Abstract
Ovarian cancer is one of the most common types of gynecological malignancy worldwide, and is the fourth leading cause of cancer-associated mortality among women. Despite improvements in therapeutic treatments, the prognosis for epithelial ovarian cancer (EOC) remains poor, mainly due to the rapid growth and metastasis of ovarian cancer tumors. An increasing number of studies have indicated that microRNAs (miRNAs) are involved in the carcinogenesis and progression of human cancer, suggesting that miRNAs may be used in clinical prognosis and as a therapeutic target in EOC. The aim of the present study was to investigate the expression levels of miRNA-494 in EOC tissues and cell lines. The clinical significance of miRNA-494 in patients with EOC was also evaluated. The results demonstrated that miRNA-494 was significantly downregulated in EOC tissues and cell lines. Low expression levels of miRNA-494 were associated with poor prognostic features, including International Federation of Gynecology and Obstetrics stage, tumor size and lymph node metastasis. In vitro functional studies demonstrated that overexpression of miRNA-494 inhibited proliferation, migration and invasion in EOC cells. By contrast, knockdown of miRNA-494 enhanced cell growth, migration and invasion in EOC cells. Notably, sirtuin 1 (SIRT1) was identified as a direct target of miRNA-494 in EOC. Furthermore, MTT, cell migration and invasion assays verified that EOC cell proliferation, migration and invasion were completely restored with forced miRNA-494 expression and SIRT1 restoration. Together, these findings suggest that miRNA-494 is a potential prognostic marker, and may provide novel therapeutic regimens of targeted therapy for EOC.
Keywords: microRNA-494, ovarian cancer, sirtuin 1, prognostic, metastasis
Introduction
Ovarian cancer is one of the most common types of gynecological malignancy worldwide and is the fourth leading cause of cancer-associated mortality among women (1). In 2015, it was estimated that there would be 21,290 new cases and 14,180 mortalities from ovarian cancer in the USA (2). Epithelial ovarian cancer (EOC), a major subtype accounting for ~90% cases of ovarian cancer, includes serous adenocarcinoma, endometrial adenocarcinoma and clear cell carcinoma (3). Among the gynecological cancers, EOC has the highest mortality rate, and contributes to >50% of gynecological cancer mortalities (4). At present, the primary therapeutic strategies for EOC are surgical resection of the visible disease, followed by platinum-based chemotherapy (5). Despite improvements in therapeutic treatments, the prognosis for EOC remains poor, mainly due to the rapid growth and metastasis of the tumor (6,7). Furthermore, due to the lack of symptoms in the early stage, EOC is usually diagnosed in an advanced stage (8). The 5-year survival rate for EOC at International Federation of Gynecology and Obstetrics (FIGO) stage (9) I–II was ~90%, while it was <30% for patients with EOC diagnosed at advanced FIGO stage (III–IV). It is therefore essential to elucidate the molecular mechanisms underlying the rapid growth and metastasis of EOC cells, since this may provide a potential therapeutic target for the treatment of EOC.
An increasing number of studies have suggested that the initiation and development of EOC may be associated with multistep changes in the genome, in particular abnormal expression of microRNA (miRNA/miR) (10–12). miRNAs are a large family of non-protein-coding short RNA molecules with 18–25 nucleotides that were first identified in 1993 by Lee et al (13). Accumulated computational and experimental studies have demonstrated that messenger RNAs (mRNAs) are regulated by miRNAs, indicating that miRNAs may act as master regulators in numerous diverse physiological and pathological processes (14–16). miRNAs negatively regulate gene expression by directly binding to the 3 untranslated region (UTR) of their target mRNA in a complementary base-pairing manner, thus causing mRNA stability and/or translation inhibition (17).
Previous studies have demonstrated that miRNAs perform important roles in numerous biological processes, including cellular proliferation, apoptosis, cell cycle, development, differentiation, migration and invasion (18,19). Currently, it is well known that certain miRNAs are dysregulated in various types of human cancer (8). Upregulated miRNAs may function as oncogenes by negatively regulating tumor-suppressor genes, whereas downregulated miRNAs may act as tumor-suppressor genes via the blockade of oncogenes (20). Therefore, the identification of the targets of miRNAs is essential for understanding the functions of miRNAs in cancer initiation and progression.
The present study aimed to investigate the expression of miR-494 in EOC tissues and cell lines, and the biological roles of miR-494 in EOC cells. Furthermore, the molecular mechanism underlying its tumor suppressive roles was evaluated. The results of the present study may contribute towards identifying a novel therapeutic target for the treatment of patients with EOC.
Materials and methods
Tissue samples
The present study was approved by the Medical Ethics Committee of the Affiliated Hospital of Jining Medical University (Jining, China). Written informed consent was obtained from patients for the use of their tissue samples. All EOC tissues and matched NATs were obtained from 96 EOC patients who underwent surgery at the Affiliated Hospital of Jining Medical University between July 2013 and January 2015. None of these EOC patients had received other therapeutic treatments prior to surgery. All tissues samples were immediately snap-frozen in liquid nitrogen and transferred to a −80°C freezer.
Cell culture
Human EOC OVCAR3, SKOV3, ES-2 and CAOV-3 cell lines, human normal ovarian epithelial NOEC cell line and HEK293T cell line were purchased from the American Type Culture Collection (Manassas, VA, USA). NOEC cells were maintained in Ham's F-12 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), containing 20% fetal bovine serum (FBS), 120 mg/ml streptomycin and 120 mg/ml penicillin (all from Gibco; Thermo Fisher Scientific, Inc.). All other cells were cultured in RPMI-1640 medium supplemented with 10% FBS, 100 mg/ml penicillin and 100 mg/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc.) in a humidified 5% CO2 cell incubator at 37°C.
