Abstract
Gefitinib resistance and relapse of the disease were the greatest challenges facing clinical therapy of non-small-cell lung cancer (NSCLC). Of note, regarding the hypoxia condition in solid tumor tissues in vivo, roles of hypoxia in gefitinib adaptive resistance and its association with lung cancer stem cells (LCSCs) have not been fully elucidated. In the present study, the role of hypoxia in gefitinib adaptive resistance and its association with aldehyde dehydrogenase (ALDH)-based LCSC gefitinib resistance were comparatively studied using RNA-sequencing (RNA-seq) technology. Co-treatment of PC9 cells with gefitinib and hypoxia (1% O2) significantly enhanced adaptive resistance compared with gefitinib or hypoxia treatment alone. An ALDEFLUOR assay demonstrated that there was a significant increase of ALDH expression level in hypoxia and gefitinib co-treated PC9 cells, in addition to a higher ratio of G0/G1 quiescent cell enrichment and acquisition of epithelial-mesenchymal transition. RNA-seq analysis revealed that interleukin-6 (IL-6) is an important common factor in hypoxia and ALDH-based gefitinib resistance, supported by inflammation-associated tumor necrosis factor, nuclear factor-κB and Janus kinase-signal transducer and activator of transcription signaling pathway enrichment. Furthermore, exposure of PC9 and HCC827 cells to IL-6 increased gefitinib adaptive resistance. Consequently, IL-6 expression level was a poor prognostic marker for patients with NSCLC and adenocarcinoma. Thus, targeting IL-6 combined with tyrosine kinase inhibitor treatment may be promising in NSCLC clinical therapy in the future.
Keywords: gefitinib, hypoxia, aldehyde dehydrogenase, adaptive resistance, interleukin-6
Introduction
Lung cancer is currently the leading cause of mortality among all types of cancer worldwide (1). It may be divided into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), which account for ~80 and ~20% of cases, respectively. On the basis of oncogene mutation profiles, epidermal growth factor receptor (EGFR) mutations and Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations account for ~50 and ~10% of patients with NSCLC in Asians and 10–15% and 30% in Caucasians (1). Owing to its promising therapeutic efficacy and low toxicity, targeted therapy, particularly tyrosine kinase inhibitors, including gefitinib and erlotinib, are being widely used in clinical therapy for patients with NSCLC (2); however, patients using tyrosine kinase inhibitors (TKIs) acquire drug resistance within 7–11 months of administration. The mechanisms of TKI resistance may be attributed to second-site mutations within the EGFR kinase domain (T790M), met proto-oncogene (MET) gene amplification, epithelial-mesenchymal transition (EMT) and histological transformation from NSCLC to SCLC. Cancer stem cells (CSCs) were also revealed to be responsible for drug resistance and disease recurrence; however, previous studies revealed that EMT induction and CSC emergence were associated (3,4). In addition, disturbed microcirculation and structure induced a hypoxic microenvironment in inner solid tumor tissues in vivo (5). Sustained hypoxia in a growing tumor induced deterioration of the disease and it was also reported to serve important roles in TKI resistance, EMT induction and CSC generation (6,7). A low-oxygen culture condition may induce non-stem cancer cells to a more CSC-like phenotype (7). Hypoxia-inducible factor expression levels under hypoxic conditions were suggested induce a dynamic state of stemness in pathological conditions (8).
In the present study, in order to investigate the synergistic effects of hypoxia and gefitinib, PC9 cells were treated with gefitinib and/or hypoxia (1% O2). Aldehyde dehydrogenase (ALDH), a detoxifying metabolic enzyme, was used as a functional marker for sorting lung cancer stem cells (LCSCs). ALDH serves roles in early differentiation of stem cells via oxidization of retinol to retinoic acid, and has been used for isolation of stem cell populations from a variety of tissues and cell lines (9). The expression and phenotypes of ALDH cells were investigated in various NSCLC cell lines with different genetic backgrounds, which demonstrated consistent enhanced tumorsphere-forming capacities in vitro. Furthermore, compared with gefitinib or hypoxia treatment only, co-treatment of PC9 cells with gefitinib and hypoxia significantly increased the percentages of ALDH-positive cells, along with significantly increased half-maximal inhibitory concentrations (IC50) to gefitinib. Cell cycle analysis demonstrated G0/G1 cell cycle phase enrichment upon gefitinib or/and hypoxia treatment. Subsequently, ALDH-positive LCSCs were sorted by fluorescence-activated cell sorting (FACs). Genome-wide RNA-sequencing (seq) technology was performed to analyze the transcriptome status among PC9 cells, gefitinib or/and hypoxia-treated PC9 cells and ALDH-positive PC9 cells. By comparison, data mining combined with cancer pathway enrichment analysis revealed that interleukin-6 (IL-6) was an important factor in gefitinib resistance mechanisms. Signaling transduction enrichment analysis demonstrated that inflammation-associated tumor necrosis factor (TNF), nuclear factor-κB (NF-κB) and the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling pathway upregulation, indicating the critical role of inflammation signaling pathways in maintaining the core transcription networks in gefitinib resistance. Western blotting demonstrated co-treatment of PC9 cells with hypoxia and gefitinib enhanced EMT acquisition. In addition, exposure of PC9 and HCC827 cells to IL-6 increased the IC50 of gefitinib, indicating the important role of IL-6 in gefitinib resistance. Clinical evidences revealed that IL-6 was a poor prognostic marker in a cohort of 719 patients with adenocarcinoma in a KM plotter database (10); however, the investigations were performed in patients with general treatment guidelines rather than in gefitinib-resistant patients. The role of IL-6 will be elucidated if the clinical studies were performed specifically in patients with adaptive resistant and recurrent NSCLC following initial gefitinib treatment. In summary, the findings of the present study suggested that combined targeting of IL-6 signaling pathways with TKI administration may be a promising strategy for the treatment of patients with NSCLC.
Materials and methods
Cell lines, cell culture and chemotherapeutic reagents
The PC9 (EGFRE746_A750del), H1975 (EGFRT790M-L858R), HCC827 (EGFRE746_A750del) and H358 (KRASG12C) human lung adenocarcinoma cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured according to the manufacturer's protocol. Briefly, cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% (v/v) fetal bovine serum (HyClone; GE Healthcare Logan, UT, USA) supplemented with 1% penicillin/streptomycin in humidified incubator at 37°C supplied with 5% CO2. Gefitinib was purchased from Selleck Chemicals (Houston, TX, USA).
