Abstract
The clinical outcome of children with high-risk relapsed B-cell precursor acute lymphoblastic leukemia (BCP-ALL) is poor. The present study assessed the utility and prognostic value of selected microRNA (miRNA/miR) in BCP-ALL. The changes in the expression levels of these miRNAs regarding known gene lesions affecting lymphoid development [early B-cell factor 1 (EBF1), ETS variant 6 (ETV6), IKAROS family zinc finger 1 (IKZF1), paired box 5 (PAX5), cyclin dependent kinase inhibitor (CDKN) 2A/CDKN2B, retinoblastoma 1 (RB1), pseudoautosomal region 1 (PAR1), B-cell translocation gene 1 protein (BTG1)] were analyzed. The following miRNAs were analyzed: miR-24, miR-31, miR-128, miR-542, and miR-708. The present study focused on patients with deletions of the IKAROS transcriptional factor gene IKZF1, which is currently considered to be an independent negative prognostic factor for ALL outcome. It was demonstrated that the expression level of miR-128 was significantly lower in patients with IKZF1 deletion compared with patients without IKZF1 deletion. Additionally, low expression of miR-542 was associated with CDKN2A/B and miR-31deletions, and low expression of miR-24 was associated with miR-31 deletion. Low expression of miR-31, miR-24, miR-708 and miR-128 was associated with PAX5 deletion, high expression of miR-24 and miR-542 was associated with PAR1 deletion and high expression of miR-708 was associated with ETV6 deletion. The expression of the selected miRNAs was not associated with deletions of BTG1, EBF1 and RB1. These data, by emphasizing the association of miRNAs expression level with microdeletions, may assist to elucidate ALL biology and contribute to future studies on the possible applications of the miRNA profile for diagnosis.
Keywords: pediatric acute lymphoblastic leukemia, microdeletions, transcriptional factors, IKZF1, microRNA
Introduction
Acute lymphoblastic leukemia (ALL) is the most common cancer malignancy and the leading cause of cancer-associated mortality in children (1). Assessment of the risk of relapse is crucial in modern treatment protocols, as relapses are the main cause of therapeutic failure in childhood ALL. Despite investigation of the clinical and cytogenetic predictors, there remains no sufficient marker enabling early identification of patients at high risk of relapse. Therefore, the molecular markers for risk assessment at the time of diagnosis and during the treatment course are urgently needed.
A particularly poor treatment outcome is observed in patients with B-cell precursor ALL (BCP-ALL) with t(9;22)(q34;q11) translocation, and in patients with breakpoint cluster region/ABL proto-oncogene 1, non-receptor tyrosine kinase (BCR/ABL1)-like ALL subtypes (2). BCR/ABL1-like ALL exhibits a gene expression profile similar to BCR/ABL1-positive ALL, but without t(9;22) rearrangement (3). These leukemia subtypes are most commonly characterized by genetic abnormalities affecting the following pathways: Lymphoid development [early B-cell factor 1 (EBF1), ETS variant 6(ETV6), IKAROS family zinc finger (IKZF)1, LIM domain only 2 and paired box 5 (PAX5)]; signal transduction (ABL1, ABL2, cytokine receptor-like factor 2, colony stimulating factor 1 receptor, erythropoietin receptor, interleukin 7 receptor, janus kinase 2, platelet derived growth factor receptor β); and tumor suppression and cell cycle regulation [cyclin dependent kinase inhibitor (CDKN)2A/CDKN2B, retinoblastoma 1 (RB1) and tumor protein 53] (4).
Microdeletions and point mutations of the aforementioned genes are considered important factors in lymphoblast differentiation and leukemia progression, but the final pathogenetic effect is controlled by post-transcriptional regulators (5–7). Small noncoding RNA molecules (~22 base pairs), referred to as microRNA (miRNA/miR), are types of these regulators. miRNAs recognize a seed region in an mRNA sequence and destabilize mRNA by acting in a RNA-induced silencing complex, thereby inhibiting protein production (8). Evidence from previous studies indicates that miRNAs promote neoplastic transformation. In lymphoid cells, miRNA may serve a role in hematopoiesis, and in the development of leukemia by the suppression of tumor suppressor genes or stimulation of oncogenes (9,10). Dysregulation of miRNA expression is observed in leukemic cells and is potentially linked to drug resistance and poor outcome (11,12).
As aforementioned, miRNAs are involved in numerous processes at the transcription level, therefore their signature may reflect normal homeostatic processes as well as pathological changes and leukemogenic disturbance. This suggests that miRNAs are potential biomarkers for early leukemia detection.
The present study aimed to identify miRNAs whose expression level variations may be associated with known molecular defects in childhood ALL. To preselect the miRNAs, bioinformatics tools were used for in silico miRNA analysis, such as Mircancer.ecu.edu, miRWalk data base, miRTarBase, and microRNA.org (13). Based on target prediction, a set of potential miRNA markers: miR-24, miR-31, miR-128, miR-542 and miR-708were identified, and then their expression levels were evaluated in a cohort of children with ALL and their association with known prognostic genetic lesions and clinical features of ALL.
Materials and methods
miRNA profiles
For the analysis of the miRNA profiles, 5 miRNAs (miR-128, miR-542, miR-708, miR-24 and miR-31) were selected using bioinformatics tools and databases [miRCancer (14), miRWalk database (15), miRTarBase (16), microRNA.org (17)], and were selected using literature reviews (9–12). The mirSVR score was applied for ranking microRNA target sites by a downregulation score. The mirSVR is machine learning method based on regression modelling and was calculated based on seed-site pairing, site context, conservation and free-energy of the selected miRNAs (18).
