Abstract
Colorectal cancer is currently the third most common cancer in males and the second in females worldwide. In spite of marked progress having been achieved in surgical resection, radiotherapy and chemotherapy, the prognosis for patients with colorectal cancer remains poor. Previous studies have demonstrated that the abnormal expression of microRNAs contributed to human cancer carcinogenesis and progression, suggesting miRNAs as novel therapeutic targets in colorectal cancer. The aim of the present study was to investigate the expression, functions and underlying molecular mechanisms of microRNA-184 (miR-184) in colorectal cancer. The results identified that miR-184 was significantly downregulated in colorectal cancer tissues and cell lines. In vitro functional studies demonstrated that miR-184 significantly inhibited colorectal cancer cell proliferation, migration and invasion. Notably, insulin-like growth factor 1 receptor (IGF-1R) was identified as a direct target of miR-184 in colorectal cancer. Furthermore, the functions of IGF-1R small interfering RNA were similar to those induced by miR-184 in colorectal cancer, suggesting IGF-1R as a functional target of miR-184 in colorectal cancer. The results of the present study indicated that miR-184 may be a novel therapeutic strategy regimen of targeted therapy for colorectal cancer.
Keywords: microRNA-184, insulin-like growth factor 1 receptor, proliferation, metastasis, colorectal cancer
Introduction
Colorectal cancer (CRC) is currently the third most common cancer in males and the second in females worldwide, with an estimated 93,090 novel cases and 49,700 mortalities in the USA in 2015 (1,2). The morbidity of CRC varies markedly worldwide, mainly due to differences in lifestyle, environment and genetics. Epidemiological studies have demonstrated that a number of risk factors contribute to the initiation and progression of CRC, including advanced age, hereditary components, obesity, excess alcohol and red meat, smoking and a lack of physical exercise (3–6). Over the last decades, marked progress has been achieved in surgery resection, radiotherapy and chemotherapy (7). However, the clinical outcome and prognosis of remains poor for patients with CRC with metastasis or recurrence (8,9). In total ~20% of patients with CRC were identified to already bear distant metastases. Furthermore, >50% patients eventually develop recurrent disease and metastasis following surgery with curative intent (10,11). The 5-year overall survival rate for patients with CRC is ~90%, but decreases to <5% in cases of CRC with metastases or recurrence (12). Therefore, it is crucial to understand the molecular mechanisms underlying CRC progression which may provide a potential therapeutic target for the treatment of CRC.
Previous studies have demonstrated that microRNAs (miRNAs/miRs) are deregulated in a number of human cancers, including CRC (13–15). miRNAs are a class of single-stranded non-coding endogenous small RNA molecules of between 19 and 24 nucleotides in length (16). They negatively regulate gene expression through binding to the 3′ untranslated regions (3′UTRs) of target mRNAs and inducing degradation or translational repression of target gene mRNA (16,17). miRNAs have been identified to be involved in a wide range of physiological and pathological processes, including cell proliferation, apoptosis, cell cycle, metastasis, development and differentiation (18,19). Furthermore, previous studies have demonstrated that miRNAs are able to function as tumor suppressors or oncogenes depending on their target mRNAs (20–22). Upregulated miRNAs function as oncogenes by negatively regulating tumor suppressor genes. In contrast, downregulated miRNAs act as tumor suppressors via blockade of oncogenes in tumor progression (23,24). Therefore, miRNAs have been investigated in carcinogenesis and progression as a novel therapeutic choice for patients with CRC.
In the present study, miR-184 was identified to be downregulated and behave as a tumor suppressor in CRC by inhibiting cancer cell growth, migration and invasion. Mechanistically, it was demonstrated that miR-184 exerted its tumor suppressive roles by directly targeting insulin-like growth factor 1 receptor (IGF-1R) in CRC.
Materials and methods
Tissue specimens
The procedure of human tissue specimen collection was approved by the Ethical Committee of the Affiliated Hospital of Nanjing University of Traditional Chinese Medicine (Nanjing, China). In addition, written informed consent was obtained from all patients. In the present study, all experimental protocols were performed according to the approved guidelines. In total, 32 pairs of CRC tissues and matched normal adjacent tissues (NATs) were obtained from patients (age range, 38–72 years; median age, 51; 19 males and 13 females) who had been diagnosed with CRC and underwent surgery at the Affiliated Hospital of Nanjing University of Traditional Chinese Medicine between June 2013 and February 2015. CRC tissues and matched NATs were immediately frozen in liquid nitrogen and stored at −80°C until use.
Cell culture
Human CRC cell lines (HCT116, HT29 and SW620) and human normal colon epithelium cell line (FHC) were purchased from the American Type Culture Collection (Manassas, VA, USA). The human embryonic kidney HEK293T cell line was obtained from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). All cells were cultured in Dulbecco's modified Eagle's medium (HT29 and FHC cells) or RPMI-1640 medium (HCT116 and SW620 cells) containing 10% fetal bovine serum (FBS), 100 mg/ml penicillin and 100 mg/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All cells were incubated at 37°C in a humidified incubator containing 5% CO2.
