Abstract

Stable isotope labeling (SIL) techniques have the potential to enhance different aspects of liquid chromatography–high-resolution mass spectrometry (LC-HRMS)-based untargeted metabolomics methods including metabolite detection, annotation of unknown metabolites, and comparative quantification. In this work, we present MetExtract II, a software toolbox for detection of biologically derived compounds. It exploits SIL-specific isotope patterns and elution profiles in LC-HRMS(/MS) data. The toolbox consists of three complementary modules: M1 (AllExtract) uses mixtures of uniformly highly isotope-enriched and native biological samples for selective detection of the entire accessible metabolome. M2 (TracExtract) is particularly suited to probe the metabolism of endogenous or exogenous secondary metabolites and facilitates the untargeted screening of tracer derivatives from concurrently metabolized native and uniformly labeled tracer substances. With M3 (FragExtract), tandem mass spectrometry (MS/MS) fragments of corresponding native and uniformly labeled ions are evaluated and automatically assigned with putative sum formulas. Generated results can be graphically illustrated and exported as a comprehensive data matrix that contains all detected pairs of native and labeled metabolite ions that can be used for database queries, metabolome-wide internal standardization, and statistical analysis. The software, associated documentation, and sample data sets are freely available for noncommercial use at http://metabolomics-ifa.boku.ac.at/metextractII.
Untargeted metabolomics research is the unbiased study of all low molecular weight compounds of a biological system. In contrast to targeted approaches, untargeted strategies aim at detecting all metabolites present in a sample, regardless of their identity, and subsequently comparing their relative abundances under different experimental conditions.1 However, due to the immense chemical diversity of metabolites and their wide range of concentrations, a single analytical technique is insufficient to probe the entire metabolic space of a biological sample at once. Reversed-phase (RP) liquid chromatography coupled to high-resolution mass spectrometry (LC-HRMS) is among the most commonly used analytical techniques, as it is well understood, highly customizable, robust, and requires little sample preparation. Untargeted LC-HRMS approaches provide a two-dimensional orthogonal separation [retention time and mass-to-charge ratio (m/z)] of the metabolites in a sample, allowing sensitive and selective concurrent detection of many hundreds to thousands of metabolites. Subsequent to an initial screening, selected metabolites can be further studied and characterized with the help of tandem mass spectrometry (LC-HRMS/MS), which results in metabolite-specific fragmentation patterns.2
However, unspecific signals and chemical and electronic noise complicate automated data processing. Additionally, ion suppression/enhancement in the electrospray ionization source (ESI), which results from coelution of different compounds, can distort relative metabolite abundances. These effects may vary between different samples (e.g., experimental conditions) and thus complicate statistical analysis and biological interpretation. Moreover, studying the metabolic fate of a tracer (e.g., toxin) is complicated by the fact that identification of most biotransformation products is currently not feasible.3
Stable isotope labeling (SIL) has gained much attention in the past years, reviewed for example by Klein and Heinzle.4 It utilizes low-abundance naturally occurring stable isotopes (e.g., 13C, 15N) to artificially produce labeled metabolite molecules. Compared to their nonlabeled pendants, labeled metabolites are enriched with naturally occurring low-abundance stable isotopes and thus possess a higher molecular weight. Additionally, native and labeled metabolites form unique isotope patterns in the LC-HRMS(/MS) data. With the help of appropriate experimental protocols, SIL can improve relative quantification and thus statistical analysis of the metabolomics experiment.5
Currently, only a few software tools exploiting the advantages of SIL are available for untargeted metabolomics. For gas chromatography coupled with mass spectrometry (GC-MS), Hiller et al.6 have developed NTFD software for untargeted detection of isotopically labeled primary metabolites and calculation of mass isotopomer distributions. For LC-HRMS, the programs X13CMS,7 geoRge,8 and mzMATCH-Iso9 provide methods for detecting metabolites with altered isotope patterns resulting from differential incorporation of a labeling isotope between experimental conditions. Kessler et al.10 developed the ALLocator online platform for mass isotopomer ratio analysis and compound annotation, and Leeming et al.11 designed the HiTIME algorithm for untargeted detection of drug metabolites by use of isotopic labeling.
Here, the second version of the MetExtract toolbox is presented. The originally published basic concept was restricted to the use of labeling-derived isotopologue mass signals (uniformly 13C-labeled) for untargeted MS spectrum inspection and automated metabolite detection in complex samples.12 Compared to the first version, MetExtract II has been considerably extended with metabolic feature pair detection, chromatographic peak picking, convolution of different ions contained in single mass spectra of the same metabolite, and their annotation as different ions of the same metabolite. Moreover, in-source fragments and heteroatom isotopologues (e.g., 37Cl, 34S) can be recognized in the LC-HRMS data, and the new software also supports tracer experiments to investigate the metabolism of isotopically labeled precursor substances. Finally, product ion MS/MS spectra can be elucidated, which was not possible with the previous MetExtract. MetExtract II consists of three modules. While M1 (AllExtract) facilitates the untargeted detection of all corresponding pairs of native and uniformly labeled metabolites, the new module M2 (TracExtract) supports the detection of mainly secondary metabolites derived from native and labeled forms of both endogenous and exogenous tracer substances (e.g., U-13C-labeled toxins). The third module M3 (FragExtract) supports the processing of LC-HRMS/MS fragmentation spectra of native and labeled metabolites, thereby allowing annotation of their fragment ions and cleaning of the spectra from unspecific signals.
Materials and Methods
AllExtract and TracExtract
The AllExtract module (M1) is designed for detecting all metabolites of a biological system, while the TracExtract module (M2) is designed for detecting only biotransformation products of a tracer compound under investigation. Both modules work with LC-HRMS data and require native and uniformly labeled material to be analyzed in a single sample. An overview of the data processing workflow is depicted in Figure 1.
