Abstract
Cervical cancer is one of the most common types of gynecological cancer worldwide. MicroRNA-320 (miR-320) has been reported to be downregulated in a number of types of human cancer. However, the expression level and functions of miR-320 in cervical cancer remain unknown. In the present study, miR-320 was identified to be markedly downregulated in cervical cancer tissues and cell lines. For the functional studies, miR-320 mimic or miR-320 inhibitor was introduced into cervical cancer cell lines. The effects of miR-320 on cervical cancer cell viability, migration and invasion were evaluated using MTT, migration and invasion assays, respectively. The results of the present study identified that overexpression of miR-320 suppressed the viability, migration and invasion of cervical cancer cells. In contrast, underexpression of miR-320 improved the viability, migration and invasion of cervical cancer cells. Bioinformatics analysis, dual-luciferase reporter assay and western blot analysis were adopted to investigate the underlying molecular mechanism of the suppressive functions of miR-320 in cervical cancer. The results of the present study demonstrated that miR-320 negatively regulated forkhead box M1 (FOXM1) expression by directly targeting the 3′ untranslated region of FOXM1. Furthermore, the functions of FOXM1 short interfering RNA were similar to those induced by miR-320 in cervical cancer, identifying FOXM1 as a functional target of miR-320 in cervical cancer. The results of the present study indicated that miR-320 acted as a tumor suppressor in the viability, migration and invasion of cervical cancer through directly targeting FOXM1, suggesting that miR-320 may be a target for the therapeutic treatment of cervical cancer.
Keywords: microRNA-320, cervical cancer, forkhead box M1, prognosis, metastasis
Introduction
Cervical cancer is the second highest cancer associated-morbidity and mortality in females worldwide, with >500,000 novel cases and ~260,000 mortalities caused by cervical cancer annually (1,2). Cervical cancer is more prevalent in developing countries, primarily due to the lack of screening programs, diagnostic procedures and effective therapies (3,4). Patients with cervical cancer at an early stage are treated with surgery, followed by radiotherapy and/or chemotherapy (5,6). For patients with metastatic diseases, or those at a more advanced stage who are at marked risk of recurrence, concurrent chemoradiotherapy was recommended as a standard therapy (7,8). However, the overall survival rate for patients with locally advanced and metastatic cervical cancer is between 30 and 50%, and between 5 and 15%, respectively (9). Furthermore, ~30% of patients presented with cancer recurrence, lymph node recurrence and distant metastasis, and obtained an unfavorable prognosis (10). Although the mechanism of cervical carcinogenesis and progression has been hypothesized, the underlying molecular mechanism of cervical carcinogenesis and progression remains unknown (11). Thus, understanding the molecular mechanism underlying the initiation and progression of cervical cancer, and identifying effective therapeutic treatments for patients with cervical cancer is required.
MicroRNAs (miRNAs) are a group of endogenous non-protein-coding single-stranded short RNA molecules of between 19 and 25 nucleotides in length (12). miRNAs negatively regulate protein expression predominantly through binding to the 3′ untranslated regions (3′UTRs) of target mRNAs in a base-pairing manner, resulting in the degradation or translation inhibition of mRNA (13–15). Previous studies have indicated that miRNAs regulate the activity of >30% of human genes and are implicated in a variety of physiological and pathological processes, including cell viability, development, apoptosis, survival and metastasis (16–18). In addition, an association between abnormal expression of miRNAs and the initiation and development of cancers has been identified (19,20). Furthermore, miRNAs may function as oncogenes or as tumor suppressors, depending on the nature of their target mRNAs (21,22). Therefore, targeting miRNAs may be used to identify a novel therapeutic treatment for patients with cervical cancer.
The aim of the present study was to investigate the expression, functions and underlying molecular mechanism of miR-320 in cervical cancer. miR-320 was identified to be markedly downregulated in cervical cancer tissues and cell lines. In vitro functional studies revealed that overexpression of miR-320 inhibited the viability, migration and invasion of cervical cancer, whereas the inhibition of miR-320 enhanced these processes. Additionally, forkhead box M1 (FOXM1) was identified as a direct functional target gene of miR-320 in cervical cancer. The results of the present study revealed a novel underlying molecular mechanism involved in the regulation of FOXM1 and cervical carcinogenesis and development. Thus, miR-320/FOXM1-based targeted therapy may be a therapeutic strategy for patients with cervical cancer.
