Abstract

We show that sequential protein deposition is possible by photodeprotection of films formed from a tetraethylene-glycol functionalized nitrophenylethoxycarbonyl-protected aminopropyltriethoxysilane (NPEOC-APTES). Exposure to near-UV irradiation removes the protein-resistant protecting group, and allows protein adsorption onto the resulting aminated surface. The protein resistance was tested using proteins with fluorescent labels and microspectroscopy of two-component structures formed by micro- and nanopatterning and deposition of yellow and green fluorescent proteins (YFP/GFP). Nonspecific adsorption onto regions where the protecting group remained intact was negligible. Multiple component patterns were also formed by near-field methods. Because reading and writing can be decoupled in a near-field microscope, it is possible to carry out sequential patterning steps at a single location involving different proteins. Up to four different proteins were formed into geometric patterns using near-field lithography. Interferometric lithography facilitates the organization of proteins over square cm areas. Two-component patterns consisting of 150 nm streptavidin dots formed within an orthogonal grid of bars of GFP at a period of ca. 500 nm could just be resolved by fluorescence microscopy.
Introduction
Patterned assemblies of biomolecules have attracted a great deal of interest for applications in biosensors and in clinical diagnostics.1−4 For example, arrays of antibodies are selected to bind antigens that are markers for disease. Patterned assemblies of proteins have also been used to investigate a variety of interfacial biological phenomena including inflammation,5 cellular attachment,6−9 and thrombogenesis,10,11 which are regulated by the presentation of proteins at interfaces. The spatial arrangement of proteins is often important in biological mechanisms, for example in photosynthetic membranes where protein organization determines mechanisms of photon capture and energy transfer.12−14 By forming spatially organized assemblies of proteins on chips, it is possible to explore the relationship between their function and their spatiotemporal organization.15,16
Proteins are irreversibly adsorbed to many surfaces, and the most basic criterion for achieving effective spatial organization is to prepare a protein-resistant substrate into which binding sites may be introduced selectively. The most widely used approaches have utilized poly(ethylene glycol) and derivatives,6,17−21 although other polymers have been found to provide resistance to protein adsorption, including poly(sulfo betaines)22,23 and zwitterionic polymers such as poly(2-(methacryloyloxy)ethylphosphorylcholine)24,25 and poly(cysteine methacrylate).26 Preferably, protein immobilization should be achieved via a site-specific binding mechanism (for example, histidine tags bind strongly to nitrilotriacetic acid (NTA) groups27−31). Finally, it is desirable to be able to deposit multiple different proteins in a spatially organized array. In analytical devices, some degree of multiplexing is usually desirable, as it also is in fundamental studies, because biological systems typically utilize multiple different molecular interactions.
While there has been significant success in achieving these goals at micrometer length-scales, protein patterning remains challenging on smaller length scales. A variety of approaches have been explored, including dip-pen nanolithography,32−34 electron beam lithography,35−38 nanoimprint lithography,39,40 near-field optical methods,41−43 microcontact printing44 and interferometric lithography.26,45 Despite significant effort, the challenge of forming submicrometer scale patterns that consist of multiple different protein components remains largely unsolved. Perhaps the best results to date have been achieved by Maynard and co-workers.37,38 They utilized electron beam lithography to pattern biotin, maleimide, aminoxy or nitrilotriacetic acid groups onto protein-resistant surfaces, and then used these to bind proteins with biotin binding sites, a free cysteine, an N-terminal α-oxoamide, and a histidine tag, respectively.37 However, a limitation of such an approach is that there is a finite range of suitable affinity tags. In order to eliminate the need for affinity labels, they described an alternative approach, based on multiple electron beam exposures of films protected by spin-coating with trehalose,38 which protects proteins from the damaging effects of high vacuum.
Here we describe an alternative approach to label-free multiple-component protein patterning, based on photochemistry. Photochemical methods are an attractive route to the patterning of biological interactions at surfaces,46−55 offering the capacity to execute specific chemical transformations through strategies such as the use of nitrophenyl protecting groups,56−64 and they have been found to facilitate the formation of multiple-component biological assemblies at micrometer length scales.51 Photopatterning at the nanometer scale is feasible through the use of near-field optical methods31,41,42,65 and through the use of interferometric lithography.26,66 In the present study we demonstrate that both methods may be applied to enable selective deprotection of (methoxyheptaethylene glycol)nitrophenylethoxycarbonyl-protected aminopropyltriethoxysilane (henceforth OEG-NPEOC-APTES), an aminosilane bearing a photoremovable nitrophenyl group derivatized with an oligo(ethylene glycol) adduct.61 Intact OEG-NPEOC-APTES has been shown previously to be highly protein resistant.31,61 However, on exposure to UV light, the nitrophenyl protecting group is removed, exposing an amine group (Scheme 1). The deprotected surface is thus no longer protein-resistant. In the present work we describe how sequential deprotection of OEG-NPEOC-APTES using both interferometric and near-field methods followed by protein adsorption facilitates the fabrication of multiple component submicrometer protein patterns.
Scheme 1. Photodeprotection of OEG-NPEOC-APTES.