Recruitment of patients
Children (<18 years old) with B-cell lineage ALL (B-ALL) and T-cell lineage (T-ALL) were recruited from the Caucasian Polish population at the Department of Pediatrics, Oncology, Hematology and Diabetology, Medical University of Łódź (Łódź, Poland), and cooperating Centres of Pediatric Oncohematology in the cities of Bialystok, Bydgoszcz, Gdansk, Katowice, Kielce, Olsztyn, Poznan, Szczecin, and Zabrze (Poland). Ethics committee approval was obtained from the Institutional Review Board of the Medical University of Lodz (number, RNN/226/11/KE//KE). All patients were diagnosed between May 2004 and March 2014 and treated according to ALL-IC 2002 or 2009 protocols (19). Patients with a set of microdeletions, particularly IKZF1 defects, were preferentially enrolled into the study.
Minimal residual disease (MRD) monitoring
The bone marrow samples from patients with BCP-ALL were collected at three time points: At diagnosis and at 15 and 33 days of treatment protocol. The bone marrow samples from patients with BCP-ALL were analyzed with 8-color flow cytometry according to the protocols of the EuroFlow Consortium (20). These samples were processed according to the manufacturer's lyse and wash protocol (BD Biosciences, San Jose, CA, USA), with 1X FACS Lyse used for erythrocyte lysis (BD Biosciences) and analyzed with FACSCanto II flow cytometer (BD Biosciences). To ensure a sensitivity level of ≥104, low-cellular samples were first subjected to erythrocyte lysis in 1X ammonium chloride solution (Pharm Lyse; BD Biosciences) in a bulk lysis protocol. Subsequently, the suspension of leukocytes was stained in a single 8-color tube with antibodies adequate for the blasts'phenotype at diagnosis. The reproducibility of the obtained results was ensured by complying with the standard operating procedures developed by EuroFlow based on daily quality assessment with fluorescent beads (Sphero Rainbow Calibration Particles; Spherotech, Lake Forest, IL, USA). For data analysis, FACSDiva 6.1 software (BD Biosciences) was used. Patients with number of blasts in peripheral blood >1,000 on the 8th day following steroid administration were identified as steroid resistant.
DNA and RNA extraction
A total of 90 bone marrow samples were collected at the time of diagnosis (B-ALL, n=66; T-ALL, n=24). For DNA and RNA isolation, 300 µl bone marrow stored in TRIzol® reagent was used (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The isolation procedure was performed according to the manufacturer's protocol.
Reverse transcription and expression
The RNA samples, isolated from bone marrow, were used for miRNA expression analysis, namely reverse transcription (RT) of miRNA to complementary (c)DNA, amplification of cDNA and detection by TaqMan probes in quantitative polymerase chain reaction (qPCR). Each RT reaction was performed according to the manufacturer's protocol (TaqMan® MicroRNA Reverse Transcription kit; Thermo Fisher Scientific, Inc.) in 15 µl [7 µl mix, 3 µl primer (5X RT), 5 µl RNA sample]. The sequences of Stem-Loop RT primer target sequences and miRNA assay IDs are described in Table I. The amount of RNA used for each reaction was 40 ng/well (RNA concentration, 8 ng/µl). RT-PCR was conducted under the following conditions: 30 min at 16°C, 30 min at 42°C, 5 min 85°C and hold at 4°C.
Table I.
miRNA assay identities and target sequences.
| miRNA | Assay identity | Target sequence |
|---|---|---|
| hsa-miR-31 | TM:002279 | 5′-AGGCAAGAUGCUGGCAUAGCU-3′ |
| hsa-miR-24–2 | TM:002441 | 5′-UGCCUACUGAGCUGAAACACAG-3′ |
| hsa-miR-542-5p | TM:002240 | 5′-UCGGGGAUCAUCAUGUCACGAGA-3′ |
| hsa-miR-708 | TM:002341 | 5′-AAGGAGCUUACAAUCUAGCUGGG-3′ |
| hsa-miR-128A | TM:002216 | 5′-UCACAGUGAACCGGUCUCUUU-3′ |
| U6 snRNA | TM:001973 | 5′-GTGCTCGCTTCGGCAGCACATATACTAAAATTGGAACGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTT-3′ |
miRNA/miR, microRNA.
Each qPCR reaction was performed in duplicate, in 18 µl volumes (TaqMan® Gene Expression Master Mix 10 µl; 20X assay 1 µl; H2O 4 µl) with 3 µl of cDNA sample (8 ng). TaqMan® microRNA assays (Thermo Fisher Scientific, Inc.) were used to analyze specific miRNAs (hsa-miR-31, TM:002279; hsa-miR-24-2, TM:002441; hsa-miR-542-5p, TM:002240; hsa-miR-708, TM:002341; hsa-miR-128a, TM:002216). For normalization of PCR, as an internal control, U6 snRNA (TM:001973) was used. PCRs were conducted in duplicate under the following protocol: 2 min at 50°C, 10 min at 95°C, then 40 cycles of: 95°C for 30 sec and 60°C for 1 min.
The expression levels are presented as 2−∆∆Cq values, where ∆Cq = Cq (reference) - Cq (miRNA of interest), which produces a higher value for higher miRNA expression, facilitating its use and interpretation as a biomarker (21).
Multiplex ligation-dependent probe amplification (MLPA)
Copy number variations (CNVs) were identified by MLPA. Samples were screened for selected CNVs using P202-B1 and P335-B1 SALSA MLPA kits (MRC-Holland, Amsterdam, The Netherlands). These assays enable analysis of all exons of CDKN2A/B and IKZF1 genes, and selected exons of PAX5, BTG anti-proliferation factor 1 (BTG1), EBF1, ETV6, RB1, immunoglobulin heavy locus, IKZF2, IKZF3, metastasis associated 1 genes and the pseudoautosomal region 1 (PAR1) region. The MLPA procedure was performed according to the manufacturer's protocols.