Materials and methods
Tissue samples
The present study was approved by the Medical Ethics Committee of Beijing Chao-Yang Hospital (Beijing, China), in accordance with The Declaration of Helsinki. In addition, written informed consent was obtained from all patients in the present study, in order for patient information to be stored in the hospital database and used for the present study. A total of 36 human cervical cancer tissue samples and corresponding healthy cervical epithelial tissues were obtained from patients with cervical cancer who underwent surgery at Beijing Chao-Yang Hospital. None of the patients received chemotherapy and radiotherapy, or other therapeutic treatments prior to surgery. All tissue specimens were immediately frozen in liquid nitrogen following surgery and stored at −80°C.
Cell lines and culture conditions
An immortalized human papillomavirus (HPV)-negative skin keratinocyte line (HaCaT), four human cervical cancer cell lines (HeLa, CaSki, C33A and SiHa) and the HEK293T cell line, used for the luciferase reporter assay, were obtained from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM) or RPMI-1640 medium and supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), at 37°C in a humidified atmosphere containing 5% CO2.
miRNA/short interfering RNA (siRNA) transfection
Human miR-320 mimic, a corresponding negative control (NC), miR-320 inhibitor and NC inhibitor were obtained from GenePharma Co., Ltd. (Shanghai, China). FOXM1 siRNA and the scramble control siRNA were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China), and were used to knock down FOXM1 in cervical cancer cells. The miR-320 mimic sequence was 5′-AAAAGCUGGGUUGAGAGGGCGA-3′; the NC, 5′-UUCUCCGAACGUGUCACGUTT-3′; miR-320 inhibitor, 5′-CCUCUCAACCCAGCUUUU-3′; NC inhibitor, 5′-CAGUACUUUUGUGUAGUACAA-3′; FOXM1 siRNA, 5′-GGACCACUUUUCCCUACUUUTT-3′; scramble control siRNA, 5′-UUCUUCCGAACGUGUCACGUTT-3′. Cells were transfected with 50 pmol/ml miR-320 mimic, NC, miR-320 inhibitor, NC inhibitor, FOXM1 siRNA or scramble control siRNA using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The medium was replaced with culture medium at 6 h after transfection.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA extraction from tissues or cells was carried out using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). To determine mature miR-320 and U6 expression, reverse transcription was conducted using a TaqMan MicroRNA Reverse Transcription kit, followed by RT-qPCR using a TaqMan miRNA assay (all from Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 40 cycles of denaturation at 95°C for 15 sec and annealing/extension at 60°C for 60 sec. U6 was used as an internal control for miR-320 expression. The primers were designed as follows: miR-320, 5′-ACACTCCAGCTGGGAAAAGCTGGGTTGAGA-3′ (forward) and 5′-TGGTGTCGTGGAGTCG-3′ (reverse); U6, 5′-CTCGCTTCGGCAGCACATATACT-3′ (forward) and 5′-ACGCTTCACGAATTTGCGTGTC-3′ (reverse). All samples were performed in triplicate. Relative expression level was calculated using the 2−ΔΔCq method (23).
MTT assay
Cell viability was determined using an MTT assay (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). At 24 h after transfection, cells were collected and seeded into 96-well plates at a density of 4,000 cells/well and cultured for 24, 48, 72 and 96 h. A total of 20 µl MTT solution was added to each well prior to incubation for an additional 4 h. Subsequently, 200 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was used to dissolve the formazan precipitates. The absorbance at 490 nm was determined using an automatic multi-well spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All experiments were carried out in triplicate.
Transwell cell migration and invasion assay
Transfected cells (1×105) in 200 µl serum-free DMEM were placed into the upper Transwell chamber (8 µm; Millipore, Billerica, MA, USA), which were pre-coated with or without Matrigel (BD Biosciences, San Jose, CA, USA). The lower compartment was filled with 500 µl DMEM medium containing 20% FBS to act as a chemoattractant. Following incubation at 37°C for 24 h, cells that had not migrated or invaded to the lower surface of the filters were removed using cotton swabs. Subsequently, the cells that had migrated or invaded to the bottom surface of the Transwell chambers were fixed with 100% methanol for 10 min, stained with 0.5% crystal violet solution for 10 min, and washed with PBS, at room temperature. The capacity for cell migration and invasion was determined by counting 5 fields per membrane with an optical microscope (magnification, ×200; Olympus IX53; Olympus Corporation, Tokyo, Japan). Each experiment was performed in triplicate.
miR-320 target prediction
The potential targets of miR-320 were predicted using the PicTar (pictar.mdc-berlin.de), TargetScan (www.targetscan.org) and miRanda (www.microrna.org/microrna) databases.
Dual-luciferase reporter assay
pGL3-FOXM1-3′UTR wild-type (Wt) or pGL3-FOXM1-3′UTR mutant (Mut) were purchased from GenePharma Co., Ltd. HEK293T cells were seeded into 24-well plates. Following incubation at 37°C overnight, cells were transfected with miR-320 mimic or NC, and co-transfected with pGL3-FOXM1-3′UTR Wt or PGL3-FOXM1-3′UTR Mut using Lipofectamine 2000 reagent. Luciferase activities were determined consecutively 48 h post-transfection using Dual-Luciferase Reporter assays (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. The Renilla luciferase activities were normalized to the firefly luciferase activities. All experiments were carried out in triplicate.
