (G) MCF-7 and (H) MDA-MB-231 cell line (*P<0.05).
Hsa-miR-59 inhibited the breast cancer progression in vivo
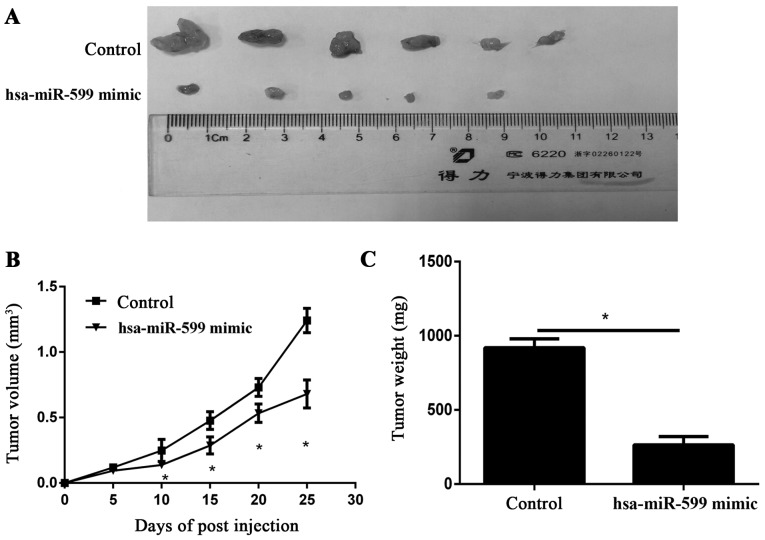
To study the relevance of hsa-miR-599 to breast cancer progression in vivo, we subcutaneously transplanted hsa-miR-599 mimics or miR-NC MCF-7 cells into nude mice (n=6/group). Twenty five days after cell implantation, the tumours were removed and photographed (Fig. 3A). The tumour volumes in the hsa-miR-599 mimic of the tumour xenograft mice were smaller than those of the mock tumour xenograft mice. Furthermore, the periodic measurements of the tumour volume and the final weight of the excised tumours demonstrated that the hsa-miR-599 mimics disrupted tumour growth at termination compared with the control group (Fig. 3B and C).
Figure 3.
Hsa-miR-599 inhibited breast cancer growth in vivo. (A) The hsa-miR-599 mimic tumor xenografts were smaller than the control tumor xenografts (*P<0.05). (B) Hsa-miR-599 tumor growth was obviously slower than that in the mock tumor xenografts after 25 days (*P<0.05). (C) The average weight of the resected tumors (*P<0.05).
Hsa-miR-599 suppressed the migration and invasion potential of breast cancer cells
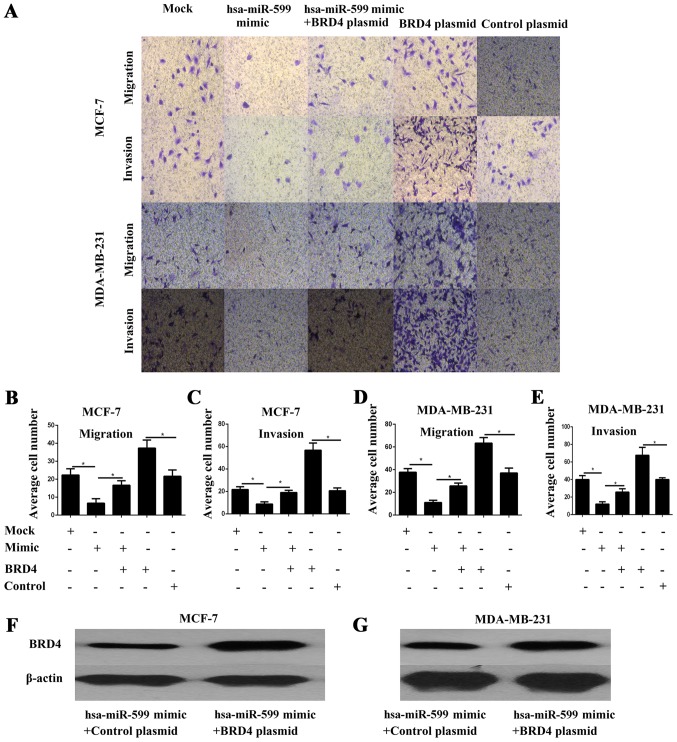
Cell migration and invasion are critical events in tumour metastasis. Thus, the effects of hsa-miR-599 on the migration and invasion of breast cancer cells were explored in vitro. The cell migration and invasion of MCF-7 and MDA-MB-231 were investigated through a transwell assay. The results showed that the hsa-miR-599 mimics significantly inhibited cell migration and invasion (P<0.05; Fig. 4A-E).
