Abstract
Liver cancer primarily accounts for the majority of malignancies of the liver. MicroRNAs (miRNAs) are endogenous non-coding RNAs, which are important in tumorigenesis. Abnormal expression of microRNA-346 (miR-346) has been demonstrated in various types of human cancer, however, its expression and potential molecular mechanism in liver cancer remains to be elucidated. Expression levels of miR-346 in liver cancer cell lines were determined by quantitative polymerase chain reaction. The effect of miR-346 on proliferation was evaluated by an MTT assay; cell migration and invasion were evaluated by Transwell migration and invasion assays and target protein expression was determined by western blotting. The present study observed that miR-346 was upregulated in liver cancer cell lines. miR-346 overexpression promoted cell proliferation, migration and invasion in liver cancer cells and conversely, inhibition of miR-346 resulted in the opposite effects. Furthermore, F-Box and leucine rich repeat protein (FBXL)2 was identified as a direct target of miR-346. miR-346 promoted proliferation, migration and invasion of liver cancer via FBXL2. Overall, these findings demonstrated that miR-346 may act as a potential prognostic marker and therapeutic target against liver cancer in the future.
Keywords: miRNA-346, proliferation, migration, invasion, hepatocellular carcinoma
Introduction
Liver cancer is responsible for primary malignancy of the liver (1–3) and is currently the third pre-eminent cause of cancer deaths around the world, with over 500,000 people affected by the disease (4,5). Liver cancer cases mostly occur in Africa and Asia, where the high number of hepatitis B and hepatitis C cases strongly predisposes to the development of liver disease and the subsequent development of liver cancer (6,7). Consequently, comprehensive elucidation of the mechanisms governing recurrence of liver cancer and metastasis are urgently needed.
MicroRNAs (miRNAs) are endogenous non-coding RNAs, which play vital roles in tumor development and tumorigenesis (8–10). In some human cancers, miRNAs are often unregulated and have an oncogenic function, while most miRNAs are downregulated and may possess a tumor-suppressive activity (11–13). Emerging evidence suggests that the abnormal expression of miRNAs is involved in the invasion and metastasis, during the progression of various human cancers (14,15). miR-346 has been reported as an oncomir in various human cancers, including cutaneous squamous cell carcinoma (16), cervical cancer (17), prostate cancer (18), nasopharyngeal carcinoma (19), lung cancer (20) and breast cancer (21).
The present study aimed to investigate the expression and the molecular regulatory mechanism of miR-346 in liver cancer. We demonstrated that miR-346 is upregulated in liver cancer cell lines. Furthermore, ectopic expression or inhibition of miR-346 could accelerate or block proliferation, migration, and invasion abilities of liver cancer cells, respectively. Furthermore, we further identified FBXL2 as a functional target of miR-346 and demonstrated FBXL2 involve in the effects of increased miR-346 on promoting proliferation, migration, and invasion. Our findings, for the first time, suggest a fundamental role for miR-346 in liver cancer.
Materials and methods
Cell lines and cell culture
Human liver cancer cell lines MHCC-97H, SMMC-7221 (22), HepG2, Huh-7, and Hep3B, the normal liver cell lines THLE-2 and THLE-3 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cell lines were cultivated in Dulbecco's modified Eagle's medium (Gibco, USA) supplied with 10% fetal bovine serum (FBS, Gibco, USA) at 37°C in a humidified atmosphere containing 5% CO2.
RNA extraction, reverse transcription and real-time PCR
Total RNA from cultured cells was extracted using the Trizol reagent (Takara, Japan) according to the manufacturer's instructions. TaqMan microRNA assays (Applied Biosystems, Foster City, CA, USA) were used to determine the expression levels of miR-346 after reverse transcribing by sequence-specific primers (Applied Biosystems), and U6 small nuclear RNA was used as an internal control. Three independent experiments were performed to analyze the relative gene expression.