Western blot analysis
Western blot analysis was used to evaluate the expression level of FOXM1. Cells were collected and lysed in ice-cold radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China) 72 h post-transfection. The concentration of total protein was measured using a Bicinchoninic Acid Protein assay kit (Thermo Fisher Scientific, Inc.). Equal amounts of protein (30 µg) were separated using SDS-PAGE (10% gel) and subsequently transferred onto polyvinylidene fluoride membranes (Millipore). Following incubation with Tris-buffered saline-Tween-20 (TBST) containing 5% non-fat dry milk at room temperature for 1 h, the membranes were probed with the following primary antibodies: Mouse anti-human monoclonal FOXM1 (1:1,000 dilution; cat. no. sc-166709; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and β-actin (1:1,000 dilution; cat. no. sc-130301; Santa Cruz Biotechnology, Inc.). Following incubation at 4°C overnight, the membranes were washed with TBST three times, followed by incubation with corresponding horseradish peroxidase-conjugated secondary antibody (1:1,000 dilution; cat. no. A0216; Beyotime Institute of Biotechnology) for 1 h at room temperature. The protein bands were determined using the an enhanced chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc.).
Statistical analysis
Data are presented as the mean ± standard deviation and compared with Student's t-test, or one-way analysis of variance followed by the SNK multiple comparison test, using SPSS software (version 13.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-320 is downregulated in cervical cancer tissue samples and cell lines
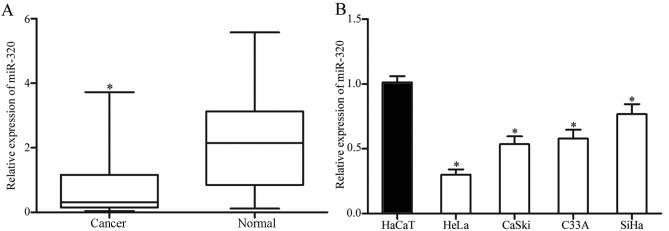
To explore whether miR-320 was involved in the carcinogenesis and progression of cervical cancer, the expression levels of miR-320 in human cervical cancer tissues and corresponding healthy cervical epithelial tissues were determined. As presented in Fig. 1A, miR-320 was markedly downregulated in cervical cancer tissue samples, compared with that in corresponding healthy cervical epithelial tissues. Consistent with the results identified in cervical cancer tissues, the expression level of miR-320 was decreased in HeLa, CaSki, C33A and SiHa cell lines, compared with that in an immortalized HPV-negative skin keratinocyte line (HaCaT; Fig. 1B). Thus, miR-320 was identified to be downregulated in cervical cancer.
Figure 1.
miR-320 is markedly downregulated in cervical cancer tissues and cell lines. (A) RT-qPCR analysis of miR-320 expression levels in human cervical cancer tissues and corresponding healthy cervical epithelial tissues. (B) RT-qPCR analysis of miR-320 expression in four cervical cancer cell lines (HeLa, CaSki, C33A and SiHa) and an immortalized human papillomavirus-negative skin keratinocyte line (HaCaT). *P<0.05 vs. respective controls. miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Overexpression of miR-320 suppresses viability, migration and invasion of cervical cancer cells
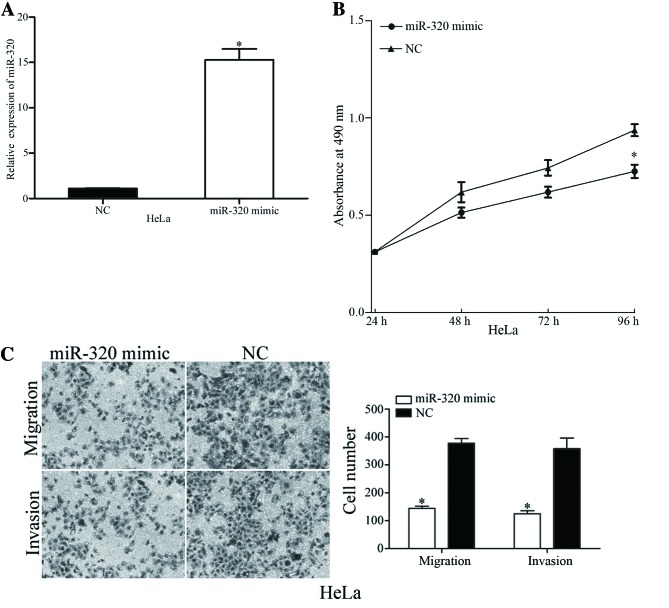
To identify the functions of miR-320 in cervical cancer, miR-320 mimic or NC was transfected into HeLa cells, which exhibited decreased expression levels of miR-320. Following transfection, miR-320 expression was markedly overexpressed in HeLa cells (Fig. 2A). The viability, migration and invasion of miR-320-overexpressing cells was evaluated using MTT, migration and invasion assays, respectively. The MTT assay results revealed that overexpression of miR-320 suppressed the viability of HeLa cells, compared with the NC (Fig. 2B). Furthermore, the migration and invasion assay demonstrated that upregulation of miR-320 decreased the migratory and invasive potential of HeLa cells (Fig. 2C). The results of the present study indicated that miR-320 exhibited a suppressive role in the viability and metastasis of cervical cancer cells.