ALDEFLUOR assay
The ALDEFLUOR assay was performed using an ALDEFLUOR™ kit (Stemcell Technologies, Inc., Vancouver, BC, USA), according to the manufacturer's protocol. Briefly, BODIPY™-aminoacetaldehyde (BAAA) was used as a non-toxic fluorescent substrate for ALDH. BAAA is able to freely diffuse into intact and viable cells and ALDH is able to convert BAAA into BODIPY™-aminoacetate and remain inside the cells. The expression level of fluorescence is proportional to the ALDH activity. Viable ALDH−bright cells may be evaluated or sorted using a flow cytometer. Diethylaminobenzaldehyde, a specific inhibitor of ALDH, was used as the control for background fluorescence. The ALDH signal was detected in the green fluorescence channel (FL1, 530±15 nm) using an Accuri C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). A FACSAria flow cytometer (BD Biosciences) was used for ALDH-positive cell sorting.
Tumorsphere formation
The ALDH-positive cells were sorted from PC9, H358 and H1975 cells using flow cytometry and plated at density of 1×105 cells/ml in a AggreWell™ 400 microplate (Stemcell Technologies, Inc.), which contained ~1,200 microwells with a diameter of 400 µm. The medium for the tumorsphere analysis was StemXVivo™ Serum-Free Tumorsphere Medium (R&D Systems, Inc., Minneapolis, MN, USA), supplemented with heparin (2 U/ml) and hydrocortisone (0.5 µg/ml). A total of 20 ng/ml epidermal growth factor (Gibco; Thermo Fisher Scientific, Inc.) and basic fibroblast growth factor (Invitrogen; Thermo Fisher Scientific, Inc.) were added to the culture medium every other day. After 7 days, the tumorspheres formed were observed and manually counted under a light microscope.
Cell treatment and cell growth curves
PC9 cells were seeded in 10 cm culture dishes (Corning Incorporated, Corning, NY, USA) at the density of 1×106 cells/ml, treated with gefitinib or/and hypoxia at 37°C with 5% CO2 for 1 week. The concentration of gefitinib was 0.1 µM and the hypoxia condition was controlled in a hypoxic cell culture chamber (ProOx 110; BioSpherix, Ltd., Lacona, IA, USA) with 1% O2. The cell morphology was observed under an inverted phase-contrast microscope. The cells were counted every day to create growth curves.
Half-maximal inhibitory concentration (IC50) calculation
For IC50 determinations, serial dilutions of compounds [10 mM stock concentration in dimethyl sulfoxide (DMSO)] were performed in 100% DMSO with a three-fold dilution factor. PC9 cells were seeded at a density of 5,000 cells per well in 96-well plates. After 24 h at 37°C, the cells were treated with dose concentrations of gefitinib for 72 h; the concentration started at 100 µM and increased 3-fold each time for a series of 11 concentrations. The IC50 of gefitinib was investigated using a Cell Counting Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology, Haimen, China). Following a 72-h treatment of cells with a dose concentration of gefitinib as previously described, 20 µl CCK-8 solution was added to each well and the plates were incubated for an additional 4 h at 37°C. Optical density was evaluated at 450 nm using SkanIt software version 2.4.3 analysis program interfaced with a VarioskanFlash Microplate Reader (Thermo Fisher Scientific, Inc.). For each concentration, the percentage inhibition values were determined and the IC50 values and Hill slopes were evaluated using a four-parameter dose-response (variable slope) equation with GraphPad Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
Cell cycle analysis
PC9 cells were harvested and washed in PBS. Subsequently, 75% pre-cold ethanol was added dropwise to the cell pellet for fixation and cells were fixed at −20°C overnight. Subsequently, cells were washed in PBS to remove ethanol and were treated with 25 µg/ml RNase (Cell Signaling Technology, Inc., Danvers, MA, USA) at 37°C for 20 min. Finally, propidium iodide (PI) solution (50 µg/ml) was added for DNA staining. The cells were incubated at room temperature for 30 min in the dark and put on ice. The PI signal was detected in the PI fluorescence channel (FL2, 585±20 nm) using an Accuri C6 flow cytometer (BD Biosciences). BD Biosciences Accuri C6 software (version, 1.0.264.21) was used for data acquisition and analysis.
RNA-seq analysis
Total RNA was extracted from each group of PC9 cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The RNA quality and quantity was determined using an Agilent 2100 Bioanalyzer instrument (Agilent Technologies, Inc., Santa, Clara, CA, USA). Subsequently, mRNA libraries were generated using a TruSeq RNA Sample Preparation kit (Illumina, Inc., San Diego, CA, USA) and high-throughput RNA sequencing studies were performed using a HiSeq 2500 System (Illumina, Inc.). Paired-end reads 125 bp in length were mapped to the human reference transcriptome using Ensembl gene database (ftp.ensembl.org/pub/release-75/fasta/homo_sapiens/dna) and were aligned using Bowtie/Tophat software (ccb.jhu.edu/software/tophat/index.shtml). RNA expression levels were evaluated as=total exon reads/mapped reads (millions) × exon length (kb). Differentially expressed genes were analyzed using Deseq (version, 1.28.0; Bioconductor, http://bioconductor.org/) and evaluated at log2 fold-change (FC) threshold (|log2FC|>1). The annotation and functional enrichment of differentially expressed genes in Gene Ontology (GO; http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg/) were performed as GO and KEGG databases provided arranged genes of specific informative groups. P<0.05 was considered to indicate a statistically significant difference. The RNA-seq and data analysis were performed by Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). All RNA-seq data have been deposited in the GEO database (GEO accession no. GSE69599).