Western blot analysis
Western blot was performed according to the method described previously (23). The following antibodies were used for analysis: anti-FBXL2 (1:1,000, ab17018, Abcam, Cambridge, MA, USA); GAPDH (1:5,000, HRP-60004, Proteintech Group, Chicago, IL, USA) was served as the loading control.
MiR-346 mimics and inhibitors
MiRNA-346 mimics, miR-346 inhibitors, small interference RNA for FBXL2, and their negative control RNA and their negative control RNA oligonucleotides were purchased from GenePharma (Shanghai, China). For overexpression experiments, the human FBXL2 cDNA were cloned into the pcDNA3.1 vector (Clonetech). The liver cancer cell line cells were transfected using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instruction. Scramble siRNA were used as the control.
Cell proliferation assay
1×104 cells per well were incubated in 96-well culture plates in 100 µl of medium. CCK-8 (Dojindo Laboratories, Japan) was used according to the manufacturer's instructions. Plates were incubated at 37°C for 2 h, and the absorbance at 450 nm was then measured. Proliferation rates were determined at days 1, 2, 3, 4 and 5 post-transfection.
Migration and invasion assay
Cells were seeded into Transwell chambers (Corning, Corning, NY, USA) in the upper chambers in medium containing 10% FBS. 16 h later, the filters were stained with crystal violet. For invasion assay, chambers were coated with Matrigel (BD Biosciences, San Jose, CA, USA). Cell migration and invasion were assessed by counting the number of cells that had penetrated through the filter. These experiments were repeated three times.
Statistical analysis
All statistical analysis were performed using SPSS17.0 (SPSS Statistics, Chicago, IL, USA) software. Data were expressed as mean ± SD and analyzed using the Student's t-test. Paired t-test was used for paired samples. P<0.05 was considered to indicate a statistically significant difference.
Results
MiR-346 is highly expressed in human liver cancer cell lines
To determine the role of miR-346 in liver cancer, real-Time PCR was performed to show that miR-346 was overexpressed in five liver cancer cell lines, including MHCC-97H, SMMC-7221, HepG2, Huh-7, and Hep3B, compared with the normal liver cell lines THLE-2 and THLE-3 (Fig. 1). This result demonstrated that, compared with normal human liver cells, miR-346 was significantly upregulated in human liver cancer cell lines.
Figure 1.
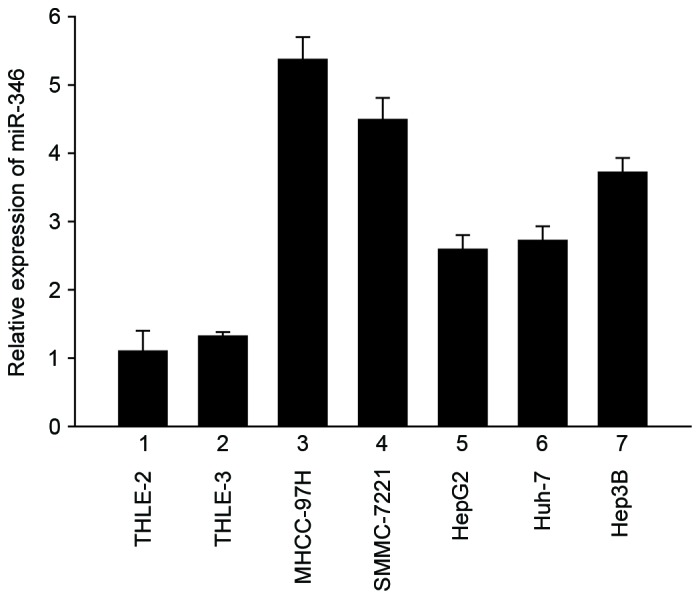
MiR-346 was significantly upregulated in human liver cancer cell lines. Expression of miR-346 in five liver cancer cell lines (3–7) compared with two normal liver cell lines (1–2). Data are presented as mean ± SD from three independent experiments.