Figure 4.
Hsa-miR-599 mimics inhibited the migration and invasion of breast cancer cells, while forced expression of bromodomain containing 4 (BRD4) reversed the migration and invasion of breast cancer cells reduced by hsa-miR-599 mimics. (A) MCF-7 and MDA-MB-231 cells were co-transfected with either hsa-miR-599 mimics or mock and BRD4 plasmid or control plasmid. Transwell assays were conducted and quantified. Migrant and invasion cells were fixed, stained and counted and images were captured (magnification, ×100). (B) Quantification of migration MCF-7 cells in the lower chamber (*P<0.05). (C) Quantification of invasion MCF-7 cells in the lower chamber (*P<0.05). (D) Quantification of migration MDA-MB-231 cells in the lower chamber (*P<0.05). (E) Quantification of invasion MDA-MB-231 cells in the lower chamber (*P<0.05). Western blot analysis was performed to confirm the re-expression of BRD4 in (F) MCF-7 and (G) MDA-MB-231 cells.
BRD4 is a direct target gene of hsa-miR-599 in vitro
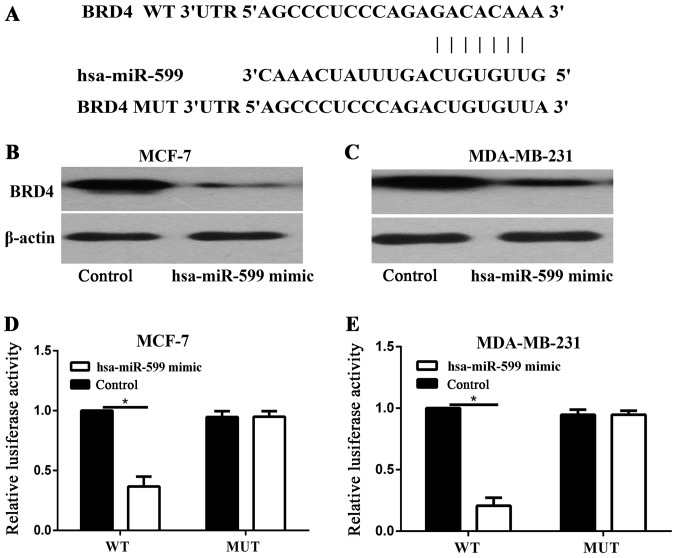
TargetScan was used to identify the target of hsa-miR-599. As BRD4 was predicted to be a target of hsa-miR-599 (Fig. 5A), Western blot analysis was performed to determine whether BRD4 was downregulated at the protein level in breast cancer cells after it was transfected with hsa-miR-599. As shown in Fig. 5B and C, the expression levels of BRD4 was significantly reduced in the MCF-7 and MDA-MB-231 cells after BRD4 was transfected with hsa-miR-599 (P<0.05). Luciferase assays were then performed to determine whether hsa-miR-599 directly target BRD4. As shown in Fig. 5D and E, hsa-miR-599 significantly inhibited the luciferase activity of BRD4 WT, but not of BRD4 MUT, in the MCF-7 and MDA-MB-231 cells. Overall, these results indicated that BRD4 was a direct target gene of hsa-miR-599 in vitro.
Figure 5.
Bromodomain containing 4 (BRD4) was a direct target of hsa-miR-599. (A) Sequence alignment of human hsa-miR-599 with 3′-untranslated region (3′-UTR) of BRD4 predicted by TargetScan. (B and C) BRD4 protein expression in breast cancer cell lines was determined through western blot analysis at 72 h post-transfection. β-actin was used as a control (*P<0.05). Luciferase assay results for (D) MCF-7 and (E) MDA-MB-231 cells showed that hsa-miR-599 upregulation decreased luciferase activity.
Forced BRD4 expression partially rescued the migration and invasion capability of hsa-miR-599-transfected breast cancer cells
To further investigate whether the effects of hsa-miR-599 on cell migration and invasion are mediated by BRD4, we performed transwell assays in MCF-7 and MDA-MB-231 cells co-transfected with hsa-miR-599 and BRD4 overexpressed plasmid. BRD4 protein expression was confirmed by Western blot analysis (Fig. 4F-G). The results showed that forced BRD4 expression increased the migration and invasion of breast cancer cells (Fig. 4A-E). More importantly, the partial but significant restoration of BRD4 rescued the migration and invasion of the hsa-miR-599 cells (Fig. 4A-E). Overall, these results illustrated that the effects of hsa-miR-599 on cell migration and invasion were partly mediated by BRD4.