Western blotting
The cells were lysed in radioimmunoprecipitation buffer (Cell Signaling Technology, Inc.) supplemented with a protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 5 min on ice. A bicinchoninic acid assay (Pierce; Thermo Fisher Scientific, Inc.) was used for the determination of protein concentration, and 20–30 µg of total protein was loaded per well. The lysates were separated by SDS-PAGE (10% gel) and transferred onto polyvinylidene difluoride membranes. Blocking was performed for 1 h at room temperature using PBS-Tween-20 (PBST) buffer supplemented with 5% milk and incubated with primary antibodies overnight at 4°C. Primary antibodies used include antibodies specific for epithelial (E-)cadherin (cat. no, 3195), vimentin (cat. no. 5741), slug (cat. no. 9585), NF-κB p65 (cat. no. 8242), phosphorylated NF-κB p65 (cat. no. 3033) and GAPDH (cat. no. 5174; all Cell Signaling Technology, Inc.) at a dilution of 1:1,000. The membranes were washed three times in PBST and incubated with anti-rabbit horseradish-linked secondary antibody (dilution, 1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.) for 1 h. Finally, following washing with PBST, the membranes were developed using enhanced chemiluminescence (Pierce; Thermo Fisher Scientific, Inc.) and images were captured using an Odyssey SA Imaging system (LI-COR Biosciences, Lincoln, NE, USA). Densitometry of western blot data relative to GAPDH was performed Image Lab 6.0 software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
Statistical analysis was performed using GraphPad Prism software. Results are presented as the mean ± standard deviation (n=3) and comparisons were performed using Student's t-test between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
ALDH is a cancer stem cell biomarker for NSCLC cells with various genetic backgrounds
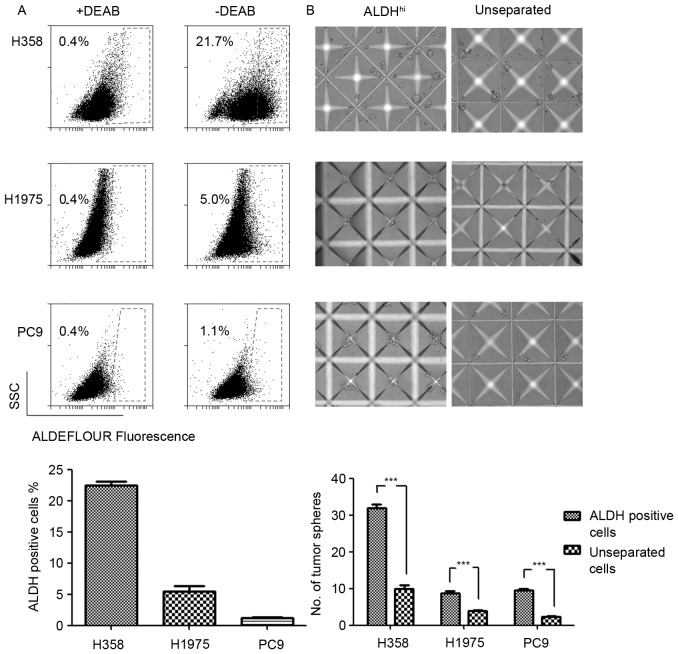
The expression levels and phenotypes of LCSC markers varied due to distinct genetic backgrounds (11). The results of the present study revealed differential expression levels of ALDH in EGFR and KRAS mutated NSCLC cell lines. The ALDH expression levels in H358 (KRASG12C), H1975 (EGFRT790M-L858R) and PC9 (EGFRE746_A750del) cell lines were evaluated using an ALDEFLUOR assay. The results demonstrated increased ALDH expression levels in the KRAS mutated H358 cell line and decreased expression levels in the EGFR mutated H1975 and PC9 cell lines (Fig. 1A). Considering the poor prognosis of patients with KRAS mutations with NSCLC compared with patients with EGFR mutations, increased ALDH expression levels may explain the poor prognosis of patients with NSCLC with KRAS mutations to a certain extent.
Figure 1.
ALDH is a reliable biomarker of lung cancer stem cells in non-small cell lung cancer cell lines. (A) Percentages of ALDH-positive cells detected by ALDEFLUOR assay. DEAB, and inhibitor of ALDH enzyme, was used as the background control in the experiment. Results are expressed as the ratios of the ALDH-positive cells and presented as the mean ± standard deviation (n=3). (B) ALDH-positive cells exhibited enhanced tumorsphere-forming capacities compared with the unseparated cells cultured in an AggreWell™ 400 microplate. Original magnification, ×10. Results are presented as the mean ± standard deviation. ***P<0.001 vs. the unseparated cells. ALDH, aldehyde dehydrogenase; DEAB, diethylaminobenzaldehyde.
Tumorsphere formation assay has previously been typically used to evaluate the functional capacity of CSCs and involves three-dimensional culture systems. The present study sorted the ALDH-positive LCSCs in H358, H1975 and PC9 NSCLC cell lines by FACS. The sorted ALDH-positive cells were cultured in AggreWell™ 400 microplates to generate consistently sized and shaped tumorspheres. As the control, the unseparated cells formed significantly fewer tumorspheres compared with the ALDH-positive cells. Three NSCLC cell lines were analyzed and demonstrated consistent ALDH-positive cells phenotypes in the tumorsphere formation assay (Fig. 1B). This result indicated that ALDH was a reliable LCSCs marker in the experimental system used.
Hypoxia and gefitinib exert synergistic resistance effects in NSCLC cells
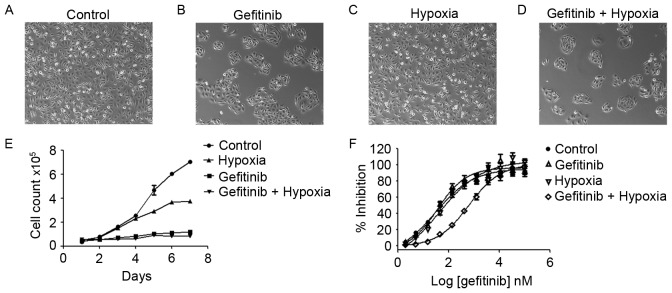
To investigate the treatment effects of PC9 cells with gefitinib or/and hypoxia, PC9 cells were pretreated with gefitinib (0.1 µM) or/and hypoxia (1% O2) for 1 week (Fig. 2A-D). Treatment of PC9 cells in normoxia (21% O2) with gefitinib significantly inhibited the cell proliferation rate, whereas co-treatment with hypoxia and gefitinib further inhibited PC9 cell growth (Fig. 2E). PC9 cells clustered under gefitinib pretreatment. Gefitinib or hypoxia pretreatment alone slightly increased the IC50 of gefitinib compared with the PC9 control cells; however, PC9 cells co-treated with gefitinib and hypoxia revealed a markedly increased IC50 value, as presented in Fig. 2F.
Figure 2.