Transfection
miR-184 mimic, negative control (NC), IGF-1R small interfering RNA (siRNA) and NC siRNA were purchased from Guangzhou RiboBio Co., Ltd (Guangzhou, China). The miR-184 mimic sequence was 5′-GGCAUUCUGUAUACAUCGGAG-3′ and the NC sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. The IGF-1R siRNA sequence was 5′-CACCGCGGCTGGAAACTCTTCTACACGAATGTAGAAGAGTTTCCAGCCGC-3′ and the NC siRNA sequence was 5′-CACCGCTCACCGGCTCCAGATTTATCGAAATAAATCTGGAGCCGGTGAGC-3′. Luciferase reporter plasmids, PGL3-IGF-1R-3′UTR wild-type (WT) and PGL3-IGF-1R-3′UTR mutant (Mut), were obtained from GenePharma (Shanghai, China). Cell transfections were performed using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from tissues or cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The expression levels of miR-184 in CRC tissues and cell lines were determined using a TaqMan miRNA assay (Applied Biosystems; Thermo Fisher Scientific, Inc.). For determination of miRNA expression, cDNA was synthesized from total RNA and amplified using a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The cycling conditions for amplification were as follows: 40 cycles of denaturation at 95°C for 15 sec; and annealing/extension at 60°C for 60 sec. To quantify IGF-1R mRNA expression, cDNA was synthesized using the PrimeScript RT-PCR kit (Takara Bio, Inc., Otsu, Japan). IGF-1R expression was assessed using a SYBR-Green PCR kit (Takara Bio, Inc.) on an ABI 7500 Real-Time PCR Detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions for IGF-1R quantification were as follows: 95°C for 10 min; 40 cycles of 95°C for 15 sec and 60°C for 1 min. miR-184 expression was normalized to that of U6 RNA, and GADPH was used as an internal control for IGF-1R expression. The primers were designed as follows: miR-184 forward, 5′-GCATGCCTAAATGTTGACAGCC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse, 5′-ACGCTTCACGAATTTGCGTGTC-3′; IGF-1R forward, 5′-GGAGGCTGAATACCGCAAAGTC-3′ and reverse, 5′-AAAGACGAAGTTGGAGGCGCT-3′; and GAPDH forward, 5′-ATCTGGAGTTTACCGCTGG-3′ and reverse, 5′-TACCGATGTCTGGTAGACGAT-3′. Relative expression levels were calculated using the 2−ΔΔCq method (25).
Cell proliferation assay
Cell proliferation was determined using the Cell Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Cells were seeded into 96-well plates at a density of 3,000 cells/well in 150 µl culture medium. Following incubation at 37°C overnight, cells underwent transfection. Following various incubation times (24, 48, 72 or 96 h), cell proliferation assay was conducted. A 10 µl volume of CCK-8 assay solution was added to each well, and the 96-well plates were incubated at 37°C for a further 2 h. The absorbance at 450 nm was determined using an automatic multiwell spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All experiments were performed in triplicate.
Transwell assay
To evaluate cell migration and invasion abilities, Transwell chambers (Costar; Corning Incorporated, Corning, NY, USA) with 8 µm diameter pores were used. For analysis of cell invasive ability, Matrigel (BD Biosciences, San Jose, CA, USA) was added into the Transwell apparatus. A total of 5×104 transfected cells in 200 µl serum-free culture medium was placed in the upper chamber and 500 µl culture medium containing 20% FBS was added to the lower chamber as a chemoattractant. Following incubation at 37°C for 24 h, Transwell chambers were stained with 0.5% crystal violet and washed with PBS three times. Non-migrating and non-invading cells were carefully wiped from the upper chambers with cotton wool. Values for cell migration and invasion were obtained by counting 5 fields per membrane using a light microscope.
Bioinformatic analysis
Bioinformatic analysis was performed to predict the potential targets of miR-184 using TargetScan (http://www.targetscan.org/) and PicTar (http://pictar.mdcberlin.de/).
Western blot analysis
Transfected cells were harvested and lysed in radioimmunoprecipitation buffer (Thermo Fisher Scientific, Inc.). Total protein concentration was quantified using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.). Equal amounts of protein were separated using SDS-PAGE (10%). Proteins were transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA), followed by incubation at room temperature with 5% non-fat dry milk in TBST for 1 h. The membranes were incubated with primary antibodies at 4°C overnight, including mouse anti-human monoclonal IGF-1R (1:500 dilution; cat. no. sc-81464; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and GADPH (1:500 dilution; cat. no. sc-59540; Santa Cruz Biotechnology, Inc.). On the second day, the membranes were incubated with a corresponding secondary antibody conjugated to horseradish peroxidase (1:1,000 dilution; cat. no. sc-2005; Santa Cruz Biotechnology, Inc.). Each blot was visualized using enhanced chemiluminescence (Pierce; Thermo Fisher Scientific, Inc.), and densitometry analysis was performed using ImageJ 1.49 (National Institutes of Health, Bethesda, MD, USA).
Luciferase reporter assay
HEK293T cells were seeded in a 24-well plates. Cells were co-transfected with miR-184 mimic or NC, and co-transfection with PGL3-IGF-1R-3′UTR WT or PGL3-IGF-1R-3′UTR Mut, using Lipofectamine 2000. At 48 h after transfection, luciferase activity was determined using the Dual-Luciferase Reporter assay system (Promega Corporation, Madison, WI, USA), and normalized to the activity of Renilla luciferase. Each assay was performed three times.
Statistical analysis
All results are expressed as the mean ± standard deviation. Results were compared using SPSS software (version 17.0; SPSS Inc., Chicago, IL, USA). P<0.05 (two-tailed) was considered to indicate a statistically significant difference.