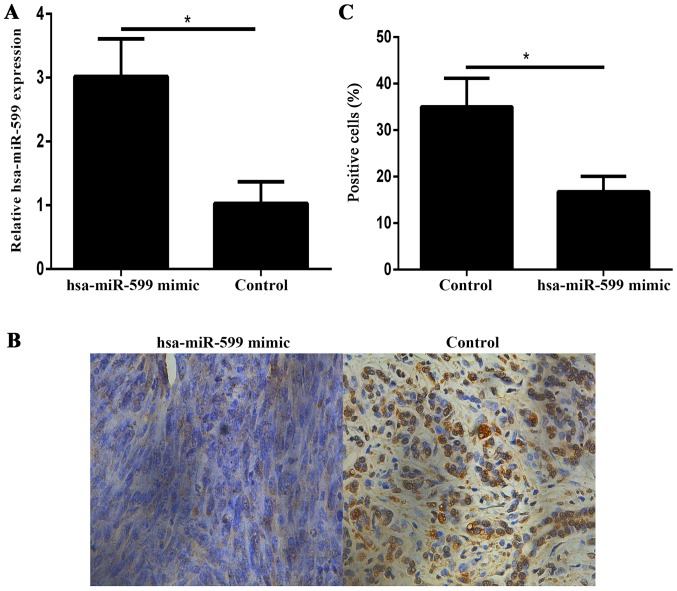
Hsa-miR-599 downregulates BRD4 expression in vivo
Immunohistochemistry showed that the BRD4 expression levels in the hsa-miR-599 mimic tumour xenografts were lower than those in the control xenografts (Fig. 6A-C).
Figure 6.
Hsa-miR-599 downregulated BRD4 expression in tumor xenografts. (A) qPCR analysis of Hsa-miR-599 expression levels in the hsa-miR-599 mimics tumor xenograft mice and control tumor xenograft mice. (B) Representative images (×200 magnification) of breast tumor sections of the control and hsa-miR-599 mimics MCF-7 tumors stained for BRD4. (C) Quantification of the percentage of bromodomain containing 4 (+) cells in the breast tumor sections (*P<0.05).
Discussion
Increasing evidence has shown that miRNAs are abnormally expressed in breast cancers, and changes in miRNA expression affect the onset, development and metastasis of breast cancers (6,12). However, miRNAs affecting breast cancer progression require further exploration. Recent study has identified that hsa-miR-599 served as tumour suppressor in hepatocellular carcinoma. However, the function of hsa-miR-599 in breast cancer has yet to be elucidated, and the mechanism through which hsa-miR-599 inhibits tumour migration and invasion remains unclear.
In the present study, we found that hsa-miR-599 is downregulated in breast cancer tissues and breast cancer cell lines. The colony formation assay showed that cells transfected with hsa-miR-599 grew markedly slower, and the results of the transwell assays revealed that hsa-miR-599 transfection inhibited cell migration and invasion. Tumour xenografting revealed that hsa-miR-599 transfection inhibited tumour volume and growth speed. These results illustrated that hsa-miR-599 functioned as a tumour suppressor during the proliferation and metastasis of breast cancer.
Identifying hsa-miR-599 target genes is essential for understanding their functions in the carcinogenesis and progression of breast cancer. It is also important for the investigation of novel targeted therapies for breast cancer. In the present study, a molecular link between hsa-miR-599 and BRD4 was identified. BRD4 is an epigenome reader and a member of the bromodomain and extra-terminal (BET) family of proteins, which consist of two bromodomains in tandem and an extra terminal domain. Recent studies showed that BRD4 promotes cell cycle progression and regulates cell growth and transcription (13). Subsequent studies demonstrated that BRD4 played an important role in tumour proliferation and growth in pancreatic cancer (14), melanoma (15), neuroblastoma (16), glioblastoma (17), hepatocellular carcinoma (18), lung adenocarcinoma (19), malignant peripheral nerve sheath tumours (20) and breast cancer (21). However, the relation between miRNA and BRD4 were not illustrated in these studies. In the present study, BRD4 was confirmed as a direct target gene of hsa-miR-599, as indicated by the results of luciferase assay and Western blot analysis. Moreover, rescue experiments indicated that the most important effect exerted by hsa-miR-599 on breast cancer cells invasion and migration, which was partially reversed when co-transfected with BRD4. These results demonstrated that BRD4 was a functional target gene of hsa-miR-599 in breast cancer. Notably, each miRNA can regulate numerous genes, and multiple miRNAs may regulate the same gene, and these characteristics affect the activities of pathways (22). In fact, hsa-miR-599 regulates biological functions of different diseases by targeting various genes, such as TGFB2 (23) and MYC (11). In our study, although increased expression of hsa-miR-599 inhibited the migration and invasion of breast cancer cells, these capabilities were partially rescued by BRD4 overexpression. Thus, other target genes were possibly involved in the suppressive effects of hsa-miR-599.