Treatment of PC9 cells with gefitinib (0.1 µM) and/or hypoxia (1% O2). (A) Morphology of PC9 cells, (B) PC9 cells treated with gefitinib in normoxia (21% O2), (C) PC9 cells under hypoxia, and (D) PC9 cells treated with gefitinib and hypoxia (magnification, ×10). (E) Growth curves of each group of PC9 cells. (F) IC50 curves revealed an increased IC50 in gefitinib and hypoxia co-treated PC9 cells (612.7±102 nM) compared with gefitinib (35.47±12.45 nM) or hypoxia (42.07±20.14 nM) treatment alone or control PC9 cells under normoxia (28.76±10.12 nM). Co-treatment of PC9 cells with gefitinib and hypoxia revealed an increase in the IC50 value compared with PC9 cells and either treatment alone. Results are presented as the mean ± standard deviation (n=3). IC50, half-maximal inhibitory concentration.
Hypoxia augments the gefitinib-induced ALDH expression
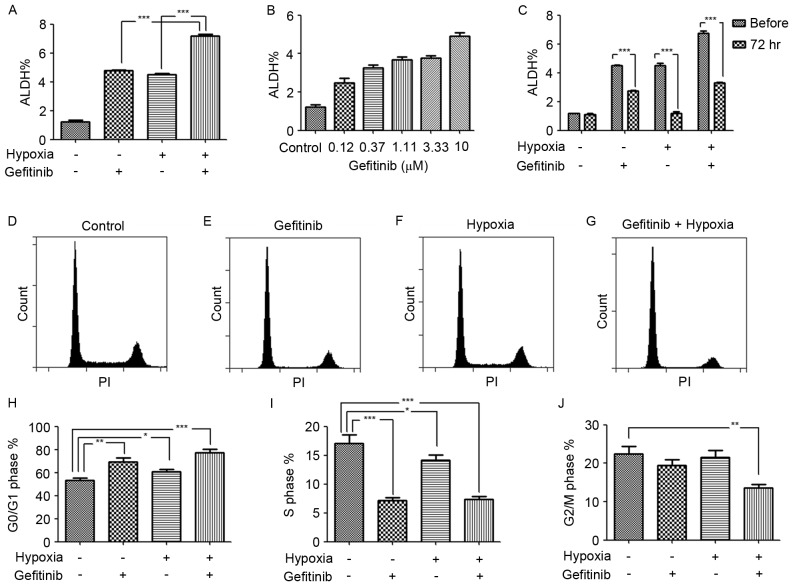
The drug resistance and disease recurrence in patients with NSCLC following long-term gefitinib administration in clinics suggested that gefitinib induced cancer cell reprograming to resist apoptosis and may also activate stemness related pathways for recurrence. Of note, an increased ALDH expression level in gefitinib or/and hypoxia treated PC9 cells was observed in the present study. In particular, hypoxia enhanced the role of gefitinib in the promotion of the ALDH expression level, as revealed in gefitinib- and hypoxia-co-treated PC9 cells (Fig. 3A). This result indicated that a hypoxic environment in inner solid tumor tissues was the ‘accomplice’ of gefitinib in cancer malignancy by enrichment of LCSCs or transforming of lung cancer cells to LCSCs. In order to verify the results, the dose response of gefitinib with regard to the ALDH expression level was investigated. It was revealed that, as the concentration of gefitinib increased, the ALDH expression level also increased (Fig. 3B). This result was in accordance with the hypothesis of therapy-induced enrichment of LCSCs in a previous study (12). However, the expression level of ALDH was unstable in PC9 cells treated with gefitinib or/and hypoxia, as revealed by the decrease in ALDH expression 3 days after passaging and cessation of treatment (Fig. 3C). Correspondingly, short-term hypoxia or/and gefitinib treated PC9 cells may survive in an incomplete reprograming of stemness pathway network status.
Figure 3.
Increased ALDH expression level following gefitinib and/or hypoxia treatment and enrichment of quiescent G0/G1 phase cells. (A) Ratios of ALDH-positive cells in PC9 cells, gefitinib- and/or hypoxia-treated PC9 cells. Gefitinib- and hypoxia-co-treated PC9 cells demonstrated significantly increased ratio of ALDH-positive cells compared with gefitinib or hypoxia treatment alone. Results are presented as the mean ± standard deviation (n=3). ***P<0.001. (B) An ALDEFLUOR assay revealed increased ALDH expression levels following treatment of PC9 cells with dose concentrations of gefitinib for 72 h. Results are presented as the mean ± standard deviation (n=3). (C) Unstable ALDH expression levels in short-term gefitinib- and/or hypoxia-treated PC9 cells after passaging and treatment cancellation for 72 h. Results are presented as the mean ± standard deviation (n=3). ***P<0.001. Flow cytometric cell cycle analysis. Results are from a typical cell cycle experiment of (D) PC9 cells under normoxia, (E) PC9 cells treated with gefitinib under normoxia, (F) PC9 cells under hypoxia and (G) PC9 cells treated with gefitinib under hypoxia. Statistics are presented as the percentages of total cells in the (H) G0/G1 phase, (I) S phase and (J) G2/M phase. Co-treatment of PC9 cells with hypoxia and gefitinib significantly increased G0/G1 cell cycle phase quiescent cells compared with hypoxia or gefitinib treatment alone, and revealed decreased proportions of S and G2/M phase cells. Results are presented as the mean ± standard deviation (n=3). *P<0.05, **P<0.01 and ***P<0.001. ALDH, aldehyde dehydrogenase.
Hypoxia enhances the enrichment of G0/G1 phase cells in gefitinib-treated PC9 cells
The LCSCs typically exhibit a quiescent slow-cycling phenotype, which partially explains an inherent mechanism of chemotherapy resistance and disease recurrence in clinics. Mizuno et al (13) identified that ALDH−high cells in ovarian carcinoma exhibited a more quiescent characteristic (G0/G1 cell cycle phase) compared with ALDH−low cells. Furthermore, Brennan et al (14) identified that ALDH-positive cells in mantle cell lymphoma remained in a relatively quiescent state and were resistant to a wide variety of agents. In the present study, following treatment with hypoxia (1% O2) or gefitinib for 7 days, quiescent G0/G1 phase cell enrichment was identified following gefitinib or/and hypoxia treatment. Compared with PC9 control cells and gefitinib or hypoxia-treated only PC9 cells, the ratio of G0/G1 phase cells increased mostly in gefitinib- and hypoxia-co-treated PC9 cells, indicating the synergistic role of hypoxia in promoting gefitinib-induced G0/G1 phase cell enrichment (Fig. 3D-J).