Figure 2.
Overexpression of miR-320 suppresses the cell viability, migration and invasion of HeLa cells. (A) Reverse transcription-quantitative polymerase chain reaction analysis of miR-320 expression levels in HeLa cells, following transfection with miR-320 mimic or NC. (B) An MTT assay revealed that overexpression of miR-320 resulted in a significant decrease in the viability of HeLa cells. (C) Transwell cell migration and invasion assays identified that migratory and invasive capabilities were decreased by the overexpression of miR-320 in HeLa cells. *P<0.05 vs. respective controls. miR, microRNA; NC, negative control.
Underexpression of miR-320 enhances viability, migration and invasion of cervical cancer cells
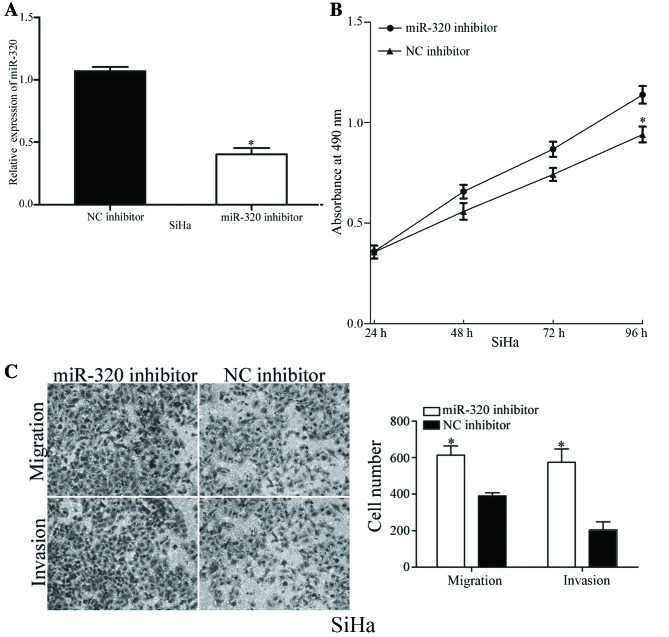
To validate the functions of miR-320 in the viability and metastasis of cervical cancer cells, SiHa cells, which exhibited increased expression levels of miR-320, were transfected with an miR-320 inhibitor or NC inhibitor. As presented in Fig. 3A, the expression level of miR-320 was markedly decreased in miR-320 inhibitor-transfected SiHa cells. The viability, migration and invasion of miR-320-underexpressing cells was analyzed using MTT, migration and invasion assays, respectively. Decreasing the expression levels of miR-320 resulted in an increased viability rate and improved cell migratory and invasive abilities of SiHa cells (Fig. 3B and C). The results of the present study demonstrated the inhibitory functions of miR-320 in the viability, migration and invasion of cervical cancer cells.
Figure 3.
Underexpression of miR-320 increases the cell viability, migration and invasion in HeLa cells. (A) Relative expression levels of miR-320 in SiHa cells were determined using the reverse transcription-quantitative polymerase chain reaction, following transfection with a miR-320 inhibitor or NC inhibitor. (B) Underexpression of miR-320 increased the viability of SiHa cells. (C) Downregulation of miR-320 increased the in vitro cell migratory and invasive abilities of SiHa cells. *P<0.05 vs. respective controls. miR, microRNA; NC, negative control.
