Abstract
NMR metabolomics are primarily conducted with 1D nuclear Overhauser enhancement spectroscopy (NOESY) presat for water suppression and Carr–Purcell–Meiboom–Gill (CPMG) presat as a T2 filter to remove macromolecule signals. Others pulse sequences exist for these two objectives but are not often used in metabolomics studies, because they are less robust or unknown to the NMR metabolomics community. However, recent improvements on alternative pulse sequences provide attractive alternatives to 1D NOESY presat and CPMG presat. We focus this perspective on PURGE, a water suppression technique, and PROJECT presat, a T2 filter. These two pulse sequences, when optimized, performed at least on par with 1D NOESY presat and CPMG presat, if not better. These pulse sequences were tested on common samples for metabolomics, human plasma, and urine.
NMR-based metabolomics has become established over the past 2 decades, with a great amount of work done to standardize the acquisition parameters and analysis of the data.1−3 While 2D NMR metabolomics are sometimes used,4−6 the majority of NMR-based metabolomics are conducted using 1D pulse sequences, with most of these studies relying on two pulse sequences: 1D nuclear Overhauser enhancement spectroscopy (NOESY) presat7,8 (solvent suppression) and Carr–Purcell–Meiboom–Gill (CPMG) presat9,10 (T2 filter).
Both pulse sequences have become gold standards for NMR-based metabolomics, thanks to their effectiveness at removing unwanted signals and general robustness. Despite their utility, these pulse sequences have limitations, as we will show below. For these cases, recently developed alternatives are worth considering.
For water suppression, several pulse sequences have been already designed, like presaturation,11 composite pulses,12 WET,13,14 WATERGATE,15−18 or PURGE.19 Despite all these variants, the 1D NOESY presat pulse sequence (noesypr1d on Bruker spectrometers) has emerged as the leading choice for NMR-based metabolomics. One of the main reasons for the success of noesypr1d is its ability to greatly suppress the water peak without intensity losses for most of the other peaks (except those close to the water resonance, like glucose and carnitine) and not needing gradients to have adequate water suppression, so gradient imperfections do not affect the final spectra, as seen in Figure 1. However, the 1D NOESY presat does have some shortcomings, the main one being the noncomplete suppression of “faraway water”,20 i.e., water outside of the homogeneous part of the NMR volume, which does not experience the same frequency as the rest of the water.
Figure 1.
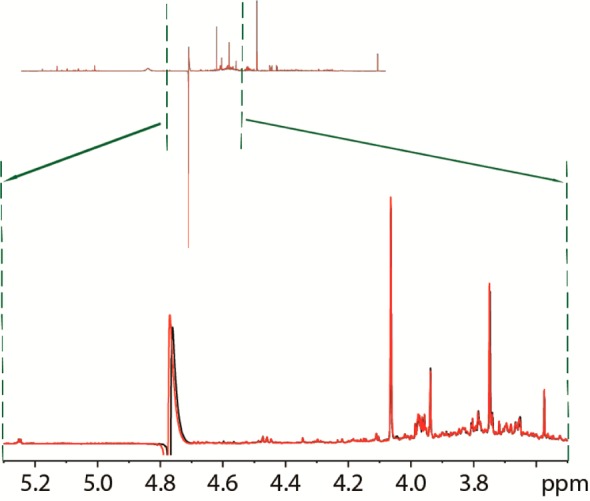
Zoom of the 1D NOESY presat spectra overlay of pooled urine without (black) and with (red) gradients, along with the complete spectrum shown above, to show the difference in residual water peak intensity.
Alternate Water Suppression Techniques and Avoiding Some of Their Shortcomings
Some of the water suppression techniques cited above are more efficient at suppressing faraway water, along with pulse sequence variants aimed at suppressing faraway water, like PRESAT18021 or WET180.22 However, they either use shaped pulses or gradients, which limits the robustness and thus reproducibility of the pulse sequence, an important requirement for metabolomics applications. Among these alternatives, PURGE19 was developed for applications requiring extensive solvent suppression (LC–NMR, metabolomics, protein study, etc.).
Since its first publication, the PURGE pulse sequence has been used on a regular basis by the team that created it,23−35 along with several other groups,36−46 and even suggested in a Nature protocol.47 Despite this visibility, the PURGE pulse sequence has not become routine in metabolomics. The relative lack of popularity may come from a lack of familiarity by a majority of NMR-based metabolomics researchers and/or an important shortcoming of the PURGE pulse sequence, namely, sensitivity to gradient imperfections.
This sensitivity to gradient imperfections is apparent in our relatively new 600 MHz AVIII-HD, as shown on a sample of glucose in water (Figure 2b). This spectrometer is equipped with a cryoprobe and is currently the main spectrometer for our high-throughput metabolomics studies, which means the PURGE pulse sequence is not the best fit for most of our metabolomics studies due to its poor line shape performance. This issue has also been mentioned in at least one other paper.44 However, a recent study48 showed that pulse sequences sensitive to gradient imperfections could still give good line shape by alternating gradient signs between scans. In that case, the WATERGATE water suppression scheme was used, but as we show here the scheme seems to be useful for other pulse sequences, including PURGE.