Figure 1.
Illustration of implemented data processing steps in the presented software modules AllExtract, TracExtract, and FragExtract.
Nomenclature
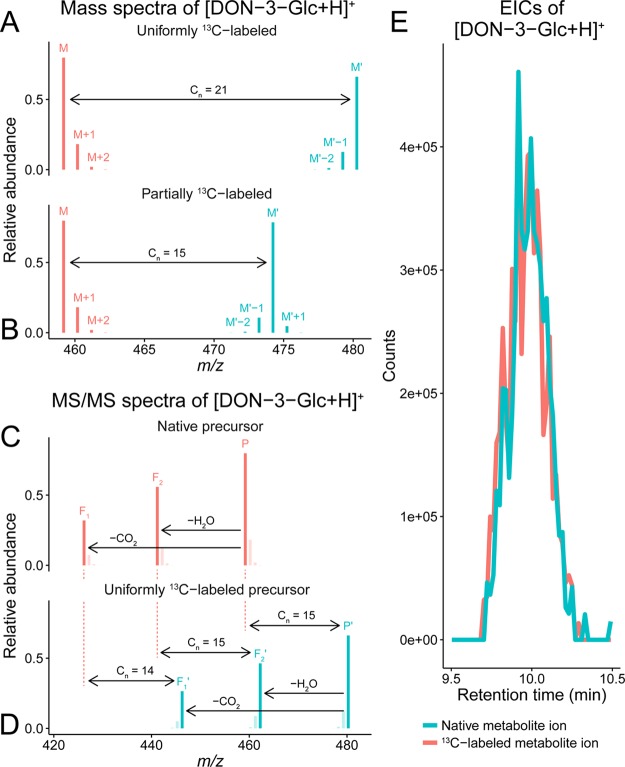
A monoisotopic, native metabolite-derived ion only consisting of the principal isotopes of its respective elements (1H, 12C, 16O, etc.) is termed M. M′ denotes the most intense ion of the labeled isotopologues: In the case of uniformly (U-) labeled metabolites [all atoms of the labeling element have an equal isotopic enrichment with the labeling isotope (e.g., 98.6% 13C), which is usually less than 100%],13 as required by AllExtract, this is the fully labeled isotopologue consisting only of the labeling isotope (e.g., 13C). In TracExtract, M′ denotes the partly labeled isotopologue, in which all atoms of the labeling element originating from the studied tracer are labeled (e.g., 13C) while any atom of the labeling element originating from the investigated native organism are the element’s natural principal isotope (e.g., 12C). M + i denotes the ith isotopologue of the native metabolite with i heavier isotopes (e.g., 13C instead of 12C), while M′ – i denotes the ith isotopologue of M′ having i atoms of the labeling element’s principal isotope (e.g., 12C instead of 13C). The m/z difference between the labeling isotope (e.g., 13C) and the principal isotope of the labeling element (e.g., 12C) is denoted as Δm (e.g., 1.00335 for 13C labeling). The charge number of an ion is denoted with z. Figure 2 panels A and B depict two MS scans of a U-13C-labeled metabolite and a partly 13C-labeled biotransformation product.
Figure 2.
LC-HRMS(/MS) data illustrating deoxynivalenol 3-glucoside (DON-3-Glc, C21H30O11). (A) Theoretical isotopologue patterns of native (M, [12C211H3016O11 + 1H]+; m/z 459.1862, native 12C enrichment of 98.93%) and U-13C-labeled (M′, [13C211H3016O11 + 1H]+; m/z 480.2566, uniform 13C enrichment of 99.1%) metabolite ions. Other isotopologue signals (e.g., 18O or 2H) are not depicted as their abundance is too low. (B) Isotope patterns of native and partly 13C-labeled (M′, [13C1512C61H3016O11 + 1H]+; m/z 474.2365) biotransformation product ions. In M′, only the 15 carbon atoms of DON are 13C, while the remaining six carbon atoms of Glc are 12C. The m/z difference (Δm) between M and M′ corresponds to the total number of labeled atoms in the respective ions (Cn). (C, D) Section of simulated MS/MS spectra of native and U-13C-labeled [DON-3-Glc + H]+ precursor ions. Mass increments between corresponding fragment ions (Fx and Fx′) reflect the number of 13C atoms per MS/MS fragment. Isotopologue signals of native and labeled precursor ions, as well as their fragments, will be present only if a corresponding broad mass isolation window is used in MS/MS analysis. (E) Coeluting chromatographic peaks of native and partly 13C-labeled [DON-3-Glc + H]+ (data provided by Kluger et al.).22
In the following subsections, the consecutive data-processing steps of MetExtract II are described. Steps 1–5 are performed separately for positive and negative ionization modes and each LC-HRMS file.
Step 1: Matching of Corresponding Mass Peaks from Native and Labeled Metabolite Ions in Each Scan
The first data-processing step of AllExtract and TracExtract detects pairs of native and labeled metabolite ion signals. For each data file, the following steps are successively performed for all recorded LC-HRMS scans. Each mass peak is initially considered to represent the monoisotopic ion M of a native metabolite or biotransformation product. This assumption is verified with the following criteria:
(i) An isotopologue M′ from the labeled metabolite or biotransformation product must be present in the same MS scan. As the charge (z) and the number of labeling isotopes (Xn) cannot be deduced from the single mass peak M, MetExtract II tests several user-defined combinations of Xn and z. For each combination, an m/z value for the putatively labeled isotopologue M′ is calculated [m/z(M′) = m/z(M) + (XnΔm)/z] and searched for in the same MS scan. If a mass peak with such an m/z value is present within a user-defined tolerance window (i.e., intrascan mass accuracy of the HRMS instrument used), it is considered a putative labeled signal M′. Together with Xn and z, M and M′ represent a putative ion pair. However, if no mass peak is found for any combination of Xn and z values, the current mass peak M is rejected.