Statistical analysis
Statistical analysis was performed using Statistical 12.5PL software (StatSoft, Inc., Tulsa, OK, USA). The distribution of variables was tested with the Shapiro-Wilk test and Kolmogorov-Smirnov test with Lilliefors correction. Due to the non-normal distribution of all analyzed variables, non-parametric tests were used and the results are presented as medians followed by interquartile ranges (IQRs). P<0.05 was considered to indicate a statistically significant difference. Categorical variables were compared using the χ2 test or Fisher's exact test. Continuous variables were compared using the U Mann-Whitney test to analyze differences between two groups and the Kruskal-Wallis along with post hoc (Dunn's) test to analyze differences between more than two groups. The R Spearman's correlation was used to analyze correlations between continuous variables.
Results
Study group characteristics
A total of 90 children were recruited, of which 24 (26.67%) were diagnosed with T-ALL and 66 (73.33%) were diagnosed with B-ALL (Table II). In the B-ALL group, 2 patients (3.23%) exhibited a BCR/ABL fusion, 1 (1.67%) exhibited a mixed lineage leukemia (MLL)/AF4 fusion and 11 (19.64%) exhibited ETV6/runt related transcription factor 1 (RUNX1) fusions. BCR/ABL, MLL/AF4 and ETV6/RUNX1 fusions were not identified among the patients with T-ALL.
Table II.
Clinical characteristics of the study population.
| Variable | Total | T-ALL | B-ALL | P-value |
|---|---|---|---|---|
| Total | 90 | 24 | 66 | |
| Sex | 0.16519 | |||
| Male, n (%) | 59 (65.56) | 19 (79.17) | 40 (60.61) | |
| Female, n (%) | 31 (34.44) | 5 (20.83) | 26 (39.39) | |
| Age at diagnosis, years(IQR) | 8.36 (3.62–12.88) | 8.87 (3.22–13.05) | 8.05 (3.79–12.73) | 0.98908 |
| WBC, ×103/µl (IQR) | 26.65 (6.36–88.10) | 108.85 (29.74–279.01) | 14.13 (4.45–53.00) | 0.00008 |
| MRD at day 15, % (IQR) | 1.06 (0.13–9.50) | 4.2 (0.83–29.30) | 0.61 (0.07–5.25) | 0.00740 |
| Resistance to steroids, % | 0.01456 | |||
| Positive (IQR) | 14 (15.55) | 8 (33.33) | 6 (9.09) | |
| Negative (IQR) | 76 (84.44) | 16 (66.66) | 60 (90.91) |
Nominal variables are presented as numbers with percentages in brackets and compared using the χ2 test, while continuous variables are presented as medians with interquartile ranges in brackets and compared using U Mann-Whitney test. Steroid resistance was defined as a peripheral blood blast count>1,000 in 8th day of treatment. WBC, white blood cells; MRD, minimal residual diseases (measured as percentage of leukemic cells in bone marrow); T-ALL, T-cell lineage acute lymphoblastic leukemia; B-ALL, B-cell lineage acute lymphoblastic leukemia.
Microdeletions were determined in 83 (92.22%) patients (Table III). IKZF1 deletions and ETV6 deletions were more common in patients with B-ALL compared with patients with T-ALL (40.63% vs. 10.53%, P=0.0147 and 24.28% vs. 0%, P=0.029). No other significant differences in the microdeletion profiles between T-ALL and B-ALL were observed.
Table III.
Microdeletion profiles among included patients.
| Microdeletion locus | All | T-ALL | B-ALL | P-value |
|---|---|---|---|---|
| IKZF1 | 28 (33.73) | 2 (10.52) | 26 (40.62) | 0.01468 |
| CDKN2A/B | 51 (61.44) | 13 (68.42) | 38 (59.37) | 0.65777 |
| MIR31 | 18 (23.07) | 3 (18.75) | 15 (24.19) | 0.75102 |
| PAX5 | 16 (20.00) | 2 (11.76) | 14 (22.22) | 0.50036 |
| PAR1 | 19 (26.76) | 3 (23.07) | 16 (27.58) | 1.00000 |
| ETV6 | 17 (24.28) | 0 (0) | 17 (29.82) | 0.02876 |
| BTG1 | 5 (7.14) | 0 (0) | 5 (8.77) | 0.57571 |
| EBF1 | 5 (7.14) | 0 (0) | 5 (8.77) | 0.57571 |
| RB1 | 10 (14.08) | 2 (15.38) | 8 (13.79) | 1.00000 |
Categorical variables were compared using the χ2 test. All data presented at the number of deletions with percentages in brackets. IKZF1, IKAROS family zinc finger 1; CDKN2A/B, cyclin dependent kinase inhibitor 2A/B; MIR31, microRNA 31; PAX5, paired box 5; PAR1, pseudoautosomal region 1; ETV6, ETS variant 6; BTG1, B-cell translocation gene 1 protein; EBF1, early B-cell factor 1; RB1, retinoblastoma 1.
The expression levels of all selected miRNAs were measurable in 76 (84.44%) of selected samples. A comparison of the expression levels of the selected miRNAs between T-ALL and B-ALL patients revealed that patients with B-ALL were characterized by significantly elevated level of miR-708 and decreased level of miR-542 (P=0.0144 and P=0.0380, respectively; Fig. 1).
Figure 1.