Cell transfection
miR-494 mimics, 5′-UGAAACAUACACGGGAAACCUC-3′; miR-494 inhibitor, 5′-GGUUUCCCGUGUAUGUUUCAUU-3′; negative control (NC), 5′-UUCUCCGAACGUGUCACGU-3′; and NC inhibitor, 5′-ACGUGACACGUUCGGAGAA-3′ were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). SIRT1 vectors and empty vector were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Prior to transfection, the cell culture medium was replaced with antibiotic-free medium. Cell transfection was performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
RT-qPCR
RT-qPCR was performed to detect miR-494 expression and to determine the expression level of SIRT1. Total RNA was isolated from tissues and cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. A TaqMan MicroRNA assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) was adopted to analyze miR-494 expression on an Applied Biosystems 7300 instrument (Thermo Fisher Scientific, Inc.). U6 was used as an endogenous control for miR-494 expression. The cycling conditions for this stage were as follows: 40 cycles of denaturation at 95°C for 15 sec; and annealing/extension at 60°C for 60 sec. SIRT1 mRNA expression was measured by RT of total RNA into complementary DNA using M-MLV Reverse Transcriptase (Promega Corporation, Madison, WI, USA), followed by qPCR with SYBR Green Master Mix (Takara Biotechnology Co. Ltd., Dalian, China). The thermocycling conditions of qPCR were as follows: 95°C for 10 min; 40 cycles of 95°C for 15 sec; and 60°C for 1 min. GAPDH was used as an internal control for SIRT1 mRNA expression. Each sample was analyzed in triplicate.
The primer sequences for miR-494 were as follows: Forward, 5′-CATAGCCCGTGAAACATACACG-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 forward, 5′-CGCTTCGGCAGCACATATACTA-3′ and reverse, 5′-GCGAGCACAGAATTAATACGAC-3′; SIRT1 forward, 5′-TCAGTGTCATGGTTCCTTTGC-3′ and reverse, 5′-AATCTGCTCCTTTGCCACTCT-3′; and GAPDH forward, 5′-CCCCTGGCCAAGGTCATCCA-3′ and reverse, 5′-CGGAAGCCATGCCAGTGAG-3′. Relative expression was calculated using the 2−ΔΔCq method (21).
MTT assay
Cellular viability was assessed using an MTT assay (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Briefly, cells were harvested and seeded into 96-well plates at a density of 4,000 cells/well. Following an overnight incubation at room temperature, cells were transfected with the corresponding vectors as described above. At different time points (24–96 h), 20 µl MTT solution (5 mg/ml) was added into each well and the 96-well plates were incubated at 37°C for an additional 4 h. The culture medium was then removed carefully and the formazan precipitates were dissolved in 150 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA). The absorbance at 490 nm was detected using an automatic multi-well spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All samples were analyzed in triplicate.
In vitro cell migration and invasion assays
Transwell chambers with a pore size of 8 µm (Costar; Corning Life Sciences, Cambridge, MA, USA) were adopted to measure cell migration and invasion abilities. For the in vitro cell invasion assay, Transwell chambers coated with Matrigel (BD Biosciences, San Jose, CA, USA) were used. For the migration and invasion assays, transfected cells were harvested and 5×104 cells in 200 µl serum-free RPMI-1640 medium was seeded onto the upper chambers. Culture medium supplemented with 20% FBS in the lower chamber acted as a chemoattractant. Following a 24-h incubation at 37°C for migration and invasion assays, the Transwell chambers were fixed in 100% methanol at room temperature, stained with 0.5% crystal violet and washed with PBS three times. The cells that had not migrated or invaded through the pores were removed with cotton swabs. Cells on the lower surface of the membranes were then counted with an inverted microscope (Olympus Corporation, Tokyo, Japan).
miR-494 targets prediction
The target genes of miR-494 were predicted using the following TargetScan (http://www.targetscan.org/index.html) and miRanda (http://www.microrna.org).
Western blot analysis
Total protein was extracted using radioimmunoprecipitation assay lysis buffer containing protease inhibitors (both Beyotime Institute of Biotechnology, Haimen, China). The total protein concentration was evaluated using a Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Equal quantities of protein (30 µg/lane) were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat dry milk in PBS plus 0.05% Tween-20 (PBST), followed by incubation at 37°C with primary antibodies overnight. Subsequent to washing with PBST three times, membranes were incubated with the corresponding horseradish peroxidase-conjugated secondary antibody (dilution, 1:5,000; cat. no. sc-2005; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for 1 h at room temperature. The protein bands were visualized with an enhanced chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The primary antibodies used in the present study were mouse anti-human monoclonal SIRT1 (dilution, 1:500; cat. no. sc-74504; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and β-actin (dilution, 1:500; cat. no. sc-47778; Santa Cruz Biotechnology, Inc.) antibodies. β-actin was used as a loading control.
Dual-luciferase reporter assay
Dual-luciferase reporter assays were performed to explore whether miR-494 was directly targeted by SIRT1-3′UTR. PGL3-SIRT1-3′UTR wild type (Wt) and PGL3-SIRT1-3′UTR mutant (Mut) were purchased from Shanghai GenePharma Co., Ltd. HEK293T cells were seeded into 24-well plates at a density of 1.5×105 cells/well and incubated overnight at 37°C. Cells were transfected with miR-494 mimics or NC, or with miR-494 inhibitor or NC inhibitor, in addition to PGL3-SIRT1-3′UTR Wt or PGL3-SIRT1-3′UTR Mut, using Lipofectamine 2000. Following 48 h of transfection, transfected cells were harvested. Firefly and Renilla luciferase activities were measured using a Dual-Luciferase Reporter (DLR) Assay System (Promega Corporation). Normalized luciferase activity was reported as Firefly luciferase activity/Renilla luciferase activity. Each assay was replicated three times.