Results
miR-184 is downregulated in CRC tissues and cell lines
To investigate the roles of miR-184 in CRC, the miR-184 expression level was examined in CRC tissues and matched NATs using RT-qPCR. The results indicated that miR-184 was significantly downregulated in CRC tissues compared with matched NATs (P<0.05; Fig. 1A).
Figure 1.
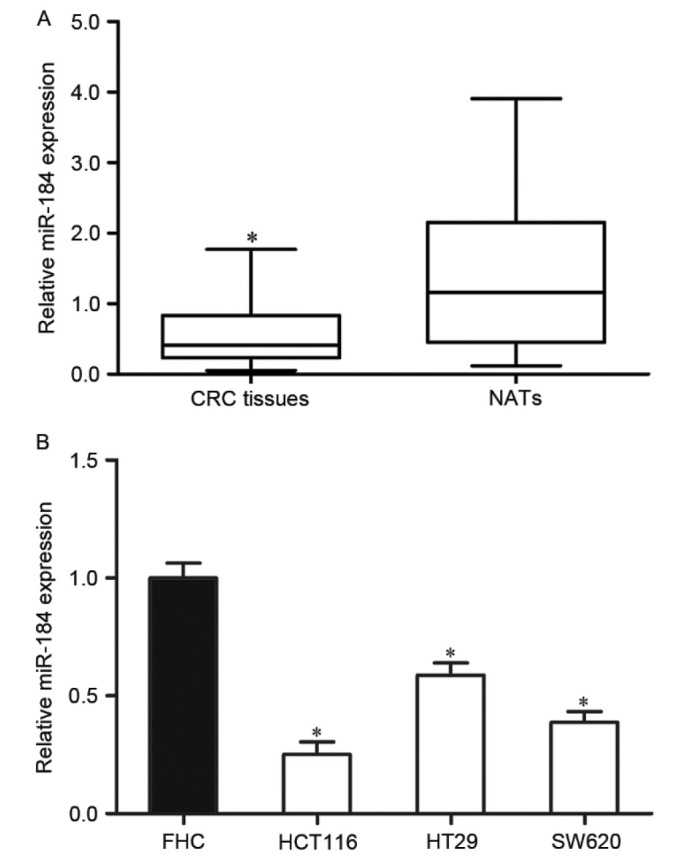
miR-184 is downregulated in CRC tissues and cell lines. (A) Expression of miR-184 was significantly downregulated in CRC tissues. (B) The miR-184 expression level was significantly decreased in the three CRC cell lines compared with in the human normal colon epithelium cell line FHC. *P<0.05 vs. the respective controls. miR-184, microRNA-184; CRC, colorectal cancer; NAT, normal adjacent tissue.
In addition, the expression level of miR-184 in human CRC cell lines was also determined. The results indicated that the miR-184 expression level was significantly decreased in the three CRC cell lines compared with that of the human normal colon epithelium cell line FHC (P<0.05; Fig. 1B). These results indicated that miR-184 may serve an important role in CRC.
miR-184 inhibits CRC cell proliferation, migration and invasion
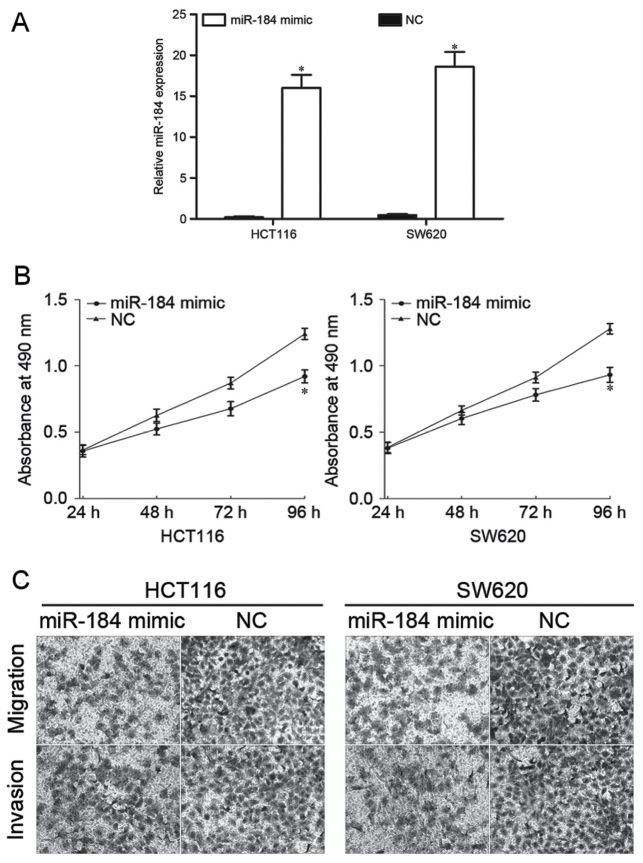
To investigate the effect of miR-184 in CRC proliferation, migration and invasion, miR-184 mimic or NC was transfected into HCT116 and SW620 cells. Following transfection, RT-qPCR was performed to detect miR-184 expression. RT-qPCR analysis demonstrated that miR-184 was significantly upregulated in HCT116 and SW620 cells (Fig. 2A). The effect of overexpression of miR-184 on cell proliferation using an MTT assay was subsequently evaluated. The results indicated that the proliferation of CRC cells was markedly inhibited following transfection with miR-184 mimic compared with cells transfected with NC (Fig. 2B). Furthermore, Transwell assays were used to assess the effect of upregulated miR-184 in CRC cell migration and invasion. The results identified that overexpression of miR-184 significantly decreased the migratory and invasive abilities of HCT116 and SW620 cells (Fig. 2C). These results suggested that miR-184 inhibited the proliferation, migration and invasion of CRC cells in vitro.