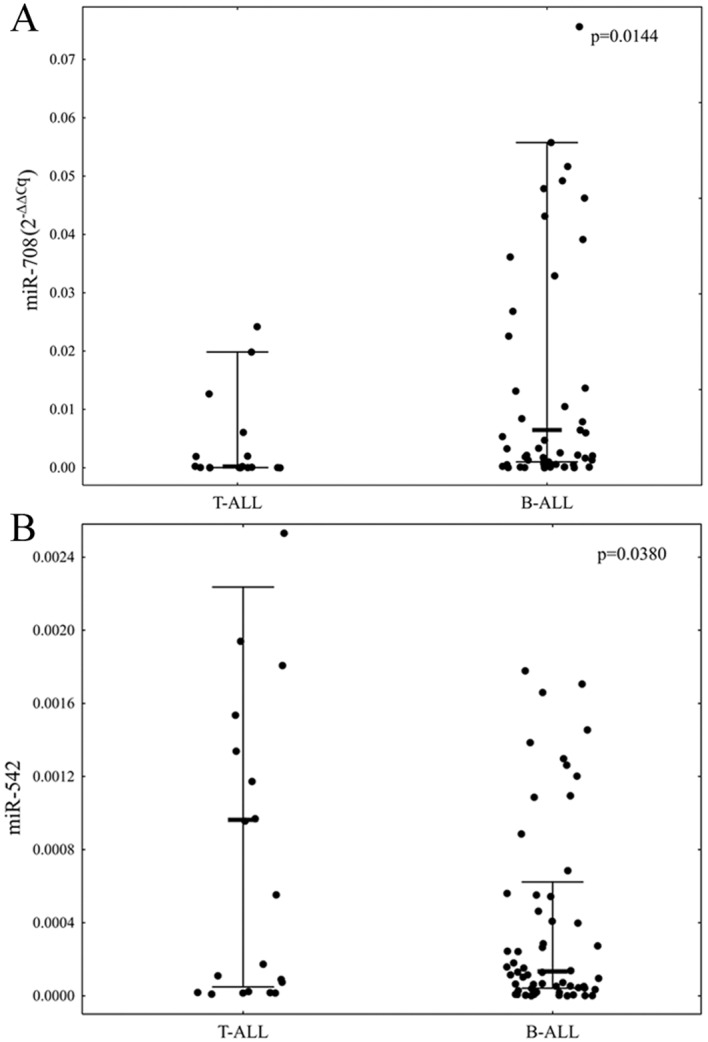
A comparison of the miR-708 and miR-542 expression levels between T-ALL and B-ALL patients. (A) P-value refers to the statistically significant difference in the expression level of miR-708 between patients with T-ALL and B-ALL. (B) P-value refers to the statistically significant differences in the expression level of miR-542 between patients with T-ALL and B-ALL. miRNA expression levels are presented as 2−∆∆Cq value. Bold line represents median, whiskers represent interquartile range and dots represent raw data. miRNA, microRNA; T-ALL, T-cell lineage acute lymphoblastic leukemia; B-ALL, B-cell lineage acute lymphoblastic leukemia.
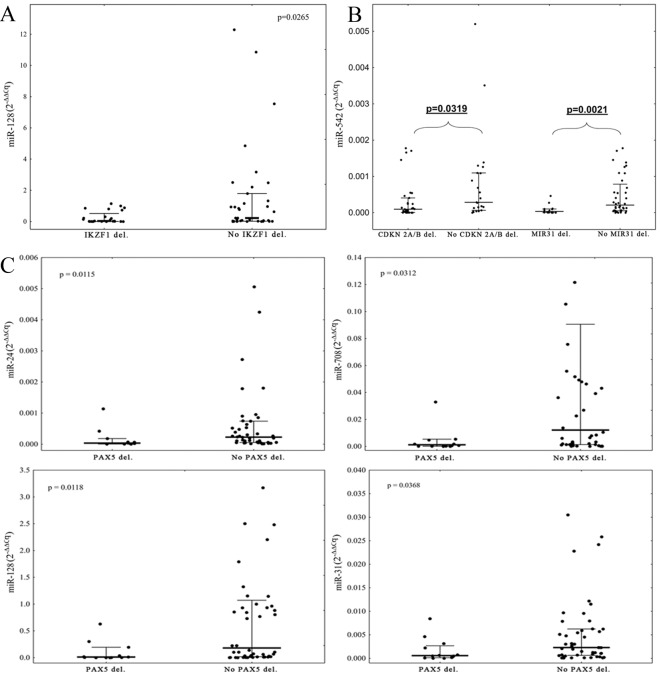
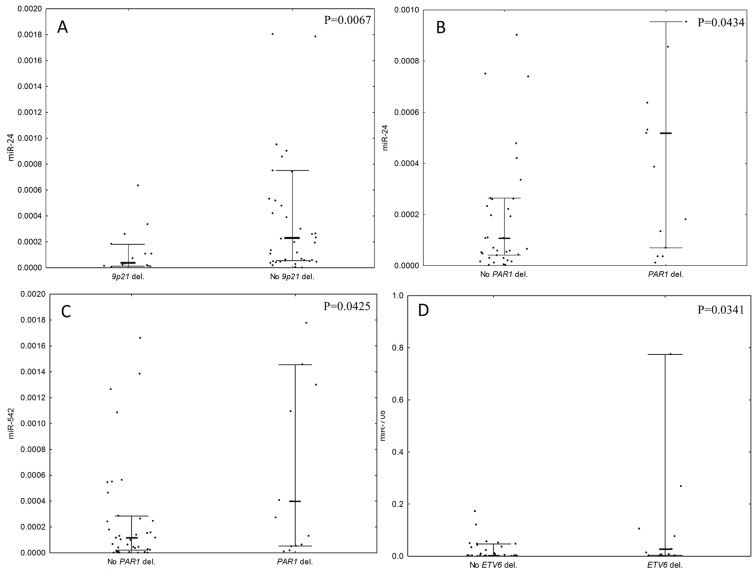
Due to the small number of patients with T-ALL with microdeletions, the expression levels of miRNAs with the microdeletion status were compared only within the group of patients with B-ALL. It was identified that the expression level of miR-128 was lower in patients with IKZF1 deletions compared with patients without IKZF1 deletions (P=0.0265, Fig. 2A). Deletions of CDKN2A/B and MIR31 were associated with low expression of miR-542 in comparison with patients without deletions (P=0.0319 and P=0.0021, respectively; Fig. 2B). Patients with PAX5 deletions exhibited significantly lower expression levels of miR-31, miR-24, miR-708 and miR-128 in comparison with patients without deletions of PAX5 (P=0.0368, 0.0115, 0.0312 and 0.0118, respectively; Fig. 2C). Large deletions in the 9p21 chromosomal region, encompassing CDKN2A, CDKN2B and MIR31 deletions, were also associated with low expression of miR-24 (P=0.0067; Fig. 3A). Deletions in the PAR1 region were associated with high expression levels of miR-24 and miR-542 (P=0.0434 and 0.0425; Fig. 3B and C, respectively). Deletions of ETV6 were associated with a high expression of miR-708 (P=0.0341, Fig. 3D). There was no association between the expression levels of selected miRNAs and deletions of BTG1, EBF1, or RB1 observed.