Experimental Section
Film Preparation
Sulfuric acid (1.83 S.G. 95+ %), hydrogen peroxide solution (100 volumes 30+ %), ammonia solution (S. G. 0.88, 35%), and toluene (HPLC grade) were supplied by Fisher Chemical and used as received. Ethanol (absolute) and glutaraldehyde (GA) solution (Grade II, 50% in water) were obtained from VWR international S.A.S. Phosphate buffered saline (PBS) tablets, ammonium acetate and HEPES were supplied by Sigma-Aldrich and prepared into buffer solutions (pH 7.4). Oligo (ethylene glycol) modified 2-nitrophenylethoxycarbonyl protected aminopropyltriethoxysilane (OEG-NPEOC-APTES) was synthesized by AF ChemPharm Ltd. Streptavidin and NeutrAvidin were supplied by Life Technologies and used as received.
Silicon wafers (reclaimed, p-type, < 100>) were supplied by Compart Technology and coverslips (22 × 50 mm # 1, 5) were supplied by Menzel-Gläser. Quartz slides (50 mm × 25 mm × 1 mm), 1000 mesh and 2000 mesh copper grids with diameter of 3.05 mm for micrometer-scale patterning were supplied by Agar Scientific Limited. Water was deionized using an ELGA Veolia water system (PURELAB Ultra). Silicon wafers, quartz or glass slides were cleaned with piranha solution, a mixture of 30% hydrogen peroxide and 95% concentrated sulfuric acid in the ratio of 3:7 (caution: piranha solution is a strong oxidizing agent and reacts violently with organic matter) and RCA solution, a mixture of water, 30% hydrogen peroxide, and 35% ammonia solution in the ratio of 5:1:1. After cleaning, the substrates were rinsed with copious amounts of deionized water and dried overnight in an oven. Clean, dry substrates were immersed in a 0.1% (v/v) solution of OEG-NPEOC-APTES in toluene for 48 h. After reaction, the substrates were washed by rinsing with toluene and ethanol several times and dried under a stream of nitrogen. Finally, the samples were annealed by heating to 120 °C for 1 h in a vacuum oven.
Proteins
The gene sequence of yellow fluorescent protein (YFP) was amplified by PCR from pCS2-Venus vector (a kind gift from Dr. Atsushi Miyawaki, RIKEN Brain science institute, Japan). The resulting Nde I /BamHI fragment was cloned into a pET14b expression vector (Novagen). Introducing the combined F64L, S65T, V68L, S72A, M153T, V163A, S175G, and A206K mutations into the YFP gene resulted in enhanced green fluorescent protein (GFP) gene.67 Both His6-YFP and His6-GFP proteins were produced by heterologous expression in Escherichia coli (BL21); cells were grown to an OD680 of 0.6 at 37 °C then induced using isopropyl β-d-1-thiogalactopyranoside (IPTG; 0.4 mM) for 12 h at 25 °C. Pelleted cells (19000g/20 min) were lysed by sonication, and the resulting lysate was clarified by a further spin (33000g/30 min). Both His-tagged fluorescent proteins were purified to homogeneity from clarified lysate using a Chelating Sepharose Fast Flow Ni-NTA gravity flow column (GE Healthcare) as detailed in the manufacturer’s instructions. Protein purity was assessed by gel electrophoresis (SDS-PAGE).
FITC-conjugated antisheep IgG, Neutravidin, and streptavidin-Alexa Fluor 750 were obtained from Life Technologies. Streptavidin-Atto 488 and streptavidin-Atto 655 were obtained from Sigma-Aldrich (Poole, UK).
Photolithography
Glass slides modified by OEG-NPEOC-APTES were cut into small pieces. For micropatterning, samples were exposed to light from a Coherent Innova 300 FreD frequency-doubled argon ion laser emitting at 244 nm through a copper grid mask with a variable power in the range 1–100 mW.
For interferometric lithography, a Lloyd’s mirror interferometer, consisting of a sample and mirror set at an angle 2θ relative to each other, was used in conjunction with the same laser.
Near-field lithography was carried out using two different systems. A home-built optical fiber scanning near-field optical microscope was used for patterning at 244 nm. The instrument was constructed using a Digital Instruments Multimode atomic force microscope (AFM) base with a NanoScope IIIa controller and Basic extender. A home-built, tuning fork based shear-force detection system68 was used in place of the AFM head. The probe is attached to one leg of a quartz crystal tuning fork (32768 Hz resonant frequency, 12.5 pF capacitance, Farnell) using UV curing epoxy (Norland 81). The tuning fork is mounted on a small shaker piezo, driven by the tapping drive signal from the AFM. The response of the tuning fork is measured by passing the current from each contact through a transimpedance amplifier (Analog Devices AD823) with a gain of 108 V/A, placed as close as possible to the tuning fork to reduce stray capacitance. The resulting voltage signals are subtracted using an instrumentation amplifier (Analog Devices AMP02) with a gain of 100 to give a signal approximately proportional to the differential displacement of the tuning fork legs. This amplitude signal is fed into the AFM feedback channel using a homemade patch cable connected to the extender module. The shear force system was controlled through the AFM software, operating in tapping mode. The amplitude sensitivity of this system, measured in the AFM software, is approximately 1 Å/V, and scans were typically performed with a free amplitude of 1 V and a set point maintained at 80–90% of this throughout the scan. Probes were prepared by etching optical fiber in HF, followed by deposition of 20 nm Al by thermal evaporation. Apertures were created by colliding the probe with a surface.