Statistical analysis
Data are presented as the mean ± standard deviation and were compared with Student's t-tests or one-way analysis of variance (ANOVA) plus multiple comparisons using SPSS software (version 17; SPSS Inc., Chicago, IL, USA). Student-Newman-Keuls test was used as a post hoc test following ANOVA. A two-tailed P<0.05 was considered to indicate a statistically significant difference.
Results
Expression level of miR-494 in ECO tissues and cell lines
The present study evaluated the expression of miR-494 in EOC tissues and matched NATs. RT-qPCR analysis indicated that the relative expression of miR-494 was decreased in EOC tissues in comparison with that in matched NATs (Fig. 1A, P<0.05). The expression of miR-494 in the human NOEC and the EOC VCAR3, SKOV3, ES-2 and CAOV-3 cell lines was then examined. As determined by RT-qPCR in EOC tissues, miR-494 was downregulated in the four EOC cell lines compared with its expression in NOEC cells (Fig. 1B, P<0.05). These results indicated that miR-494 expression was impaired in EOC.
Figure 1.
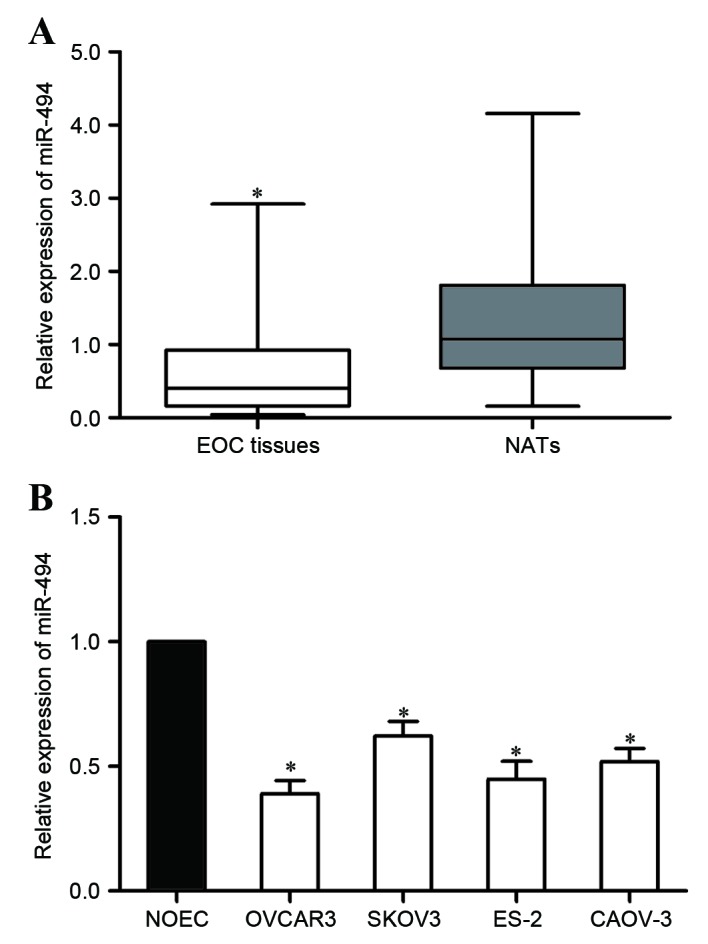
Expression of miR-494 in EOC tissues and cell lines. (A) miR-494 was significantly downregulated in EOC tissues compared with its expression in matched NATs. (B) The expression of miR-494 was lower in the four EOC cell lines investigated than that in the human NOEC cell line. *P<0.05 compared with their respective controls. Data represent the mean ± or + standard deviation. EOC, epithelial ovarian cancer; NOEC, normal ovarian epithelial cell; miR, microRNA.
Clinical significance of miR-494 in EOC patients
To further explore the role of miR-494 in EOC, the association between miR-494 expression level and clinicopathological variables of patients with EOC was studied. As presented in Table I, low expression levels of miR-494 were significantly associated with FIGO stage (P=0.003), tumor size (P=0.010) and lymph node metastasis (P=0.037) in patients with EOC. However, no significant associations were identified between miR-494 expression level and other clinicopathological variables, including age, differentiation and histological subtype (all P>0.05).
Table I.
Association between the expression of microRNA-494 and clinicopathological features in patients with epithelial ovarian cancer.
| MicroRNA-494 expression | ||||
|---|---|---|---|---|
| Clinical features | Case number | Low (n=59) | High (n=37) | P-value |
| Age, years | 0.516 | |||
| <50 | 35 | 23 | 12 | |
| ≥50 | 61 | 36 | 25 | |
| FIGO stage | 0.003 | |||
| I–II | 39 | 17 | 22 | |
| III–IV | 57 | 42 | 15 | |
| Differentiation | 0.964 | |||
| 1/2 | 62 | 38 | 24 | |
| 3 | 34 | 21 | 13 | |
| Tumor size, cm | 0.010 | |||
| <5 | 34 | 15 | 19 | |
| ≥5 | 62 | 44 | 18 | |
| Histological subtype | 0.995 | |||
| Serous | 83 | 51 | 32 | |
| Non-serous | 13 | 8 | 5 | |
| LN metastasis | 0.037 | |||
| No | 66 | 35 | 31 | |
| Yes | 30 | 24 | 6 | |
FIGO, International Federation of Gynecology and Obstetrics; LN, lymph node.