Differential expression transcripts and biological pathway analysis by genome-wide RNA-seq technology identifies IL-6 as a candidate gene in hypoxia- and ALDH-based gefitinib resistance mechanisms
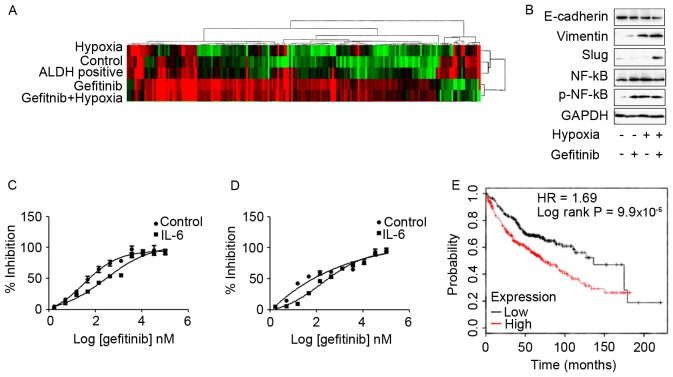
Subsequently, genome-wide transcriptome analysis to investigate the effects of gefitinib or/and hypoxia in PC9 cells was performed in the present study. High-throughput RNA sequencing of each group of PC9 cells was performed using a HiSeq 2500 System. Differentially expressed transcripts were analyzed using Deseq and evaluated at log2 fold-change thresholds (|log2FC|>1). The hierarchical clustering and heat-map analysis of the RNA-seq data are presented in Fig. 4A. In the present study, the common upregulated transcripts in PC9 cells that exhibited high IC50 values to gefitinib, including PC9 cells co-treated with gefitinib and hypoxia and ALDH-positive PC9 cells, were screened. Mapping of common differential transcripts to the KEGG cancer signaling pathway revealed that, with the exception of candidate genes also highly expressed in low gefitinib IC50 PC9 cells, fibroblast growth factor 22, Axin2, IL-6 and Fms-related tyrosine kinase-3 ligand are candidate genes in gefitinib resistance (Table I). Among these genes, IL-6 was reported to confer acquired TKI resistance in EGFR-driven NSCLC (15–17). IL-6 expression was associated with EMT, and it was revealed that IL-6 was a common gefitinib resistance gene shared in hypoxia and ALDH-based mechanisms. Furthermore, the KEGG biological signaling pathway enrichment analysis demonstrated that inflammation-associated TNF, NF-κB and JAK-STAT signaling pathways were highly expressed in PC9 cells that revealed increased expression levels of IL-6, as presented in Table II. Enrichment of TNF, NF-κB and the JAK-STAT signaling pathways may be due to IL-6 upregulation, since IL-6 activates the downstream NF-κB signaling pathway to enhance drug resistance in lung cancer cells (18). Concerning the role of IL-6 in LCSCs, previous studies revealed that IL-6 was indispensable for LCSC generation, proliferation and phenotype maintenance (19,20).
Figure 4.
(A) Hierarchical clustering and heatmap analysis of the RNA-seq data in each group of PC9 cells. Differential expression transcripts and biological pathway analysis by genome-wide RNA-seq technology identified IL-6 as an important gefitinib-resistant gene in hypoxia- and gefitinib-co-treated PC9 cells and ALDH-positive PC9 cells. (B) Treatment of PC9 cells with gefitinib and/or hypoxia activated epithelial-mesenchymal transition pathway expression levels. In particular, co-treatment of PC9 cells with gefitinib and hypoxia had significantly increased expression levels of mesenchymal markers compared with cell treated with gefitinib or hypoxia alone. Exposure of (C) PC9 and (D) HCC827 cells to 10 ng/ml IL-6 for 72 h significantly enhanced the resistance to gefitinib. (E) A cohort of 719 patients with adenocarcinoma from the KM plotter database revealed decreased overall survival times in patients with high IL-6 expression levels. RNA-seq, RNA-sequencing; IL-6, interleukin 6; ALDH, aldehyde dehydrogenase; NF-κB, nuclear factor-κB; p, phosphorylated; E-cadherin, epithelial cadherin; HR, hazard ratio.
Table I.
Differentially expressed reference pathway (KO) analysis of pathways in cancer.
| Genes overexpressed in group 4 and 5 compared with group 1 | Genes overexpressed in group 2, 3, 4 and 5 compared with group 1 | ||||
|---|---|---|---|---|---|
| KO | Gene name | Description | KO | Gene name | Description |
| K04385 | AXIN2 | Axin2 | K06625 | CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| K06625 | CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | K06236 | COL11A2 | Collagen, type XI, α2 |
| K06236 | COL11A2 | Collagen, type XI, α2 | K06237 | COL4A1 | Collagen, type IV, α1 |
| K06236 | COL1A2 | Collagen, type I, α2 | K04547 | GNG13 | Guanine nucleotide-binding protein (G-protein), γ13 |
| K06237 | COL4A1 | Collagen, type IV, α1 | K08006 | MMP28 | Matrix metallopeptidase 28 |
| K04358 | FGF22 | Fibroblast growth factor 22 | K01403 | MMP9 | Matrix metallopeptidase 9 (gelatinase B, 92 kDa gelatinase, 92 kDa type IV collagenase) |
| K05454 | FLT3LG | Fms-related tyrosine kinase 3 ligand | K11987 | PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| K07826 | GNG2 | Guanine nucleotide-binding protein (G-protein), γ2 | K01357 | WNT10B | Wingless-type MMTV integration site family, member 10B |
| K04548 | GNGT1 | Guanine nucleotide-binding protein (G-protein), γ transducing activity polypeptide 1 | K01384 | WNT11 | Wingless-type MMTV integration site family, member 11 |
| K05405 | IL6 | Interleukin 6 (interferon, β2) | |||
| K11987 | PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | |||
| K01357 | WNT10B | Wingless-type MMTV integration site family, member 10B | |||
1, PC9 cells under normoxia; 2, PC9 cells treated with gefitinib under normoxia; 3, PC9 cells under hypoxia; 4, PC9 cells treated with gefitinib under hypoxia; 5, Aldehyde dehydrogenase-positive PC9 cells. MMTV, mouse mammary tumor virus.
Table II.