For patterning at 325 nm, a HeCd laser (IK 3202R-D, Kimmon, Tokyo, Japan) was coupled to a WiTec AlphaSNOM scanning near-field optical microscope (WiTec, Ulm, Germany). The laser power was 11 mW. WiTec AlphaSNOM cantilever-type probes were used (WiTec, Ulm, Germany), with a writing rate of 1 μm s–1. These probes have pyramidal tips with apertures at their apexes. The resolution is defined by the aperture size, which was ca. 150 nm. After exposure, the samples were immersed into PBS solution (pH 7.4) for 15 min then transferred to a solution of the appropriate protein. After protein adsorption, the samples were rinsed with PBS solution (pH 7.4) or HEPES buffer (pH 7.4) and stored in PBS solution or HEPES buffer at 4 °C in a fridge prior to analysis. For sequential protein patterning by near-field lithography, it was necessary to locate the same position on the sample after immersion in a protein solution. To aid this, a “finder grid” was formed on the sample surface prior to commencing lithography, by shadow deposition of metal through a suitable mask.
Friction force microscopy (FFM) and tapping-mode AFM images were acquired on a Veeco Multimode atomic force microscope with a Nanoscope IV controller (Veeco, Cambridge, UK), using Si3N4 cantilevers (NP-10, nominal spring constant 0.06 N m–1, Veeco) for FFM measurements and silicon tapping-mode cantilevers (Veeco MPP- 11100-W; spring constant between 20 and 50 N m–1).
Confocal Microscopy
Confocal microscopy was carried out using an LSM 510 laser scanning confocal microscope (Carl Zeiss, Welwyn Garden City, UK). Lasers emitting at 488 nm (GFP) and 543 nm (CpcA-PEB) were used for excitation. A 40× or 63× magnification oil immersion lens was used for imaging the samples, which were mounted in an antifade reagent (glycerol-PBS solution, AF1) (Citifluor Ltd., London, United Kingdom). The captured images were analyzed using Zeiss LSM image browser software. Samples stored in HEPES buffer (pH 7.4) were rinsed with 100 mM ammonium acetate solution (pH 7.4) and dried by nitrogen gas, to eliminate deposits from the buffer remaining at the surface after protein adsorption. Ammonium acetate is volatile and leaves no residue, and effectively displaces materials deposited from buffer solutions.
Results and Discussion
The methods used for protein nanopatterning are shown schematically in Figure 1. Near-field lithography allows the fabrication of arbitrary patterns, but patterning large areas can be slow because the method is serial. A parallel near-field lithography device, the “Snomipede”, has been described, which allows patterning over areas 1 mm wide.69 However, in the present work patterning is confined to a single probe. By contrast, interferometric lithography does not permit arbitrary pattern formation, but does facilitate rapid patterning over macroscopic areas (cm2). Sequential exposures enable the fabrication of a variety of different morphologies (Figure 1c).
Figure 1.
Schematic diagram showing the fabrication methods used in the present work. (a) Near-field lithography with a fiber or cantilever probe leads to selective photoremoval of OEG-NPEOC protecting groups, exposing a protein-adhesive aminated surface onto which protein is adsorbed. (b) In two-beam interferometric lithography using a Lloyd’s mirror apparatus, a laser beam is directed at a sample and mirror set at an angle 2θ to each other. Half the beam falls on the sample, while the other half falls on the mirror from where it is reflected onto the sample to interfere with the first half of the beam. (c) The resulting interferogram has a sinusoidal cross-section with a pitch λ/2 nsin θ, where n is the refractive index of the medium. By carrying out two exposures, and rotating the sample through an angle ϕ between exposures, a variety of pattern morphologies can be fabricated.
Proof of Concept Using Micropatterned Structures
To demonstrate the general efficacy of the patterning methodology utilized here, patterns were formed first by using an electron microscope grid as a mask. An OEG-NPEOC-APTES coated substrate was exposed to UV light from a frequency-doubled argon ion laser (244 nm) through the mask, causing localized photodeprotection. The kinetics of the photodeprotection process have been described elsewhere;31,61 a dose of 3 J cm–2 is required for full deprotection of OEG-NPEOC-APTES. The sample was immersed in a solution of Alexa Fluor 750-labeled streptavidin, which emits red fluorescence. Figure 2(a) shows a confocal fluorescence image of the resulting micropattern. Bright red fluorescence is observed from the square regions that were exposed to UV light, but dark contrast is observed elsewhere. Flooding the sample with light from an HeCd laser (λ = 325 nm) leads to deprotection of the dark bars, which were masked during the first exposure. A longer wavelength that is not damaging to the adsorbed protein is used for this second step, but the first step is carried out at 244 nm because patterning is more rapid with these more energetic photons. This enables adsorption of polymer nanoparticles coated with yellow-green fluorescent Neutravidin. Yellow-green fluorescence is observed only from the bars, because streptavidin adsorbed during the first patterning step blocks the square regions against adsorption of Neutravidin-coated particles.
Figure 2.