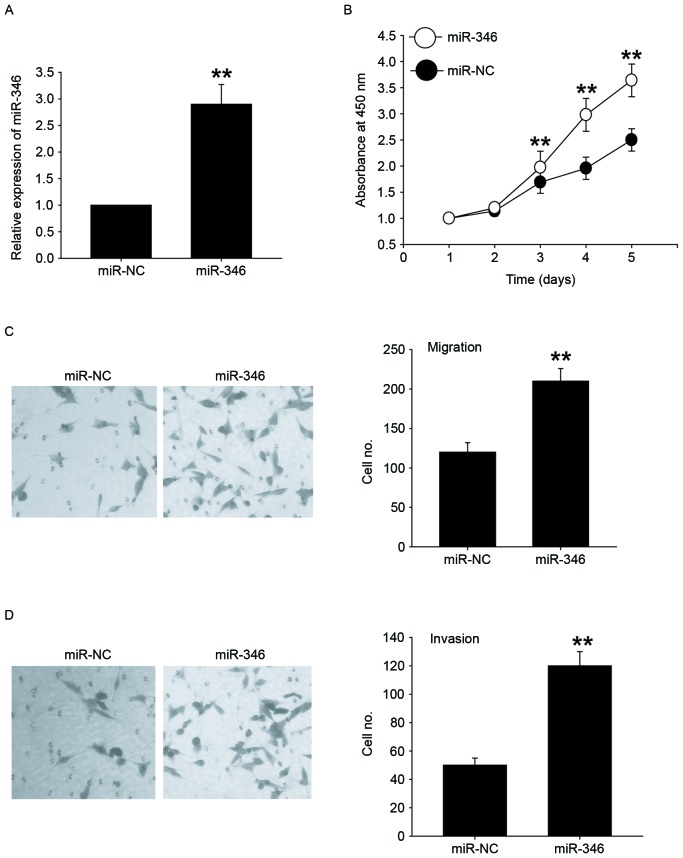
Overexpression of miR-346 promotes the proliferation, migration, and invasion of liver cancer cells
We transfected miR-346 mimics into HepG2 cells with lower expression of miR-346 to investigate the role of miR-346 (Fig. 2A). CCK-8 analysis showed that overexpression of miR-346 significantly promoted the cell proliferation (Fig. 2B). Next, we explored the potential roles of miR-346 in migration and invasion of HepG2 cells by using Transwell migration and invasion assays. miR-346 overexpression significantly increased cell migration (Fig. 2C) and invasion (Fig. 2D) in HepG2 cell line. Together, these results indicated that miR-346 overexpression promoted cell proliferation, migration, and invasion.
Figure 2.
Overexpression of miR-346 promotes the proliferation, migration, and invasion of liver cancer cells. (A) Confirmation of miR-346 overexpression by qRT-PCR in HepG2 cells transfected with miR-346 mimics. (B) CCK-8 assay was performed to evaluate the effects of miR-346 mimics on cell proliferation in HepG2 cells. (C) Transwell-migration assay was performed in HepG2 cells transfected with miR-346 mimics or miR-NC. (D) Transwell-invasion assay was performed in HepG2 cells transfected with miR-346 mimics or miR-NC. All data are shown as mean ± SD from three independent experiments. **P<0.01 vs. NC.