In conclusion, hsa-miR-599 inhibited breast cancer cell progression and metastasis by targeting BRD4. Hsa-miR-599 restoration may provide new therapeutic methods for breast cancer treatment.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Pivot X. Adjuvant chemotherapy for local relapse breast cancer. Lancet Oncol. 2014;15:125–126. doi: 10.1016/S1470-2045(13)70610-7. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 5.Shen J, Stass SA, Jiang F. MicroRNAs as potential biomarkers in human solid tumors. Cancer Lett. 2013;329:125–136. doi: 10.1016/j.canlet.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asghari F, Haghnavaz N, Baradaran B, Hemmatzadeh M, Kazemi T. Tumor suppressor microRNAs: Targeted molecules and signaling pathways in breast cancer. Biomed Pharmacother. 2016;81:305–317. doi: 10.1016/j.biopha.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Peng F, Xiong L, Tang H, Peng C, Chen J. Regulation of epithelial-mesenchymal transition through microRNAs: Clinical and biological significance of microRNAs in breast cancer. Tumour Biol. 2016;37:14463–14477. doi: 10.1007/s13277-016-5334-1. [DOI] [PubMed] [Google Scholar]
- 8.Guo L, Yuan J, Xie N, Wu H, Chen W, Song S, Wang X. miRNA-411 acts as a potential tumor suppressor miRNA via the downregulation of specificity protein 1 in breast cancer. Mol Med Rep. 2016;14:2975–2982. doi: 10.3892/mmr.2016.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao L, Zheng XY. MicroRNA-490 inhibits tumorigenesis and progression in breast cancer. Onco Targets Ther. 2016;9:4505–4516. doi: 10.2147/OTT.S100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao H, Kang X, Xia X, Wo L, Gu X, Hu Y, Xie X, Chang H, Lou L, Shen X. miR-145 suppresses breast cancer cell migration by targeting FSCN-1 and inhibiting epithelial-mesenchymal transition. Am J Transl Res. 2016;8:3106–3114. [PMC free article] [PubMed] [Google Scholar]
- 11.Tian J, Hu X, Gao W, Zhang J, Chen M, Zhang X, Ma J, Yuan H. Identification a novel tumor-suppressive hsa-miR-599 regulates cells proliferation, migration and invasion by targeting oncogenic MYC in hepatocellular carcinoma. Am J Transl Res. 2016;8:2575–2584. [PMC free article] [PubMed] [Google Scholar]
- 12.Joyce DP, Kerin MJ, Dwyer RM. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int J Cancer. 2016;139:1443–1448. doi: 10.1002/ijc.30179. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang YH, Sui YN, Yan K, Wang LS, Wang F, Zhou JH. BRD4 promotes pancreatic ductal adenocarcinoma cell proliferation and enhances gemcitabine resistance. Oncol Rep. 2015;33:1699–1706. doi: 10.3892/or.2015.3774. [DOI] [PubMed] [Google Scholar]
- 15.Segura MF, Fontanals-Cirera B, Gaziel-Sovran A, Guijarro MV, Hanniford D, Zhang G, González-Gomez P, Morante M, Jubierre L, Zhang W, et al. BRD4 sustains melanoma proliferation and represents a new target for epigenetic therapy. Cancer Res. 2013;73:6264–6276. doi: 10.1158/0008-5472.CAN-13-0122-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, Nekritz EA, Zeid R, Gustafson WC, Greninger P, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce LA, Thompson RC, Muller S, Knapp S, Wang J. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang YH, Sui XM, Sui YN, Zhu QW, Yan K, Wang LS, Wang F, Zhou JH. BRD4 induces cell migration and invasion in HCC cells through MMP-2 and MMP-9 activation mediated by the Sonic hedgehog signaling pathway. Oncol Lett. 2015;10:2227–2232. doi: 10.3892/ol.2015.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci USA. 2012;109:19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel AJ, Liao CP, Chen Z, Liu C, Wang Y, Le LQ. BET bromodomain inhibition triggers apoptosis of NF1-associated malignant peripheral nerve sheath tumors through Bim induction. Cell Rep. 2014;6:81–92. doi: 10.1016/j.celrep.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrieu G, Tran AH, Strissel KJ, Denis GV. BRD4 regulates breast cancer dissemination through Jagged1/Notch1 signaling. Cancer Res. 2016;76:6555–6567. doi: 10.1158/0008-5472.CAN-16-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: Approaches and considerations. Nat Rev Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie B, Zhang C, Kang K, Jiang S. miR-599 inhibits vascular smooth muscle cells proliferation and migration by targeting TGFB2. PLoS One. 2015;10:e0141512. doi: 10.1371/journal.pone.0141512. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]