Abstract
MicroRNAs (miRNAs/miRs) are important molecules that are able to regulate multiple cellular processes in cancer cells. miR-7 has been previously identified as a tumor suppressive miRNA in several types of cancer. The aim of the present study was to investigate whether miR-7 is able to regulate autophagy in hepatocellular carcinoma (HCC) cells. It was identified that miR-7 was significantly downregulated in tumor tissues compared with adjacent normal tissues. Overexpression of miR-7 inhibited cell proliferative activity, which was partially reversed by miR-7 inhibitor. In addition, overexpression of miR-7 significantly induced an increasen in autophagic activity, and luciferase activity assay and western blot analysis identified that mammalian target of rapamycin (mTOR) was a direct target of miR-7. In addition, inhibition of autophagy by 3-methyladenine resulted in a marked enhancement of the proliferation inhibition effect of miR-7. In conclusion, miR-7 was identified to induce proliferation inhibition and autophagy in HCC cells by targeting mTOR, and inhibition of autophagy may be utilized to enhance the antitumor activity of miR-7.
Keywords: hepatocellular carcinoma, microRNA-7, autophagy, mammalian target of rapamycin
Introduction
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-associated mortality worldwide (1). HCC is often detected at an advanced stage and exhibits poor prognosis (2). The pathogenesis of HCC is incompletely understood, and an effective treatment for HCC remains a requirement. As such, it is of great importance to elucidate the molecular mechanisms underlying the biological behavior of HCC cells.
Autophagy is a catabolic process that is induced under various conditions of stress (3). Despite its diverse roles proposed by several studies, evidence suggests that autophagy is critical for cell survival (4), and aberrant autophagic activity is implicated in the development, progression and drug resistance of HCC (5–7). Mammalian target of rapamycin (mTOR) tethers several upstream signals to exert an inhibitory effect on autophagy (8). Inhibition of mTOR signaling is one of the potential strategies for HCC therapy (9,10); however, the role of autophagy in this context is not fully understood.
MicroRNAs (miRNAs/miRs) have emerged as a class of key regulators in multiple types of cancers (11,12). mRNAs canonically match with the 3′untranslated region (UTR) of targeted mRNAs, which consequently results in gene silencing (13). A broad spectrum of miRNAs have been implicated in the diagnostic and therapeutic strategies for HCC (14). It has been reported that miR-7 is downregulated in various types of cancer and functions as a tumor suppressor (15,16). Previous studies have identified a number of oncogenes to be targeted by miR-7 (17–20). In HCC, it exhibits anticancer activity by causing cell cycle arrest via targeting critical genes in cell cycle progression (17). However, whether miR-7 regulates other important cell machinery, such as autophagy, remains unknown. In particular, miR-7 contributes to the inhibition of tumor growth and metastasis by modulating the phosphoinositide 3-kinase/protein kinase B (Akt)/mTOR signaling pathway (21), a major signaling pathway involved in the regulation of autophagy induction, indicating its potential involvement in autophagy regulation.
In the present study it was identified that miR-7 is significantly downregulated in HCC tissues compared with the normal adjacent tissue. Supplementation of miR-7 in HepG2 cancer cells inhibited the proliferation of the cells. In addition to the proliferation inhibition effect of miR-7, autophagy was identified to be induced by miR-7 at the same time, and inhibition of mTOR was responsible for the autophagy induction. Most notably, suppression of autophagy enhanced the proliferation inhibition effect of miR-7. Therefore, the results of the present study revealed a novel mechanism by which miR-7 regulates autophagy, and modulation of autophagy may be a feasible approach to enhance the antitumor activity of miR-7.
Materials and methods
Clinical samples
A total of 17 HCC tissues (age, 32–65; 9 males and 8 females) and paired adjacent tissues were obtained under the supervision of the ethics committee of the local hospital (Yantai City Hospital for Infectious Diseases, Shandong, China) between March 2015 and June 2015. All the patients did not receive any chemotherapy prior to surgery. The diagnoses of HCC were histologically confirmed by three independent pathologists, and the negative resection margin was confirmed during the surgery. Tissues were snap-frozen in liquid nitrogen. All patients provided written informed consent for the study.
Cell culture and 3-methyladenine (3-MA) treatment
The human HCC HepG2 and Huh-7 cell lines and human embryonic kidney HEK293-T cell line used in the present study were obtained from the American Type Culture Collection (Manassas, VA, USA). The cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Hyclone; GE Healthcare, Logan, UT, USA) with 10% fetal bovine serum (FBS; Hyclone; GE Healthcare). Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2, and the medium was changed every 3 days. 3-MA was purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany) and dissolved in the culture medium, and a 5 mM final concentration was applied to cells to inhibit autophagy. For autophagy flux assay, co-treatment with 20 µM chloroquine (C6628; Sigma-Aldrich; Merck KGaA) was applied.