(ii) The observed abundances of both mass peaks M and M′ must exceed a certain, user-defined intensity threshold. If any do not, the ion pair is rejected.
(iii) Depending on the experimental setup, the user can optionally define an intensity ratio of M:M′ (e.g., the ratio of native to labeled tracer applied in the biological experiment). If the ratio is not within the specified tolerance window, the ion pair is rejected.
(iv) The observed isotopologue patterns originating from native and labeled metabolite ions must match with their respective theoretical patterns. This is tested by comparing the observed isotopologue ratios by using the intensity ratio of M + 1 to M [I(M + 1)/I(M)] as well as M′ – 1 to M′ [I(M′ – 1)/I(M′)] with the expected ratios for a compound having the assigned number of labeling isotopes (Xn) as well as the isotopic abundance with either the principal isotope or the labeling isotope. Theeoretical ratios for such isotopologues are calculated by use of eq 1 with a = Xn, s = 1, and e = relative abundance of the principal isotope in nature (e.g., for 12C, 98.93%) or the isotopic enrichment with the labeling isotope (the 13C isotopic enrichment used).
| 1 |
Labeled biotransformation products may partly consist of native (nonlabeled) structure units (any conjugated moiety from the native biological system), which do not contribute to the m/z shift between M and M′. These moieties need to be accounted for in the native ion forms by the TracExtract module when the observed isotopologue ratio I(M + 1)/I(M) is compared to the theoretical ratio for Xn labeling atoms as their presence increases the theoretical ratio I(M + 1)/I(M). Thus, the observed ratio I(M + 1)/I(M) is corrected for the ratio of nonlabeled atoms I(M′ + 1)/I(M′) (corresponding to all atoms of the labeling element in the native moiety but not in the tracer itself) before it is tested against its corresponding theoretical ratio. This corrected ratio I(M + 1)/I(M) – I(M′ + 1)/I(M′) represents only the number of atoms of the labeling-element originating from the studied tracer. The ratio I(M′ – 1)/I(M′), however, is derived solely from the Xn labeling isotope atoms and must not be calculated differently than in the AllExtract module. If the observed and calculated theoretical isotopologue ratios deviate by less than a user-defined tolerance window (the expected relative isotopologue abundance error of the used HRMS instrument), the ion pair is accepted. If either of the two isotopologue ratio tests exceeds the maximum allowed tolerance, the ion pair is rejected. Any MS signal pair passing these verification criteria is considered to be an ion of a native and a corresponding labeled metabolite or biotransformation product.
Step 2: Clustering of Detected Ion Pairs
The second data-processing step is to cluster the detected ion pairs of the same ions with hierarchical clustering (HC). The purpose of this is to cluster similar ion pairs of the same metabolite ions (i.e., all signals recorded in different MS scans within a chromatographic peak) and determine average m/z values of the native and labeled isotopologues. For each assigned number of labeled atoms Xn and charge z, a separate hierarchical dendrogram (Euclidean distance, average linkage) is calculated with the m/z values of M of all corresponding ion pairs. This dendrogram is then split (top-down) and (sub)clusters with an m/z deviation between their highest and lowest values of less than a user-defined value (interscan mass deviation of the MS instrument) and within the putative chromatographic peak widths (user-set time interval) are kept as ion clusters and not split.
Step 3: Deconvolution of Chromatographic Peaks
In the next data-processing step, ion chromatograms of both isotopologues M and M′ are extracted from the raw chromatogram with the mean values of M and M′ of all ion pairs in a respective ion cluster and a user-defined mass-tolerance window (the instrument’s resolving power). Each of the thereby generated extracted ion chromatograms (EICs) is inspected separately for chromatographic peaks with the R-package MassSpecWavelet.14 Only chromatographic peak pairs (denoted as feature pairs) that (a) are present in the EICs of the corresponding M and M′ ions at approximately the same retention time (user-defined tolerance window) and (b) have a similar peak profile (Pearson correlation coefficient; user-defined minimal correlation) are kept. Other chromatographic peaks with no corresponding chromatographic peak in the respective native or labeled ion-derived EIC, or pairs of chromatographic peaks not coeluting sufficiently, are discarded. Then, previously detected ion pairs are assigned to their respective chromatographic peak pairs. Any feature pair detected in fewer mass scans than a user-defined minimum number is rejected. Each remaining chromatographic peak pair is considered to represent a pair of a native and a corresponding labeled metabolite ion (feature pair) that show coelution in the LC-HRMS data. Additionally, each feature pair is assigned an average m/z value of M, a determined number of labeling atoms Xn, and a charge z. Figure 2E illustrates a typical feature pair.
Step 4: Removal of Incorrectly Matched Isotopologue Pairs
Subsequent to their extraction, feature pairs that originate from pairings of M + 1 with M′ or M with M′ – 1 isotopologues, which do not represent the desired pairing of the true M and M′ isotopologues, are removed. This is achieved by inspecting coeluting feature pairs for either an m/z offset or a reduced number of assigned labeling isotopes. If their m/z values of M differ by Δm or Xn differs by 1, the feature pair with the higher m/z value of M or the lower value of Xn is flagged as an incorrect pairing and discarded. All remaining feature pairs represent correct pairings of the isotopologues M and M′.
Step 5: Annotation with Putative Heteroatoms
The following data-processing step annotates detected feature pairs with putative heteroatoms that have a distinct isotopologue pattern in LC-HRMS data if present in a metabolite (e.g., 37Cl, 54Fe). Depending on the mass increment of the heteroatom’s isotopologue Δm relative to its principal isotope, it is either searched for at the isotopologue pattern of the native (negative mass offset, e.g., 54Fe −1.9944) or the labeled metabolite form (positive mass offset, e.g., 37Cl +1.9971). The search is performed in each scan of the chromatographic peaks of a feature pair by calculating the mass increment expected for the heteroatom under investigation. If a peak with the predicted m/z value is present within a certain tolerance window and the intensity ratio of the isotopologue relative to M or M′ is within a user-defined window, the feature pair is considered to contain the respective heteroatom. The feature pair is then annotated with this heteroatom if it was detected in a minimal number of scans per EIC peak (user-defined value).