miR-320 regulates FOXM1 expression by directly targeting 3′UTR of FOXM1
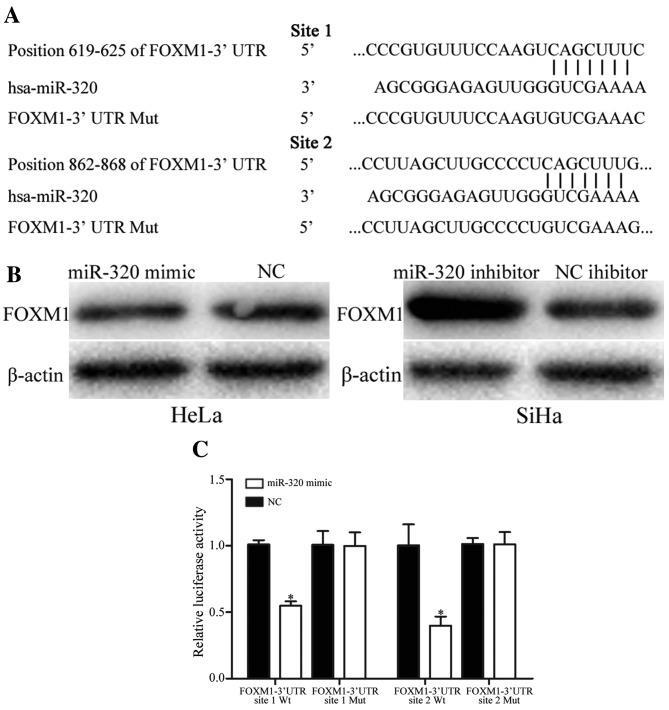
To identify the molecular mechanism underlying the suppressive functions of miR-320 in cell viability, migration and invasion, PicTar, TargetScan and miRanda databases were used to predict the target genes of miR-320. Bioinformatics analysis predicated that FOXM1 was a direct target gene of miR-320 (Fig. 4A). Western blot analysis revealed that overexpression of miR-320 decreased the expression level of FOXM1 protein in HeLa cells, whereas underexpression of miR-320 increased the expression level of FOXM1 in SiHa cells (Fig. 4B).
Figure 4.
miR-320 regulates FOXM1 expression levels by directly targeting the 3′UTR of FOXM1 in cervical cancer. (A) miR-320-binding site in the 3′UTR of FOXM1 identified using PicTar, TargetScan and miRanda databases, and the FOXM1 3′UTR mutant sequence. (B) Western blot analysis of FOXM1 protein expression in HeLa cells following transfection with the miR-320 mimic or NC, and SiHa cells transfected with miR-320 inhibitor or NC inhibitor. β-actin was used as a loading control. (C) miR-320 decreased the FOXM1-3′UTR site 1 Wt and FOXM1-3′UTR site 2 Wt luciferase activity, but not the FOXM1-3′UTR site 1 Mut and FOXM1-3′UTR site 2 Mut luciferase activity in HEK293T cells. *P<0.05 vs. respective controls. miR, microRNA; FOXM1, forkhead box M1; UTR, untranslated region; NC, negative control; Wt, wild-type; Mut, mutant; hsa, human.
Furthermore, the binding of miR-320 on the 3′UTR of FOXM1 was evaluated using a dual-luciferase reporter assay in HEK293T cells. The results demonstrated that HEK293T cells co-transfected with miR-320 mimic and FOXM1-3′UTR Wt markedly decreased the luciferase activities, whereas no effect was observed when the 3′UTR of FOXM1 was mutated (Fig. 4C). The results of the present study suggested that miR-320 negatively regulated FOXM1 expression by directly targeting the 3′UTR of FOXM1.
miR-320 suppresses the viability, migration and invasion of cervical cancer cells via the regulation of FOXM1
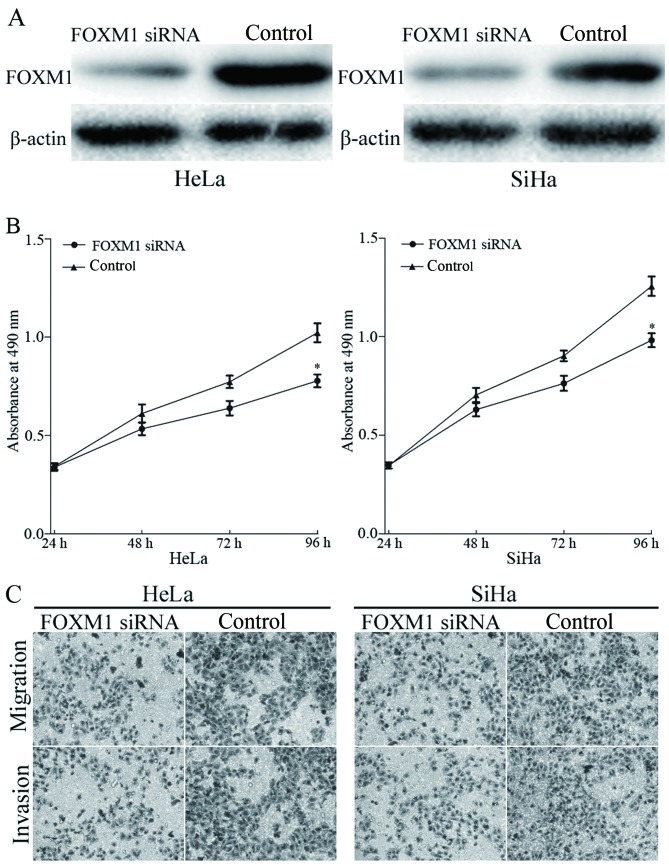
FOXM1 was identified as a direct target of miR-320 in cervical cancer. To elucidate whether miR-320 functions through FOXM1, FOXM1 siRNA was transfected into HeLa and SiHa cells. Transfection with FOXM1 siRNA resulted in marked downregulation of FOXM1 expression in HeLa and SiHa cells (Fig. 5A). The effects of FOXM1 siRNA on cell viability, migration and invasion was evaluated using MTT, migration and invasion assays, respectively. The results identified that knockdown of FOXM1 inhibited viability (Fig. 5B), migration and invasion (Fig. 5C) of HeLa and SiHa cells, compared with the control groups. The results indicated that miR-320, by knocking down FOXM1 expression, inhibited the viability, migration and invasion of cervical cancer cells.