Transfection
The miRNA mimic for miR-7 (miR10004553-1-5) and its exact antisense inhibitor (miR200004553-1-5) were obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The nucleotides were transfected using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at a final concentration of 50 nM, according to the manufacturer's protocol. miRNAs were diluted in DMEM and incubated with a proper amount (10 µl for one 6-well plate) of Lipofectamine 2000 at 37°C for 20 min. The Lipofectamine 2000-miRNA mixture was then added to each well of a 6-well plate; after 12 h of incubation in serum-free conditions, the medium was refreshed with complete medium containing 10% FBS.
Cell proliferation assay
To determine the cell proliferation, a Cell Counting Kit-8 (CCK-8; Wuhan Boster Biological Technology, Ltd., Wuhan, China) was used. The experiment was conducted according to the manufacturer's protocol. The absorbance value at 450 nm was determined using a plate reader as a measure of proliferative activity.
Reverse transcription-polymerase chain reaction (RT-PCR)
Following treatment, cells were collected and lysed with 1 ml TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Chloroform (0.2 ml) was then added to the tube and mixed vigorously with TRIzol reagent. RNA was precipitated using isopropyl alcohol from the upper phase and washed using 75% ethanol. The RNA samples were resolved in nuclease-free double-distilled water (Promega Corporation, Madison, WI, USA). miR-7 was then reverse-transcribed using a specific stem-loop primer, followed by a SYBR Green-based amplification protocol. Reverse transcription was performed with the GoScript reverse transcription kit (Promega Corporation), at 25°C for 5 min, and then 42°C for 1 h. PCR was performed using the GoTaq aPCR Master Mix (Promega Corporation), which included the DNA polymerase. The thermocycling conditions were as follows: 95°C for 20 sec, 58°C for 20 sec and 72°C for 20 sec, for 40 cycles. Small nuclear RNA U6 was used as an internal control. The primer sets for miR-7 and U6 were all obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China; cat. no., miRQ0000252-1-2). All experiments were repeated three times. Quantification was performed using the 2−ΔΔCq method (22).
Western blot analysis
Total protein from the treated cells was obtained by homogenizing cells with radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Inc.). The protein concentration was quantified by a bicinchoninic acid kit (Thermo Fisher Scientific, Inc.). Protein (~40 µg) was loaded on 15% SDS-PAGE gels. Following electrophoresis, the protein was blotted onto a polyvinylidene fluoride membrane. The membrane was blocked with non-fat milk at 37°C for 1 h, and incubated with primary antibodies [LC3 (cat. no., 3868; dilution, 1:1,000), mTOR (cat. no., 2983; dilution, 1:1,000), p62 (cat. no., 8025; dilution, 1:1,000), p-mTOR (cat. no., 5536; dilution, 1:1,000) and β-actin (cat. no., TA310155; dilution, 1:2,000)] at 4°C for 16 h. Following 5 washes of 5 min with PBS containing 0.5% Tween-20, membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (goat anti-rabbit HRP (Santa Cruz Biotechnology, Inc., Dallas, TX, USA; cat. no., sc-2005; dilution, 1:5,000) and goat anti-mouse HRP (Santa Cruz Biotechnology, Inc.; cat. no., sc-2004; dilution, 1:5,000) at room temperature for 1 h, followed by visualization using an enhanced chemiluminescence detection kit (GE Healthcare Life Sciences, Little Chalfont, UK), according to the manufacturer's protocol. Antibodies against 1A/1B-light chain 3 (LC3), mTOR, phosphorylated mTOR (p-mTOR) and p62 were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Antibody against β-actin was purchased from OriGene Technologies, Inc. (Beijing, China).
Luciferase activity assay
A 500 bp fragment containing the putative binding site of miR-7 was amplified from the cDNA of mTOR, the fragment of mTOR 3′UTR was then inserted into the 3′UTR of PGL-3 firefly reporter (Promega Corporation). HEK293-T cells grown in 24-well plates were transfected with promoter Renilla luciferase-thymidine kinase (internal control), PGL3-mTOR 3′UTR and miR-7 or miR-7 + miR-7 inhibitor, using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). The luciferase activity was detected after 36 h using the substrates provided in the Dual Luciferase Reporter Assay kit (Promega Corporation), according to the manufacturer's protocol. The firefly luciferase activity was normalized to Renilla luciferase activity, and the relative luciferase activity in each group was obtained by normalizing the value of (firefly luciferase activity)/(Renilla luciferase activity) to that of the negative control (NC)-transfected group.