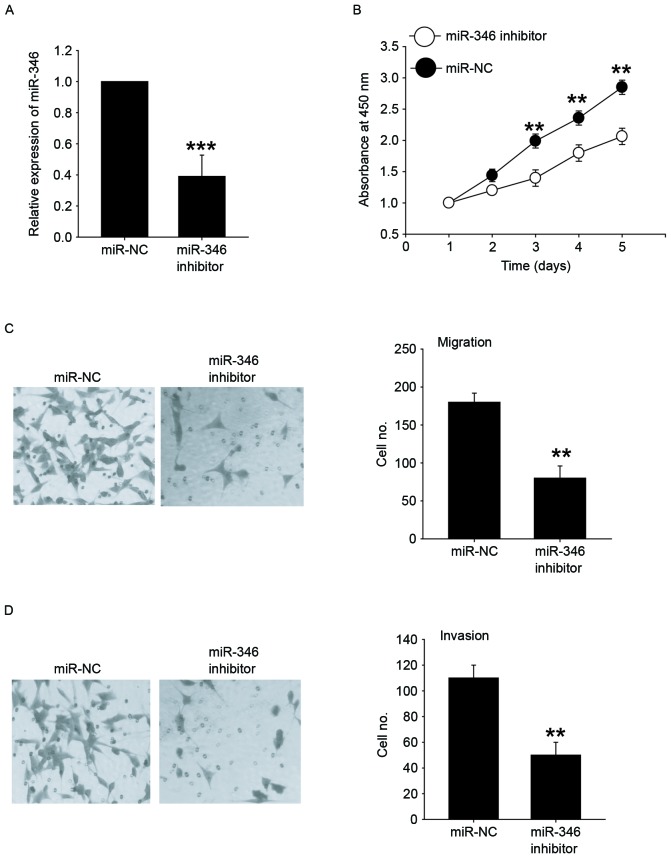
Downregulation of miR-346 inhibits the proliferation, migration, and invasion of liver cancer cells
Next, MHCC-97H cells with higher expression of miR-346 were successfully transfected with the miR-346 inhibitors (Fig. 3A). The CCK-8 assay demonstrated that inhibition of miR-346 significantly reduced the cellular growth of MHCC-97H cells (Fig. 3B). Furthermore, MHCC-97H cells were transfected with miR-346 inhibitors and then analyzed for their metastatic potential using Transwell assays. Our results showed that the migration and invasion abilities in the miR-346 downexpression group was significantly reduced when compared with the control group (Fig. 3C and D). These results demonstrated that downregulation of miR-346 could inhibit the proliferative and metastatic potential of liver cancer cells.
Figure 3.
Downregulation of miR-346 inhibits the proliferation, migration, and invasion of liver cancer cells. (A) Confirmation of miR-346 inhibition by qRT-PCR in MHCC-97H cells transfected with miR-346 inhibitors. (B) CCK-8 assay was performed to evaluate the effects of miR-346 inhibitors on cell proliferation in MHCC-97H cells. (C) Transwell-migration assay was performed in MHCC-97H cells transfected with miR-346 inhibitors or miR-NC. (D) Transwell-invasion assay was performed in MHCC-97H cells transfected with miR-346 inhibitors or miR-NC. All data are shown as mean ± SD from three independent experiments. **P<0.01; ***P<0.001 vs. NC.
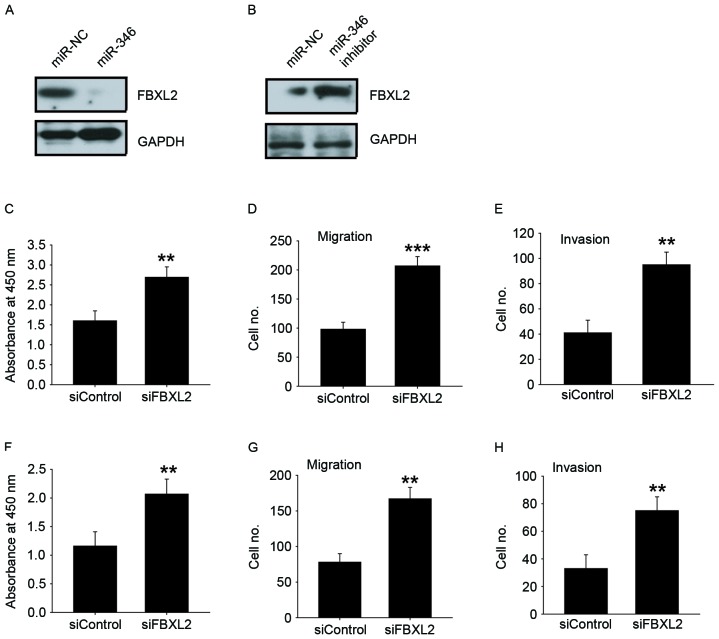
FBXL2 was a direct and functional target of miR-346 in liver cancer cells
FBXL2 was the theoretical target gene of miR-346, determined by analysis using publicly available algorithm Target Scan. Western blot analysis indicated that overexpression of miR-346 inhibited FBXL2 expression in HepG2 cells with lower expression of miR-346 (Fig. 4A); in contrast, knockdown of miR-346 increased FBXL2 expression in MHCC-97H cells with higher expression of miR-346 (Fig. 4B). We found that FBXL2 silencing could promote cell proliferation, migration, and invasion in HepG2 (Fig. 4C-E) and in Huh-7 cells (Fig. 4F-H). These results showed that FBXL2 has an opposite effect on cell proliferation, migration, and invasion, compared to miR-346, suggesting that it may be a functional target of miR-346 in liver cancer cells.