miR-494 inhibits EOC cellular viability, migration and invasion in vitro
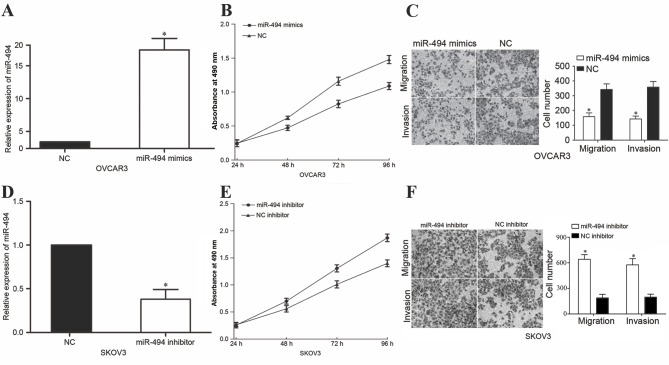
To investigate the functions of miR-494 in EOC, miR-494 was upregulated in OVCAR3 cells by transfection with miR-494 mimics. Following transfection for 48 h, miR-494 was markedly overexpressed in OVCAR3 cells (Fig. 2A, P<0.05). MTT assays were performed to evaluate the effect of altering miR-494 levels on EOC cellular viability. The present study revealed that upregulation of miR-494 resulted in a significant reduction in the proliferation of OVCAR3 cells (Fig. 2B, P<0.05). In vitro cell migration and invasion assays were used to determine the effect of overexpression of miR-494 in cell migration and invasion. The results demonstrated that enforced miR-494 expression led to suppression of migration and invasion in OVCAR3 cells (Fig. 2C, P<0.05).
Figure 2.
miR-494 inhibited EOC cells proliferation, migration and invasion. (A) OVCAR3 cells that were transfected with miR-494 mimics or NC were subjected to RT-qPCR to detect miR-494 expression. (B) MTT assays demonstrated that upregulation of miR-494 resulted in a significant reduction in cellular proliferation in OVCAR3 cells. (C) Cell migration and invasion were suppressed by the overexpression of miR-494 in OVCAR3 cells as compared with that in NC-transfected cells. (D) SKOV3 cells that were transfected with miR-494 inhibitor or NC inhibitor were subjected to RT-qPCR for evaluation of miR-494 expression. (E) Downregulation of miR-494 enhanced the proliferation of SKOV3 cells. (F) Knockdown of miR-494 improved the in vitro cell migration and invasion abilities of SKOV3 cells. *P<0.05 compared with their respective controls. Data represent the mean ± or + standard deviation. EOC, epithelial ovarian cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NC, negative control; miR, microRNA.
miR-494 was also downregulated in SKOV3 cells via transfection with a miR-494 inhibitor. As measured by RT-qPCR, miR-494 was significantly downregulated in SKOV3 cells (Fig. 2D, P<0.05). As expected, downregulation of miR-494 enhanced cell growth, migration and invasion in SKOV3 cells (Fig. 2E and F, P<0.05). Together, these findings suggest that miR-494 acts as a tumor suppressor in EOC.
SIRT1 is a direct target of miR-494 in EOC
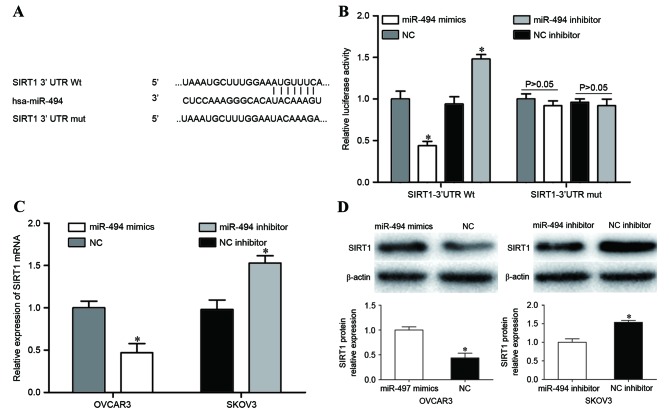
To explore the molecular mechanism by which miR-494 inhibited cellular viability, migration and invasion in EOC, bioinformatic predication algorithms algorithms (TargetScan, Whitehead Institute for Biomedical Research, Cambridge, MA, USA; miRanda, Memorial Sloan-Kettering Cancer Center, NY, USA) were performed. As shown in Fig. 3A, SIRT1 contained a miR-494 7-mer seed match at position 765–771 of the SIRT1-3UTR. To explore whether SIRT1 was directly targeted by miR-494 in EOC, dual-luciferase reporter assays were performed in HEK293T cells. miR-494 markedly suppressed the luciferase activity of SIRT1-3′UTR Wt, but did not inhibit the luciferase activity of SIRT1-3′UTR Mut. When the miR-494 inhibitor was transfected, an increase in luciferase activity of SIRT1-3′UTR Wt was observed. However, co-transfection of miR-494 inhibitor and SIRT1-3′UTR Mut did not cause a decrease in luciferase activity (Fig. 3B, P<0.05).
Figure 3.
SIRT1 was identified as a direct target of miR-494 in EOC. (A) Potential miR-494 binding sequence of SIRT1-3′UTR and the SIRT1-3′UTR Mut sequence at the binding site. (B) miR-494 decreased the luciferase activity caused by SIRT1-3′UTR Wt but not by SIRT1-3′UTR Mut. miR-494 inhibitor improved the luciferase activity of SIRT1-3′UTR Wt. (C) Reverse transcription-quantitative polymerase chain reaction was performed to measure SIRT1 mRNA expression level in OVCAR3 and SKOV3 cells following transfection. (D) Western blot analysis was used to detect SIRT1 protein level in OVCAR3 and SKOV3 cells following transfection. *P<0.05 compared with their respective controls. Data represents the mean + standard deviation. SIRT1, sirtuin 1; miR, microRNA; EOC, epithelial ovarian cancer; UTR, untranslated region; Mut, mutant; Wt, wild type; NC, negative control; mRNA, messenger RNA; hsa, Homo sapiens.