Figure 5.
FOXM1 siRNA suppressed the viability, migration and invasion of HeLa and SiHa cells. (A) Western blot analysis of FOXM1 protein expression in HeLa and SiHa cells following transfection with the FOXM1 siRNA. β-actin was used as a loading control. (B) An MTT assay demonstrated that FOXM1 siRNA suppressed the cellular viability in HeLa and SiHa cells, compared with that in control groups. (C) Transwell migration and invasion assays identified that FOXM1 siRNA significantly decreased the cell migratory and invasive capabilities of HeLa and SiHa cells. *P<0.05, vs. respective controls. FOXM1, forkhead box M1; siRNA, short interfering RNA.
Discussion
Cervical cancer is one of the most common types of gynecological cancer worldwide (24). Currently, the standard therapeutic treatments for patients are surgical resection, followed by chemotherapy and radiotherapy. However, the overall survival rate remains low (25). Therefore, the development of novel therapeutic strategies by targeting the molecules involved in carcinogenesis and development of cervical cancer are required to improve the prognosis for patients with cervical cancer. In the present study, miR-320 was identified to be downregulated in cervical cancer tissues and cell lines. Functional studies revealed that the overexpression of miR-320 markedly decreased the viability, migration and invasion of cervical cancer cells, whereas the underexpression of miR-320 led to the opposite results. In addition, FOXM1 was demonstrated to be a functional target of miR-320 in cervical cancer. The results of the present study suggested that miR-320 may be a therapeutic target for patients with cervical cancer.
miR-320 has been reported to be downregulated in a number of types of human cancer, and serves important functions in carcinogenesis and cancer progression. Sun et al (26) identified that miR-320 was markedly downregulated in glioma tissues, compared with that in healthy tissues, and overexpression of miR-320 inhibited cell viability and metastasis by the downregulation of E2F transcription factor 1. Cheng et al (27) reported that miR-320 expression levels were decreased in human osteosarcoma tissues, and increased miR-320 expression in osteosarcoma cells suppressed viability in vitro and in vivo by directly targeting fatty acid synthase. Furthermore, Wu et al (28) demonstrated that miR-320, which was decreased in oral squamous cell carcinoma tissues and cell lines, suppressed the tumorigenicity of oral squamous cell carcinoma cells in vitro and tumor angiogenesis in vivo, via inhibition of neuropilin 1. In addition, Wu et al (28) reported that miR-320 was downregulated in prostate cancer, and that the upregulation of miR-320 inhibited carcinogenesis and progression of prostate cancer through the downregulation of the Wnt/β-catenin signaling pathway. The overexpression of miR-320 in prostate stem-like tumor-initiating cells markedly inhibited the stem cell-like properties of prostate cancer cells, including tumorsphere formation, chemoresistance and tumorigenic abilities (29). In the present study, it was revealed that miR-320 was downregulated in cervical cancer and functioned as a tumor suppressor in cervical cancer by inhibiting the viability, migration and invasion, via directly targeting FOXM1.
miRNAs typically exert biological functions through the negative regulation of target mRNAs, via directly binding to the 3′UTRs of target mRNAs (30). In the present study, three bioinformatics algorithms were used to predict the target genes of miR-320. FOXM1 was identified to contain two miR-320 seed matches at positions between 619 and 625, and between 862 and 868 of the 3′UTR of FOXM1. The dual-luciferase reporter assay validated that miR-320 directly targeted the 3′UTR of FOXM1. Western blot analysis revealed that the overexpression of miR-320 decreased FOXM1 expression levels in cervical cancer, whereas underexpression of miR-320 increased FOXM1 expression levels. Furthermore, the functions of FOXM1 siRNA were similar to those induced by miR-320 in cervical cancer, suggesting that FOXM1 was a functional target of miR-320 in cervical cancer. The identification of target genes of miR-320 is required to elucidate the functions of miR-320 in the carcinogenesis and progression of cervical cancer, and may provide novel therapeutic targets for the treatment of patients with cervical cancer.