Figure 4.
FBXL2 was a direct and functional target of miR-346 in liver cancer cells. (A) Western blot analysis of FBXL2 expression in miR-346-transfected HepG2 cells. (B) Western blot analysis of FBXL2 expression in miR-346 inhibitors-transfected MHCC-97H cells. CCK-8 assay C), Transwell-migration assay (D), Transwell-invasion assay (E) were performed to evaluate the effects of FBXL2 siRNA on cell proliferation, migration, and invasion in HepG2 cells. CCK-8 assay (F), Transwell-migration assay (G), Transwell-invasion assay (H) were performed to evaluate the effects of FBXL2 siRNA on cell proliferation, migration, and invasion in Huh-7 cells. All data are shown as mean ± SD from three independent experiments. **P<0.01; ***P<0.001 vs. siControl.
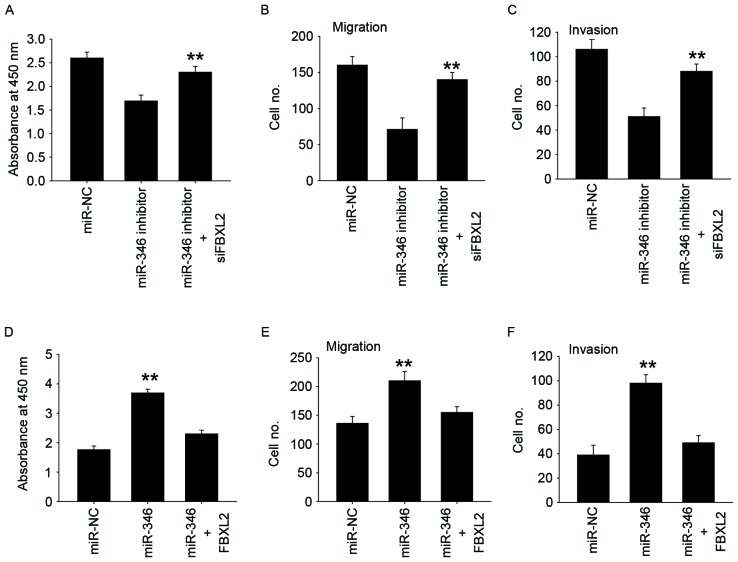
miR-346 promotes the proliferation, migration, and invasion of liver cancer cells via FBXL2
To verify whether miR-346 promotes proliferation and metastasis by targeting FBXL2, gain-and-loss assays were performed. We found that simultaneous inhibition of FBXL2 expression in MHCC-97H cells with miR-346 inhibition can restore their proliferation and metastatic capacity (Fig. 5A-C), while simultaneous overexpression of both FBXL2 expression in HepG2 cells with miR-346 overexpression eliminated the role of miR-346 in proliferation and metastasis (Fig. 5D-F). These results indicate that miR-346 may promote proliferation, migration, and invasion via FBXL2.