Immunofluorescent staining
Cells (2×104/ml) were seeded onto coverslips and cultured in 24-well plates. Immediately after transfection, cells were incubated at 37°C for 48 h. The cells on coverslips were then fixed with PBS containing 4% paraformaldehyde, and quickly penetrated with 0.3% Triton X-100 on ice for 5 min. The coverslips were blocked with 5% bovine serum albumin (Sigma-Aldrich; Merck KGaA) for 1 h at room temperature. Primary antibody against LC3 (Cell Signaling Technology, Inc.; cat. no., 3868; dilution, 1:200) was then added to the coverslips. Cells were incubated with primary antibody in a humid chamber overnight at 4°C, and the coverslips were then washed with PBS containing Triton X100 (0.5%). Alexa Fluor® 594-conjugated anti-rabbit secondary antibody (cat. no., R37117; Thermo Fisher Scientific, Inc.) was added and incubated at room temperature for 1 h to label the primary antibody for LC3. Cells were counterstained with DAPI and observed by fluorescence microscopy in five fields of view (Nikon Corporation, Tokyo, Japan; magnification, ×400).
Statistical analysis
Data are expressed as the mean ± standard deviation. A paired Student's t-test was used to determine the differential expression of miR-7 in tumor tissues and normal tissues. One-way analysis of variance followed by Dunnett's test was used for multiple comparisons. P<0.05 (two-tailed) was considered to indicate a statistically significant difference. Statistical analysis was performed using SPSS version 19 software (IBM SPSS, Armonk, NY, USA).
Results
miR-7 expression is downregulated in HCC
Previous studies have demonstrated that miR-7 functions as a tumor suppressor and is downregulated in various types of cancer, including lung cancer and gastric cancer (19,20,23). The present study compared the expression of miR-7 in tumor samples and paired normal tissue. As presented in Fig. 1, a significant decrease in miR-7 was observed in tumor tissue compared with normal tissue, suggesting the potential antitumor role for miR-7 in HCC.
Figure 1.
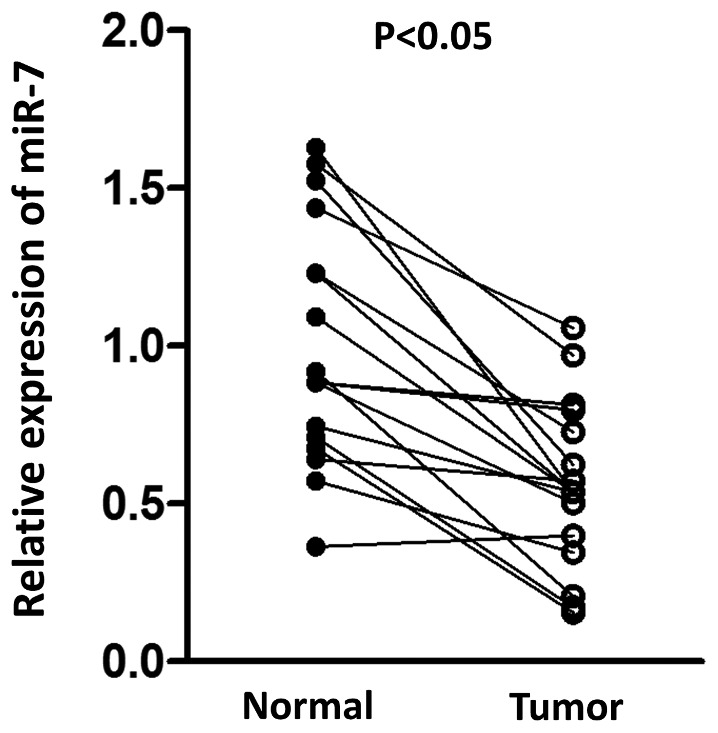
Differential expression of miR-7 in hepatocellular carcinoma tissues and paired adjacent normal tissues. A significant decrease in miR-7 expression was observed in tumor tissues compared with normal tissues (P<0.05). n=17. miR-7, microRNA-7.