FOXM1 serves a function in tumor initiation and progression (31). FOXM1 has been reported to be upregulated in a number of types of tumor, including lung, breast, liver, pancreatic and cervical cancer, and glioblastoma (32,33). FOXM1 serves a function in the regulation of a number of biological processes, including cell viability, cell cycle progression, cell differentiation, DNA damage repair, tissue homeostasis, angiogenesis and apoptosis (34). In cervical cancer, FOXM1 was markedly overexpressed in cervical cancer tissues, and increased expression levels of FOXM1 was associated with the tumor late stage and cell viability marker Ki67 (35).
In functional studies, enforced FOXM1 expression increased the migration and invasion of cervical cancer cells, whereas the knockdown of FOXM1 exhibited the opposite effect in cervical metastasis (33). Furthermore, Chen et al (36) reported that the underexpression of FOXM1 suppressed the viability, invasion and angiogenesis of cervical cancer cells in vitro and in vivo (36). Therefore, the overexpression of FOXM1 protein in cervical cancer was associated with the carcinogenesis and progression of cervical cancer, and may be investigated as a novel therapeutic target for cervical cancer. In the present study, overexpression of miR-320 was identified to target FOXM1, and inhibit viability and metastasis of cervical cancer, suggesting that miR-320/FOXM1-based targeted therapy may be a novel therapeutic strategy for the treatment of patients with cervical cancer.
The results of the present study demonstrated that miR-320 was downregulated in cervical cancer tissues and cell lines. Additionally, it was revealed that miR-320 acted as a tumor suppressor and exhibited a marked suppressive effect on the viability, migration and invasion of cervical cancer in vitro. Furthermore, FOXM1 was identified as a direct target gene of miR-320 in cervical cancer. The overexpression of miR-320 may be a novel therapeutic target for the treatment of cervical cancer.
References
- 1.Arbyn M, Castellsagué X, de Sanjosé S, Bruni L, Saraiya M, Bray F, Ferlay J. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–2686. doi: 10.1093/annonc/mdr015. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Su SY, Huang JY, Ho CC, Liaw YP. Evidence for cervical cancer mortality with screening program in Taiwan, 1981–2010: Age-period-cohort model. BMC Public Health. 2013;13:13. doi: 10.1186/1471-2458-13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Yao D, Li Y, Chen H, He C, Ding N, Lu Y, Ou T, Zhao S, Li L, Long F. Serum microRNA expression levels can predict lymph node metastasis in patients with early-stage cervical squamous cell carcinoma. Int J Mol Med. 2013;32:557–567. doi: 10.3892/ijmm.2013.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yee GP, de Souza P, Khachigian LM. Current and potential treatments for cervical cancer. Curr Cancer Drug Targets. 2013;13:205–220. doi: 10.2174/1568009611313020009. [DOI] [PubMed] [Google Scholar]
- 6.Wang F, Liu M, Li X, Tang H. MiR-214 reduces cell survival and enhances cisplatin-induced cytotoxicity via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett. 2013;587:488–495. doi: 10.1016/j.febslet.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Rose PG, Bundy BN, Watkins EB, Thigpen JT, Deppe G, Maiman MA, Clarke-Pearson DL, Insalaco S. Concurrent cisplatin-based radiotherapy and chemotherapy for locally advanced cervical cancer. N Engl J Med. 1999;340:1144–1153. doi: 10.1056/NEJM199904153401502. [DOI] [PubMed] [Google Scholar]
- 8.Thomas GM. Improved treatment for cervical cancer-concurrent chemotherapy and radiotherapy. N Engl J Med. 1999;340:1198–1200. doi: 10.1056/NEJM199904153401509. [DOI] [PubMed] [Google Scholar]