Step 6: Convolution of Feature Pairs into Feature Groups
Next, different feature pairs originating from the same metabolite are convoluted into feature groups. The Pearson correlation coefficient is again utilized for assessing whether the chromatographic peaks of two closely eluting feature pairs are similar. If two feature pairs have a high correlation, they are put into the same feature group. This convolution is performed for all feature pairs regardless of the ionization mode in which they were detected. After all possible pairwise correlations have been calculated, feature groups are inspected for erroneously linked feature pairs. To this end, a hierarchical clustering dendrogram using the determined correlation of all feature pairs in a group is calculated. If the number of feature pairs with a low correlation exceeds a certain threshold, the dendrogram is split into two subclusters and two new hierarchical dendrograms are calculated by using the remaining feature pairs in each subcluster. This step is repeated until clusters need not be split further or until a cluster consists only of a single feature pair (a single ion species was detected).
After feature group convolution, each group is inspected for ion species frequently observed in ESI spectra (user-defined adducts; e.g., [M + H]+, [M – H]−, [M + Na]+). To this end, for all feature pairs the 12C ions are used for pairwise comparison of mass increments. If the m/z difference corresponds to that between two known ion species (e.g., [M + H]+ and [M + Cl]−, Δm/z = 33.96213) and if their numbers of labeling isotopes (Xn) are identical, the two feature pairs are annotated accordingly. If two feature pairs could not be annotated with common adducts, their m/z value difference is used to calculate putative neutral losses. For this, the determined number of labeled isotopes is used and possible sum formulas are generated with the Seven Golden Rules.15 Feature pair convolution and annotation is performed jointly for the positive and negative ionization modes (in case fast-polarity switching has been used for sample measurement) but separately for each LC-HRMS file.
Step 7: Bracketing of Detected Feature Pairs across All LC-HRMS Files
After assignment of feature groups and annotation of ion species, detected feature pairs of the data set are combined into a two-dimensional data matrix (feature pairs and analyzed samples). This bracketing is performed by use of the ionization mode, the determined number of labeling isotopes Xn, and the charge number z as well as the retention time and m/z value of each detected feature pair. Feature pairs from all analyzed samples are first bracketed in the m/z domain with hierarchical clustering. Again, only subclusters differing by a maximal value for the m/z range are not split further. Then, for all feature pairs in a remaining cluster, an optional chromatographic alignment is calculated on the EICs of the labeled isotopologue M′. For this, the R package PolynomialTimeWarping (PTW)16 is utilized, and the retention times of the feature pairs are clustered with hierarchical clustering. All feature pairs in a subcluster with similar retention time (user-defined maximum deviation) are assumed to represent the same ion of a metabolite in the different LC-HRMS files and are bracketed and saved in one row of the final data matrix.
Following this processing step, the feature group annotation from the different LC-HRMS files is used to calculate an overall feature pair graph, and a majority vote system is used for annotation. If a link between two or more feature pairs in the data matrix was detected in at least n files (user-defined cutoff), the link is also retained in the final data matrix.
Step 8: Reintegration of Originally Missed Extracted Ion Chromatogram Peaks
The last data-processing step is aimed at reintegrating all M and M′ features that were not successfully detected with these rather strict data-processing criteria. To this end, feature pairs from the final data matrix missing in certain samples are searched for in a targeted manner. Peak areas of chromatographic peaks for the feature pairs detected in the EICs of M or M′ are inserted into the data matrix without verifying the presence of any of their isotopologues (M + 1 and M′ – 1) or the accuracy of their isotopologue intensity ratios [I(M + 1)/I(M) or I(M′ – 1)/I(M′)]. Consequently, this step fills missing values mostly originating from low-abundant metabolites in the inspected samples.
FragExtract
The FragExtract module17 is designed for studying native and labeled metabolites with LC-HRMS/MS. It requires separate LC-HRMS/MS spectra of the native and labeled metabolite. Steps 1–3 are carried out for each of the defined precursors.
Nomenclature
Any fragment peak observed in the MS/MS spectrum of the native precursor (P) that has a corresponding fragment peak in the MS/MS spectrum of the labeled precursor ion is termed F. Vice versa, any fragment peak in the MS/MS spectrum of the labeled precursor (P′) that has a corresponding peak in the MS/MS spectrum of the native precursor ion is termed F′. Figure 2 panels C and D show two successive fragmentation spectra of a native and a uniformly 13C-labeled precursor ion.
Step 1: Selection of Tandem Mass Spectrometric Scans of Native and Labeled Precursors
The first data-processing step in FragExtract is to determine the apex of the chromatographic LC-HRMS full-scan peak of P and then select the two successive MS/MS scans (termed S and S′), one for the native precursor ion and one for the labeled precursor ion. Then, all mass peaks that have an m/z value higher than their respective precursor ions are removed, and the intensity values of the product ion mass spectra are normalized to the base peak.
Step 2: Matching of Corresponding Fragment Peaks
In the next data-processing step, corresponding native and labeled fragment peaks in S and S′ are matched. Each mass peak in S is initially considered to represent a monoisotopic fragment F. This assumption is verified by searching for a respective labeled fragment peak F′ in S′. For this, FragExtract iterates over a predefined number of the labeling isotopes per fragment and calculates its corresponding m/z value. If a peak with this m/z value is detected in S′ within a user-defined error window and if their normalized intensities in S and S′ are within a user-defined tolerance window, the fragment pair F and F′ is accepted. Any such fragment pair represents a fragment of the investigated metabolite precursor and is annotated with the determined number of labeling isotopes. All other peaks recorded in either S or S′ that are not matched with a peak of the respective other MS/MS spectrum are discarded.