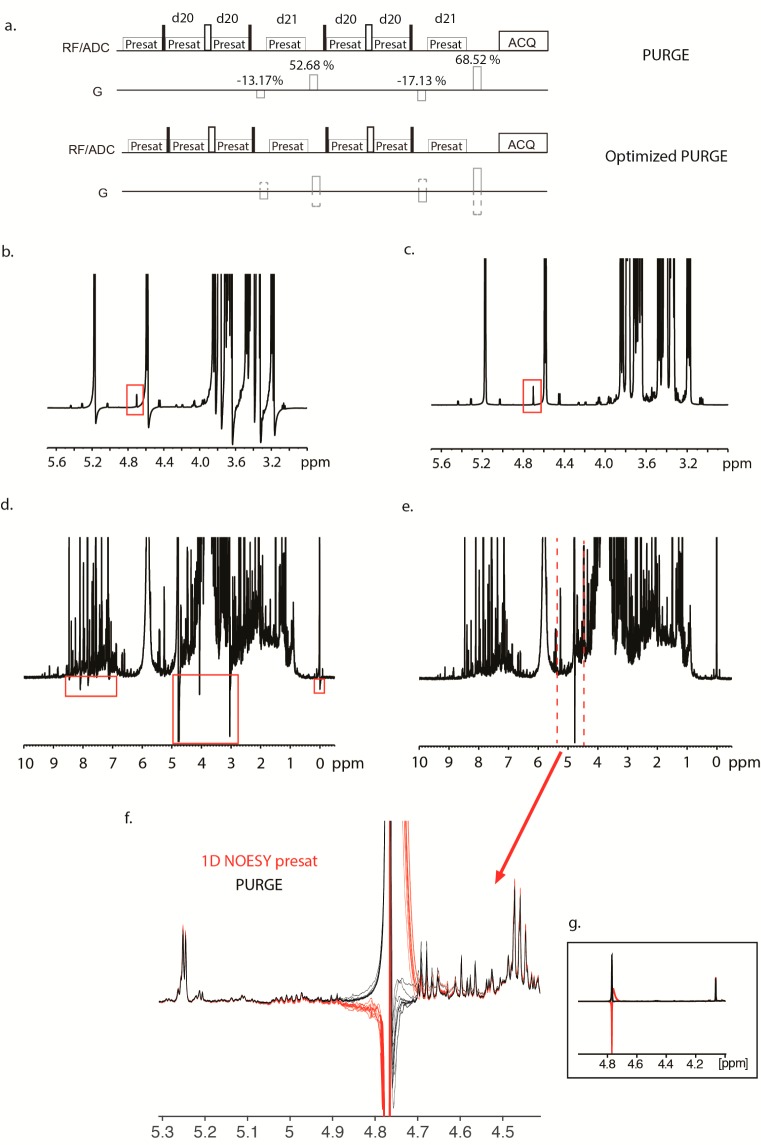
Figure 2.
Pulse sequences for the original PURGE and the optimized PURGE (a), where gradient signs are inverted between every scan. The gradient levels themselves are shown for the original PURGE. d20 and d21 are the delays used within the pulse sequence for short presaturation times and were set to 200 μs, the recommend values. 1D 1H spectra of glucose with the original PURGE (b) and the optimized PURGE (c) pulse sequences, with the water peak framed in red, showing the nonexistent impact of alternating gradients on the water peak. 1D 1H spectra of a urine sample with the original PURGE (d) and the optimized PURGE (e) pulse sequences. An expansion of part e is shown in part f, where both the 1D NOESY presat (red) and the PURGE (black) spectra of 10 urine samples are superimposed. A more global view of the water peak is shown in the framed inset (g).
We modified the standard PURGE to an “optimized” PURGE sequence by applying gradients that alternated in sign between scans (Figure 2a), similar to the ROBUST-5 approach of Aguilar and co-workers.48 While we started by including both alternating gradients and prefocusing gradients, we found an increase in the water resonance using prefocusing gradients (data not shown). Therefore, we kept only the alternating gradients. On a concentrated glucose sample (100 mM), the original PURGE shows sensitivity to gradient imperfections, which causes line shape distortions (Figure 2b), while the optimized PURGE shows good line shape and baseline without modification of the water peak (Figure 2c).
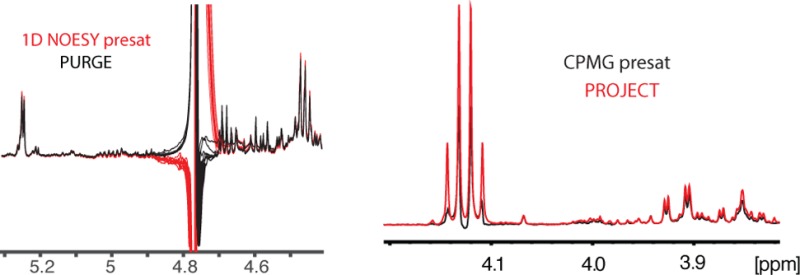
After verifying the optimized PURGE on the glucose sample, we compared it to 1D NOESY preset using 10 replicates of pooled human urine samples containing a high concentration of salts and 90% H2O. In these conditions, the optimized PURGE still showed good line shape and baseline compared to the original PURGE (Figure 2d,e). Spectra from each pulse sequence are shown in Figure 2f. This figure shows that the water peak in 1D NOESY presat distorted the baseline over a bigger chemical shift range than for PURGE. This property is probably caused by an incomplete suppression of “faraway water”, as seen in the framed expansion in Figure 2g. In addition, the resonances closest to the water have a higher intensity in the PURGE spectrum, despite using the same presaturation power for both experiments. Our results confirm the utility of the PURGE pulse sequence, especially with the rather simple solution to compensate for gradient imperfections.48 It should be noted that it is possible to adjust the pre-emphasis on most gradients. However, this is time-consuming and also specific for each probe. Alternating the sign of the gradients provides a much simpler and robust solution to the problem.