To further investigate the potential effects of miR-494 in the regulation of SIRT1, RT-qPCR and western blot analysis were used to measure SIRT1 mRNA and protein expression. SIRT1 mRNA and protein levels were significantly downregulated in miR-494 mimics-transfected OVCAR3 cells compared with those in the NC group. However, SIRT1 mRNA and protein levels were increased by the downregulation of miR-494 in SKOV3 cells (Fig. 3C and D, P<0.05). Overall, these results identified SIRT1 as a direct target gene of miR-494 in EOC.
miR-494 regulates EOC cellular proliferation, migration and invasion via SIRT1 pathway
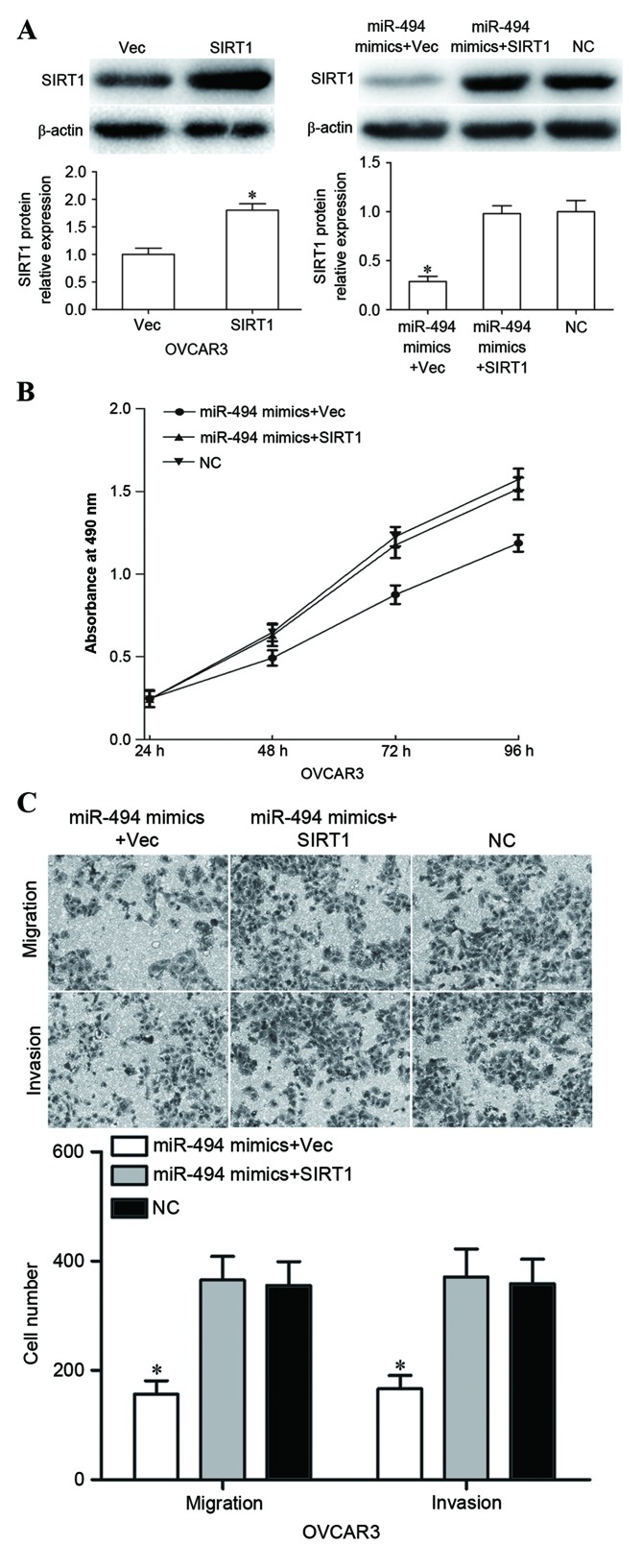
The present study next explored whether SIRT1 could counteract the tumor-suppressor functions of miR-494 in EOC. SIRT1 vectors or empty vector were co-transfected with miR-494 mimics into OVCAR3 cells. Subsequent to transfection, western blot analysis was used to measure the SIRT1 expression level. As expected, SIRT1 was significantly upregulated in SIRT1 vectors-transfected OVCAR3 cells (Fig. 4A, P<0.05). Enforced SIRT1 expression reversed the miR-494-mediated growth-suppression effect in OVCAR3 cells (Fig. 4B). In vitro cell migration and invasion assays demonstrated that upregulated SIRT1 expression abrogated the migration- and invasion-inhibitory effects exerted by miR-494 in OVCAR3 cells (Fig. 4C). Collectively, these results indicate that SIRT1 mediated the tumor-suppressor functions of miR-494 in EOC.
Figure 4.
Overexpression of SIRT1 counteracted the miR-494-mediated tumor-suppressing effects in OVCAR3 cells. (A) Upregulation of SIRT1 in OVCAR3 cells was evaluated by western blot analysis, with β-actin used as a control. (B) Overexpression of SIRT1 reversed the inhibitory effect in proliferation mediated by miR-494 in OVCAR3 cells. (C) OVCAR3 cells' migration and invasion abilities were restored with a forced miR-494 expression and SIRT1 restoration. *P<0.05 compared with their respective controls. Data represents the mean ± or + standard deviation. SIRT1, sirtuin 1; miR, microRNA; Vec, vector.