Figure 2.
Overexpression of miR-184 inhibits CRC cell proliferation, migration and invasion. (A) The reverse transcription-quantitative polymerase chain reaction was performed to detect the expression level of miR-184 in HCT116 and SW620 cells following transfection with miR-184 mimic or NC, respectively. (B) Cell proliferation was determined using an MTT assay in HCT116 and SW620 cells following transfection with miR-184 mimic or NC. Overexpression of miR-184 was identified top inhibit cell proliferation in HCT116 and SW620 cells. (C) Cell migration and invasion was determined using a Transwell assay in HCT116 and SW620 cells following transfection with miR-184 mimic or NC. Upregulation of miR-184 suppressed CRC cell migration and invasion compared with NC-transfected cells. *P<0.05 vs. NC. miR-184, microRNA-184; CRC, colorectal cancer; NC, negative control.
miR-184 negatively regulates IGF-1R expression
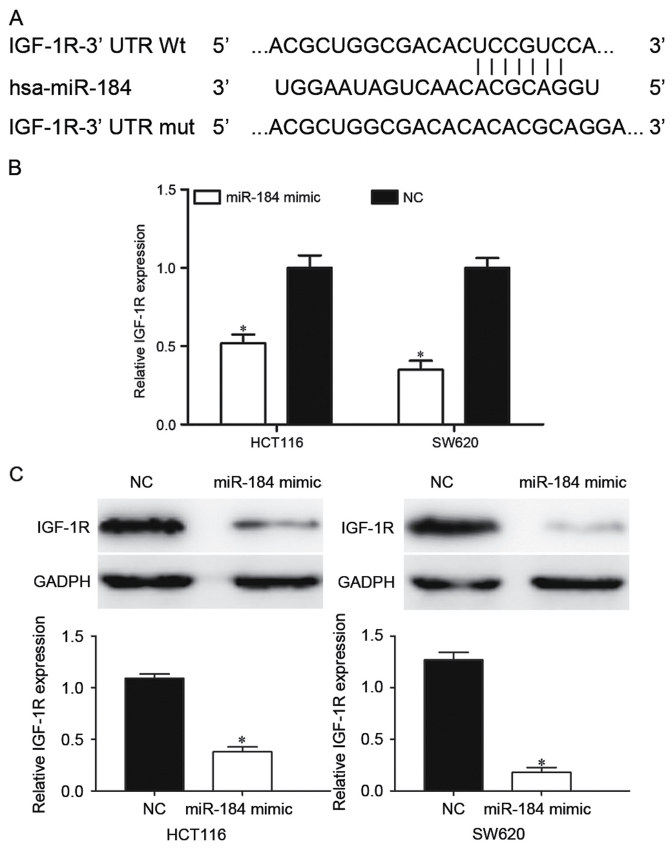
Bioinformatic databases were used to predicted potential targets of miR-184. Among these predicted targets, IGF-1R was a clear candidate since it was predicted by TargetScan and PicTar. As presented in Fig. 3A, IGF-1R contained a miR-184 seed match of the IGF-1R 3′-UTR. To investigate the effect of miR-184 in the regulation of IGF-1R expression, RT-qPCR and western blot analysis were performed. It was identified that the IGF-1R mRNA was significantly downregulated in miR-184 mimic-transfected HCT116 and SW620 cells (P<0.05; Fig. 3B). Western blot analysis results indicated that miR-184 significantly decreased IGF-1R protein expression in HCT116 and SW620 cells compared with the NC groups (P<0.05; Fig. 3C).
Figure 3.
miR-184 suppresses IGF-1R expression in CRC cells. (A) The potential miR-184-binding sequence of IGF-1R-3′UTR and the mutant IGF-1R-3′UTR sequence at the binding site. (B) Determination of IGF-1R mRNA expression levels in HCT116 and SW620 cells using the reverse transcription-quantitative polymerase chain reaction following transfection with the miR-184 mimic or NC. IGF-1R mRNA expression levels were significantly decreased in miR-184 mimic-transfected HCT116 and SW620 cells. (C) Determination of IGF-1R protein expression levels in HCT116 and SW620 cells by western blot analysis following transfection with the miR-184 mimic or NC. GAPDH was used as a loading control. miR-184 significantly inhibited IGF-1R protein expression in HCT116 and SW620 cells. *P<0.05 vs. NC. miR-184, microRNA-184; IGF-1R, insulin-like growth factor 1 receptor; CRC, colorectal cancer; UTR, untranslated region; NC, negative control.
IGF-1R is a direct target of miR-184
To verify whether IGF-1R was a direct target of miR-184 in CRC, PGL3-IGF-1R-3′UTR WT and PGL3-IGF-1R-3′UTR Mut along with miR-184 mimic or NC were transfected into HEK293T cells. Following transfection for 48 h, luciferase reporter assays were performed. The results identified that miR-184 significantly inhibited the luciferase activity of PGL3-IGF-1R-3′UTR WT (P<0.05; Fig. 4). However, the activity of PGL3-IGF-1R-3′UTR Mut was not affected (Fig. 4), which suggested that IGF-1R was a direct target of miR-184.