Figure 5.
miR-346 promotes the proliferation, migration, and invasion of liver cancer cells via FBXL2. CCK-8 assay (A), Transwell-migration assay (B), Transwell-invasion assay (C) were performed to evaluate the reversal by FBXL2 siRNA of miR-346 inhibitors induced inhibition of cell proliferation, migration, and invasion in MHCC-97H cells. CCK-8 assay (D), Transwell-migration assay (E), Transwell-invasion assay (F) were performed to evaluate the reversal by FBXL2 overexpression of miR-346 mimics induced increase of cell proliferation, migration, and invasion in HepG2 cells. All data are shown as mean ± SD from three independent experiments. **P<0.01.
Discussion
The molecular mechanisms underlying liver cancer development, however, remain largely unknown (24,25). Increasing data findings have indicated miRNAs to be critical regulators of cancer-related processes, but it is still unknown the molecular mechanisms by which miRNAs modulate the behavior of cancer cells (26,27). miR-346 has been reported as an oncomir because it facilitates cell growth and metastasis in various human cancers (16–21). This is the first study that indicates that ectopically expressed miR-346 can promote migration, proliferation, and invasion of liver cancer cells.
In the present study, we found that miR-346 is upregulated in liver cancer cells; this suggests that it might contribute to the development and ultimately progression of liver disease. It was also found that overexpression of miR-346 promoted, migration, proliferation, and invasion of the cells, whereas the knockdown of miR-346 inhibited cell migration, proliferation, and invasion. miR-346 promotes the proliferation, migration, and invasion of liver cancer cells via FBXL2 which is a tumor suppressor and is also a direct target of miR-346.
FBXL2, which is an SCF (Skp1-Cullin-F-box) E3 ligase component, was initially found as maintenance of cellular homeostasis (28). Then, Chen et al suggested that FBXL2 impaired cell proliferation by mediating cyclin D3 polyubiquitination and degradation (29). Accumulating evidence has demonstrated that FBXL2 plays the tumor suppressor role in different kinds of cancers. For example, FBXL2 exerted human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest (30). It also inhibited leukemic cell proliferation by targeting cyclin D2 (31). FBXL2 also decreased gastric cancer proliferation by promoting degradation of forkhead box M1 (32).
The present study, therefore, demonstrates that miR-346 is markedly upregulated in liver cancer cells. Also, FBXL2 is a direct target gene of miR-346 and overexpression of miR-346 reduced the expression of FBXL2 and promoted the migration, proliferation, and invasion of liver cancer cells, whereas the downregulation of miR-346 had the opposite effect. Further investigation is required to fully characterize the biological function of miR-346 and its clinical relevance in the development of liver cancer. Although the particular mechanisms are not yet fully understood, the present study suggests that miR-346 may play a significant role in regulating the migration, proliferation, and invasion of liver cancer cells, and indicates that miR-346 may represent a potential therapeutic target for liver cancer.
Acknowledgments
The present study was supported by the Department of Hepatobiliary Surgery, Chinese PLA General Hospital.