Figure 2.
A comparison of the microRNAs expression levels with respect to IKZF1, CDKN2A/B, MIR31 or PAX5 deletion status. P-values refer to the statistically significant difference in the expression level of: (A) miR-128 with respect to IKZF1 deletion status; (B) miR-542 with respect to CDKN2A/B and MIR31 deletion status; and (C) miRNAs expression level with respect to PAX5 deletion status in patients with B-ALL. Expression levels are presented as 2−∆∆Cq value. Bold line represents median, whiskers represent interquartile range and dots represent raw data. miRNA, microRNA; B-ALL, B-cell lineage acute lymphoblastic leukemia.
Figure 3.
A comparison of the microRNA's expression levels with respect to 9p21, PAR1 and ETV6 deletion status. P-values refer to the statistically significant difference in the expression level of: (A) miR-24 with respect to 9p21 large deletion status; (B) miR-24 with respect to PAR1 deletion status and (C) miR-542 with respect to PAR1 deletion status in patients with B-ALL (D) miR-708 with respect to ETV6 deletion status. Expression levels are presented as 2−∆∆Cq value. Bold line represents the median, whiskers represent interquartile range and dots represent raw data. miRNA, microRNA; B-ALL, B-cell lineage acute lymphoblastic leukemia; PAR1, pseudoautosomal region 1; ETV6, ETS variant 6.
Correlation analyses revealed a negative correlation between age at diagnosis and miR-542 (R2=−0.26, P=0.043), and between MRD at day 15 and miR-31 (−0.32, P=0.017) and miR-708 (−0.32, P=0.016) in patients with B-ALL (Table IV). There were no statistically significant associations with sex, WBC or resistance to steroid therapy observed.
Table IV.
Correlation analyses.
| Variable | miR-31 | miR-24 | miR-708 | miR-542 | miR-128 |
|---|---|---|---|---|---|
| Age at diagnosis | −0.11 | −0.19 | −0.16 | -0.26a | −0.16 |
| WBC at diagnosis | −0.08 | 0.09 | −0.05 | −0.03 | 0.09 |
| MRD_15 | -0.32a | −0.11 | -0.32a | −0.11 | −0.19 |
| MRD_33 | −0.05 | 0.11 | 0.02 | 0.16 | −0.01 |
Quantitative variables were compared using Spearmann's correlation analyses. We present all correlation coefficients bolded and followed by asterisks if significant.
b and cstand for P=0.043, P=0.016, P=0.017, respectively. WBC, white blood cells; MRD, minimal residual disease.
Discussion
The data of the present study indicate that selected miRNAs: miR-128, miR-542, miR-708, miR-24 and miR-31, are associated with the ALL immunophenotype and microdeletion status of crucial genes affecting lymphoid development and cell cycle regulation.
Firstly, it was identified that the expression of miR-128 was lower in patients with IKZF1 deletions in comparison with patients without these deletions. IKZF1 encodes Ikaros protein, which has a key role as a transcription factor in lymphocyte differentiation (22). Ikaros influences more than half of the known genes associated with hematopoiesis and early B-cell development (23). IKZF1 deletions are associated with a poor outcome in high-risk groups of pediatric patients (24,25). In a comparison between normal controls and patients with AML, an increased expression of miR-128 in patients with ALL has been suggested (26). Altered miR-128 expression may discriminate ALL from adult myeloid leukemia and normal bone marrow samples (27–30). Notably, Mi et al (27) revealed that the differential expression of miR-128 in acute leukemia is not a consequence of an altered genomic DNA copy number, and suggested that this miRNA is also under other types of DNA epigenetic regulation. The association between the lower expression of miR-128 and steroid resistance was not evaluated in the present study. In these and other previous studies, miR-128 expression did not correlate with other biological and clinical features (28,31). The role of miR-128 insufficiency in the pathogenesis and response to treatment in patients with IKZF1-deletion ALL should be additionally examined with large-scale pediatric patients with ALL.
Lower miR-128 expression was detected in patients with deletion of another transcriptional factor gene, PAX5. Patients with PAX5 deletions exhibited significantly lower expression levels of miR-31, miR-24 and miR-708 in comparison with patients without PAX5 deletions.
Compared to patients with T-ALL, a high expression of miR-708 and a low expression of miR-542 was observed in patients with B-ALL. miR-708 was overexpressed in patients with ETV6 deletions, and this gene is involved in lymphoid development (32). These miRNAs were negatively correlated with MRD status at day 15 of the therapeutic protocol. Higher MRD was associated with lower miR-708. The investigation of miR-708 performed by de Oliveira et al (33) revealed lower miR-708 expression in normal bone marrow samples compared with ALL samples. In their study, miR-708 expression was not associated with clinical features, such as white blood cell level, bone marrow blast infiltration, steroid resistance, or central nervous system or testis involvement. Han et al (34) revealed a correlation between miR-708 and the response to glucocorticoid therapy, and suggested that low miR-708 expression was associated with higher risk of leukemogenesis or relapse. Higher miR-708 expression has been identified in patients with transcription factor ETV6-acute myeloid leukemia 1 protein, breakpoint cluster region-abelson tyrosine-protein kinase 1 (BCR-ABL1), 3-pre-B-cell leukemia transcription factor 1, hyperdiploid and ‘B-other’ ALL (B-cell ALL patients without aforementioned defects) compared with MLL-rearranged B-ALL and T-ALL cases (12,35). In another study, Li et al (36) demonstrated that miR-708 was overexpressed in small groups of Chinese common precursor patients with B-ALL: Expression of miR-708 was higher in the high-risk group compared with the intermediate-risk group.