Figure 4.
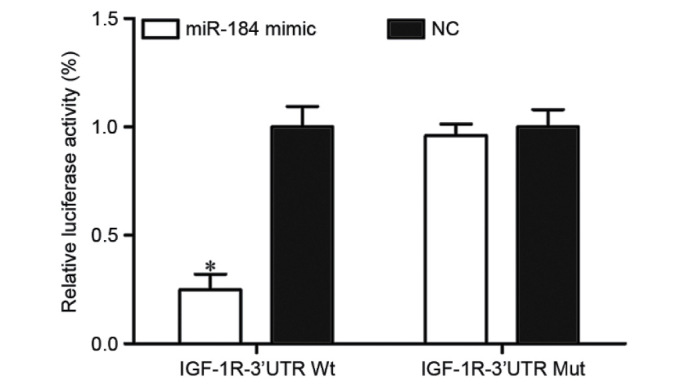
miR-184 directly targets the 3′UTR of IGF-1R. Luciferase assay in HEK293T cells co-transfected with miR-184 or NC and PGL3-IGF-1R-3′UTR WT or PGL3-IGF-1R-3′UTR Mut. miR-184 significantly suppressed the luciferase activity of PGL3-IGF-1R-3′UTR WT. However, the activity of the PGL3-IGF-1R-3′UTR Mut was not affected. *P<0.05 vs. NC. miR-184, miRNA-184; UTR, untranslated region; IGF-1R, insulin-like growth factor 1 receptor; NC, negative control; WT, wild-type; Mut, mutant.
IGF-1R is involved in miR-184-mediated proliferation, migration and invasion in CRC cells
To explore further whether IGF-1R acts as a downstream effector in miR-184-mediated suppressive effects in CRC cells, IGF-1R siRNA or NC siRNA was transfected into HCT116 and SW620 cells. Following transfection, western blot analysis was performed to determine the expression of IGF-1R protein. As presented in Fig. 5A, IGF-1R was significantly downregulated in IGF-1R siRNA-transfected HCT116 and SW620 cells compared with NC siRNA-transfected cells (P<0.05).
Figure 5.
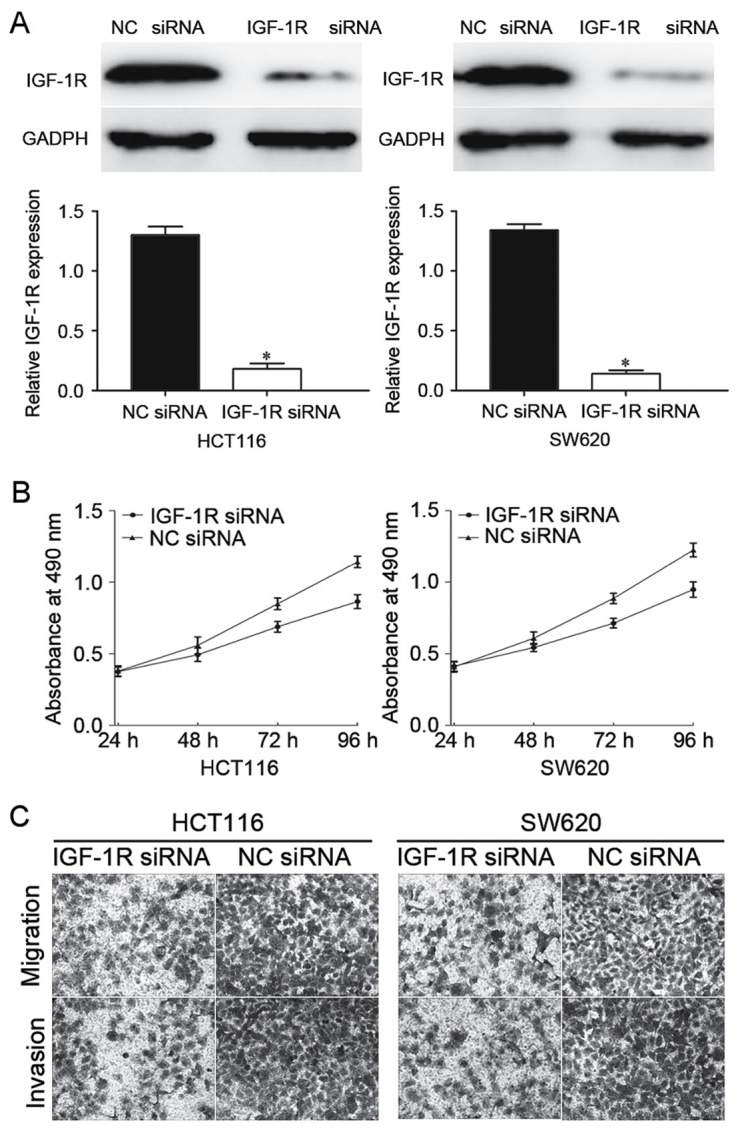
IGF-1R is involved in the miR-184-mediated tumor suppressing effect in CRC cells. (A) Determination of IGF-1R protein expression levels in HCT116 and SW620 cells by western blot analysis following transfection with the IGF-1R siRNA or NC siRNA. GAPDH was used as a loading control. (B) IGF-1R siRNA inhibited HCT116 and SW620 cell proliferation. (C) IGF-1R siRNA suppressed HCT116 and SW620 cell migration and invasion. *P<0.05 vs. NC siRNA. IGF-1R, insulin-like growth factor 1 receptor; miR-184, microRNA-184; CRC, colorectal cancer; siRNA, small interfering RNA; NC, negative control.