T2 Filter
For samples with a large quantity of macromolecules (like proteins or lipids), T2 filters are useful in order to remove the broad unwanted resonances, as seen in Figure 3b. The pulse sequence mainly used for T2 filters is the CPMG,10 which has been used for decades. However, a variant called PROJECT has been developed recently, which has some distinct advantages with no significant downsides (Figure 3a).49 This variant uses perfect echoes instead of spin echoes, an approach developed in 198950 but recently reintroduced after it was shown that it can avoid J-modulation for most spin systems, in contrast to CPMG.49
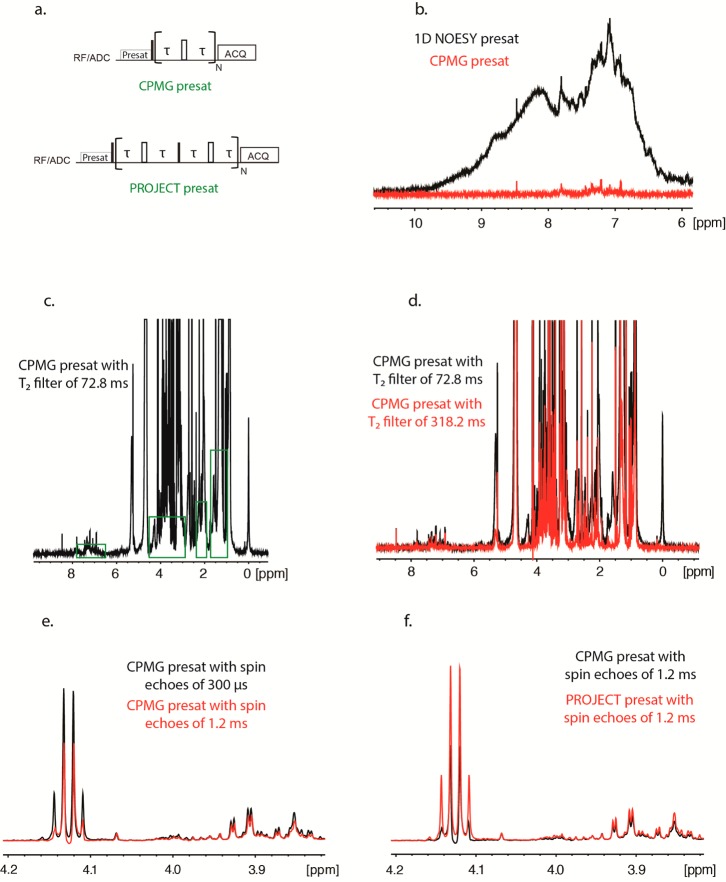
Figure 3.
1D 1H spectrum of whole plasma with 1D NOESY presat and CPMG presat, using standardized parameters3 (a), along with a close-up of the CPMG presat spectrum, showing in green frame regions where there is still some significant signal from macromolecules (b). The use of a longer T2 filter allows reducing further signals from macromolecules (c), but the use of longer spin echoes (without changing the length of the T2 filter) decreases the sensitivity of signals and even distorts the line shape of some of them (d). The use of the PROJECT pulse sequence as shown in (e) and compared to the CPMG pulse sequence, allows retaining sensitivity when using longer spin echoes (f).
Since the first publication of PROJECT, a few other papers have shown its utility for T2 measurements or as a T2 filter.51−57 However, a large number of the references to PROJECT use it as an alternative spin echo for other pulse sequences,58−65 like WATERGATE, HSQC, INEPT, etc. To our knowledge there has been no mention of PROJECT for metabolomics. For that reason, we were interested to compare PROJECT with CPMG, especially for the quantification of plasma or serum, along with intact tissues with HR-MAS, as shown elsewhere.54 It should be noted that no new parameters had to be introduced in the PROJECT pulse sequence, so it should be as robust and simple as CPMG.
We compared intact plasma with methanol/water extracted plasma66,67 using the same pooled Red Cross plasma that is used for quality control in the Southeast Center for Integrated Metabolomics (SECIM). Intact plasma allows for the comparison between CPMG presat and PROJECT presat, while extracted plasma was used as the standard in terms of quantification.67 It should be noted that, as described by Gowda and co-workers, extracted plasma or serum produce excellent results. However, it adds extra steps to the sample preparation and also removes the lipoproteins, which can be analyzed directly on the same sample to obtain additional information.68−70 We first applied CPMG presat with standardized parameters for metabolomics with plasma (Figure 3b).3 While most of the macromolecular signals are already suppressed, a close examination of the baseline shows that some broad signal remains (Figure 3c). These residual signals can interfere with quantification, so we first tried to increase the length of the T2 filter.