In the present study, deletions in the 9p21 chromosomal region (CDKN2A and CDKN2B, MIR31) were associated with a lower expression of miR-542 in comparison with patients without deletions. miR-542 regulates the DNA repair/notch pathway in epithelial prostate cancer, and functions as a tumor suppressor in neuroblastoma (37,38). This miRNA may positively regulate tumor protein 53 (p53) activity (39). The homozygous loss of the miR-31 gene is described in 9p instability in ALL (40). The 9p instability is detected in 19% of patients with ALL, and always includes the homozygous loss of CDKN2A along with loss of CDKN2B, which are associated with BCR/ABL1 and IKZF1 dysfunction (40–43). A lower expression of miR-542 is associated with a deletion of the 9p region. Schotte et al (12) demonstrated that miR-542 was 1 of 6 significantly upregulated miRNAs in T-ALL compared with normal thymocytes. Reduced expression of miR-542 and its role in B-ALL biology requires additional examination, as the current literature regarding miR-542 expression in ALL and lymphoproliferative diseases is limited. The present study also identified that deletions of MIR31were associated with a low miR-24 expression. miR-24 serves a role in the response to DNA damage, and may enhance apoptosis by targeting B-cell lymphoma 2 (44,45). miR-24 potentially reduces cellular viability and induces apoptosis in combination with docetaxel (46). High expression of miR-24 also correlates with the response to DNA damage and apoptosis in cells treated with etoposide (47).
The present study observed altered miR-24 expression in patients with ALL with deletions in PAR1. PAR1 deletions are frequently observed in patients who are positive for IKZF1 deletion (40). High expressions of miR-24 and miR-542 in patients who are positive for PAR1 deletion were detected. The expression level of miR-24 in patients with acute leukemia is higher compared with the healthy individuals, and is associated with shorter overall survival, high risk of relapse and poor survival (48).
Notably, the present study demonstrated that the lower expression of miR-31 and miR-708 was associated with a high MRD level and positive PAX5 mutation status. It should be emphasized that those two investigated miRNAs (miR-31 and miR-708) negatively influence the NF-κB signaling pathway, which has a role in B-cell receptor activation (49). Aberrant B-cell receptor activation leads to a number of lymphoid malignancies (50). At present, targeted therapies are focused on the inhibition of BCR signaling (51,52). miR-708 strongly represses NF-κB signaling in chronic lymphocytic leukemia (53). miR-31 in adult T-ALL negatively regulates the NF-κB pathway by targeting NF-κB inducing kinase (54). Based on this, it is hypothesized that the loss of activity of these miRNAs may serve a role in the pathogenesis of ALL, and in treatment failure.
Certain issues must be taken into consideration when interpreting the present data. One of the most conspicuous points is the mutation profile of the patients. The mutation rate in the study population is inconsistent with that of the general population, which is a consequence of the enrollment criteria. Patients with IKZF1 deletions were enrolled with priority as the present study was designed to focus on high-risk patients such as patients with BCR/ABL1-like ALL. Another important issue is the underrepresentation of patients with T-ALL, and these patients were excluded from the majority of additional analyses. However, differences in the biological features between B-ALL and T-ALL were demonstrated. Data from another study suggesting that patients with B-ALL, in comparison to T-ALL, were characterized by lower white blood cell counts at diagnosis, lower MRD at day 15 of the therapeutic protocol and greater sensitivity to steroid treatment were also confirmed by the present study. Finally, it was not possible to elaborate the activity of all selected miRNAs in all the investigated samples. Despite this, the expression levels of at least 4 selected miRNAs were successfully measured in 84 patients (93.33%). To the best of our knowledge, this is the first study estimating selected miRNA expression, genetic defects and clinical features in pediatric ALL subtypes. These data indicate that an aberrant miRNA pattern is associated with ALL types, and that microdeletions are commonly observed in high-risk patients with B-ALL.
As demonstrated in the present study, identifying the miRNA expression status in patients with ALL may be useful for elucidating the mechanisms of the disease. miRNAs may be used as molecular markers to assess risk of leukemia relapse, at the time of diagnosis, and to tailor specific therapies to the individual patient. Additional studies with larger cohorts are needed to examine the potential use of these miRNAs (miR-128, miR-542, miR-708, miR-24 and miR-31) as markers of genetic lesions.
Acknowledgements
The present study was supported by the National Science Centre, Poland (grant no. 2011/01/B/NZ4/03345). The study was performed within the project Centre for Innovative Research in Medical and Natural Sciences realized by the University of Rzeszow, and co-financed by Regional Operational Programme for the Podkarpackie Province for the years 2007–2013 (contract no. UDA-RPPK.01.03.00–18-004/12-00). Ms J. Madzio was supported by The National Science Centre (grant no. PRELUDIUM 2015/17/N/NZ5/00669). Dr M. Braun was supported by Polish Ministry of Science and Higher Education under the Diamond Grant Program (grant no. DI2012017042). Ms J. Madzio, Dr M. Braun and A.P. were also supported by National Centre of Research and Development (grant no. LIDER 031/635/l-5/13/NCBR/2014).