Discussion
It has been demonstrated that the expression of miR-494 is altered in several types of human cancer; it is upregulated in the cancer tissues of hepatocellular carcinoma (22), colorectal (23) and non-small-cell lung cancer (24), while it is downregulated in various types of human cancer, including gastric carcinoma (25), prostate cancer (26), esophageal squamous cell carcinoma (ESCC) (27), oral cancer (28), breast cancer (29), pancreatic cancer (30), chondrosarcoma (31) and cervical cancer (32). These conflicting studies indicate that the expression of miR-494 in cancers has tissue specificity. The present study identified that miR-494 was downregulated in EOC tissues and cell lines. In addition, low expression levels of miR-494 was demonstrated to be associated with FIGO stage, tumor size and lymph node metastasis in EOC. This indicates that miR-494 may possess tumor-suppressive roles in the carcinogenesis and progression of ovarian cancer.
miR-494 has been identified as an oncogene in cancers. Liu et al (22) reported that the upregulation of miR-494 in hepatocellular carcinoma was positively associated with tumor differentiation, tumor node metastasis (TNM) stage and lymph node metastasis. Furthermore, miR-494 enhanced cellular proliferation and metastasis, decreased cell apoptosis and cell cycle arrest at G1 stage, and conferred sorafenib resistance to hepatocellular carcinoma (22). In colorectal cancer, the overexpression of miR-494 occurred more frequently in tissue specimens with adverse clinical stage and presence of distant metastasis. In addition, multivariate survival analyses demonstrated that the upregulation of miR-494 was an independent prognostic factor for progression-free and overall survival rates (23). A previous functional study demonstrated that miR-494 enhanced the migration and invasion of colorectal cancer cells by negative regulation of phosphatase and tensin homologue (23). In non-small cell lung cancer (NSCLC), miR-494 expression level was positively associated with pathological TNM stage and lymph node metastasis. The expression of miR-494 was negatively associated with the grade of differentiation. In addition, patients with upregulation of miR-494 had a shorter overall survival rate (24). Overexpression of miR-494 improved angiogenesis in NSCLC.
Functions of miRNAs are tissue-type dependent. To date, the vast majority of studies have demonstrated that miR-494 acts as a tumor suppressor. For example, in prostate cancer, the overexpression of miR-494 suppressed cancer cell proliferation, migration and invasion, and enhanced cell apoptosis by post-transcriptional regulation of C-X-C chemokine receptor type 4 (CXCR4) (26). Zhang et al (27) identified that miR-494 expression level was associated with lymph node metastases in ESCC. Upregulation of miR-494 decreased ESCC cell growth, migration and invasion, and induced apoptosis by directly targeting the CLPTM1-like gene (27). In breast cancer, ectopic expression of miR-494 significantly inhibited cellular proliferation and invasion via blockade of CXCR4 through the Wnt/β-catenin signaling pathway (29). In pancreatic cancer, decreased miR-494 expression level was associated with larger tumor size, late TNM stage, lymphatic invasion, distant metastasis and poor prognosis (30). Functional experiments revealed that miR-494 suppressed growth, invasion and chemoresistance in pancreatic cancer through downregulation of SIRT1 and c-Myc (30). The present study demonstrated that miR-494 suppressed ovarian cancer cellular viability, migration and invasion. These findings indicated that upregulating miR-494 or providing analogous pharmaceutical compounds exogenously could be effective cancer therapies for tumors resulting from the activation or overexpression of these oncogenes.
Identification of miR-494 target mRNAs is important for understanding its roles in ovarian cancer initiation and progression. It is also important for exploring new targeted therapies for ovarian cancer. The present study demonstrated that SIRT1 was the direct target gene of miR-494 in ovarian cancer. SIRT1, a highly conserved nicotinamide adenine dinucleotide+-dependent protein deacetylase, is the best-characterized sirtuin (33). SIRT1 has a large variety of substrates, including p53, myogenic differentiation 1, forkhead box O3 and nuclear factor kB (34). Previous studies have shown that SIRT1 was upregulated in numerous types of cancer, including human epithelial, breast, colon and prostate cancer, as well as mouse pulmonary adenocarcinoma, sarcoma and lymphoma, indicating that SIRT1 is involved in cancer carcinogenesis and progression (35–38). SIRT1 performs important functions in a wide spectrum of biological processes, including cellular survival, cellular stress response, energy metabolism, migration, invasion and apoptosis (39–42).
SIRT1 was also observed to be upregulated in malignant ovarian epithelial tumors compared with its expression in benign or borderline tumors. Furthermore, SIRT1 expression level was associated with increased overall survival rate in SIRT1-positive serous carcinoma (43). A previous study demonstrated that SIRT1 was upregulated in EOC tissues compared with is expression in matched NATs (44). This study revealed that SIRT1 contributed to EOC cell growth and metastasis. These findings suggest that SIRT1 was involved in EOC initiation and progression. Therefore, it is worthwhile to investigate novel targeted therapies against SIRT1. Previous studies have demonstrated that SIRT1 was regulated by miRs in numerous human cancer types (45–47). To the best of our knowledge, the present study demonstrates for the first time that SIRT1 was negatively regulated by miR-494 in EOC, which indicated that miR-494 could be investigated as a targeted therapy for EOC.
In conclusion, the present study identified that miR-494 was downregulated in EOC tissues and cell lines. In addition, low expression levels of miR-494 were associated with poor prognostic features, including FIGO stage, tumor size and lymph node metastasis. In vitro studies demonstrated that miR-494 suppressed cellular proliferation, migration and invasion in EOC cells. Mechanistically, the present study demonstrated that miR-494 inhibited EOC cell growth, migration and invasion through directly targeting SIRT1. The present results demonstrate that miR-494 may be a potential clinical biomarker and could also be a therapeutic target in EOC.