In order to increase the length of T2 and avoid heating the sample, it is preferable to increase the length of each echo rather than the number of echoes. This strategy allows a further reduction of intensity of macromolecules compared to the standardized CPMG presat (Figure 3d). However, when compared to a T2 filter with the same total duration with shorter spin echoes, the CPMG presat with longer spin echoes leads to sensitivity losses for most of the peaks (from 10% to more than 50% in our data set), along with line shape distortion for some peaks that is caused by J-modulation that is not completely suppressed by CPMG presat (Figure 3e). These shortcomings of CPMG presat can be resolved by using the PROJECT presat pulse sequence, which produces spectra almost identical to the one obtained with the CPMG presat using short spin echoes (Figure 3f).
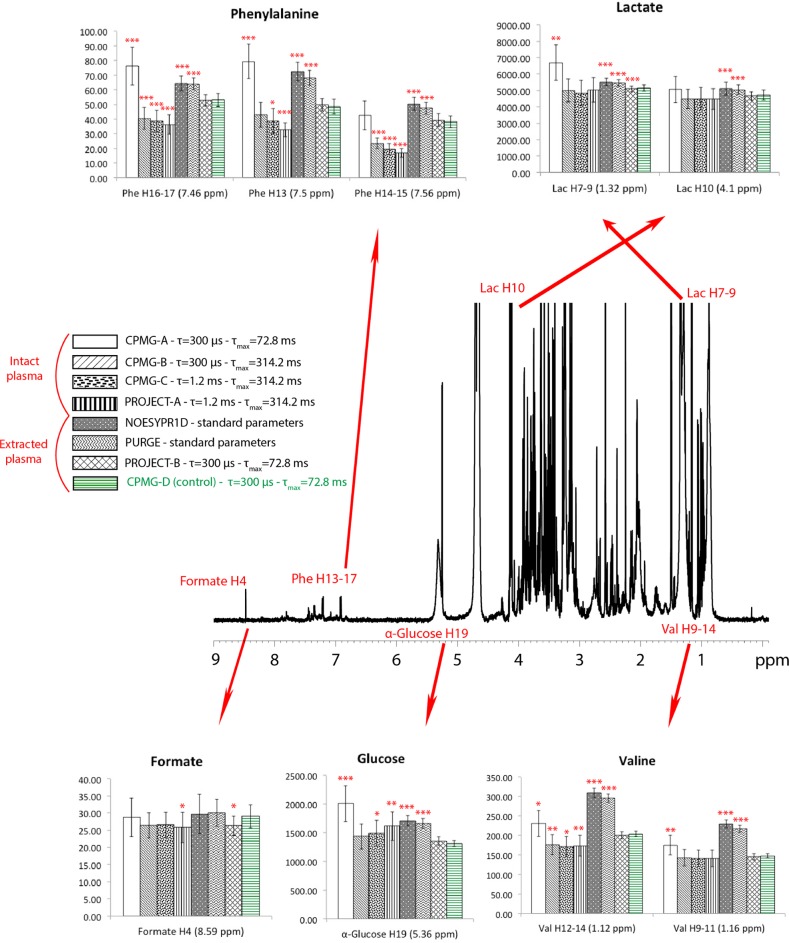
Along with this qualitative overview of the differences in the spectra, we also wanted to determine the effect of the changes in parameters for quantification, results shown in Figure 4. Peaks from 5 different molecules were selected for quantification. These had different properties: isolated peaks (lactate H10 formate H4), peaks with some overlap with macromolecule signals (valine H9-11 and H12-14, α-glucose H19) and peaks with major overlap with macromolecules (lactate H7-9, phenylalanine H13, H14-15, and H16-17). All atom labeling comes from the recently developed ALATIS, which creates a unique and atom-specific InChI string.71
Figure 4.
CPMG presat spectrum of methanol/water extracted plasma. Assigned peaks used for quantification are displayed. Ten replicate pooled aliquots were prepared for intact and extracted samples (20 total), and each reported pulse sequence was applied to each sample. The bar plots are the mean concentration (in μM) and standard deviation of 10 replicates of each condition, each measured from standard addition, using 3 spectra: one of the extract alone and two after 2 consecutive standard additions of each quantified metabolite. For this figure, the different variants correspond to different parameters (designated in the legend) and not different pulse sequences. CPMG-A, CPMG-B, CPMG-C, and PROJECT-A were used on intact plasma, while NOESYPR1d, PURGE, PROJECT-B, and CPMG-D were used on extracted plasma. τ, spin echo delay between two pulses; τmax, total duration of the spin echo. The parameters for CPMG-A are considered standard for analysis of intact plasma. For each peak, t tests were done, comparing the results of the CPMG presat of extracted plasma (CPMG-D) to the 7 other spectra. *, p < 0.05; **, p < 0.005; ***, p < 0.0005.