- 9.Mayr NA, Huang Z, Wang JZ, Lo SS, Fan JM, Grecula JC, Sammet S, Sammet CL, Jia G, Zhang J, et al. Characterizing tumor heterogeneity with functional imaging and quantifying high-risk tumor volume for early prediction of treatment outcome: Cervical cancer as a model. Int J Radiat Oncol Biol Phys. 2012;83:972–979. doi: 10.1016/j.ijrobp.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kogo R, How C, Chaudary N, Bruce J, Shi W, Hill RP, Zahedi P, Yip KW, Liu FF. The microRNA-218~Survivin axis regulates migration, invasion and lymph node metastasis in cervical cancer. Oncotarget. 2015;6:1090–1100. doi: 10.18632/oncotarget.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castellsaguè X, Díaz M, de Sanjosè S, Muñoz N, Herrero R, Franceschi S, Peeling RW, Ashley R, Smith JS, Snijders PJ, et al. Worldwide human papillomavirus etiology of cervical adenocarcinoma and its cofactors: Implications for screening and prevention. J Natl Cancer Inst. 2006;98:303–315. doi: 10.1093/jnci/djj067. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 13.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- 14.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 15.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: Microrna biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 16.Aigner A. MicroRNAs (miRNAs) in cancer invasion and metastasis: Therapeutic approaches based on metastasis-related miRNAs. J Mol Med (Berl) 2011;89:445–457. doi: 10.1007/s00109-010-0716-0. [DOI] [PubMed] [Google Scholar]
- 17.Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho WC. MicroRNAs: Potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42:1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Chang L, Li Z, Gao Q, Cai D, Tian Y, Zeng L, Li M. miR-99a and −99b inhibit cervical cancer cell proliferation and invasion by targeting mTOR signaling pathway. Med Oncol. 2014;31:934. doi: 10.1007/s12032-014-0934-3. [DOI] [PubMed] [Google Scholar]
- 20.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 21.Babashah S, Soleimani M. The oncogenic and tumour suppressive roles of microRNAs in cancer and apoptosis. Eur J Cancer. 2011;47:1127–1137. doi: 10.1016/j.ejca.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 25.de Freitas AC, Gomes Leitão Mda C, Coimbra EC. Prospects of molecularly-targeted therapies for cervical cancer treatment. Curr Drug Targets. 2015;16:77–91. doi: 10.2174/1389450116666141205150942. [DOI] [PubMed] [Google Scholar]
- 26.Sun JY, Xiao WZ, Wang F, Wang YQ, Zhu YH, Wu YF, Miao ZL, Lin YC. MicroRNA-320 inhibits cell proliferation in glioma by targeting E2F1. Mol Med Rep. 2015;12:2355–2359. doi: 10.3892/mmr.2015.3657. [DOI] [PubMed] [Google Scholar]
- 27.Cheng C, Chen ZQ, Shi XT. MicroRNA-320 inhibits osteosarcoma cells proliferation by directly targeting fatty acid synthase. Tumour Biol. 2014;35:4177–4183. doi: 10.1007/s13277-013-1546-9. [DOI] [PubMed] [Google Scholar]
- 28.Wu YY, Chen YL, Jao YC, Hsieh IS, Chang KC, Hong TM. miR-320 regulates tumor angiogenesis driven by vascular endothelial cells in oral cancer by silencing neuropilin 1. Angiogenesis. 2014;17:247–260. doi: 10.1007/s10456-013-9394-1. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ, Lee KH, Yeh SD, Hong TM, Chen YL. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis. 2013;34:530–538. doi: 10.1093/carcin/bgs371. [DOI] [PubMed] [Google Scholar]
- 30.Siciliano V, Garzilli I, Fracassi C, Criscuolo S, Ventre S, di Bernardo D. MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat Commun. 2013;4:2364. doi: 10.1038/ncomms3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 32.Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta. 2012;1819:28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 33.He SY, Shen HW, Xu L, Zhao XH, Yuan L, Niu G, You ZS, Yao SZ. FOXM1 promotes tumor cell invasion and correlates with poor prognosis in early-stage cervical cancer. Gynecol Oncol. 2012;127:601–610. doi: 10.1016/j.ygyno.2012.08.036. [DOI] [PubMed] [Google Scholar]
- 34.Hou Y, Li W, Sheng Y, Li L, Huang Y, Zhang Z, Zhu T, Peace D, Quigley JG, Wu W, et al. The transcription factor Foxm1 is essential for the quiescence and maintenance of hematopoietic stem cells. Nat Immunol. 2015;16:810–818. doi: 10.1038/ni.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan DW, Yu SY, Chiu PM, Yao KM, Liu VW, Cheung AN, Ngan HY. Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis. J Pathol. 2008;215:245–252. doi: 10.1002/path.2355. [DOI] [PubMed] [Google Scholar]
- 36.Chen H, Zou Y, Yang H, Wang J, Pan H. Downregulation of FoxM1 inhibits proliferation, invasion and angiogenesis of HeLa cells in vitro and in vivo. Int J Oncol. 2014;45:2355–2364. doi: 10.3892/ijo.2014.2645. [DOI] [PubMed] [Google Scholar]