Two-component patterning. An OEG-NPEOC-APTES film was exposed at 244 nm through an electron microscope grid mask and immersed in a solution of Alexa Fluor 750-labeled streptavidin. Subsequently, the sample was exposed at 325 nm and immersed in a solution of yellow-green fluorescent Neutravidin-coated polymer particles. (a) Pattern of red fluorescence from Alexa Fluor 750-labeled streptavidin. (b) Fluorescence from Neutravidin-coated polymer particles. (c) Overlay of micrographs in (a) and (b).
It is known that during photoremoval of nitrophenyl protecting groups a side reaction occurs that leads to the formation of a nitroso derivative. Nevertheless, it was determined for OEG-NPEOC-APTES using XPS that ∼70% of adsorbates undergo complete deprotection to expose the amine group.31 Xia et al. demonstrated that this side reaction did not impede surface functionalization of deprotected films with nitrilotriaceticacid (NTA) or subsequent site-specific attachment of histidine-tagged proteins. Protein binding was reversed by treatment with imidazole, suggesting that the presence of nitroso byproducts did not have a significant impact on the utilization of the films form protein binding.31 The data presented in Figure 2 demonstrate that if the side reaction does occur, it does not inhibit the physical adsorption of protein either.
To provide a more rigorous test for the efficacy of this patterning method, microspectroscopy was used to characterize patterns consisting of discrete regions to which green fluorescent protein (GFP) and yellow fluorescent protein (YFP) had been adsorbed. After an initial exposure of the sample through a mask, GFP was adsorbed onto the exposed regions (squares). The sample was then flooded with near-UV light from an HeCd laser, to deprotect the remaining intact OEG-NPEOC-APTES regions and immersed in a solution of YFP. GFP and YFP have distinct emission spectra, with emission maxima at 509 and 527 nm, respectively. Consequently, if the integrity of masked OEG-NPEOC-APTES is not preserved during the adsorption of GFP, the masked regions (bars) would contain both proteins, leading to the observation of two peaks in spectra recorded from those regions.
Figure 3 shows a fluorescence image of a patterned sample consisting of GFP (squares) and YFP (bars). There is a small difference in the intensity of fluorescence observed from regions covered with the different adsorbed proteins. Spectra acquired from the square regions yielded a single maximum at 510 nm, corresponding to the emission maximum of GFP, and spectra acquired from the bars exhibited a single emission maximum at 527 nm corresponding to the emission maximum of YFP. These data provide very strong evidence that the methodology described here is effective in controlling the regions of the surface to which the different proteins are adsorbed.
Figure 3.

Two-component patterning. Fluorescence microscopy of pattern consisting of GFP (squares) surrounded by YFP (bars), together with spectra acquired at selected locations as shown.
Near-field lithography
It is possible to write arbitrary nanostructures using a near-field probe, while interferometric lithography (see below) enables the fabrication of periodic structures over macroscopic areas. Using a commercial cantilever-based near-field microscope, coupled to an HeCd laser (325 nm), features were written into an OEG-NPEOC-APTES film and imaged using friction force microscopy70−72 (Figure 4a). The exposed regions (lines) exhibit bright contrast relative to the unmodified regions of the film. The feature size is limited by the aperture size of the probe. Using commercial cantilever probes, with aperture sizes of ca. 200 nm, feature sizes of ca. 200 nm were achieved, as determined from friction force microscopy images such as the one in Figure 4a. Figure 4b shows a confocal fluorescence image of two geometric structures (overlaid triangles) formed by near-field lithography followed by immersion of the sample in a solution of FITC-labeled IgG in buffer. There is a clear contrast difference between the bright features formed in the lithographic process and the dark background, confirming the selectivity of attachment of protein to regions exposed to the near-field probe.
Figure 4.

(a) Friction force microscopy image of a pattern fabricated by near-field exposure of an OEG-NPEOC-APTES film. (b) Fluorescence microscopy image showing bright contrast from geometric shapes formed by near-field lithography followed by adsorption of FITC-labeled IgG. (c) Tapping-mode phase image of YFP adsorbed to nanolines fabricated using an optical fiber probe in shear-force mode to modify OEG-NPEOC-APTES. (d) Tapping mode height image of the lines shown in panel c.
The smallest feature sizes were achieved using a custom-built shear-force near-field microscope fitted with an optical fiber probe. Figure 4c shows a tapping mode phase image of three lines formed by using a near-field probe to expose a PEG-NPEOC-APTES film followed by immersion of the sample in a solution of YFP in buffer. The YFP exhibits brighter contrast, indicative of a higher rate of energy dissipation in the tip–sample contact than is the case for the silane film. The mean value of the full width and half-maximum height (fwhm) of these lines is 80 nm.
A distinctive feature of near-field microscopy is that the processes of reading and writing can be decoupled. By switching off the excitation source, it is possible to image the surface in shear-force mode, or in a cantilever-type system by operating the probe in the same way that an AFM is used. This makes it possible to fabricate a pattern, remove the sample from the microscope for functionalization of the pattern, return it to the microscope, and then relocate the same region in a sample, before proceeding to fabricate a second structure. To aid relocation of the precise same point on the surface, a metallic finder structure was first deposited by shadow deposition onto the substrate, prior to formation of the OEG-NPEOC-APTES film.