For each selected resonance, quantification was done on both intact and extracted plasma67 by integrating the area under each resonance and by using standard addition to build the calibration curves. The spiking buffer was identical to that used for the original samples. The single spiking solution contained 55 mM lactate, 57 mM glucose, 10 mM valine, 11 mM phenylalanine, and 0.55 mM formate. We added 20 μL of this solution and recorded all 1D experiments for each spike; this was done twice. For extracted plasma, all pulse sequences mentioned in this paper (noesypr1d, PURGE, cpmgpr1d, and PROJECTpr1d) were tested, using the standard parameters.3,67
t tests for each selected resonance were performed to statistically evaluate the changes in quantification between the different pulse sequences, using a threshold of 0.05 for the p-value. The concentrations from the CPMG presat spectra of the extracted plasma were used as the reference standards. These analyses show two trends. First, the pulse sequences that show the greatest differences with the CPMG presat of extracted plasma are the ones without T2 filters (noesypr1d, PURGE), even though these all used the extracted plasma. This shows that the remaining proteins after extraction are still concentrated enough to interfere with 1D quantification, demonstrating the need for a T2 filter.67 This difference can be mainly explained by the influence of protein signals in the total area under some of the peaks, especially lactate H7-9, both valine peaks, and α-glucose H19. For phenylalanine, in addition to some interfering protein signal, the different results can be explained by low signal-to-noise (S/N) and the longer T2 filters that further reduce the S/N.
Second, increasing the length of the T2 filter in intact plasma tends toward the values from the CPMG of the extracted plasma. The main exceptions are the phenylalanine peaks, as mentioned above. It can also be noted that for quantification, the length of the T2 filters is more important than the length of the spin echoes or the variant of T2 filter.
Results show that while both CPMG presat and PROJECT presat can be used for quantification, samples with high amount of proteins need long T2 filters, and PROJECT gave better results in these conditions, due to better overall sensitivity and line shape.
In summary, we have shown that there are useful alternatives to the most commonly used 1D NMR metabolomics pulse sequences. Should everyone adopt these in their metabolomics workflow? The answer to this question is complicated by the opposing need for the field to move toward standardization so that data can be compared across studies and between different laboratories. One could argue that it is more important to keep protocols like pulse sequences constant than to further optimize, but this argument is countered by each new generation of instrumentation that is available, which will undoubtedly have a significant impact on the spectra or we would not be willing to pay large amounts of money to buy them. The good news is that the sequences described here produce similar results to their currently used counterparts. In our hands, the optimized PURGE pulse sequence improved the water region when compared with noesypr1d for all samples tested, but other regions of the spectra were essentially unchanged. For plasma or serum, things are more complicated, because every combination of pulse sequence and sample preparation method gives different quantitative values. We think that we understand these differences and have attempted to describe them here. Our recommendation is to use PROJECT presat in place of CPMG presat for intact or extracted samples, because there is no change in parameter optimization and no change for most resonances. However, there is a great improvement when longer T2 filters need to be applied, because PROJECT does not cause J-coupling distortions that arise in CPMG. As the field advances and new techniques are introduced, the best way to establish back-compatibility with older data sets and methods will be to employ a reference standard, such as the NIST plasma (SRM 1950).72
Acknowledgments
This work was supported by the Southeast Center for Integrated Metabolomics (Grant NIH/NIDDK U24DK097209) and the Georgia Research Alliance.