Acknowledgements
The present study was supported by the Natural Science Foundation Projects of Shandong Province (grant no. ZR2012HL002) and Research Projects of Jining Medical University (grant no. JY2013KJ016).
References
- 1.Knutson KL, Karyampudi L, Lamichhane P, Preston C. Targeted immune therapy of ovarian cancer. Cancer Metastasis Rev. 2015;34:53–74. doi: 10.1007/s10555-014-9540-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Suh DH, Kim JW, Kim K, Kim HJ, Lee KH. Major clinical research advances in gynecologic cancer in 2012. J Gynecol Oncol. 2013;24:66–82. doi: 10.3802/jgo.2013.24.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y, Wang M, Wu J, Jie Z, Chang S, Shuang T. The clinicopathological significance of miR-1307 in chemotherapy resistant epithelial ovarian cancer. J Ovarian Res. 2015;8:23. doi: 10.1186/s13048-015-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aletti GD, Gallenberg MM, Cliby WA, Jatoi A, Hartmann LC. Current management strategies for ovarian cancer; Mayo Clin Proc; 2007; pp. 751–770. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz PE. Current diagnosis and treatment modalities for ovarian cancer. Cancer Treat Res. 2002;107:99–118. doi: 10.1007/978-1-4757-3587-1_4. [DOI] [PubMed] [Google Scholar]
- 7.Ji K, Ye L, Mason MD, Jiang WG. The Kiss-1/Kiss-1R complex as a negative regulator of cell motility and cancer metastasis (Review) Int J Mol Med. 2013;32:747–754. doi: 10.3892/ijmm.2013.1472. [DOI] [PubMed] [Google Scholar]
- 8.Fan Y, Fan J, Huang L, Ye M, Huang Z, Wang Y, Li Q, Huang J. Increased expression of microRNA-196a predicts poor prognosis in human ovarian carcinoma. Int J Clin Exp Pathol. 2015;8:4132–4137. [PMC free article] [PubMed] [Google Scholar]
- 9.Rutten MJ, van de Vrie R, Bruining A, Spijkerboer AM, Mol BW, Kenter GG, Buist MR. Predicting surgical outcome in patients with International Federation of Gynecology and Obstetrics stage III or IV ovarian cancer using computed tomography: A systematic review of prediction models. Int J Gynecol Cancer. 2015;25:407–415. doi: 10.1097/IGC.0000000000000368. [DOI] [PubMed] [Google Scholar]
- 10.Luo J, Zhou J, Cheng Q, Zhou C, Ding Z. Role of microRNA-133a in epithelial ovarian cancer pathogenesis and progression. Oncol Lett. 2014;7:1043–1048. doi: 10.3892/ol.2014.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katz B, Tropé CG, Reich R, Davidson B. MicroRNAs in Ovarian Cancer. Hum Pathol. 2015;46:1245–1256. doi: 10.1016/j.humpath.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Lu Z, Unruh AK, Ivan C, Baggerly KA, Calin GA, Li Z, Bast RC, Jr, Le XF. Clinically relevant microRNAs in ovarian cancer. Mol Cancer Res. 2015;13:393–401. doi: 10.1158/1541-7786.MCR-14-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 14.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah AA, Leidinger P, Blin N, Meese E. miRNA: Small molecules as potential novel biomarkers in cancer. Curr Med Chem. 2010;17:4427–4432. doi: 10.2174/092986710794182980. [DOI] [PubMed] [Google Scholar]
- 16.Jankovic R, Radulovic S, Brankovic-Magic M. siRNA and miRNA for the treatment of cancer. J BUON. 2009;14(Suppl 1):S43–S49. [PubMed] [Google Scholar]
- 17.Li X, Abdel-Mageed AB, Mondal D, Kandil E. MicroRNA expression profiles in differentiated thyroid cancer, a review. Int J Clin Exp Med. 2013;6:74–80. [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40–R44. [PubMed] [Google Scholar]
- 19.Zhu W, Zhu D, Lu S, Wang T, Wang J, Jiang B, Shu Y, Liu P. miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med Oncol. 2012;29:384–391. doi: 10.1007/s12032-010-9797-4. [DOI] [PubMed] [Google Scholar]
- 20.Dong LL, Chen LM, Wang WM, Zhang LM. Decreased expression of microRNA-124 is an independent unfavorable prognostic factor for patients with breast cancer. Diagn Pathol. 2015;10:45. doi: 10.1186/s13000-015-0257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Liu K, Liu S, Zhang W, Jia B, Tan L, Jin Z, Liu Y. miR-494 promotes cell proliferation, migration and invasion and increased sorafenib resistance in hepatocellular carcinoma by targeting PTEN. Oncol Rep. 2015;34:1003–1010. doi: 10.3892/or.2015.4030. [DOI] [PubMed] [Google Scholar]
- 23.Sun HB, Chen X, Ji H, Wu T, Lu HW, Zhang Y, Li H, Li YM. miR-494 is an independent prognostic factor and promotes cell migration and invasion in colorectal cancer by directly targeting PTEN. Int J Oncol. 2014;45:2486–2494. doi: 10.3892/ijo.2014.2665. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Chen H, Liao Y, Chen N, Liu T, Zhang H, Zhang H. Expression and clinical evidence of miR-494 and PTEN in non-small cell lung cancer. Tumour Biol. 2015;36:6965–6972. doi: 10.1007/s13277-015-3416-0. [DOI] [PubMed] [Google Scholar]