miR-7 inhibits the proliferation of HCC cells
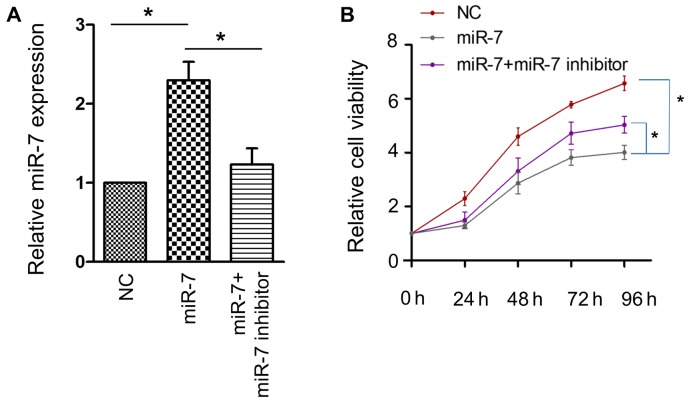
To ascertain the role of miR-7 in HCC, a CCK-8 assay was employed to examine whether miR-7 affects the proliferative activity of the HCC HepG2 cell line. The efficacy of transfection was confirmed using RT-PCR (Fig. 2A). A significant attenuation of cell proliferative activity was observed when miR-7 was overexpressed, and this effect was partially reversed by miR-7 inhibitor (Fig. 2B). These results confirmed that miR-7 serves an antitumor role in HCC.
Figure 2.
miR-7 inhibits the proliferative activity of HepG2 cells. (A) Quantification of miR-7 expression following transfection with miR-7 or miR-7 + miR-7 inhibitor. (B) Effect of miR-7 and miR-7 + miR-7 inhibitor on cell viability. *P<0.05; n=5 in each group. miR-7, microRNA-7; NC, negative control.
Overexpression of miR-7 enhances autophagy
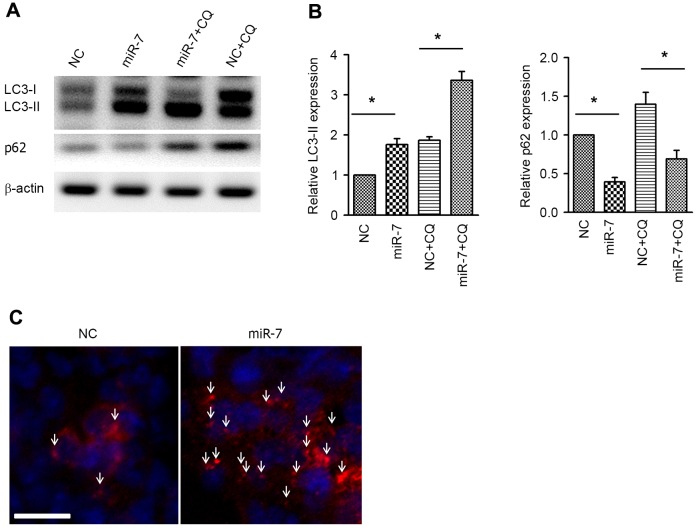
Next, the autophagic activity was examined by western blot analysis of the autophagy markers LC3 and p62. Overexpression of miR-7 resulted in increased expression of LC3-phosphatidylethanolamine conjugate (LC3-II), and, accordingly, p62, which is a specific substrate of autophagy (24), was downregulated (Fig. 3A and B). Co-administration of chloroquine (CQ) was used to conduct the autophagy flux assay. Similar to the aforementioned results, the LC3-II level was also increased in the miR-7 group in the presence of CQ, indicating the increased autophagy flux (Fig. 3A and B). Immunofluorescent staining of LC3 was performed to visualize the autophagosome, and it was revealed that the autophagosome was aggregated in miR-7-transfected cells (Fig. 3C). These results suggested that overexpression enhances autophagic activity in HCC cells.
Figure 3.
Overexpression of miR-7 induces autophagy in HepG2 cells. (A) Effect of miR-7 on autophagy marker LC3 and p62 with or without treatment with CQ. (B) Quantification of the band density of LC3-II and p62. (C) The autophagosome as demonstrated by LC3 immunofluorescent staining. Arrows indicate the autophagosome dots. Scale bar, 25 µm. *P<0.05; n=5 in each group. LC3, 1A/1B-light chain 3; LC3-II, LC3-phosphatidylethanolamine conjugate; miR-7, microRNA-7; CQ, chloroquine; NC, negative control.