Subsequently, MTT assays and Transwell assays were performed. The results identified that knockdown of IGF-1R inhibited CRC cell proliferation, migration and invasion (Fig. 5B and C), indicating that IGF-1R was involved in the miR-184-mediated proliferation, migration and invasion of CRC cells.
Discussion
Previous studies have demonstrated that the expression of miR-184 differs between normal tissues and tumors. It has been identified to be upregulated in hepatocellular carcinoma (26) and squamous cell carcinoma of the tongue (27). However, downregulation of miR-184 has also been identified in various types of human cancer, including renal cell carcinoma (28), nasopharyngeal carcinoma (29), neuroblastoma (30), non-small cell lung cancer (31), glioma (32,33) and breast cancer (32). The contradictory results of these studies indicated that the expression levels of miR-184 in cancer exhibit tissue-specificity. In the present study, it was identified that miR-184 was downregulated in CRC tissues and cell lines. The results suggested that miR-184 may serve tumor suppressive functions in the carcinogenesis and progression of CRC.
miR-184 was demonstrated to be a tumor suppressor in cancer. Lin et al (31) demonstrated that miR-184 was downregulated in non-small cell lung cancer. Furthermore, a low expression level of miR-184 may be a predictor of poor prognosis in patients with non-small cell lung cancer. Upregulation of miR-184 decreased cell proliferation and invasion by targeting cell division cycle 25A and c-Myc (31). In renal cell carcinoma, overexpression of miR-184 suppressed cell proliferation and migration, and enhanced cell apoptosis in vitro (28). Cheng et al (33) demonstrated that miR-184-targeted tumor necrosis factor α-induced protein 2 decreased glioma cell proliferation and migration in vitro, and the xenografted tumor size in vivo (33). Taken together, the results of these studies suggested a possible role for miR-184 in regulating cancer carcinogenesis and progression.
However, miR-184 has also been verified as an oncogene in certain types of cancer. For example, in squamous cell carcinoma of the tongue, miR-184 increased cell proliferation and inhibited apoptosis (27). In addition, surgical removal of the primary squamous cell carcinoma of tongue markedly suppressed miR-184 expression in plasma (34). In hepatocellular carcinoma, overexpression of miR-184 significantly increased cell proliferation, tumorigenicity and cell cycle progression by directly targeting sex-determining region Y box 7 and inositol polyphosphate phosphatase-like 1 (26,35). These results appear contradictory in that miR-184 was demonstrated to be an oncogene in certain types of cancer and a tumor suppressor in others. This contradiction may be explained by the ‘imperfect complementarity’ of the interactions between miRNAs and target genes (36).
In the present study, the functions of miR-184 in CRC cell proliferation, migration and invasion were analyzed by transfecting miR-184 mimic and NC into HCT116 and SW620 CRC cells. It was identified that miR-184 significantly inhibited the proliferation, migration and invasion of CRC cells. These results provided novel insight into the functions of miR-184 in the initiation and development of CRC. Subsequently, the potential underlying molecular mechanism involved in miR-184-mediated suppressive functions of CRC cell proliferation, migration and invasion were sought. In the present study, an important molecular link between miR-184 and IGF-1R was identified in CRC. First, bioinformatic analysis predicted that IGF-1R contained a miR-184 seed match at position 701–708 of the IGF-1R 3′-UTR. Secondly, RT-qPCR and western blot analysis demonstrated that miR-184 decreased IGF-1R expression at the mRNA and protein level. Thirdly, a luciferase reporter assay demonstrated that miR-84 directly targeted IGF-1R 3′-UTR. Finally, IGF-1R siRNA also decreased CRC cell proliferation, migration and invasion. These results indicated that miR-184 targeted IGF-1R to inhibit CRC cell proliferation and metastasis in vitro. Identification of the targets of miR-184 is essential for understanding its role in CRC, and developing novel targeted therapies for CRC.
IGF-1R, a transmembrane tyrosine kinase receptor of the insulin receptor family, contains two extracellular α subunits with the ligand-binding site and two transmembrane β subunits with intracellular tyrosine kinase activity (37). Overexpression of IGF-1R has been demonstrated to serve important functions in CRC, hepatocellular carcinoma, osteosarcoma, non-small cell lung cancer and prostate cancer (38–41). IGF-1R was identified to be involve in a range of biological processes, including cell proliferation, apoptosis, cell cycle regulation and metastasis (42–44).
Furthermore, IGF-1R expression level has been identified to be associated with an aggressive phenotype, tumor progression, drug resistance and poor outcome in several types of tumor, including colorectal, ovary, prostate, endometrial, gastric, bladder, sarcoma, glioblastoma, leukemia, myeloma, gastrointestinal stromal and breast (45). This indicated that inhibition of IGF-1R expression may serve as a novel therapeutic method for the treatment of cancer. A number of agents targeting IGF-1R have been developed or are in development, and certain agents are in clinical trials for cancer treatment (46). In the present study, miR-184 was identified for the first time, to the best of our knowledge, to negatively regulate IGF-1R expression and therefore function as a tumor suppressor in CRC. It may be investigated as a targeted therapy for the treatment of CRC.