References
- 1.Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64:83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 2.Schwab CJ, Chilton L, Morrison H, Jones L, Al-Shehhi H, Erhorn A, Russell LJ, Moorman AV, Harrison CJ. Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: Association with cytogenetics and clinical features. Haematologica. 2013;98:1081–1088. doi: 10.3324/haematol.2013.085175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven LJ, Beverloo HB, Van der Spek PJ, Escherich G, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: A genome-wide classification study. Lancet Oncol. 2009;10:125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullighan CG. The genomic landscape of acute lymphoblastic leukemia in children and young adults. Hematology Am Soc Hematol Educ Program. 2014;2014:174–180. doi: 10.1182/asheducation-2014.1.174. [DOI] [PubMed] [Google Scholar]
- 5.Figueroa ME, Chen SC, Andersson AK, Phillips LA, Li Y, Sotzen J, Kundu M, Downing JR, Melnick A, Mullighan CG. Integrated genetic and epigenetic analysis of childhood acute lymphoblastic leukemia. J Clin Invest. 2013;123:3099–3111. doi: 10.1172/JCI66203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshida T, Landhuis E, Dose M, Hazan I, Zhang J, Naito T, Jackson AF, Wu J, Perotti EA, Kaufmann C, et al. Transcriptional regulation of the Ikzf1 locus. Blood. 2013;122:3149–3159. doi: 10.1182/blood-2013-01-474916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mavrakis KJ, Van Der Meulen J, Wolfe AL, Liu X, Mets E, Taghon T, Khan AA, Setty M, Rondou P, Vandenberghe P, et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL) Nat Genet. 2011;43:673–678. doi: 10.1038/ng.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 10.Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. Investigation of microRNA alterations in leukemias and lymphomas. Methods Enzymol. 2007;427:193–213. doi: 10.1016/S0076-6879(07)27011-9. [DOI] [PubMed] [Google Scholar]
- 12.Schotte D, De Menezes RX, Akbari Moqadam F, Khankahdani LM, Lange-Turenhout E, Chen C, Pieters R, Den Boer ML. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica. 2011;96:703–711. doi: 10.3324/haematol.2010.026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie B, Ding Q, Han H, Wu D. miRCancer: A microRNA-cancer association database constructed by text mining on literature. Bioinformatics. 2013;29:638–644. doi: 10.1093/bioinformatics/btt014. [DOI] [PubMed] [Google Scholar]
- 15.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: Prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell M, Castillo L, Janic D, Jazbec J, Kaiserova E, Konja J, Kovas G, Kowalczyk J, Soycan LY. ALL IC-BFM 2009 - A Randomized Trial of the I-BFM-SG for the Management of Childhood non-B Acute Lymphoblastic Leukemia. http://tphd.org.tr/5th_hematoloji_sempozyumu/Lebriz_Yuksel_ALLIC_BFM_2009 Final Version of Therapy Protocol from August-14-2009. [Google Scholar]
- 20.van Dongen JJ, Lhermitte L, Böttcher S, Almeida J, van der Velden VH, Flores-Montero J, Rawstron A, Asnafi V, Lécrevisse Q, Lucio P, et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia. 2012;26:1908–1975. doi: 10.1038/leu.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Schwickert TA, Tagoh H, Gültekin S, Dakic A, Axelsson E, Minnich M, Ebert A, Werner B, Roth M, Cimmino L, et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol. 2014;15:283–293. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreirós-Vidal I, Carroll T, Taylor B, Terry A, Liang Z, Bruno L, Dharmalingam G, Khadayate S, Cobb BS, Smale ST, et al. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood. 2013;121:1769–1782. doi: 10.1182/blood-2012-08-450114. [DOI] [PubMed] [Google Scholar]
- 24.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dörge P, Meissner B, Zimmermann M, Möricke A, Schrauder A, Bouquin JP, Schewe D, Harbott J, Teigler-Schlegel A, Ratei R, et al. IKZF1 deletion is an independent predictor of outcome in pediatric acute lymphoblastic leukemia treated according to the ALL-BFM 2000 protocol. Haematologica. 2013;98:428–432. doi: 10.3324/haematol.2011.056135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu YD, Wang L, Sun C, Fan L, Zhu DX, Fang C, Wang YH, Zou ZJ, Zhang SJ, Li JY, Xu W. Distinctive microRNA signature is associated with the diagnosis and prognosis of acute leukemia. Med Oncol. 2012;29:2323–2331. doi: 10.1007/s12032-011-0140-5. [DOI] [PubMed] [Google Scholar]
- 27.Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci USA. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Oliveira JC, Scrideli CA, Brassesco MS, Morales AG, Pezuk JA, Queiroz Rde P, Yunes JA, Brandalise SR, Tone LG. Differential miRNA expression in childhood acute lymphoblastic leukemia and association with clinical and biological features. Leuk Res. 2012;36:293–298. doi: 10.1016/j.leukres.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Luo XQ, Zhang P, Huang LB, Zheng YS, Wu J, Zhou H, Qu LH, Xu L, Chen YQ. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One. 2009;4:e7826. doi: 10.1371/journal.pone.0007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Li Z, He C, Wang D, Yuan X, Chen J, Jin J. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44:191–197. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang LC, Swat W, Fujiwara Y, Davidson L, Visvader J, Kuo F, Alt FW, Gilliland DG, Golub TR, Orkin SH. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev. 1998;12:2392–2402. doi: 10.1101/gad.12.15.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Oliveira JC, Scrideli CA, Brassesco MS, Yunes JA, Brandalise SR, Tone LG. MiR-708-5p is differentially expressed in childhood acute lymphoblastic leukemia but not strongly associated to clinical features. Pediatr Blood Cancer. 2015;62:177–178. doi: 10.1002/pbc.25222. [DOI] [PubMed] [Google Scholar]