Step 3: Generation of Sum Formulas for Each Fragment Peak
For each verified fragment pair as well as the used precursor ion, possible sum formulas are calculated by use of the determined number of labeling isotopes and the Seven Golden Rules.15 Additionally, sum formulas for the neutral losses between the parent precursor ion and the fragment pairs are also calculated. Any combination of annotated sum formulas, neutral losses, and parent sum formulas that cannot be valid (e.g., atoms present in the fragment but not the parent) are discarded.
Implementation of MetExtract II
MetExtract II is implemented in the Python programming language (2.6, http://www.python.org/, accessed September 2015) and uses the R Project for Statistical Computing v2.15.2.18 It imports LC-HRMS(/MS) files in the mzXML or mzML formats19 and supports parallel processing of multiple data files. Processing results are saved as tab-delimited files (.tsv) and as graphical depictions (.pdf). If the mzXML format is used, MetExtract II can also save all detected feature pairs to a new mzXML file. Additionally, the software has a graphical user interface for processing the LC-HRMS(/MS) data as well as to review the extracted metabolites graphically (Figure S1).
Sample Data Sets
Several sample data sets were recorded on an LTQ Orbitrap XL instrument operated in positive ESI mode and an Orbitrap Exactive Plus instrument operated in fast-polarity-switching mode. A summary of the data sets is provided in Table 1. More details on the biological and analytical experiments, as well as data-processing results with MetExtract II, are available in Supporting Information.
Table 1. Data Sets Used for Evaluation of MetExtract II.
| data seta | instrument | description |
|---|---|---|
| AE_Std | LTQ Orbitrap XL | LC-HRMS data of different native and U-13C-labeled mycotoxin standards12 |
| AE_Wheat | Orbitrap Exactive Plus | LC-HRMS data of native and U-13C-labeled wheat |
| TE_DiW | Orbitrap Exactive Plus | LC-HRMS data of wheat treated with native and U-13C-labeled deoxynivalenol22 |
| ATE_Blanks | Orbitrap Exactive Plus | LC-HRMS data of native nonlabeled wheat |
| FE_PPAs | LTQ Orbitrap XL | LC-HRMS/MS data from three native and U-13C-labeled PPAs |
AE, AllExtract; ATE, AllExtract and TracExtract; FE, FragExtract; TE, TracExtract; DiW, DON in wheat; std, standards; PPAs, phenylpropanoid amides.
Results and Discussion
A revised and updated software toolbox, named MetExtract II, for SIL-assisted and LC-HRMS(/MS)-based untargeted metabolomics is presented. It supports biological experiments that use highly stable isotope-enriched samples/metabolites (e.g., 13C, 15N, or 34S). Carbon-13 is suggested as the main labeling element since it is present in any organic metabolite. Additionally, labeling with 13C produces highly characteristic isotope patterns in the LC-HRMS data (Figure 2A–D). Other isotopes such as 15N or 34S are also supported.
MetExtract II consists of three complementary modules for the detection of ions in LC-HRMS(/MS) data derived from native and labeled metabolites:
Module 1, AllExtract, allows fully automated, reliable detection of the global metabolic composition of a single biological sample under investigation. Assignment of the total number of labeling atoms per metabolite and metabolomewide internal standardization improve annotation and relative quantification of metabolites compared to labeling-free methods.
Module 2, TracExtract, facilitates the comprehensive untargeted screening of tracer-derived biotransformation products of concurrently metabolized native and labeled tracer substances, thus efficiently probing the secondary metabolism of both endogenous (e.g., aromatic amino acids or hormones) as well as exogenous (e.g., pesticides, drugs, or toxins) tracer compounds.
Module 3, FragExtract, addresses the challenge of investigating unknown metabolites based on LC-HRMS/MS spectra of native and labeled precursor ions of the same metabolite in highly complex biological samples. Each fragment is annotated with its total number of labeling atoms, and unrelated peaks and noise are efficiently filtered, resulting in pure MS/MS spectra of the compound under investigation.
The associated biological and analytical workflows, which are required for producing the respective biological material and the LC-HRMS(/MS) data, are summarized in Bueschl et al.20 (AllExtract), Kluger et al.21 (TracExtract), and Neumann et al.17 (FragExtract). Briefly summarized, the respective biological workflows are as follows.
AllExtract: In the case of 13C-labeling, the biological system under investigation is grown in parallel either with a native carbon source (e.g., nonlabeled glucose) or with a highly isotope-enriched carbon source (e.g., 13C-labeled glucose in the case of fungi, 13CO2 for plants) under identical environmental conditions. Then the native and labeled samples are combined, so that the samples contain both native and labeled metabolites. All biological metabolites will show corresponding isotope patterns of native and uniformly labeled metabolite ions in the sample’s LC-HRMS data. For experimentwide internal standardization, all labeled samples are pooled to produce a labeled reference sample from which equal aliquots are transferred to native experimental samples.
TracExtract: In the case of an exogenous tracer, the compound under investigation (e.g., toxin, drug) is applied to the studied biological system as both the native and labeled form, while an endogenous tracer (e.g., Phe) may be applied as the labeled form only. After a defined incubation period, a sample is taken and analyzed with LC-HRMS. Any tracer derivative will be present in native and partly labeled form.
FragExtract: Metabolites of interest (e.g., detected with AllExtract or TracExtract) are selected as MS/MS targets. Separate LC-HRMS/MS spectra of the compound’s native (M) and labeled (M′) precursor ions are then recorded in the same LC-HRMS/MS run.