Signal transduction enrichment analysis compared with PC9 parental cells.
| Signal transduction | No. of genes expressed at increased levels compared with in PC9 parental cells | ||||||
|---|---|---|---|---|---|---|---|
| KEGG orthology | Signaling pathway | 1 vs. 1 | 2 vs.1 | 3 vs. 1 | 4 vs. 1 | 5 vs. 1 | |
| ko04350 | TGF-β | 0 | −4 | 0 | 4 | 1 | |
| ko04630 | JAK-STAT | 0 | −2 | 0 | 3 | 1 | |
| ko04064 | NF-κB | 0 | −1 | 1 | 1 | 3 | |
| ko04668 | TNF | 0 | −2 | 0 | 3 | 6 | |
1, PC9 cells under normoxia; 2, PC9 cells treated with gefitinib under normoxia; 3, PC9 cells under hypoxia; 4, PC9 cells treated with gefitinib under hypoxia; 5, ALDH-positive PC9 cells. TNF, tumor necrosis factor; NF-κB, nuclear factor-κB; JAK, Janus kinase; STAT, signal transducer and activator of transcription 3; TGF-β; transforming growth factor-β.
Co-treatment of PC9 cells with hypoxia and gefitinib induced EMT- and IL-6-enhanced gefitinib resistance in TKI-sensitive EGFR mutated PC9 cells and HCC827 cells
In order to investigate the effects of hypoxia and gefitnib on PC9 cells, PC9 cells were treated with 1 µM gefitinib and hypoxia for 2 weeks. Treatment of PC9 cells with gefitinib or/and hypoxia activated the EMT signaling pathway expression. In particular, co-treatment of PC9 cells with gefitinib and hypoxia revealed significantly increased expression levels of mesenchymal markers, including vimentin and slug, whereas decreased expression levels of the epithelial marker E-cadherin compared with gefitinib or hypoxia treatment alone (Fig. 4B). Meanwhile, co-treatment of PC9 cells with gefitinib and hypoxia or either treatment alone was enough to activate phosphorylated NF-κB expression.
The function of IL-6 in promoting gefitinib resistance was confirmed by exposure of PC9 and HCC827 cells to 10 ng/ml IL-6 for 72 h, and subsequently resistance to gefitinib was analyzed in the presence of IL-6. IL-6 treatment significantly enhanced the resistance of PC9 (Fig. 4C) and HCC827 (Fig. 4D) cells to gefitinib. Furthermore, analyzing clinical data from a KM plotter database in 719 patients with adenocarcinoma revealed unfavorable clinical outcomes in patients with high expression of IL-6 with decreased overall survival times (10). The patient samples were divided into two groups with high and low expression levels of IL-6; subsequently, the two patient cohorts were compared using a Kaplan-Meier estimator survival plot and the hazard ratio (HR) with 95% confidence intervals and log rank P-value were determined. The results demonstrated that HR=1.69 (1.34–2.14) and log rank P=9.9×10−6 (Fig. 4E).
Discussion
Initial symptom relief, TKI drug resistance and disease recurrence during clinical therapy of NSCLC suggest the enrichment of LCSCs in this process. Treatment with gefitinib markedly inhibited PC9 cancer cell activities in many aspects, including proliferation and metabolism, whereas hypoxia serves critical roles in assisting the adaptation of PC9 cells to gefitinib resistance. Murakami et al (6) reported that hypoxic culture conditions are able to increase the LCSC ratio and enhance gefitinib resistance in NSCLC cells harboring TKI-sensitive EGFR mutations following exposure of these cells to gefitinib. Considering the disturbed microcirculation and structure due to the hypoxic environment in solid tumor tissues in vivo, hypoxia and gefitinib treatment may be the main cause of drug resistance, and, in particular, disease recurrence at a late stage, as confirmed by the increase in the ALDH-positive LCSC ratio in the present study. CSCs are thought to be responsible for tumor initiation, therapy resistance and particularly disease recurrence. The challenges in studying the roles of LCSCs in adaptive resistance to TKIs include controversies over LCSC biomarkers, the scarcity of LCSCs and their unstable phenotypes in vitro (21). ALDH has been validated as a reliable biomarker in numerous types of CSC, and the stemness and quiescent nature of ALDH-positive LCSCs was confirmed by determining the tumorsphere-forming capacity and enrichment of G0/G1 phase cells in the present study.
Transcriptome profiling by RNA-seq technology indicates that IL-6 is an important gefitinib-resistance gene. T790M, a secondary EGFR kinase domain mutation, may cause steric hindrance to impair the binding of gefitinib/erlotinib (22). MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling (23). There are differences in acquired resistance mechanisms associated with T790M and MET amplification; adaptive resistance to TKIs occurs early in therapy with a number of alterations to cancer cell signaling (24). Previously, Yao et al (17) revealed that the transforming growth factor β (TFG-β)-IL-6 axis mediates the adaptive resistance of lung cancer to TKIs. This study demonstrated that the inflammatory response-induced IL-6 secretion released lung cancer cells from EGFR dependency, decreased sensitivity to erlotinib and that IL-6 was indispensable for elotinib-resistant cell survival. In accordance with these results, the results of the present study revealed that exposure of PC9 and HCC827 cells harboring TKI-sensitive EGFR mutations to IL-6 increased gefitinib resistance. Increased expression levels of IL-6 in hypoxia- and gefitinib-co-treated PC9 cells was associated with increased expression levels of EMT signaling pathways. Furthermore, IL-6 was reported to enhance TKI resistance primarily by induction of EMT activation (17,18,25), and was also revealed to be indispensable for LCSC enrichment and phenotype maintenance (19,20). Accordingly, KEGG biological signaling pathway enrichment analysis confirmed that there was inflammation-associated pathways (TNF, NF-κB, JAK-STAT) enrichment in gefitinib- and hypoxia-co-treated PC9 cells and ALDH-positive PC9 cells. Of note, previously published studies have described the importance of these inflammation pathways in EMT and ALDH phenotypes maintenance. Kumar et al (26) demonstrated that TNF- and TGFβ-stimulated NSCLC spheroid cells exhibited increased EMT-associated core transcription factors, along with constitutive NF-κB expression levels. Inhibition of NF-κB decreased the expression levels of EMT-associated genes and resulted in EMT phenotype inhibition (27). Of note, Akunuru et al (28) demonstrated that TGFβ-induced EMT in NSCLC cells was accompanied by an increased ALDH expression level, which indicated a dynamic transition balance between EMT and LCSCs. These results are consistent with those of the present study that indicated that ALDH expression levels and EMT activation were associated. Additionally, these results also suggested an indispensable role of the NF-κB signaling pathway in maintaining EMT and the stemness-associated cell program (29). The stemness state of cancer cells induces resistance to TKIs therapy and drives tumorigenesis and metastasis (30). For the JAK-STAT signaling pathway, previous functional studies revealed that ALDH-positive LCSCs expressed increased levels of activated STAT3 compared with ALDH-negative cells, and inhibition of STAT3 decreased the number of ALDH-positive cells (31,32). Similar studies also revealed increased expression levels of STAT3 in ALDH-positive breast cancer cells and inhibition of STAT3 decreased the ALDH-positive cell population, suggesting an important role for STAT3 in ALDH-positive breast cancer stem cells (31,33–35).