- 34.Han BW, Feng DD, Li ZG, Luo XQ, Zhang H, Li XJ, Zhang XJ, Zheng LL, Zeng CW, Lin KY, et al. A set of miRNAs that involve in the pathways of drug resistance and leukemic stem-cell differentiation is associated with the risk of relapse and glucocorticoid response in childhood ALL. Hum Mol Genet. 2011;20:4903–4915. doi: 10.1093/hmg/ddr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schotte D, Chau JC, Sylvester G, Liu G, Chen C, van der Velden VH, Broekhuis MJ, Peters TC, Pieters R, den Boer ML. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:313–322. doi: 10.1038/leu.2008.286. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Li D, Zhuang Y, Shi Q, Wei W, Ju X. Overexpression of miR-708 and its targets in the childhood common precursor B-cell ALL. Pediatr Blood Cancer. 2013;60:2060–2067. doi: 10.1002/pbc.24583. [DOI] [PubMed] [Google Scholar]
- 37.Rane JK, Ylipää A, Adamson R, Mann VM, Simms MS, Collins AT, Visakorpi T, Nykter M, Maitland NJ. Construction of therapeutically relevant human prostate epithelial fate map by utilising miRNA and mRNA microarray expression data. Br J Cancer. 2015;113:611–615. doi: 10.1038/bjc.2015.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bray I, Tivnan A, Bryan K, Foley NH, Watters KM, Tracey L, Davidoff AM, Stallings RL. MicroRNA-542-5p as a novel tumor suppressor in neuroblastoma. Cancer Lett. 2011;303:56–64. doi: 10.1016/j.canlet.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Huang JW, Castella M, Huntsman DG, Taniguchi T. p53 is positively regulated by miR-542-3p. Cancer Res. 2014;74:3218–3227. doi: 10.1158/0008-5472.CAN-13-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Usvasalo A, Ninomiya S, Räty R, Hollmén J, Saarinen-Pihkala UM, Elonen E, Knuutila S. Focal 9p instability in hematologic neoplasias revealed by comparative genomic hybridization and single-nucleotide polymorphism microarray analyses. Genes. Chromosomes Cancer. 2010;49:309–318. doi: 10.1002/gcc.20741. [DOI] [PubMed] [Google Scholar]
- 41.Sherborne AL, Hosking FJ, Prasad RB, Kumar R, Koehler R, Vijayakrishnan J, Papaemmanuil E, Bartram CR, Stanulla M, Schrappe M, et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet. 2010;42:492–494. doi: 10.1038/ng.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsson L, Albitar F, Castor A, Behrendtz M, Biloglav A, Paulsson K, Johansson B. Cooperative genetic changes in pediatric B-cell precursor acute lymphoblastic leukemia with deletions or mutations of IKZF1. Genes Chromosomes Cancer. 2015;54:315–325. doi: 10.1002/gcc.22245. [DOI] [PubMed] [Google Scholar]
- 43.Braun M, Pastorczak A, Fendler W, Madzio J, Tomasik B, Taha J, Bielska M, Sedek L, Szczepanski T, Matysiak M, et al. Biallelic loss of CDKN2A is associated with poor response to treatment in pediatric acute lymphoblastic leukemia. Leuk Lymphoma. 2016;18:1162–1171. doi: 10.1080/10428194.2016.1228925. [DOI] [PubMed] [Google Scholar]
- 44.Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, Bentwich Z, Lieberman J, Chowdhury D. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srivastava N, Manvati S, Srivastava A, Pal R, Kalaiarasan P, Chattopadhyay S, Gochhait S, Dua R, Bamezai RN. miR-24-2 controls H2AFX expression regardless of gene copy number alteration and induces apoptosis by targeting antiapoptotic gene BCL-2: A potential for therapeutic intervention. Breast Cancer Res. 2011;13:R39. doi: 10.1186/bcr2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manvati S, Mangalhara KC, Kalaiarasan P, Srivastava N, Bamezai RN. miR-24-2 regulates genes in survival pathway and demonstrates potential in reducing cellular viability in combination with docetaxel. Gene. 2015;567:217–224. doi: 10.1016/j.gene.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Brunner S, Herndler-Brandstetter D, Arnold CR, Wiegers GJ, Villunger A, Hackl M, Grillari J, Moreno-Villanueva M, Bürkle A, Grubeck-Loebenstein B. Upregulation of miR-24 is associated with a decreased DNA damage response upon etoposide treatment in highly differentiated CD8(+) T cells sensitizing them to apoptotic cell death. Aging Cell. 2012;11:579–587. doi: 10.1111/j.1474-9726.2012.00819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Organista-Nava J, Gómez-Gómez Y, Illades-Aguiar B, Del Carmen Alarcón-Romero L, Saavedra-Herrera MV, Rivera-Ramírez AB, Garzón-Barrientos VH, Leyva-Vázquez MA. High miR-24 expression is associated with risk of relapse and poor survival in acute leukemia. Oncol Rep. 2015;33:1639–1649. doi: 10.3892/or.2015.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talab F, Thompson V, Allen JC, Lin K, Slupsky JR. Characterisation of B cell receptor-induced NF-κB activation in chronic lymphocytic leukaemia cells. Blood. 2015;116:3765. [Google Scholar]
- 50.Niemann CU, Wiestner A. B-cell receptor signaling as a driver of lymphoma development and evolution. Semin Cancer Biol. 2013;23:410–421. doi: 10.1016/j.semcancer.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lannutti BJ, Meadows SA, Herman SE, Kashishian A, Steiner B, Johnson AJ, Byrd JC, Tyner JW, Loriaux MM, Deininger M, et al. CAL-101, a p110delta selective phosphatidylinositol-3-kinase inhibitor for the treatment of B-cell malignancies, inhibits PI3K signaling and cellular viability. Blood. 2011;117:591–594. doi: 10.1182/blood-2010-03-275305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, Grant B, Sharman JP, Coleman M, Wierda WG, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baer C, Oakes CC, Ruppert AS, Claus R, Kim-Wanner SZ, Mertens D, Zenz T, Stilgenbauer S, Byrd JC, Plass C. Epigenetic silencing of miR-708 enhances NF-κB signaling in chronic lymphocytic leukemia. Int J Cancer. 2015;137:1352–1361. doi: 10.1002/ijc.29491. [DOI] [PubMed] [Google Scholar]
- 54.Yamagishi M, Nakano K, Miyake A, Yamochi T, Kagami Y, Tsutsumi A, Matsuda Y, Sato-Otsubo A, Muto S, Utsunomiya A, et al. Polycomb-mediated loss of miR-31 activates NIK-dependent NF-κB pathway in adult T cell leukemia and other cancers. Cancer Cell. 2012;21:121–135. doi: 10.1016/j.ccr.2011.12.015. [DOI] [PubMed] [Google Scholar]