mTOR is a target of miR-7
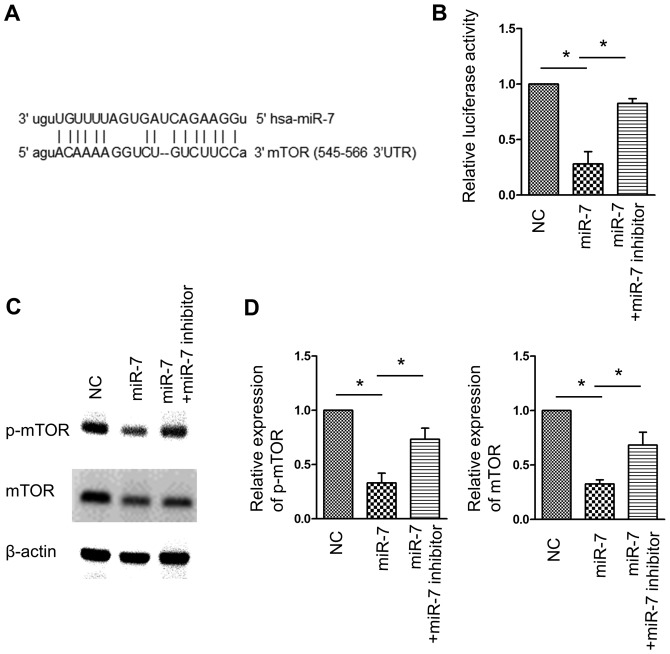
As mTOR is a canonical negative regulator of autophagy, it was hypothesized that miR-7-associated autophagy induction was associated with this pathway. A region in the 3′UTR of the mRNA of mTOR was identified to comprise the seed sequence matching miR-7 (Fig. 4A). A luciferase reporter assay demonstrated that miR-7 overexpression inhibited the luciferase activity of the reporter carrying mTOR 3′UTR, whereas co-transfection of miR-7 and miR-7 inhibitor significantly reversed this effect (Fig. 4B). Western blot analysis confirmed the inhibition of mTOR expression by miR-7, and consequently, p-mTOR, which represents the kinase activity of mTOR, was also inhibited by miR-7 (Fig. 4C and D). These data indicated that mTOR is a target of miR-7, and inhibition of mTOR may contribute to miR-7-activated autophagy.
Figure 4.
Identification of mTOR as a target of miR-7. (A) Schematic diagram of the base pair match between miR-7 and mTOR 3′UTR. (B) Effect of miR-7 and miR-7 inhibitor on luciferase activity. *P<0.05, n=5 in each group. (C) Effect of miR-7 and miR-7 inhibitor on the expression of p-mTOR and mTOR. β-actin served as an internal control. (D) Statistical analysis of the band density of mTOR and p-mTOR. *P<0.05; n=5 in each group. miR-7, microRNA-7; mTOR, mammalian target of rapamycin; p-mTOR, phosphorylated mTOR; NC, negative control; UTR, untranslated region.
Inhibition of autophagy enhances the proliferation inhibition activity of miR-7
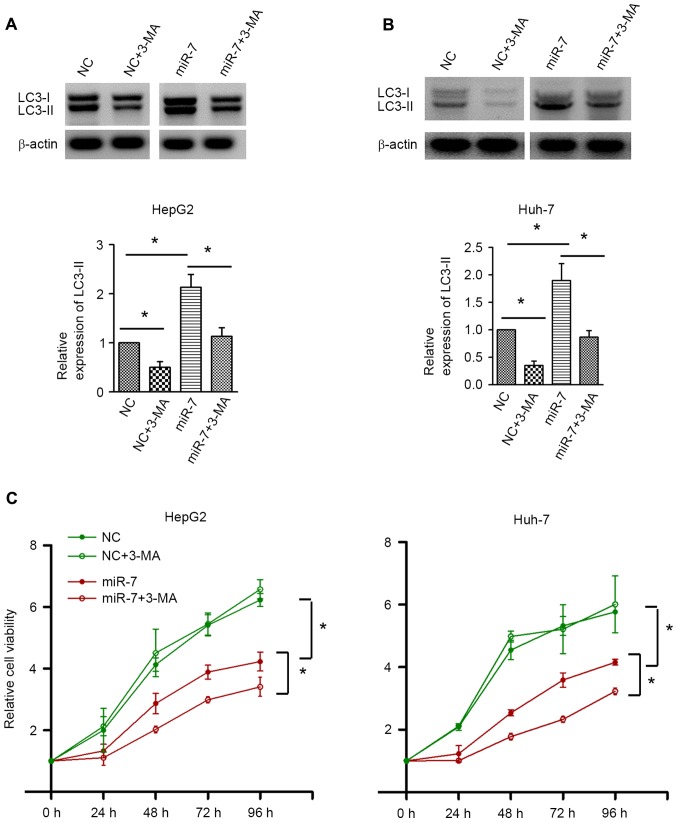
To investigate the exact role of autophagy in HCC, autophagy was inhibited by 3-MA. Western blot analysis confirmed the autophagic inhibition by 3-MA in HepG2 and Huh-7 cells (Fig. 5A and B). 3-MA treatment did not result in significant alterations in cell proliferation in the NC-transfected group. By contrast, inhibition of autophagy by 3-MA significantly decreased the cell proliferative activity upon miR-7 overexpression (Fig. 5C). These data indicated that autophagy induced by miR-7 counteracts the proliferation inhibition effect of miR-7, and inhibition of autophagy may enhance the antitumor effect of miR-7.