The results of the present study indicate that miR-184 is downregulated in CRC. Furthermore, miR-184 inhibited CRC cell proliferation, migration and invasion. Mechanistically, it was verified that miR-184 directly targeted IGF-1R. miR-184 may therefore be a therapeutic target in the treatment of CRC.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Andrews L. Dietary flavonoids for the prevention of colorectal cancer. Clin J Oncol Nurs. 2013;17:671–672. doi: 10.1188/13.CJON.671-672. [DOI] [PubMed] [Google Scholar]
- 4.Altobelli E, Lattanzi A, Paduano R, Varassi G, di Orio F. Colorectal cancer prevention in Europe: Burden of disease and status of screening programs. Prev Med. 2014;62:132–141. doi: 10.1016/j.ypmed.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Sugarbaker PH. Colorectal cancer: Prevention and management of metastatic disease. Biomed Res Int. 2014;2014:782890. doi: 10.1155/2014/782890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan DS, Lau R, Aune D, Vieira R, Greenwood DC, Kampman E, Norat T. Red and processed meat and colorectal cancer incidence: Meta-analysis of prospective studies. PLoS One. 2011;6:e20456. doi: 10.1371/journal.pone.0020456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu M, Xu Y, Ge M, Gui Z, Yan F. Regulation of UHRF1 by microRNA-9 modulates colorectal cancer cell proliferation and apoptosis. Cancer Sci. 2015;106:833–839. doi: 10.1111/cas.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta GP, Massague J. Cancer metastasis: Building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Spano D, Heck C, De Antonellis P, Christofori G, Zollo M. Molecular networks that regulate cancer metastasis. Semin Cancer Biol. 2012;22:234–249. doi: 10.1016/j.semcancer.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Xiong Y, Zhang YY, Wu YY, Wang XD, Wan LH, Li L, Zhou LM. Correlation of over-expressions of miR-21 and Notch-1 in human colorectal cancer with clinical stages. Life Sci. 2014;106:19–24. doi: 10.1016/j.lfs.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 11.Manfredi S, Bouvier AM, Lepage C, Hatem C, Dancourt V, Faivre J. Incidence and patterns of recurrence after resection for cure of colonic cancer in a well defined population. Br J Surg. 2006;93:1115–1122. doi: 10.1002/bjs.5349. [DOI] [PubMed] [Google Scholar]
- 12.Van Cutsem E, Cervantes A, Nordlinger B, Arnold D, ESMO Guidelines Working Group Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):iii1–9. doi: 10.1093/annonc/mdu260. [DOI] [PubMed] [Google Scholar]
- 13.Yuan W, Sui C, Liu Q, Tang W, An H, Ma J. Up-regulation of microRNA-145 associates with lymph node metastasis in colorectal cancer. PLoS One. 2014;9:e102017. doi: 10.1371/journal.pone.0102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Chen L, Xu Y, Li R, Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400:236–240. doi: 10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 15.Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, Ren C, et al. Identification of microRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology. 2014;60:598–609. doi: 10.1002/hep.27118. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.O'Hara SP, Mott JL, Splinter PL, Gores GJ, LaRusso NF. MicroRNAs: key modulators of posttranscriptional gene expression. Gastroenterology. 2009;136:17–25. doi: 10.1053/j.gastro.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croce CM, Calin GA. miRNAs, cancer and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 19.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 20.Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA-novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013;340:201–211. doi: 10.1016/j.canlet.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Zhang X, Dhakal IB, Beggs M, Kadlubar S, Luo D. Induction of cell proliferation and survival genes by estradiol-repressed microRNAs in breast cancer cells. BMC Cancer. 2012;12:29. doi: 10.1186/1471-2407-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanokura M, Banno K, Kobayashi Y, Kisu I, Ueki A, Ono A, Masuda K, Nomura H, Hirasawa A, Susumu N, Aoki D. MicroRNA and endometrial cancer: Roles of small RNAs in human tumors and clinical applications (Review) Oncol Lett. 2010;1:935–940. doi: 10.3892/ol.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ventura A, Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang T, Thakur A, Chen T, Yang L, Lei G, Liang Y, Zhang S, Ren H, Chen M. MicroRNA-15a induces cell apoptosis and inhibits metastasis by targeting BCL2L2 in non-small cell lung cancer. Tumour Biol. 2015;36:4357–4365. doi: 10.1007/s13277-015-3075-1. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Gao B, Gao K, Li L, Huang Z, Lin L. miR-184 functions as an oncogenic regulator in hepatocellular carcinoma (HCC) Biomed Pharmacother. 2014;68:143–148. doi: 10.1016/j.biopha.