- 25.He W, Li Y, Chen X, Lu L, Tang B, Wang Z, Pan Y, Cai S, He Y, Ke Z. miR-494 acts as an anti-oncogene in gastric carcinoma by targeting c-myc. J Gastroenterol Hepatol. 2014;29:1427–1434. doi: 10.1111/jgh.12558. [DOI] [PubMed] [Google Scholar]
- 26.Shen PF, Chen XQ, Liao YC, Chen N, Zhou Q, Wei Q, Li X, Wang J, Zeng H. MicroRNA-494-3p targets CXCR4 to suppress the proliferation, invasion, and migration of prostate cancer. Prostate. 2014;74:756–767. doi: 10.1002/pros.22795. [DOI] [PubMed] [Google Scholar]
- 27.Zhang R, Chen X, Zhang S, Zhang X, Li T, Liu Z, Wang J, Zang W, Wang Y, Du Y, Zhao G. Upregulation of miR-494 inhibits cell growth and invasion and induces cell apoptosis by targeting cleft lip and palate transmembrane 1-like in esophageal squamous cell carcinoma. Dig Dis Sci. 2015;60:1247–1255. doi: 10.1007/s10620-014-3433-7. [DOI] [PubMed] [Google Scholar]
- 28.Liborio-Kimura TN, Jung HM, Chan EK. miR-494 represses HOXA10 expression and inhibits cell proliferation in oral cancer. Oral Oncol. 2015;51:151–157. doi: 10.1016/j.oraloncology.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 29.Song L, Liu D, Wang B, He J, Zhang S, Dai Z, Ma X, Wang X. miR-494 suppresses the progression of breast cancer in vitro by targeting CXCR4 through the Wnt/β-catenin signaling pathway. Oncol Rep. 2015;34:525–531. doi: 10.3892/or.2015.3965. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Li X, Zhu S, Zhang JG, Yang M, Qin Q, Deng SC, Wang B, Tian K, Liu L, et al. Ectopic expression of miR-494 inhibited the proliferation, invasion and chemoresistance of pancreatic cancer by regulating SIRT1 and c-Myc. Gene Ther. 2015;22:729–738. doi: 10.1038/gt.2015.39. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Wang L, Liu Z, Zu C, Xing F, Yang P, Yang Y, Dang X, Wang K. MicroRNA-494 inhibits cell proliferation and invasion of chondrosarcoma cells in vivo and in vitro by directly targeting SOX9. Oncotarget. 2015;6:26216–26229. doi: 10.18632/oncotarget.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen B, Hou Z, Li C, Tong Y. MiRNA-494 inhibits metastasis of cervical cancer through Pttg1. Tumour Biol. 2015;36:7143–7149. doi: 10.1007/s13277-015-3440-0. [DOI] [PubMed] [Google Scholar]
- 33.Guarente L, Picard F. Calorie restriction-the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 34.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 35.Hida Y, Kubo Y, Murao K, Arase S. Strong expression of a longevity-related protein, SIRT1, in Bowen's disease. Arch Dermatol Res. 2007;299:103–106. doi: 10.1007/s00403-006-0725-6. [DOI] [PubMed] [Google Scholar]
- 36.Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 37.Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation; Proc Natl Acad Sci USA; 2005; pp. 1859–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis; Proc Natl Acad Sci USA; 2008; pp. 13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai L, Yan L, Gao S, Hu CL, Ge H, Davidow A, Park M, Bravo C, Iwatsubo K, Ishikawa Y, et al. Type 5 adenylyl cyclase increases oxidative stress by transcriptional regulation of manganese superoxide dismutase via the SIRT1/FoxO3a pathway. Circulation. 2013;127:1692–1701. doi: 10.1161/CIRCULATIONAHA.112.001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Cantó C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Zhang M, Dong H, Yong S, Li X, Olashaw N, Kruk PA, Cheng JQ, Bai W, Chen J, et al. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene. 2009;28:445–460. doi: 10.1038/onc.2008.388. [DOI] [PubMed] [Google Scholar]
- 43.Jang KY, Kim KS, Hwang SH, Kwon KS, Kim KR, Park HS, Park BH, Chung MJ, Kang MJ, Lee DG, Moon WS. Expression and prognostic significance of SIRT1 in ovarian epithelial tumours. Pathology. 2009;41:366–371. doi: 10.1080/00313020902884451. [DOI] [PubMed] [Google Scholar]
- 44.Mvunta DH, Miyamoto T, Asaka R, Yamada Y, Ando H, Higuchi S, Ida K, Kashima H, Shiozawa T. Overexpression of SIRT1 is associated with poor outcomes in patients with ovarian carcinoma. Appl Immunohistochem Mol Morphol. 2016 Feb 9; doi: 10.1097/PAI.0000000000000316. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 45.Shi Y, Huang J, Zhou J, Liu Y, Fu X, Li Y, Yin G, Wen J. MicroRNA-204 inhibits proliferation, migration, invasion and epithelial-mesenchymal transition in osteosarcoma cells via targeting Sirtuin 1. Oncol Rep. 2015;34:399–406. doi: 10.3892/or.2015.3986. [DOI] [PubMed] [Google Scholar]
- 46.Li K, Xyu Q, Liu X, Liu Q, Wang M. Growth inhibition of human hepatocellular carcinoma by miRNA-204 via down-regulation of Bcl-2 and Sirt1 expression. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2015;31:168–172. (In Chinese) [PubMed] [Google Scholar]
- 47.Yamakuchi M. MicroRNA regulation of SIRT1. Front Physiol. 2012;3:68. doi: 10.3389/fphys.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]