Figure 5.
Inhibition of autophagy enhances proliferation inhibition activity of miR-7. Western blot analysis confirmed the induction of autophagy by miR-7 and the inhibition of autophagy by 3-MA in (A) HepG2 and (B) Huh-7 cells, with quantification presented in the histograms. (C) Effect of 3-MA on miR-7-induced proliferation inhibition of HepG2 and Huh-7 cells. *P<0.05; n=5 in each group. miR-7, microRNA-7; 3-MA, 3-methyladenine; NC, negative control; LC3, 1A/1B-light chain 3; LC3-II, LC3-phosphatidylethanolamine conjugate.
Discussion
HCC is one of the leading causes of cancer-associated mortality (11). Owing to the relatively low sensitivity to chemotherapy drugs, the prognosis of advanced HCC is poor (25). Understanding the underlying molecular mechanism will assist in the development of alternative effective strategies for HCC. Recently, the pharmaceutical value of miRNAs has drawn wide attention in cancer treatment (26,27). A spectrum of miRNAs that are involved in cancer cell behavior have demonstrated therapeutic potential in HCC (16). The present study identified that miR-7, one of the downregulated miRNAs in HCC (21), functions as a tumor suppressor. Overexpression of miR-7 was able to significantly suppress the proliferation of HCC cells, and importantly, it was accompanied by increased autophagy. Furthermore, it was demonstrated that mTOR, a negative regulator of autophagy, is a direct target of miR-7. Pharmacological inhibition of autophagy using 3-MA enhanced the anti-proliferative activity of miR-7, indicating that the autophagy inhibitor may serve as the adjuvant drug to enhance the efficacy of miR-7 in the treatment of HCC.
Previous studies have depicted the crucial role of miRNAs in hepatocarcinogenesis (14,28). miRNAs have been incorporated into a number of pathways to regulate cell proliferation, apoptosis, transdifferentiation and drug resistance. miR-7 has been demonstrated in a previous study to directly impede the normal function of several oncogenes in various types of cancer (16). A number of critical oncogenes, including epidermal growth factor receptor (EGFR), B-cell lymphoma-2, Akt and Krüppel-like factor-4 have been validated as the target of miR-7 (18,20,21), suggesting that miR-7 functions as the concurrent upstream regulator of multiple pathways that regulate cell survival. Fang et al (21) reported that p70S6 kinase and mTOR are targeted by miR-7, which potentially provides the molecular basis for miR-7-based therapy for HCC. In the present study, it was confirmed that miR-7 was downregulated in HCC, and consistent with the previous study (21), miR-7 overexpression exerted a significant proliferation inhibition action. Considering that mTOR is critical for the nutrient balance and multiple cellular events of cancer cells (10), the association between mTOR and miR-7 was then investigated. Consistent with the results of Fang et al (21), the present study demonstrated that mTOR was a target of miR-7.
In addition to examining the role of miR-7 in the proliferative activity of HCC cells, the present study focused on other critical processes. Autophagy is a lysosomal degradation process, through which diverse essential substances, including amino acids, lipids and nucleotides, are recycled (3,5). It has been reported that autophagy at the physiological level is beneficial for cell survival, and uncontrolled autophagy may lead to cell death (4). Currently, studies on the role of autophagy in HCC are inconsistent. Luo et al (29) demonstrated that proteasome 26S subunit non-ATPase 10-induced autophagy promoted cell survival against starvation and chemotherapy. Tian et al (30) reported that autophagy is required for the prevention of hepatocarcinogenesis and the development of HCC. These studies suggested that autophagy may be a survival mechanism of HCC cells. By contrast, other studies also demonstrated that autophagy inhibits tumorigenesis in HCC (31–33). Therefore, the exact role of autophagy in the development and progression of HCC is possibly context- and pathway-dependent. There are a few studies demonstrating the regulation of autophagy by miRNAs (34,35). He et al (36) demonstrated that miR-21 inhibits autophagy via the Akt/mTOR signaling pathway, which is important for sorafenib resistance of HCC cells. The present study identified that autophagy was significantly activated upon miR-7 overexpression. Similar to miR-21, a significant suppression of mTOR by miR-7 was observed, suggesting that the mTOR signaling pathway may also confer the induction of autophagy by miR-7. More importantly, inhibition of autophagy by 3-MA sensitized cells to miR-7-induced proliferation inhibition. These data indicated that autophagy may function as an adaptive survival mechanism to overcome the adverse effect of miR-7 on cancer cells. Notably, Tazawa et al (37) demonstrated that genetically engineered oncolytic adenovirus kills lung cancer cells through an autophagic cell death signaling pathway, which is mediated by the E2F transcription factor1/miR-7/EGFR axis. In this study, inhibition of autophagy partially rescued the decrease in cell viability caused by miR-7 overexpression, indicating an opposite role for autophagy in their system. This discrepancy may be a result of several differences in the stimulation type and the gene expression profile of the recipient cells. However, the mechanism for this phenomenon requires clarification in the future. Nonetheless, the results of the present study revealed that miR-7-induced autophagy is cytoprotective in the scenario of HCC.