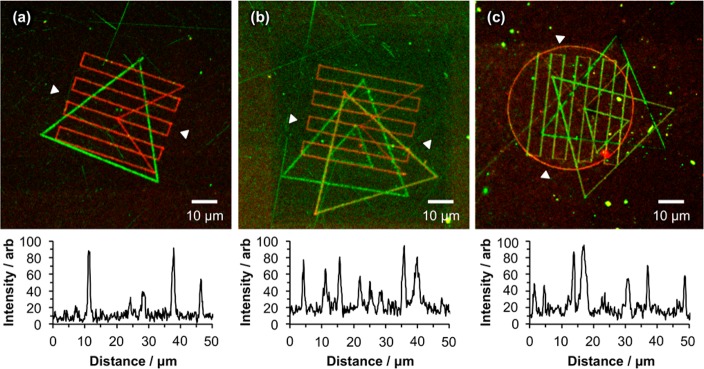
Figure 5 shows the stepwise assembly of a variety of overlaid geometrical patterns formed using a SNOM system with a cantilever probe. In Figure 5a, a triangle was first fabricated by near-field exposure of OEG-NPEOC-APTES. The sample was removed from the microscope and immersed in a solution of GFP overnight. After rinsing, the sample was dried and placed in the near-field microscope. Using the finder structure, the position at which the first lithographic process had been performed was located, and a second patterning process was implemented. A series of parallel lines was overlaid on the first pattern. The sample was then removed from the near-field microscope and placed in a solution of streptavidin labeled with Atto 655. The emission maximum of this dye lies far enough from the emission maximum of GFP that their fluorescence signals could be separated during confocal microscopy, enabling the different, discrete locations of the two protein patterns to be confirmed.
Figure 5.
Stepwise assembly of multiple component protein nanopatterns using near-field lithography. (a) GFP + streptavidin-Atto 655. (b) IgG-FITC + streptavidin-Atto 655 + streptavidin-Atto 488 (c) IgG-FITC + streptavidin-Atto 655 + streptavidin-Alexa Fluor 488 + streptavidin-Alexa Fluor 750. A representative line section is provided beneath each micrograph, measured between the white arrowheads marked on each.
Figure 5b shows a similar pattern, fabricated in the same fashion, but with a third protein component added. In this case, the IgG and streptavidin-Atto 488 have very similar emission spectra, and only the series of parallel lines (formed by the adsorption of streptavidin-Atto 655) may be distinguished using a filter. Finally Figure 5c shows a four-component pattern, consisting of a single triangle of IgG-FITC, a circle of streptavidin-Atto 655, eight lines of streptavidin-Alexa Fluor 488, and a double-triangle structure consisting of adsorbed streptavidin-Alexa Fluor 750.
These data clearly demonstrate the feasibility of executing lengthy sequences of lithographic processes using near-field methods. The use of a finder grid, combined with the decoupling of reading and writing in the near-field microscope, enables accurate registry to be achieved. Importantly for protein patterning, the unmodified OEG-NPEOC-APTES regions in Figure 5c exhibit dark contrast, indicating minimal nonspecific adsorption, even after four sequences of protein adsorption.
Two-Component Nanopatterning over Macroscopic Areas by Interferometric Lithography (IL)
Interferometric lithography does not permit arbitrary pattern formation, but it does enable the rapid fabrication of patterns over macroscopic areas. In the apparatus used in the present study, an area of ca. 1 cm2 was exposed. In a Lloyd’s mirror interferometer, two coherent laser beams interfere to produce an interferogram that consists of alternating bands of constructive and destructive interference with a sinusoidal cross-section of pitch λ/2n sin θ,73,74 where λ is the wavelength of the incident light, n is the refractive index of the medium (1 for air) and 2θ is the angle between the sample and the mirror. To identify the proteins in multiple-component nanopatterns, it is necessary to use optical microscopy; the features in the interferogram should thus have a pitch greater than the width of the point spread function of the microscope.
Figure 6 shows single-component protein patterns formed by interferometric exposure of OEG-NPEOC-APTES films. The interferometer conditions have been selected such that the features produced are likely to be just resolvable when characterized by optical microscopy. After exposure of the film, bands of alternating contrast are observed in a friction force microscopy image. Bright contrast is associated with regions where the film has been deprotected (see Figure 4a). The fwhm of these lines was 234 nm. After adsorption of GFP, a tapping-mode phase image (Figure 6b) reveals bands of alternating bright and dark contrast. The bright bands, thought to correspond to the protein, have a mean fwhm of 218 nm, indicating effective confinement of the protein to the regions that were deprotected during exposure.
Figure 6.

Patterns fabricated by IL. (a) Friction force microscopy image of parallel bands of amine functional groups produced by exposure of an OEG-NPEOC-APTES film (bright contrast). (b) Tapping mode phase image of lines of GFP after adsorption onto the sample shown in panel a. (c) Confocal fluorescence micrograph of a grid of orthogonal lines of GFP produced by carrying out a double-exposure of an OEG-NPEOC-APTES film, with a rotation of the sample through 90° between exposures. (d) Tapping-mode phase image of a region of the sample shown in panel c, revealing an array of protein-free spots (dark contrast).