MetExtract II utilizes the generic mzXML and mzML LC-HRMS(/MS) data formats and can thus be used independently of the MS instrument type or manufacturer [e.g., Orbitrap or quadrupole time-of-flight (QToF) instruments]. It supports positive and negative ionization modes and additionally supports fast-polarity-switching ESI for comprehensive metabolite coverage and annotation. Detected and convoluted metabolite ions are reported as feature pairs, each consisting of an m/z value for the native, monoisotopic metabolite ion (M), the number of labeling atoms (Xn), the number of charges (z), the retention time of its respective chromatographic peak, and the abundance values (peak areas) for the monoisotopic isotopolog M and the labeled isotopolog M′.
MetExtract II generates diagnostic plots that help the user to easily review the results and tune the data-processing parameter settings (Figure S2). Bracketed feature pairs detected in the processed LC-HRMS measurement files are reported in a comprehensive data matrix consisting of metadata as well as the relative ion abundances in the processed samples. This matrix can then be used for statistical analysis or database annotation. Individual mzXML files may also be exported as processed mzXML files that contain only the detected signals of the native and labeled metabolite ions.
Recommended Labeling Patterns To Be Used with MetExtract II
MetExtract II can be used to process data files from native and highly isotope-enriched biological samples analyzed jointly in a single LC-HRMS(/MS) run. All native and uniformly or partly labeled metabolites will produce highly characteristic isotope patterns that MetExtract II uses for metabolite detection and annotation. Thus, certain requirements must be fulfilled:
(i) Only one labeling isotope (e.g., 13C, 15N) may be used at a time.
(ii) Native and corresponding labeled metabolite ions must at least partially coelute, and no isotope exchange between metabolite or solvent molecules should occur. Thus, isotopes such as 2H or 18O should be tested for their chemical integrity in the studied metabolites.4
(iii) If the labeling is performed with a stable isotope other than 13C and the number of labeled atoms in a putative metabolite is low (e.g., 15N1 or 2H2), the facilitated HRMS instrument must be capable of resolving the isotopic fine structure of the labeling isotope and the usually dominating isotope pattern of carbon. Overlapping isotopologue signals of native carbon isotopologues and the labeling isotope may cause problems and result in incorrectly detected feature pairs. For example, a labeling experiment employing 15N will require a resolution of 220.000 (m/z 200) to separate an ion of m/z 480 with a single 15N atom from the native 13C isotopologue. If multiple atoms of the labeling element are present (e.g., 15N3 or 2H3), the resolution of the MS instrument may be set to a lower value (e.g., 70.000) since the M + 3, M + 4 carbon isotopologues can be neglected for the majority of all metabolites with a maximum of 60 carbon atoms.
(iv) If the labeling is performed with 13C, the isotope patterns of the native and the 13C-labeled ions must be clearly separated. Overlapping isotope patterns are not yet supported.
(v) TracExtract is primarily designed for studying the metabolism of secondary metabolites that do not tend to be catabolized intensively. Thus, most primary metabolites that are catabolically degraded and constantly rearranged or integrated into other metabolites result in scrambled and broad isotope patterns and are not recognized by the current version of TracExtract.
Demonstration of MetExtract II Modules
To demonstrate the MetExtract II modules, five data sets have been selected and processed. An overview of the biological experiments, LC-HRMS analysis and data processing is provided in Table 1 and Supporting Information.
The first data set (AE_Std) is used to evaluate the performance of MetExtract II. It has been taken from the previous version of MetExtract.12 The data have been generated from an LC-HRMS analysis of a mixture of 15 native and U-13C-labeled (∼99.5% 13C enrichment) fungal substances that were spiked into a complex native Fusarium graminearum culture sample. The LC-HRMS file was processed with the AllExtract module. Processing parameter settings were kept identical (where possible) to the previous MetExtract version, but several parameter settings (e.g., chromatographic separation, convolution, annotation) are new and therefore cannot be compared directly. After data processing, 72 feature pairs convoluted into 17 feature groups were detected with MetExtract II (Figure S3). These correspond to the 15 fungal standards plus three impurities of the used standards, which also clearly show the expected isotope patterns of native and U-13C-labeled substances. Metabolite ions for 13 of the 15 fungal substances were successfully convoluted into separate feature groups, and only two substances (HT-2 toxin and griseofulvin) were incorrectly convoluted since they showed coelution (Figure S3). By use of the automated heteroatom annotation, the 37Cl isotopologues of griseofulvin and ochratoxin A were annotated.
In the second data set (AE_Wheat), the metabolome of wheat ear samples was evaluated in full-scan LC-HRMS data generated in fast-polarity-switching mode on an Orbitrap Exactive plus instrument. Each sample contained a mixture of native and U-13C-labeled wheat material (∼98.6% 13C enrichment). Processing of the raw data with the AllExtract module resulted in a total of 2430 feature pairs convoluted into 506 metabolites. Each such detected feature pair is clearly derived from wheat and was verified by use of its native and corresponding U-13C-labeled ion forms. Figure S4 exemplifies one highly abundant wheat metabolite that was detected by AllExtract as 35 different ion species. Approximately one-third of all feature pairs were solely detected in the negative, and about 41% of all metabolites consisted of feature pairs from both ionization modes. Figure S5 shows a feature map of all detected feature pairs.
The third data set (TE_DiW) originates from a study presented by Kluger et al.22 It investigates the detoxification and biotransformation mechanism of native and U-13C-labeled (∼99.5% 13C enrichment) deoxynivalenol in wheat plants. The biological sample was remeasured on an Orbitrap Exactive plus instrument with fast-polarity-switching ESI. In total, 21 feature groups consisting of 84 feature pairs were detected, including the unmodified toxin tracer (Figure S6).