The prognostic value of IL-6 was confirmed in a cohort of 719 patients with adenocarcinoma, as patients with high expression levels of IL-6 revealed decreased overall survival times (10). The unfavorable clinical outcomes in patients with NSCLC and high expression levels of IL-6 were also supported by clinical evidence that the expression levels of IL-6 in 245 patients with advanced NSCLC that had received chemotherapy were negatively associated with survival time, as analyzed by Kaplan-Meier estimator and Cox's proportional hazard models (21). Additionally, patients with high IL-6 expression levels responded poorly to chemotherapy (36). However, although these studies suggested that IL-6 may be associated with poor prognosis in patients with NSCLC who received chemotherapy, the investigations were performed in patients following standard chemotherapy guidelines and not in patients who received TKI therapy only (36). TKI treatment regimens have been used as National Comprehensive Cancer Network guidelines in recent years; therefore, the duration was limited for collection of a sufficiently high number of patients for the IL-6 investigation of gefitinib resistance in clinical prognosis. Although collection of clinical data from TKI-resistant patients with NSCLC is difficult, it is crucial to investigate the expression level of IL-6 in TKI-resistant patients with NSCLC to validate the results.
In order to inhibit the IL-6 signaling pathways, the majority of the research performed to date has been with tocilizumab, the first humanized monoclonal antibody specific for the IL-6 receptor, which is currently used in the treatment of rheumatoid arthritis in clinics (37,38). Clinical trials using humanized anti-IL-6 antibodies (ALD518; NCT00866970) revealed that IL-6 appeared to be well-tolerated and ameliorates NSCLC-associated anemia and cachexia (39); however, further studies are required to elucidate its therapeutic effect in TKI resistance. Future studies of combined anti-IL-6 as a target in adjuvant with chemotherapy and/or radiotherapy may assist in overcoming NSCLC TKI resistance.
Acknowledgements
The present study was supported by the Natural Science Foundation of China (grant no. 81173084) and the Shanghai Municipal Science Foundation (grant nos. 12ZR1415700, 14YZ032 and 2013-52).
References
- 1.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 3.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graves EE, Maity A, Le QT. The tumor microenvironment in non-small-cell lung cancer. Semin Radiat Oncol. 2010;20:156–163. doi: 10.1016/j.semradonc.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murakami A, Takahashi F, Nurwidya F, Kobayashi I, Minakata K, Hashimoto M, Nara T, Kato M, Tajima K, Shimada N, et al. Hypoxia increases gefitinib-resistant lung cancer stem cells through the activation of insulin-like growth factor 1 receptor. PLoS One. 2014;9:e86459. doi: 10.1371/journal.pone.0086459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazumdar J, Dondeti V, Simon MC. Hypoxia-inducible factors in stem cells and cancer. J Cell Mol Med. 2009;13:4319–4328. doi: 10.1111/j.1582-4934.2009.00963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iida H, Suzuki M, Goitsuka R, Ueno H. Hypoxia induces CD133 expression in human lung cancer cells by up-regulation of OCT3/4 and SOX2. Int J Oncol. 2012;40:71–79. doi: 10.3892/ijo.2011.1207. [DOI] [PubMed] [Google Scholar]
- 9.Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011;7:292–306. doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- 10.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis SJ, Sinkevicius KW, Li D, Lau AN, Roach RR, Zamponi R, Woolfenden AE, Kirsch DG, Wong KK, Kim CF. Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell. 2010;7:127–133. doi: 10.1016/j.stem.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freitas DP, Teixeira CA, Santos-Silva F, Vasconcelos MH, Almeida GM. Therapy-induced enrichment of putative lung cancer stem-like cells. Int J Cancer. 2014;134:1270–1278. doi: 10.1002/ijc.28478. [DOI] [PubMed] [Google Scholar]
- 13.Mizuno T, Suzuki N, Makino H, Furui T, Morii E, Aoki H, Kunisada T, Yano M, Kuji S, Hirashima Y, et al. Cancer stem-like cells of ovarian clear cell carcinoma are enriched in the ALDH-high population associated with an accelerated scavenging system in reactive oxygen species. Gynecol Oncol. 2015;137:299–305. doi: 10.1016/j.ygyno.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Brennan SK, Meade B, Wang Q, Merchant AA, Kowalski J, Matsui W. Mantle cell lymphoma activation enhances bortezomib sensitivity. Blood. 2010;116:4185–4191. doi: 10.1182/blood-2010-02-268375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terai H, Soejima K, Yasuda H, Nakayama S, Hamamoto J, Arai D, Ishioka K, Ohgino K, Ikemura S, Sato T, et al. Activation of the FGF2-FGFR1 autocrine pathway: A novel mechanism of acquired resistance to gefitinib in NSCLC. Mol Cancer Res. 2013;11:759–767. doi: 10.1158/1541-7786.MCR-12-0652. [DOI] [PubMed] [Google Scholar]
- 16.Ware KE, Hinz TK, Kleczko E, Singleton KR, Marek LA, Helfrich BA, Cummings CT, Graham DK, Astling D, Tan AC, Heasley LE. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis. 2013;2:e39. doi: 10.1038/oncsis.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, Schlederer M, Johns C, Altorki N, Mittal V, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci USA. 2010;107:15535–15540. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan HQ, Huang XB, Ke SZ, Jiang YN, Zhang YH, Wang YN, Li J, Gao FG. Interleukin 6 augments lung cancer chemotherapeutic resistance via ataxia-telangiectasia mutated/NF-kappaB pathway activation. Cancer Sci. 2014;105:1220–1227. doi: 10.1111/cas.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu CC, Lin JH, Hsu TW, Su K, Li AF, Hsu HS, Hung SC. IL-6 enriched lung cancer stem-like cell population by inhibition of cell cycle regulators via DNMT1 upregulation. Int J Cancer. 2015;136:547–559. doi: 10.1002/ijc.29033. [DOI] [PubMed] [Google Scholar]