A grid structure was formed by carrying out a double exposure and rotating the sample through 90° between exposures. After adsorption of GFP, the grid pattern could be resolved in a fluorescence image (Figure 6c). The pitch of the interferogram was 750 nm. Tapping mode phase images revealed protein-free regions with a fwhm of 290 nm (Figure 6d, dark contrast) corresponding to regions of the OEG-NPEOC-APTES film that were not exposed during either of the two interferometric patterning steps.
The protein-free regions observed in Figure 6d can be deprotected by exposure at 325 nm. Light at this wavelength is not expected to be damaging to the proteins, especially after short exposures. Two-component patterns were thus prepared by preparing a sample like the one shown in Figure 6c, followed by flooding of the sample with near-UV light and adsorption of streptavidin-Alexa Fluor 750. The sample was imaged by confocal microscopy (Figure 7), using a filter to separate the emission from the two proteins. The GFP is clearly resolved (Figure 7a). The streptavidin-coated regions are much more challenging, because their widths are slightly less than those of the GFP lines. However, an image of the red fluorescence (Figure 7b) reveals an array of points of fluorescence corresponding to the array of GFP-free regions defined in the first lithographic process. An overlay of the fluorescence signal acquired from the two proteins (Figure 7c) confirms that the streptavidin is located at the interstices in the GFP structure.
Figure 7.
Confocal fluorescence images of two-component protein patterns formed by interferometric lithography. (a) Image of fluorescence emission from GFP. (b) Image of fluorescence emission from streptavidin-Alexa Fluor 750. (c) Overlay of the fluorescence images shown in panels a and b.
While the formation of two-component patterns is demonstrated here, it is possible to conceive of methods for the production of surfaces with larger numbers of constituent components, for example by adsorption of two different proteins to form a grid pattern, or by the introduction of surface hererogeneity (for example, self-cleaning Ti nanostructures66).
In summary, the present work has demonstrated that a variety of methods may be used to expose OEG-NPEOC-APTES films. Mask-based exposure yields micrometer-scale patterns; sequential exposure enables the adsorption of proteins to form multiple-component patterns. A number of approaches exists to the patterning of proteins at micrometer length-scales, but by contrast, there are few methods that offer a capacity for repeatable nanopatterning of proteins on submicrometer length scales, and still fewer that offer a capability for the sequential deposition of multiple different proteins. Of the published studies, Maynard’s work using electron beam lithography is the most complete.37,38 Photolithographic approaches offer an alternative approach that requires comparatively inexpensive equipment and also the possibility for scaleability to cover large areas. Interferometric lithography offers a rapid route for the organization of proteins over macroscopic (>cm2) areas, while near-field methods offer the capacity for arbitrary pattern formation. Both enable deposition of different proteins using very simple, generic strategies. In the present work, proteins are adsorbed simply by physical adsorption, which is favored following removal of the OEG-NPEOC protecting group. However, it would also be possible to introduce approaches to the site-specific binding of proteins, via elaboration of the surface chemistry as described in previous studies, to enhance the biological activity of the immobilized biomolecules.31,61 Moreover, the capacity of near-field lithography for parallel implementation over macroscopic areas under water69 presents the exciting prospect of its development into a high-throughput fabrication technology capable of producing complex biologically functional surfaces.
Conclusions
Films formed by the adsorption of OEG-NPEOC-APTES demonstrate exceptional protein-resistance, enabling the execution of multiple lithographic processing steps with little diminution of their performance. Microspectroscopy of two-component GFP/YFP patterns formed by sequential exposure and adsorption steps confirms that the proteins are found only in the desired locations, with no evidence of cross-contamination. Using near-field lithography, a series of photopatterning steps, each followed by adsorption of a different protein, may be carried out, leading to the fabrication of elaborate multiple-component nanostructures that exhibit clear fluorescence and suggest minimal adsorption to unmodified regions of the surface despite the use of up to four overnight adsorption steps. Interferometric lithography may be used to carry out multiple exposures over macroscopic areas.
Acknowledgments
The authors are grateful to EPSRC (Programme Grant EP/I012060/1) for Financial Support. C.N.H. gratefully acknowledges financial support from the Biotechnology and Biological Sciences Research Council (BBSRC UK), Award Number BB/M000265/1. C.N.H. was also supported by Advanced Award 338895 from the European Research Council. This work was also supported as part of the Photosynthetic Antenna Research Center (PARC), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC 0001035. PARC’s role was to provide partial support for C.N.H. and A.J.C.
The authors declare no competing financial interest.