References
- 1.Inokawa Y, Inaoka K, Sonohara F, Hayashi M, Kanda M, Nomoto S. Molecular alterations in the carcinogenesis and progression of hepatocellular carcinoma: Tumor factors and background liver factors. Oncol Lett. 2016;12:3662–3668. doi: 10.3892/ol.2016.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernández-Rodríguez CM, Gutiérrez-García ML. Prevention of hepatocellular carcinoma in patients with chronic hepatitis B. World J Gastrointest Pharmacol Ther. 2014;5:175–182. doi: 10.4292/wjgpt.v5.i3.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamed O, Kimchi ET, Sehmbey M, Gusani NJ, Kaifi JT, Staveley-O'Carroll K. Impact of genetic targets on cancer therapy: Hepatocellular cancer. Adv Exp Med Biol. 2013;779:67–90. doi: 10.1007/978-1-4614-6176-0_4. [DOI] [PubMed] [Google Scholar]
- 4.Niu ZS, Niu XJ, Wang WH. Genetic alterations in hepatocellular carcinoma: An update. World J Gastroenterol. 2016;22:9069–9095. doi: 10.3748/wjg.v22.i41.9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baffy G. Hepatocellular carcinoma in non-alcoholic fatty liver disease: Epidemiology, pathogenesis and prevention. J Clin Transl Hepatol. 2013;1:131–137. doi: 10.14218/JCTH.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Martel C, Maucort-Boulch D, Plummer M, Franceschi S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology. 2015;62:1190–1200. doi: 10.1002/hep.27969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakayama H, Takayama T. Management before hepatectomy for hepatocellular carcinoma with cirrhosis. World J Hepatol. 2015;7:2292–2302. doi: 10.4254/wjh.v7.i20.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahdian-Shakib A, Dorostkar R, Tat M, Hashemzadeh MS, Saidi N. Differential role of microRNAs in prognosis, diagnosis and therapy of ovarian cancer. Biomed Pharmacother. 2016;84:592–600. doi: 10.1016/j.biopha.2016.09.087. [DOI] [PubMed] [Google Scholar]
- 9.Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20:10432–10439. doi: 10.3748/wjg.v20.i30.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krauskopf J, Verheijen M, Kleinjans JC, de Kok TM, Caiment F. Development and regulatory application of microRNA biomarkers. Biomark Med. 2015;9:1137–1151. doi: 10.2217/bmm.15.50. [DOI] [PubMed] [Google Scholar]
- 11.Hsu KW, Fang WL, Huang KH, Huang TT, Lee HC, Hsieh RH, Chi CW, Yeh TS. Notch1 pathway-mediated microRNA-151-5p promotes gastric cancer progression. Oncotarget. 2016;7:38036–38051. doi: 10.18632/oncotarget.9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi RU, Miyazaki H, Takeshita F, Yamamoto Y, Minoura K, Ono M, Kodaira M, Tamura K, Mori M, Ochiya T. Loss of microRNA-27b contributes to breast cancer stem cell generation by activating ENPP1. Nat Commun. 2015;6:7318. doi: 10.1038/ncomms8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Li N, Dong Y, Li S, Xu L, Li X, Li Y, Li Z, Ng SS, Sung JJ, et al. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene. 2015;34:4142–4152. doi: 10.1038/onc.2014.348. [DOI] [PubMed] [Google Scholar]
- 14.O'Bryan S, Dong S, Mathis JM, Alahari SK. The roles of oncogenic miRNAs and their therapeutic importance in breast cancer. Eur J Cancer. 2017;72:1–11. doi: 10.1016/j.ejca.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Jiang C, Chen X, Alattar M, Wei J, Liu H. MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis of gastric cancer. Cancer Gene Ther. 2015;22:291–301. doi: 10.1038/cgt.2015.19. [DOI] [PubMed] [Google Scholar]
- 16.Chen B, Pan W, Lin X, Hu Z, Jin Y, Chen H, Ma G, Qiu Y, Chang L, Hua C, et al. MicroRNA-346 functions as an oncogene in cutaneous squamous cell carcinoma. Tumour Biol. 2016;37:2765–2771. doi: 10.1007/s13277-015-4046-2. [DOI] [PubMed] [Google Scholar]