In conclusion, the results of the present study revealed a novel mTOR-dependent mechanism by which miR-7 regulates autophagy. In addition, inhibition of autophagy enhanced the proliferation inhibition effect of miR-7. The present study suggested that modulating the levels of autophagy may be a feasible tool to enhance the therapeutic efficacy of miR-7 in the treatment of HCC.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Balogh J, Victor D, III, Asham EH, Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM, Monsour HP., Jr Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puri P, Chandra A. Autophagy modulation as a potential therapeutic target for liver diseases. J Clin Exp Hepatol. 2014;4:51–59. doi: 10.1016/j.jceh.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar A, Singh UK, Chaudhary A. Targeting autophagy to overcome drug resistance in cancer therapy. Future Med Chem. 2015;7:1535–1542. doi: 10.4155/fmc.15.88. [DOI] [PubMed] [Google Scholar]
- 7.Du H, Yang W, Chen L, Shi M, Seewoo V, Wang J, Lin A, Liu Z, Qiu W. Role of autophagy in resistance to oxaliplatin in hepatocellular carcinoma cells. Oncol Rep. 2012;27:143–150. doi: 10.3892/or.2011.1464. [DOI] [PubMed] [Google Scholar]
- 8.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bupathi M, Kaseb A, Meric-Bernstam F, Naing A. Hepatocellular carcinoma: Where there is unmet need. Mol Oncol. 2015;9:1501–1509. doi: 10.1016/j.molonc.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Jin W, Jin H, Wang X. mTOR in viral hepatitis and hepatocellular carcinoma: Function and treatment. Biomed Res Int. 2014;2014:735672. doi: 10.1155/2014/735672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inamura K. Diagnostic and therapeutic potential of MicroRNAs in lung cancer. Cancers (Basel) 2017;9 doi: 10.3390/cancers9050049. pii: E49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cătană CS, Pichler M, Giannelli G, Mader RM, Berindan-Neagoe I. Non-coding RNAs, the Trojan horse in two-way communication between tumor and stroma in colorectal and hepatocellular carcinoma. Oncotarget. 2017;8:29519–29534. doi: 10.18632/oncotarget.15706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faller M, Guo F. MicroRNA biogenesis: There's more than one way to skin a cat. Biochim Biophys Acta. 2008;1779:663–667. doi: 10.1016/j.bbagrm.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao M, Wang L, Yao Y, Gu HB, Yao DF. Biomarker-based MicroRNA therapeutic strategies for hepatocellular carcinoma. J Clin Transl Hepatol. 2014;2:253–258. doi: 10.14218/JCTH.2014.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horsham JL, Kalinowski FC, Epis MR, Ganda C, Brown RA, Leedman PJ. Clinical potential of microRNA-7 in cancer. J Clin Med. 2015;4:1668–1687. doi: 10.3390/jcm4091668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalinowski FC, Brown RA, Ganda C, Giles KM, Epis MR, Horsham J, Leedman PJ. microRNA-7: A tumor suppressor miRNA with therapeutic potential. Int J Biochem Cell Biol. 2014;54:312–317. doi: 10.1016/j.biocel.2014.05.040. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Hu S, Zhang X, Wang L, Zhang X, Yan B, Zhao J, Yang A, Zhang R. MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2014;443:1078–1084. doi: 10.1016/j.bbrc.2013.12.095. [DOI] [PubMed] [Google Scholar]
- 18.Xiong S, Zheng Y, Jiang P, Liu R, Liu X, Chu Y. MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. Int J Biol Sci. 2011;7:805–814. doi: 10.7150/ijbs.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo J, Li H, Zhang C. MicroRNA-7 inhibits the malignant phenotypes of non-small cell lung cancer in vitro by targeting Pax6. Mol Med Rep. 2015;12:5443–5448. doi: 10.3892/mmr.2015.4032. [DOI] [PubMed] [Google Scholar]