Author Contributions
A.L.G conceived the study, implemented the pulse sequences, and measured and analyzed the data. F.T. assisted with extraction protocols and data analysis. All authors contributed to writing the manuscript.
The authors declare no competing financial interest.
Notes
The raw data used in this study will be deposited in the Metabolomics Workbench.
References
- Lindon J. C.; Nicholson J. K.; Holmes E.; Everett J. R. Concepts Magn. Reson. 2000, 12, 289–320. . [DOI] [Google Scholar]
- Bernini P.; Bertini I.; Luchinat C.; Nincheri P.; Staderini S.; Turano P. J. Biomol. NMR 2011, 49, 231–243. 10.1007/s10858-011-9489-1. [DOI] [PubMed] [Google Scholar]
- Dona A. C.; Jiménez B.; Schäfer H.; Humpfer E.; Spraul M.; Lewis M. R.; Pearce J. T. M.; Holmes E.; Lindon J. C.; Nicholson J. K. Anal. Chem. 2014, 86, 9887–9894. 10.1021/ac5025039. [DOI] [PubMed] [Google Scholar]
- Robinette S. L.; Ajredini R.; Rasheed H.; Zeinomar A.; Schroeder F. C.; Dossey A. T.; Edison A. S. Anal. Chem. 2011, 83, 1649–1657. 10.1021/ac102724x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis I. A.; Schommer S. C.; Markley J. L. Magn. Reson. Chem. 2009, 47, S123–S126. 10.1002/mrc.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.; Robinette S. L.; Bruschweiler-Li L.; Brüschweiler R. Magn. Reson. Chem. 2009, 47, S118–S122. 10.1002/mrc.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J. K.; Foxall P. J. D.; Spraul M.; Farrant R. D.; Lindon J. C. Anal. Chem. 1995, 67, 793–811. 10.1021/ac00101a004. [DOI] [PubMed] [Google Scholar]
- McKay R. T. Concepts Magn. Reson., Part A 2011, 38A, 197–220. 10.1002/cmr.a.20223. [DOI] [Google Scholar]
- Carr H. Y.; Purcell E. M. Phys. Rev. 1954, 94, 630–638. 10.1103/PhysRev.94.630. [DOI] [Google Scholar]
- Meiboom S.; Gill D. Rev. Sci. Instrum. 1958, 29, 688–691. 10.1063/1.1716296. [DOI] [Google Scholar]
- Hoult D. I. J. Magn. Reson. (1969-1992) 1976, 21, 337–347. 10.1016/0022-2364(76)90081-0. [DOI] [Google Scholar]
- Bax A. J. Magn. Reson. (1969-1992) 1985, 65, 142–145. 10.1016/0022-2364(85)90383-X. [DOI] [Google Scholar]
- Ogg R. J.; Kingsley R. B.; Taylor J. S. J. Magn. Reson., Ser. B 1994, 104, 1–10. 10.1006/jmrb.1994.1048. [DOI] [PubMed] [Google Scholar]
- Smallcombe S. H.; Patt S. L.; Keifer P. A. J. Magn. Reson., Ser. A 1995, 117, 295–303. 10.1006/jmra.1995.0759. [DOI] [Google Scholar]
- Hwang T. L.; Shaka A. J. J. Magn. Reson., Ser. A 1995, 112, 275–279. 10.1006/jmra.1995.1047. [DOI] [Google Scholar]
- Liu M.; Mao X.-a.; Ye C.; Huang H.; Nicholson J. K.; Lindon J. C. J. Magn. Reson. 1998, 132, 125–129. 10.1006/jmre.1998.1405. [DOI] [PubMed] [Google Scholar]
- Piotto M.; Saudek V.; Sklenář V. J. Biomol. NMR 1992, 2, 661–665. 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Sklenar V.; Piotto M.; Leppik R.; Saudek V. J. Magn. Reson., Ser. A 1993, 102, 241–245. 10.1006/jmra.1993.1098. [DOI] [Google Scholar]
- Simpson A. J.; Brown S. A. J. Magn. Reson. 2005, 175, 340–346. 10.1016/j.jmr.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Giraudeau P.; Silvestre V.; Akoka S. Metabolomics 2015, 11, 1041–1055. 10.1007/s11306-015-0794-7. [DOI] [Google Scholar]
- Mo H.; Raftery D. J. Magn. Reson. 2008, 190, 1–6. 10.1016/j.jmr.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo H.; Raftery D. J. Biomol. NMR 2008, 41, 105–111. 10.1007/s10858-008-9246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKelvie J. R.; Yuk J.; Xu Y.; Simpson A. J.; Simpson M. J. Metabolomics 2009, 5, 84. 10.1007/s11306-008-0122-6. [DOI] [Google Scholar]
- Brown S. A. E.; Simpson A. J.; Simpson M. J. Environmental Chemistry 2009, 6, 432–440. 10.1071/EN09054. [DOI] [Google Scholar]
- Brown S. A. E.; McKelvie J. R.; Simpson A. J.; Simpson M. J. Environ. Pollut. 2010, 158, 2117–2123. 10.1016/j.envpol.2010.02.023. [DOI] [PubMed] [Google Scholar]
- Whitfield Åslund M. L.; Simpson A. J.; Simpson M. J. Ecotoxicology 2011, 20, 836–846. 10.1007/s10646-011-0638-9. [DOI] [PubMed] [Google Scholar]
- Woods G. C.; Simpson M. J.; Koerner P. J.; Napoli A.; Simpson A. J. Environ. Sci. Technol. 2011, 45, 3880–3886. 10.1021/es103425s. [DOI] [PubMed] [Google Scholar]