References
- Turner A. P. F. Biosensors: sense and sensibility. Chem. Soc. Rev. 2013, 42, 3184–3196. 10.1039/c3cs35528d. [DOI] [PubMed] [Google Scholar]
- Tamura T.; Hamachi I. Recent Progress in Design of Protein-Based Fluorescent Biosensors and Their Cellular Applications. ACS Chem. Biol. 2014, 9, 2708–2717. 10.1021/cb500661v. [DOI] [PubMed] [Google Scholar]
- Zhao W.-W.; Xu J.-J.; Chen H.-Y. Photoelectrochemical DNA Biosensors. Chem. Rev. 2014, 114, 7421–7441. 10.1021/cr500100j. [DOI] [PubMed] [Google Scholar]
- Liu Q.; Wu C.; Cai H.; Hu N.; Zhou J.; Wang P. Cell-Based Biosensors and Their Application in Biomedicine. Chem. Rev. 2014, 114, 6423–6461. 10.1021/cr2003129. [DOI] [PubMed] [Google Scholar]
- Rostam H. M.; Singh S.; Vrana N. E.; Alexander M. R.; Ghaemmaghami A. M. Impact of surface chemistry and topography on the function of antigen presenting cells. Biomater. Sci. 2015, 3, 424–441. 10.1039/C4BM00375F. [DOI] [PubMed] [Google Scholar]
- Lopez G. P.; Albers M. W.; Schreiber S. L.; Carroll R.; Peralta E.; Whitesides G. M. Convenient Methods for Patterning the Adhesion of Mammalian Cells to Surfaces using Self-Aseembled Monolayers of Alkanethiolates on Gold. J. Am. Chem. Soc. 1993, 115, 5877–5878. 10.1021/ja00066a087. [DOI] [Google Scholar]
- Chen C. S.; Mrksich M.; Huang S.; Whitesides G. M.; Ingber D. E. Geometric control of cell life and death. Science 1997, 276, 1425–1428. 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Cavalcanti-Adam A. E.; Micoulet A.; Blümmel J.; Auernheimer J.; Kessler H.; Spatz J. P. Lateral spacing of integrin ligands influences cell spreading and focal adhesion assembly. Eur. J. Cell Biol. 2006, 85, 219–224. 10.1016/j.ejcb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Cavalcanti-Adam E. A.; Volberg T.; Micoulet A.; Kessler H.; Geiger B.; Spatz J. P. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys. J. 2007, 92, 2964–74. 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie D. C.; Waterhouse A.; Berthet J. B.; Valentin T. M.; Watters A. L.; Jain A.; Kim P.; Hatton B. D.; Nedder A.; Donovan K.; Super E. H.; Howell C.; Johnson C. P.; Vu T. L.; Bolgen D. E.; Rifai S.; Hansen A. R.; Aizenberg M.; Super M.; Aizenberg J.; Ingber D. E. A bioinspired omniphobic surface coating on medical devices prevents thrombosis and biofouling. Nat. Biotechnol. 2014, 32, 1134–1140. 10.1038/nbt.3020. [DOI] [PubMed] [Google Scholar]
- Fedorov K.; Blaszykowski C.; Sheikh S.; Reheman A.; Romaschin A.; Ni H.; Thompson M. Prevention of Thrombogenesis from Whole Human Blood on Plastic Polymer by Ultrathin Monoethylene Glycol Silane Adlayer. Langmuir 2014, 30, 3217–3222. 10.1021/la500745p. [DOI] [PubMed] [Google Scholar]
- Cartron M. L.; Olsen J. D.; Sener M.; Jackson P. J.; Brindley A. A.; Qian P.; Dickman M. J.; Leggett G. J.; Schulten K.; Neil Hunter C. Integration of energy and electron transfer processes in the photosynthetic membrane of Rhodobacter sphaeroides. Biochim. Biophys. Acta, Bioenerg. 2014, 1837, 1769–1780. 10.1016/j.bbabio.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sener M.; Strumpfer J.; Singharoy A.; Hunter C. N.; Schulten K. Overall energy conversion efficiency of a photosynthetic vesicle. eLife 2016, 5, e09541. 10.7554/eLife.09541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Cartron M. L.; Mullin N.; Qian P.; Leggett G. J.; Hunter C. N.; Hobbs J. K. Direct Imaging of Protein Organization in an Intact Bacterial Organelle Using High-Resolution Atomic Force Microscopy. ACS Nano 2017, 11, 126–133. 10.1021/acsnano.6b05647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante M.; Lenferink A.; Zhao Y.; Tas N.; Huskens J.; Hunter C. N.; Subramaniam V.; Otto C. Long-Range Energy Propagation in Nanometer Arrays of Light Harvesting Antenna Complexes. Nano Lett. 2010, 10, 1450–1457. 10.1021/nl1003569. [DOI] [PubMed] [Google Scholar]
- Vasilev C.; Johnson M. P.; Gonzales E.; Wang L.; Ruban A. V.; Montano G.; Cadby A. J.; Hunter C. N. Reversible switching between nonquenched and quenched states in nanoscale linear arrays of plant light-harvesting antenna complexes. Langmuir 2014, 30, 8481–8490. 10.1021/la501483s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. M.Poly(Ethylene Glycol) Chemistry: Biochemical and Biomedical Applications; Plenum: New York, 1992. [Google Scholar]