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 28.Su Z, Chen D, Li Y, Zhang E, Yu Z, Chen T, Jiang Z, Ni L, Yang S, Gui Y, et al. microRNA-184 functions as tumor suppressor in renal cell carcinoma. Exp Ther Med. 2015;9:961–966. doi: 10.3892/etm.2015.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhen Y, Liu Z, Yang H, Yu X, Wu Q, Hua S, Long X, Jiang Q, Song Y, Cheng C, et al. Tumor suppressor PDCD4 modulates miR-184-mediated direct suppression of C-MYC and BCL2 blocking cell growth and survival in nasopharyngeal carcinoma. Cell Death Dis. 2013;4:e872. doi: 10.1038/cddis.2013.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tivnan A, Foley NH, Tracey L, Davidoff AM, Stallings RL. MicroRNA-184-mediated inhibition of tumour growth in an orthotopic murine model of neuroblastoma. Anticancer Res. 2010;30:4391–4395. [PMC free article] [PubMed] [Google Scholar]
- 31.Lin TC, Lin PL, Cheng YW, Wu TC, Chou MC, Chen CY, Lee H. MicroRNA-184 deregulated by the microRNA-21 promotes tumor malignancy and poor outcomes in non-small cell lung cancer via targeting CDC25A and c-Myc. Ann Surg Oncol. 2015;22(Suppl 3):S1532–S1539. doi: 10.1245/s10434-015-4595-z. [DOI] [PubMed] [Google Scholar]
- 32.Feng R, Dong L. Inhibitory effect of miR-184 on the potential of proliferation and invasion in human glioma and breast cancer cells in vitro. Int J Clin Exp Pathol. 2015;8:9376–9382. [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Z, Wang HZ, Li X, Wu Z, Han Y, Li Y, Chen G, Xie X, Huang Y, Du Z, et al. MicroRNA-184 inhibits cell proliferation and invasion and specifically targets TNFAIP2 in Glioma. J Exp Clin Cancer Res. 2015;34:27. doi: 10.1186/s13046-015-0142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong TS, Ho WK, Chan JY, Ng RW, Wei WI. Mature miR-184 and squamous cell carcinoma of the tongue. Scientific World Journal. 2009;9:130–132. doi: 10.1100/tsw.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu GG, Li WH, He WG, Jiang N, Zhang GX, Chen W, Yang HF, Liu QL, Huang YN, Zhang L, et al. Mir-184 post-transcriptionally regulates SOX7 expression and promotes cell proliferation in human hepatocellular carcinoma. PLoS One. 2014;9:e88796. doi: 10.1371/journal.pone.0088796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Z, Ni L, Chen D, Zhang Q, Su Z, Wang Y, Yu W, Wu X, Ye J, Yang S, et al. Identification of miR-7 as an oncogene in renal cell carcinoma. J Mol Histol. 2013;44:669–677. doi: 10.1007/s10735-013-9516-5. [DOI] [PubMed] [Google Scholar]
- 37.Hu Q, Gong JP, Li J, Zhong SL, Chen WX, Zhang JY, Ma TF, Ji H, Lv MM, Zhao JH, Tang JH. Down-regulation of miRNA-452 is associated with adriamycin-resistance in breast cancer cells. Asian Pac J Cancer Prev. 2014;15:5137–5142. doi: 10.7314/APJCP.2014.15.13.5137. [DOI] [PubMed] [Google Scholar]
- 38.Wang YH, Wang ZX, Qiu Y, Xiong J, Chen YX, Miao DS, De W. Lentivirus-mediated RNAi knockdown of insulin-like growth factor-1 receptor inhibits growth, reduces invasion and enhances radiosensitivity in human osteosarcoma cells. Mol Cell Biochem. 2009;327:257–266. doi: 10.1007/s11010-009-0064-y. [DOI] [PubMed] [Google Scholar]
- 39.Wang YH, Han XD, Qiu Y, Xiong J, Yu Y, Wang B, Zhu ZZ, Qian BP, Chen YX, Wang SF, et al. Increased expression of insulin-like growth factor-1 receptor is correlated with tumor metastasis and prognosis in patients with osteosarcoma. J Surg Oncol. 2012;105:235–243. doi: 10.1002/jso.22077. [DOI] [PubMed] [Google Scholar]
- 40.Scharf JG, Braulke T. The role of the IGF axis in hepatocarcinogenesis. Horm Metab Res. 2003;35:685–693. doi: 10.1055/s-2004-814151. [DOI] [PubMed] [Google Scholar]
- 41.Shiratsuchi I, Akagi Y, Kawahara A, Kinugasa T, Romeo K, Yoshida T, Ryu Y, Gotanda Y, Kage M, Shirouzu K. Expression of IGF-1 and IGF-1R and their relation to clinicopathological factors in colorectal cancer. Anticancer Res. 2011;31:2541–2545. [PubMed] [Google Scholar]
- 42.Werner H, LeRoith D. The role of the insulin-like growth factor system in human cancer. Adv Cancer Res. 1996;68:183–223. doi: 10.1016/S0065-230X(08)60354-1. [DOI] [PubMed] [Google Scholar]
- 43.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: An update. Nat Rev Cancer. 2012;12:159–169. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 44.King H, Aleksic T, Haluska P, Macaulay VM. Can we unlock the potential of IGF-1R inhibition in cancer therapy? Cancer Treat Rev. 2014;40:1096–1105. doi: 10.1016/j.ctrv.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hewish M, Chau I, Cunningham D. Insulin-like growth factor 1 receptor targeted therapeutics: Novel compounds and novel treatment strategies for cancer medicine. Recent Pat Anticancer Drug Discov. 2009;4:54–72. doi: 10.2174/157489209787002515. [DOI] [PubMed] [Google Scholar]
- 46.Wu J, Zhu AX. Targeting insulin-like growth factor axis in hepatocellular carcinoma. J Hematol Oncol. 2011;4:30. doi: 10.1186/1756-8722-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]