- Whitfield Åslund M.; Simpson M. J.; Simpson A. J.; Zeeb B. A.; Rutter A. Ecotoxicology 2012, 21, 1947–1956. 10.1007/s10646-012-0928-x. [DOI] [PubMed] [Google Scholar]
- Lankadurai B.; Furdui V.; Reiner E.; Simpson A.; Simpson M. Metabolites 2013, 3, 718. 10.3390/metabo3030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankadurai B. P.; Wolfe D. M.; Whitfield Åslund M. L.; Simpson A. J.; Simpson M. J. Metabolomics 2013, 9, 44–56. 10.1007/s11306-012-0427-3. [DOI] [Google Scholar]
- Lankadurai B. P.; Nagato E. G.; Simpson A. J.; Simpson M. J. Ecotoxicol. Environ. Saf. 2015, 120, 48–58. 10.1016/j.ecoenv.2015.05.020. [DOI] [PubMed] [Google Scholar]
- Marshall M. H. M.; McKelvie J. R.; Simpson A. J.; Simpson M. J. Appl. Geochem. 2015, 54, 43–53. 10.1016/j.apgeochem.2014.12.013. [DOI] [Google Scholar]
- Kovacevic V.; Simpson A. J.; Simpson M. J. Comp. Biochem. Physiol., Part D: Genomics Proteomics 2016, 19, 199–210. 10.1016/j.cbd.2016.01.004. [DOI] [PubMed] [Google Scholar]
- Nagato E. G.; Simpson A. J.; Simpson M. J. Aquat. Toxicol. 2016, 170, 175–186. 10.1016/j.aquatox.2015.11.023. [DOI] [PubMed] [Google Scholar]
- Wagner N. D.; Simpson A. J.; Simpson M. J. Environ. Toxicol. Chem. 2017, 36, 938–946. 10.1002/etc.3604. [DOI] [PubMed] [Google Scholar]
- Hölscher D.; Brand S.; Wenzler M.; Schneider B. J. Nat. Prod. 2008, 71, 251–257. 10.1021/np0705514. [DOI] [PubMed] [Google Scholar]
- Byrne C. M. P.; Hayes M. H. B.; Kumar R.; Novotny E. H.; Lanigan G.; Richards K. G.; Fay D.; Simpson A. J. Water Res. 2010, 44, 4379–4390. 10.1016/j.watres.2010.05.055. [DOI] [PubMed] [Google Scholar]
- Lesar C. T.; Decatur J.; Lukasiewicz E.; Champeil E. Forensic Sci. Int. 2011, 212, e40–e45. 10.1016/j.forsciint.2011.06.017. [DOI] [PubMed] [Google Scholar]
- Houghton J. L.; Biswas T.; Chen W.; Tsodikov O. V.; Garneau-Tsodikova S. ChemBioChem 2013, 14, 2127–2135. 10.1002/cbic.201300359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton J. L.; Green K. D.; Pricer R. E.; Mayhoub A. S.; Garneau-Tsodikova S. J. Antimicrob. Chemother. 2013, 68, 800–805. 10.1093/jac/dks497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo Torre J. C.; Schmidt G. W.; Paetz C.; Reichelt M.; Schneider B.; Gershenzon J.; D’Auria J. C. Phytochemistry 2013, 91, 177–186. 10.1016/j.phytochem.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Plaza C.; Courtier-Murias D.; Fernández J. M.; Polo A.; Simpson A. J. Soil Biol. Biochem. 2013, 57, 124–134. 10.1016/j.soilbio.2012.07.026. [DOI] [Google Scholar]
- Koehler J.; Beck Erlach M.; Crusca E.; Kremer W.; Munte C. E.; Meier A.; Kalbitzer H. R. J. Biomol. NMR 2014, 60, 45–50. 10.1007/s10858-014-9850-2. [DOI] [PubMed] [Google Scholar]
- del Campo G.; Zuriarrain J.; Zuriarrain A.; Berregi I. Food Chem. 2016, 196, 1031–1039. 10.1016/j.foodchem.2015.10.036. [DOI] [PubMed] [Google Scholar]
- Marshall D. D.; Sadykov M. R.; Thomas V. C.; Bayles K. W.; Powers R. J. Proteome Res. 2016, 15, 1205–1212. 10.1021/acs.jproteome.5b01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vučković I.; Rapinoja M.-L.; Vaismaa M.; Vanninen P.; Koskela H. Phytochem. Anal. 2016, 27, 64–72. 10.1002/pca.2600. [DOI] [PubMed] [Google Scholar]
- Kim H. K.; Choi Y. H.; Verpoorte R. Nat. Protoc. 2010, 5, 536–549. 10.1038/nprot.2009.237. [DOI] [PubMed] [Google Scholar]
- Aguilar J. A.; Kenwright S. J. Analyst 2016, 141, 236–242. 10.1039/C5AN02121A. [DOI] [PubMed] [Google Scholar]
- Aguilar J. A.; Nilsson M.; Bodenhausen G.; Morris G. A. Chem. Commun. 2012, 48, 811–813. 10.1039/C1CC16699A. [DOI] [PubMed] [Google Scholar]
- Takegoshi K.; Ogura K.; Hikichi K. J. Magn. Reson. (1969-1992) 1989, 84, 611–615. 10.1016/0022-2364(89)90127-3. [DOI] [Google Scholar]
- Leung I. K. H.; Demetriades M.; Hardy A. P.; Lejeune C.; Smart T. J.; Szöllössi A.; Kawamura A.; Schofield C. J.; Claridge T. D. W. J. Med. Chem. 2013, 56, 547–555. 10.1021/jm301583m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto L. F.; Riguera R.; Fernandez-Megia E. J. Am. Chem. Soc. 2013, 135, 11513–11516. 10.1021/ja4059348. [DOI] [PubMed] [Google Scholar]
- Sánchez-Fernández E. M.; Tarhonskaya H.; Al-Qahtani K.; Hopkinson; Richard J.; McCullagh; James S. O.; Schofield; Christopher J.; Flashman E. Biochem. J. 2013, 449, 491–496. 10.1042/BJ20121155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André M.; Dumez J.-N.; Rezig L.; Shintu L.; Piotto M.; Caldarelli S. Anal. Chem. 2014, 86, 10749–10754. 10.1021/ac502792u. [DOI] [PubMed] [Google Scholar]