- 20.Yi H, Cho HJ, Cho SM, Jo K, Park JA, Kim NH, Amidon GL, Kim JS, Shin HC. Blockade of interleukin-6 receptor suppresses the proliferation of H460 lung cancer stem cells. Int J Oncol. 2012;41:310–316. doi: 10.3892/ijo.2012.1447. [DOI] [PubMed] [Google Scholar]
- 21.Templeton AK, Miyamoto S, Babu A, Munshi A, Ramesh R. Cancer stem cells: Progress and challenges in lung cancer. Stem Cell Investig. 2014;1:9. doi: 10.3978/j.issn.2306-9759.2014.03.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godin-Heymann N, Ulkus L, Brannigan BW, McDermott U, Lamb J, Maheswaran S, Settleman J, Haber DA. The T790M ‘gatekeeper’ mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol Cancer Ther. 2008;7:874–879. doi: 10.1158/1535-7163.MCT-07-2387. [DOI] [PubMed] [Google Scholar]
- 23.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 24.Rosell R, Karachaliou N, Morales-Espinosa D, Costa C, Molina MA, Sansano I, Gasco A, Viteri S, Massuti B, Wei J, et al. Adaptive resistance to targeted therapies in cancer. Transl Lung Cancer Res. 2013;2:152–159. doi: 10.3978/j.issn.2218-6751.2012.12.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Han R, Xiao H, Lin C, Wang Y, Liu H, Li K, Chen H, Sun F, Yang Z, et al. Metformin sensitizes EGFR-TKI-resistant human lung cancer cells in vitro and in vivo through inhibition of IL-6 signaling and EMT reversal. Clin Cancer Res. 2014;20:2714–2726. doi: 10.1158/1078-0432.CCR-13-2613. [DOI] [PubMed] [Google Scholar]
- 26.Kumar M, Allison DF, Baranova NN, Wamsley JJ, Katz AJ, Bekiranov S, Jones DR, Mayo MW. NF-κB regulates mesenchymal transition for the induction of non-small cell lung cancer initiating cells. PLoS One. 2013;8:e68597. doi: 10.1371/journal.pone.0068597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min C, Eddy SF, Sherr DH, Sonenshein GE. NF-kappaB and epithelial to mesenchymal transition of cancer. J Cell Biochem. 2008;104:733–744. doi: 10.1002/jcb.21695. [DOI] [PubMed] [Google Scholar]
- 28.Akunuru S, James Zhai Q, Zheng Y. Non-small cell lung cancer stem/progenitor cells are enriched in multiple distinct phenotypic subpopulations and exhibit plasticity. Cell Death Dis. 2012;3:e352. doi: 10.1038/cddis.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang TS, Chen CL, Wu YC, Liu JJ, Kuo YC, Lee KF, Lin SY, Lin SE, Tung SY, Kuo LM, et al. Inflammation promotes expression of stemness-related properties in HBV-related hepatocellular carcinoma. PLoS One. 2016;11:e0149897. doi: 10.1371/journal.pone.0149897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medema JP. Cancer stem cells: The challenges ahead. Nat Cell Biol. 2013;15:338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- 31.Lin L, Hutzen B, Lee HF, Peng Z, Wang W, Zhao C, Lin HJ, Sun D, Li PK, Li C, et al. Evaluation of STAT3 signaling in ALDH+ and ALDH+/CD44+/CD24− subpopulations of breast cancer cells. PLoS One. 2013;8:e82821. doi: 10.1371/journal.pone.0082821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao C, Sullivan JP, Girard L, Augustyn A, Yenerall P, Rodriguez-Canales J, Liu H, Behrens C, Shay JW, Wistuba II, Minna JD. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin Cancer Res. 2014;20:4154–4166. doi: 10.1158/1078-0432.CCR-13-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Zee M, Sacchetti A, Cansoy M, Joosten R, Teeuwssen M, Heijmans-Antonissen C, Ewing-Graham PC, Burger CW, Blok LJ, Fodde R. IL6/JAK1/STAT3 signaling blockade in endometrial cancer affects the ALDHhi/CD126+ stem-like component and reduces tumor burden. Cancer Res. 2015;75:3608–3622. doi: 10.1158/0008-5472.CAN-14-2498. [DOI] [PubMed] [Google Scholar]
- 34.Han Z, Wang X, Ma L, Chen L, Xiao M, Huang L, Cao Y, Bai J, Ma D, Zhou J, Hong Z. Inhibition of STAT3 signaling targets both tumor-initiating and differentiated cell populations in prostate cancer. Oncotarget. 2014;5:8416–8428. doi: 10.18632/oncotarget.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin L, Fuchs J, Li C, Olson V, Bekaii-Saab T, Lin J. STAT3 signaling pathway is necessary for cell survival and tumorsphere forming capacity in ALDH+/CD133+ stem cell-like human colon cancer cells. Biochem Biophys Res Commun. 2011;416:246–251. doi: 10.1016/j.bbrc.2011.10.112. [DOI] [PubMed] [Google Scholar]
- 36.Chang CH, Hsiao CF, Yeh YM, Chang GC, Tsai YH, Chen YM, Huang MS, Chen HL, Li YJ, Yang PC, et al. Circulating interleukin-6 level is a prognostic marker for survival in advanced nonsmall cell lung cancer patients treated with chemotherapy. Int J Cancer. 2013;132:1977–1985. doi: 10.1002/ijc.27892. [DOI] [PubMed] [Google Scholar]
- 37.Hennigan S, Kavanaugh A. Interleukin-6 inhibitors in the treatment of rheumatoid arthritis. Ther Clin Risk Manag. 2008;4:767–775. doi: 10.2147/TCRM.S3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sebba A. Tocilizumab: The first interleukin-6-receptor inhibitor. Am J Health Syst Pharm. 2008;65:1413–1418. doi: 10.2146/ajhp070449. [DOI] [PubMed] [Google Scholar]
- 39.Bayliss TJ, Smith JT, Schuster M, Dragnev KH, Rigas JR. A humanized anti-IL-6 antibody (ALD518) in non-small cell lung cancer. Expert Opin Biol Ther. 2011;11:1663–1668. doi: 10.1517/14712598.2011.627850. [DOI] [PubMed] [Google Scholar]