The fourth data set (ATE_Blanks) consisted of solvent blanks as well as blanks containing only native wheat ear extracts without any labeled pendants. Thus, no feature pairs should be detected in these blanks. After data processing with the AllExtract and TracExtract modules, on average less than two feature pairs were detected (maximum in one sample, six pairs), which demonstrates the high selectivity of the presented approach. The detected false positives were random pairings of signals and noise artifacts.
The fifth data set (FE_PPAs) exemplifies the FragExtract module and is composed of LC-HRMS/MS data of three phenylpropanoid amides (PPAs): namely, p-coumaroylputrescine (CouPut), p-coumaroylagmatine (CouAgm), and p-coumaroylserotonin (CouSer). LC-HRMS/MS analysis of the native monoisotopic and the uniformly 13C-labeled metabolites were performed successively with collision-induced dissociation (CID) on an LTQ Orbitrap XL instrument in positive ESI mode. Acquired data files were processed with the FragExtract module, which matched fragment ions of native and U-13C-labeled precursor ions for spectral cleaning and fragment peak annotation. Corresponding signals of nine, eight, and three native and uniformly 13C-labeled metabolite fragment ions were successfully matched for the investigated CouPut, CouAgm, and CouSer precursors, respectively. Each fragment ion was annotated with its total number of carbon atoms and a unique sum formula as well as the neutral loss relative to its parent ion. The accurate masses of the predicted fragment ions deviated less than 5 parts per million (ppm) from their respective theoretical values. Moreover, signals not matching native and 13C-labeled fragment ions were removed. Figure S7 shows the processing results for the metabolite CouAgm with FragExtract.
The five data sets demonstrate the high sensitivity and selectivity of the presented MetExtract II modules. Compounds of nonbiological origin or non-tracer-derived metabolites (data set ATE_Blanks) are efficiently removed, and only biological relevant metabolites remain as detected metabolite or biotransformation product ions of the tracer compound ions (data sets AE_Std, AE_Wheat, and TE_DiW). Moreover, with the FragExtract module, LC-HRMS/MS spectra of (unknown) native and highly isotope-enriched metabolites are annotated and cleaned from nonspecific signals (data set FE_PPAs). With the help of metabolome- and experimentwide internal standardization (pooled 13C sample material) comparative quantification of different biological samples can also be improved (not detailed in this paper; the interested reader is referred to Giavalisco et al.23 and Bueschl et al.20).
In addition to these five demonstration data sets, earlier versions of the presented MetExtract II software have already been used successfully, for example, to study the detoxification mechanisms of the mycotoxins T-2 toxin and HT-2 toxin in barley,24 the effect of heat stress upon flavonoids in grapes,25 and the formation of deoxynivalenol derivatives in wheat22 and phenylalanine-derived secondary metabolites in wheat.21 Moreover, MetExtract II was used to evaluate different extraction solvents used in untargeted metabolomics research.26
Comparison to Other Software Tools
SIL-assisted metabolomics research can resort to many different software tools (e.g., X13CMS,7 geoRge,8 mzMatch-ISO9), with each tool being designed for a particular type of experiment and requiring data from respective labeling and measurement strategies. A comparison of the available software tools is therefore not straightforward. For example, to study the metabolism of certain tracer compounds, the geoRge workflow has been designed for tracer metabolism studies and requires separate samples of the native and labeled tracer under investigation, while the TracExtract workflow requires concurrent metabolization of the native and labeled tracers in a single sample. As a result, the different tools and their associated experimental workflows cannot be compared without major modifications of the tools and additional sourcecode or even additional biological experiments. Consequently, a direct comparison of MetExtract II-derived results with those of other software tools is not feasible. However, this demonstrates that MetExtract II is a complementary addition to the growing number of software tools for SIL-assisted untargeted metabolomics research.
Concluding Remarks
With SIL being increasingly used in untargeted metabolomics research, there is a constant need for novel software tools, many of which are intended for certain types of experiments and applications. The presented MetExtract II software is a versatile tool for LC-HRMS-based and SIL-assisted untargeted metabolomics. It supports experiments that use native and highly isotope-enriched biological material or tracer compounds.
Since MetExtract II uses the characteristic and unique mass increments and isotope patterns between native and highly isotope-enriched forms of the same metabolites, it can (i) efficiently discriminate biological relevant metabolites from unspecific compounds, (ii) track the metabolic fate of tracer substances under investigation, (iii) assign the total number of labeling isotopes to each detected metabolite or biotransformation product, (iv) report ion abundances of both native and labeled metabolite forms, and (v) improve MS/MS annotation and clean MS/MS product ion spectra of known and unknown metabolites.
In conclusion, MetExtract II addresses untargeted metabolomics experiments that employ native and highly isotope-enriched samples and thus perfectly complements other software in the field. MetExtract II is freely available for noncommercial use and can be downloaded from http://metabolomics-ifa.boku.ac.at/metextractII.
Acknowledgments
We thank Sylvia Lehner, Marc Lemmens, Elisabeth Varga, Franz Berthiller, Jürgen Hartler, Justyna Rechthaler, and Gerhard Adam for useful discussions. This work was supported by the Austrian Science Fund (Projects SFB-Fusarium 3706/3715 and T2/HT2 P26213), the Government of Lower Austria (Projects NovAlgo and NoBiTUM), and the Austrian Ministry of Science, Research and Economy (HSRSM Grant BioTechMed Omics Center Graz to G.G.T.).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.7b02518.