- 20.Chang YL, Zhou PJ, Wei L, Li W, Ji Z, Fang YX, Gao WQ. MicroRNA-7 inhibits the stemness of prostate cancer stem-like cells and tumorigenesis by repressing KLF4/PI3K/Akt/p21 pathway. Oncotarget. 2015;6:24017–24031. doi: 10.18632/oncotarget.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852–1862. doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Xie J, Chen M, Zhou J, Mo MS, Zhu LH, Liu YP, Gui QJ, Zhang L, Li GQ. miR-7 inhibits the invasion and metastasis of gastric cancer cells by suppressing epidermal growth factor receptor expression. Oncol Rep. 2014;31:1715–1722. doi: 10.3892/or.2014.3052. [DOI] [PubMed] [Google Scholar]
- 24.Bitto A, Lerner CA, Nacarelli T, Crowe E, Torres C, Sell C. P62/SQSTM1 at the interface of aging, autophagy, and disease. Age (Dordr) 2014;36:9626. doi: 10.1007/s11357-014-9626-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu AX. Systemic treatment of hepatocellular carcinoma: Dawn of a new era? Ann Surg Oncol. 2010;17:1247–1256. doi: 10.1245/s10434-010-0975-6. [DOI] [PubMed] [Google Scholar]
- 26.Gandellini P, Doldi V, Zaffaroni N. microRNAs as players and signals in the metastatic cascade: Implications for the development of novel anti-metastatic therapies. Semin Cancer Biol. 2017 Mar 23; doi: 10.1016/j.semcancer.2017.03.005. (Epub ahead of Print) [DOI] [PubMed] [Google Scholar]
- 27.Denaro N, Merlano MC, Russi EG, Lo Nigro C. Non coding RNAs in head and neck squamous cell carcinoma (HNSCC): A clinical perspective. Anticancer Res. 2014;34:6887–6896. [PubMed] [Google Scholar]
- 28.Karagonlar ZF, Korhan P, Atabey N. Targeting c-Met in cancer by MicroRNAs: Potential therapeutic applications in hepatocellular carcinoma. Drug Dev Res. 2015;76:357–367. doi: 10.1002/ddr.21274. [DOI] [PubMed] [Google Scholar]
- 29.Luo T, Fu J, Xu A, Su B, Ren Y, Li N, Zhu J, Zhao X, Dai R, Cao J, et al. PSMD10/gankyrin induces autophagy to promote tumor progression through cytoplasmic interaction with ATG7 and nuclear transactivation of ATG7 expression. Autophagy. 2016;12:1355–1371. doi: 10.1080/15548627.2015.1034405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian Y, Kuo CF, Sir D, Wang L, Govindarajan S, Petrovic LM, Ou JH. Autophagy inhibits oxidative stress and tumor suppressors to exert its dual effect on hepatocarcinogenesis. Cell Death Differ. 2015;22:1025–1034. doi: 10.1038/cdd.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang GM, Jiang QH, Cai C, Qu M, Shen W. SCD1 negatively regulates autophagy-induced cell death in human hepatocellular carcinoma through inactivation of the AMPK signaling pathway. Cancer Lett. 2015;358:180–190. doi: 10.1016/j.canlet.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Wu J, Lin B, Li X, Zhang H, Ding H, Chen X, Lan L, Luo H. Galangin suppresses HepG2 cell proliferation by activating the TGF-β receptor/Smad pathway. Toxicology. 2014;326:9–17. doi: 10.1016/j.tox.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, Lin YJ, Wu CT, Liu HS. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014;59:505–517. doi: 10.1002/hep.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gozuacik D, Akkoc Y, Ozturk DG, Kocak M. Autophagy-Regulating microRNAs and cancer. Front Oncol. 2017;7:65. doi: 10.3389/fonc.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, Liao JZ, He XX, Li PY. The role of autophagy in hepatocellular carcinoma: Friend or foe. Oncotarget. 2017 Apr 18; doi: 10.18632/oncotarget.17202. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He C, Dong X, Zhai B, Jiang X, Dong D, Li B, Jiang H, Xu S, Sun X. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. 2015;6:28867–28881. doi: 10.18632/oncotarget.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tazawa H, Yano S, Yoshida R, Yamasaki Y, Sasaki T, Hashimoto Y, Kuroda S, Ouchi M, Onishi T, Uno F, et al. Genetically engineered oncolytic adenovirus induces autophagic cell death through an E2F1-microRNA-7-epidermal growth factor receptor axis. Int J Cancer. 2012;131:2939–2950. doi: 10.1002/ijc.27589. [DOI] [PubMed] [Google Scholar]