- 17.Guo J, Lv J, Liu M, Tang H. miR-346 Up-regulates Argonaute 2 (AGO2) protein expression to augment the activity of other MicroRNAs (miRNAs) and contributes to cervical cancer cell malignancy. J Biol Chem. 2015;290:30342–30350. doi: 10.1074/jbc.M115.691857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Wang S, Zhang W, Qiu J, Shan Y, Yang D, Shen B. Screening key microRNAs for castration-resistant prostate cancer based on miRNA/mRNA functional synergistic network. Oncotarget. 2015;6:43819–43830. doi: 10.18632/oncotarget.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan HL, Li L, Li SJ, Zhang HS, Xu W. miR-346 promotes migration and invasion of nasopharyngeal carcinoma cells via targeting BRMS1. J Biochem Mol Toxicol. 2016;30:602–607. doi: 10.1002/jbt.21827. [DOI] [PubMed] [Google Scholar]
- 20.Sun CC, Li SJ, Yuan ZP, Li DJ. MicroRNA-346 facilitates cell growth and metastasis, and suppresses cell apoptosis in human non-small cell lung cancer by regulation of XPC/ERK/Snail/E-cadherin pathway. Aging (Albany NY) 2016;8:2509–2524. doi: 10.18632/aging.101080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Yang F, Luo LJ, Zhang L, Wang DD, Yang SJ, Ding L, Li J, Chen D, Ma R, Wu JZ, Tang JH. MiR-346 promotes the biological function of breast cancer cells by targeting SRCIN1 and reduces chemosensitivity to docetaxel. Gene. 2017;600:21–28. doi: 10.1016/j.gene.2016.11.037. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, Cao Y, Zhang J, Lei M, Deng X, Zahid KR, Liu Y, Liu K, Yang J, Xiong G, et al. Determination of glutathione in apoptotic SMMC-7221 cells induced by xylitol selenite using capillary electrophoresis. Biotechnol Lett. 2016;38:761–766. doi: 10.1007/s10529-016-2056-9. [DOI] [PubMed] [Google Scholar]
- 23.Cheng CW, Wang HW, Chang CW, Chu HW, Chen CY, Yu JC, Chao JI, Liu HF, Ding SL, Shen CY. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res Treat. 2012;134:1081–1093. doi: 10.1007/s10549-012-2034-4. [DOI] [PubMed] [Google Scholar]
- 24.Brito AF, Abrantes AM, Tralhão JG, Botelho MF. Targeting Hepatocellular Carcinoma: What did we Discover so Far? Oncol Rev. 2016;10:302. doi: 10.4081/oncol.2016.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tornesello ML, Buonaguro L, Izzo F, Buonaguro FM. Molecular alterations in hepatocellular carcinoma associated with hepatitis B and hepatitis C infections. Oncotarget. 2016;7:25087–25102. doi: 10.18632/oncotarget.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie T, Huang M, Wang Y, Wang L, Chen C, Chu X. MicroRNAs as regulators, biomarkers and therapeutic targets in the drug resistance of colorectal cancer. Cell Physiol Biochem. 2016;40:62–76. doi: 10.1159/000452525. [DOI] [PubMed] [Google Scholar]
- 27.Mohammadi A, Mansoori B, Baradaran B. The role of microRNAs in colorectal cancer. Biomed Pharmacother. 2016;84:705–713. doi: 10.1016/j.biopha.2016.09.099. [DOI] [PubMed] [Google Scholar]
- 28.Chen BB, Coon TA, Glasser JR, Mallampalli RK. Calmodulin antagonizes a calcium-activated SCF ubiquitin E3 ligase subunit, FBXL2, to regulate surfactant homeostasis. Mol Cell Biol. 2011;31:1905–1920. doi: 10.1128/MCB.00723-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen BB, Glasser JR, Coon TA, Mallampalli RK. FBXL2 is a ubiquitin E3 ligase subunit that triggers mitotic arrest. Cell Cycle. 2011;10:3487–3494. doi: 10.4161/cc.10.20.17742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen BB, Glasser JR, Coon TA, Mallampalli RK. F-box protein FBXL2 exerts human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest. Oncogene. 2012;31:2566–2579. doi: 10.1038/onc.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen BB, Glasser JR, Coon TA, Zou C, Miller HL, Fenton M, McDyer JF, Boyiadzis M, Mallampalli RK. F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. Blood. 2012;119:3132–3141. doi: 10.1182/blood-2011-06-358911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li LQ, Pan D, Chen H, Zhang L, Xie WJ. F-box protein FBXL2 inhibits gastric cancer proliferation by ubiquitin-mediated degradation of forkhead box M1. FEBS Lett. 2016;590:445–452. doi: 10.1002/1873-3468.12071. [DOI] [PubMed] [Google Scholar]