- Castañar L.; Nolis P.; Virgili A.; Parella T. J. Magn. Reson. 2014, 244, 30–35. 10.1016/j.jmr.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Walport L. J.; Hopkinson R. J.; Vollmar M.; Madden S. K.; Gileadi C.; Oppermann U.; Schofield C. J.; Johansson C. J. Biol. Chem. 2014, 289, 18302–18313. 10.1074/jbc.M114.555052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman P.; Meiri N.; Colnago L. A.; Moraes T. B.; Linder C.; Levi O.; Parmet Y.; Saunders M.; Wiesman Z. Biotechnol. Biofuels 2015, 8, 12. 10.1186/s13068-014-0194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klika K. D. Org. Lett. 2012, 14, 524–527. 10.1021/ol2031334. [DOI] [PubMed] [Google Scholar]
- Adams R. W.; Holroyd C. M.; Aguilar J. A.; Nilsson M.; Morris G. A. Chem. Commun. 2013, 49, 358–360. 10.1039/C2CC37579F. [DOI] [PubMed] [Google Scholar]
- Baishya B.; Khetrapal C. L.; Dey K. K. J. Magn. Reson. 2013, 234, 67–74. 10.1016/j.jmr.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Gambarota G.; Bondon A.; Floch M. L.; Mulkern R. V.; Saint-Jalmes H. J. Magn. Reson. 2013, 228, 76–80. 10.1016/j.jmr.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Baishya B.; Khetrapal C. L. J. Magn. Reson. 2014, 242, 143–154. 10.1016/j.jmr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Kaltschnee L.; Kolmer A.; Timari I.; Schmidts V.; Adams R. W.; Nilsson M.; Kover K. E.; Morris G. A.; Thiele C. M. Chem. Commun. 2014, 50, 15702–15705. 10.1039/C4CC04217D. [DOI] [PubMed] [Google Scholar]
- Baishya B.; Verma A. J. Magn. Reson. 2015, 252, 41–48. 10.1016/j.jmr.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Aguilar J. A.; Adams R. W.; Nilsson M.; Morris G. A. J. Magn. Reson. 2014, 238, 16–19. 10.1016/j.jmr.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Gowda G. A. N.; Raftery D. Anal. Chem. 2014, 86, 5433–5440. 10.1021/ac5005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagana Gowda G. A.; Gowda Y. N.; Raftery D. Anal. Chem. 2015, 87, 706–715. 10.1021/ac503651e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soininen P.; Kangas A. J.; Wurtz P.; Tukiainen T.; Tynkkynen T.; Laatikainen R.; Jarvelin M.-R.; Kahonen M.; Lehtimaki T.; Viikari J.; Raitakari O. T.; Savolainen M. J.; Ala-Korpela M. Analyst 2009, 134, 1781–1785. 10.1039/b910205a. [DOI] [PubMed] [Google Scholar]
- Kaess B. M.; Tomaszewski M.; Braund P. S.; Stark K.; Rafelt S.; Fischer M.; Hardwick R.; Nelson C. P.; Debiec R.; Huber F.; Kremer W.; Kalbitzer H. R.; Rose L. M.; Chasman D. I.; Hopewell J.; Clarke R.; Burton P. R.; Tobin M. D.; Hengstenberg C.; Samani N. J. PLoS One 2011, 6, e14529. 10.1371/journal.pone.0014529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen A.-K.; Stark K.; Musameh M. D.; Nelson C. P.; Römisch-Margl W.; Kremer W.; Raffler J.; Krug S.; Skurk T.; Rist M. J.; Daniel H.; Hauner H.; Adamski J.; Tomaszewski M.; Döring A.; Peters A.; Wichmann H. E.; Kaess B. M.; Kalbitzer H. R.; Huber F.; Pfahlert V.; Samani N. J.; Kronenberg F.; Dieplinger H.; Illig T.; Hengstenberg C.; Suhre K.; Gieger C.; Kastenmüller G. Hum. Mol. Genet. 2012, 21, 1433–1443. 10.1093/hmg/ddr580. [DOI] [PubMed] [Google Scholar]
- Dashti H.; Westler W. M.; Markley J. L.; Eghbalnia H. R. Sci. Data 2017, 4, 170073. 10.1038/sdata.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney K. W.; Ballihaut G.; Bedner M.; Benford B. S.; Camara J. E.; Christopher S. J.; Davis W. C.; Dodder N. G.; Eppe G.; Lang B. E.; Long S. E.; Lowenthal M. S.; McGaw E. A.; Murphy K. E.; Nelson B. C.; Prendergast J. L.; Reiner J. L.; Rimmer C. A.; Sander L. C.; Schantz M. M.; Sharpless K. E.; Sniegoski L. T.; Tai S. S. C.; Thomas J. B.; Vetter T. W.; Welch M. J.; Wise S. A.; Wood L. J.; Guthrie W. F.; Hagwood C. R.; Leigh S. D.; Yen J. H.; Zhang N.-F.; Chaudhary-Webb M.; Chen H.; Fazili Z.; LaVoie D. J.; McCoy L. F.; Momin S. S.; Paladugula N.; Pendergrast E. C.; Pfeiffer C. M.; Powers C. D.; Rabinowitz D.; Rybak M. E.; Schleicher R. L.; Toombs B. M. H.; Xu M.; Zhang M.; Castle A. L. Anal. Chem. 2013, 85, 11732–11738. 10.1021/ac402689t. [DOI] [PMC free article] [PubMed] [Google Scholar]