Four sections of additional text, with seven figures and five tables, describing biological experiments, LC-HRMS analysis, data processing, and generated results (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Patti G. J.; Yanes O.; Siuzdak G. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. Plant Mol. Biol. 2002, 48, 155–171. 10.1023/A:1013713905833. [DOI] [PubMed] [Google Scholar]
- Hegeman A. D. Briefings Funct. Genomics 2010, 9, 139–148. 10.1093/bfgp/elp053. [DOI] [PubMed] [Google Scholar]
- Klein S.; Heinzle E. Wiley Interdiscip. Rev.: Syst. Biol. Med. 2012, 4, 261–272. 10.1002/wsbm.1167. [DOI] [PubMed] [Google Scholar]
- Bueschl C.; Krska R.; Kluger B.; Schuhmacher R. Anal. Bioanal. Chem. 2013, 405, 27–33. 10.1007/s00216-012-6375-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller K.; Wegner A.; Weindl D.; Cordes T.; Metallo C. M.; Kelleher J. K.; Stephanopoulos G. Bioinformatics 2013, 29, 1226–1228. 10.1093/bioinformatics/btt119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.; Chen Y. J.; Cho K.; Nikolskiy I.; Crawford P. A.; Patti G. J. Anal. Chem. 2014, 86, 1632–1639. 10.1021/ac403384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellades J.; Navarro M.; Samino S.; Garcia-Ramirez M.; Hernandez C.; Simo R.; Vinaixa M.; Yanes O. Anal. Chem. 2016, 88, 621–628. 10.1021/acs.analchem.5b03628. [DOI] [PubMed] [Google Scholar]
- Chokkathukalam A.; Jankevics A.; Creek D. J.; Achcar F.; Barrett M. P.; Breitling R. Bioinformatics 2013, 29, 281–283. 10.1093/bioinformatics/bts674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler N.; Walter F.; Persicke M.; Albaum S. P.; Kalinowski J.; Goesmann A.; Niehaus K.; Nattkemper T. W. PLoS One 2014, 9, e113909 10.1371/journal.pone.0113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeming M. G.; Isaac A. P.; Pope B. J.; Cranswick N.; Wright C. E.; Ziogas J.; O’Hair R. A. J.; Donald W. A. Anal. Chem. 2015, 87, 4104–4109. 10.1021/ac504767d. [DOI] [PubMed] [Google Scholar]
- Bueschl C.; Kluger B.; Berthiller F.; Lirk G.; Winkler S.; Krska R.; Schuhmacher R. Bioinformatics 2012, 28, 736–738. 10.1093/bioinformatics/bts012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUPAC Compendium of Chemical Terminology (Gold Book), 2nd ed., 2006; https://goldbook.iupac.org/index.html.
- Du P.; Kibbe W. A.; Lin S. M. Bioinformatics 2006, 22, 2059–2065. 10.1093/bioinformatics/btl355. [DOI] [PubMed] [Google Scholar]
- Kind T.; Fiehn O. BMC Bioinf. 2007, 8, 105 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemberg T. G.; Gerretzen J.; Wouters H. J.; Gloerich J.; van Dael M.; Wessels H. J.; van den Heuvel L. P.; Eilers P. H.; Buydens L. M.; Wehrens R. Chemom. Intell. Lab. Syst. 2010, 104, 65–74. 10.1016/j.chemolab.2010.04.008. [DOI] [Google Scholar]
- Neumann N. K. N.; Lehner S. M.; Kluger B.; Bueschl C.; Sedelmaier K.; Lemmens M.; Krska R.; Schuhmacher R. Anal. Chem. 2014, 86, 7320–7327. 10.1021/ac501358z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R Project for Statistical Computing, v2.15.2, 2014; https://www.r-project.org/.
- Pedrioli P. G. A.; Eng J. K.; Hubley R.; Vogelzang M.; Deutsch E. W.; Raught B.; Pratt B.; Nilsson E.; Angeletti R. H.; Apweiler R.; Cheung K.; Costello C. E.; Hermjakob H.; Huang S.; Julian R. K.; Kapp E.; McComb M. E.; Oliver S. G.; Omenn G.; Paton N. W.; Simpson R.; Smith R.; Taylor C. F.; Zhu W. M.; Aebersold R. Nat. Biotechnol. 2004, 22, 1459–1466. 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- Bueschl C.; Kluger B.; Lemmens M.; Adam G.; Wiesenberger G.; Maschietto V.; Marocco A.; Strauss J.; Bodi S.; Thallinger G. G.; Krska R.; Schuhmacher R. Metabolomics 2014, 10, 754–769. 10.1007/s11306-013-0611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluger B.; Bueschl C.; Neumann N.; Stuckler R.; Doppler M.; Chassy A. W.; Waterhouse A. L.; Rechthaler J.; Kampleitner N.; Thallinger G. G.; Adam G.; Krska R.; Schuhmacher R. Anal. Chem. 2014, 86, 11533–11537. 10.1021/ac503290j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluger B.; Bueschl C.; Lemmens M.; Berthiller F.; Haubl G.; Jaunecker G.; Adam G.; Krska R.; Schuhmacher R. Anal. Bioanal. Chem. 2013, 405, 5031–5036. 10.1007/s00216-012-6483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavalisco P.; Koehl K.; Hummel J.; Seiwert B.; Willmitzer L. Anal. Chem. 2009, 81, 6546–6551. 10.1021/ac900979e. [DOI] [PubMed] [Google Scholar]
- Meng-Reiterer J.; Varga E.; Nathanail A. V.; Bueschl C.; Rechthaler J.; McCormick S. P.; Michlmayr H.; Malachova A.; Fruhmann P.; Adam G.; Berthiller F.; Lemmens M.; Schuhmacher R. Anal. Bioanal. Chem. 2015, 407, 8019–8033. 10.1007/s00216-015-8975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassy A. W.; Bueschl C.; Lee H.; Lerno L.; Oberholster A.; Barile D.; Schuhmacher R.; Waterhouse A. L. Food Chem. 2015, 166, 448–455. 10.1016/j.foodchem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Doppler M.; Kluger B.; Bueschl C.; Schneider C.; Krska R.; Delcambre S.; Hiller K.; Lemmens M.; Schuhmacher R. Int. J. Mol. Sci. 2016, 17, 1017 10.3390/ijms17071017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.