- Prime K. L.; Whitesides G. M. Self-assembled organic monolayers: model systems for studying adsorption of proteins at surfaces. Science 1991, 252, 1164–1167. 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- Pale-Grosdemange C.; Simon E. S.; Prime K. L.; Whitesides G. M. Formation of self-assembled monolayers by chemisorption of derivatives of oligo(ethylene glycol) of structure HS(CH2)11(OCH2CH2)mOH on gold. J. Am. Chem. Soc. 1991, 113, 12–20. 10.1021/ja00001a002. [DOI] [Google Scholar]
- Ma H.; Li D.; Sheng X.; Zhao B.; Chilkoti A. Protein resistant polymer brushes on silicon oxide by surface initiated atom transfer radical polymerization. Langmuir 2006, 22, 3751–3756. 10.1021/la052796r. [DOI] [PubMed] [Google Scholar]
- Hucknall A.; Rangarajan S.; Chilkoti A. In Pursuit of Zero: Polymer Brushes that Resist the Adsorption of Proteins. Adv. Mater. 2009, 21, 2441–2446. 10.1002/adma.200900383. [DOI] [Google Scholar]
- Zhang Z.; Chao T.; Chen S.; Jiang S. Superlow Fouling Sulfobetaine and Carboxybetaine Polymers on Glass Slides. Langmuir 2006, 22, 10072–10077. 10.1021/la062175d. [DOI] [PubMed] [Google Scholar]
- Yang W.; Chen S.; Cheng G.; Vaisocherová H.; Xue H.; Li W.; Zhang J.; Jiang S. Film Thickness Dependence of Protein Adsorption from Blood Serum and Plasma onto Poly(sulfobetaine)-Grafted Surfaces. Langmuir 2008, 24, 9211–9214. 10.1021/la801487f. [DOI] [PubMed] [Google Scholar]
- Feng W.; Zhu S.; Ishihara K.; Brash J. L. Adsorption of Fibrinogen and Lysozyme on Silicon Grafted with Poly(2-methacryloyloxyethyl Phosphorylcholine) via Surface-Initiated Atom Transfer Radical Polymerization. Langmuir 2005, 21, 5980–5987. 10.1021/la050277i. [DOI] [PubMed] [Google Scholar]
- Feng W.; Brash J. L.; Zhu S. Non-biofouling materials prepared by atom transfer radical polymerization grafting of 2-methacryloloxyethyl phosphorylcholine: Separate effects of graft density and chain length on protein repulsion. Biomaterials 2006, 27, 847–855. 10.1016/j.biomaterials.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Alswieleh A. M.; Cheng N.; Canton I.; Ustbas B.; Xue X.; Ladmiral V.; Xia S.; Ducker R. E.; El Zubir O.; Cartron M. L.; Hunter C. N.; Leggett G. J.; Armes S. P. Zwitterionic Poly(amino acid methacrylate) Brushes. J. Am. Chem. Soc. 2014, 136, 9404–9413. 10.1021/ja503400r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal G. B.; Bamdad C.; Barberis A.; Strominger J.; Whitesides G. M. A Self-Assembled Monolayer for the Binding and Study of Histidine-Tagged Proteins by Surface Plasmon Resonance. Anal. Chem. 1996, 68, 490–497. 10.1021/ac9504023. [DOI] [PubMed] [Google Scholar]
- Schmid E. L.; Keller T. A.; Dienes Z.; Vogel H. Reversible Oriented Surface Immobilization of Functional Proteins on Oxide Surfaces. Anal. Chem. 1997, 69, 1979–1985. 10.1021/ac9700033. [DOI] [PubMed] [Google Scholar]
- Stora T.; Dienes Z.; Vogel H.; Duschl C. Histidine-Tagged Amphiphiles for the Reversible Formation of Lipid Bilayer Aggregates on Chelator-Functionalized Gold Surfaces. Langmuir 2000, 16, 5471–5478. 10.1021/la991711h. [DOI] [Google Scholar]
- Maury P.; Escalante M.; Péter M.; Reinhoudt D. N.; Subramaniam V.; Huskens J. Creating Nanopatterns of His-Tagged Proteins on Surfaces by Nanoimprint Lithography Using Specific NiNTA-Histidine Interactions. Small 2007, 3, 1584–1592. 10.1002/smll.200700046. [DOI] [PubMed] [Google Scholar]
- Xia S.; Cartron M.; Morby J.; Bryant D. A.; Hunter C. N.; Leggett G. J. Fabrication of Nanometer- and Micrometer-Scale Protein Structures by Site-Specific Immobilization of Histidine-Tagged Proteins to Aminosiloxane Films with Photoremovable Protein-Resistant Protecting Groups. Langmuir 2016, 32, 1818–1827. 10.1021/acs.langmuir.5b04368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-B.; Park S.-J.; Mirkin C. A.; Smith J. C.; Mrksich M. Protein Nanoarrays Generated by Dip-Pen Nanolithography. Science 2002, 295, 1702–1705. 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- Lee K.-B.; Lim J.-H.; Mirkin C. A. Protein Nanostructures formed via Direct-Write Dip-Pen Nanolithography. J. Am. Chem. Soc. 2003, 125, 5588–5589. 10.1021/ja034236p. [DOI] [PubMed] [Google Scholar]
- Hyun J.; Ahn S. J.; Lee W. K.; Chilkoti A.; Zauscher S. Molecular Recognition-Mediated Fabrication of Protein Nanostructures by Dip-Pen Nanolithography. Nano Lett. 2002, 2, 1203–1207. 10.1021/nl0257364. [DOI] [Google Scholar]
- Ballav N.; Thomas H.; Winkler T.; Terfort A.; Zharnikov M. Making Protein Patterns by Writing in a Protein-Repelling Matrix. Angew. Chem., Int. Ed. 2009, 48, 5833–5836. 10.1002/anie.200900950. [DOI] [PubMed] [Google Scholar]
- Krakert S.; Ballav N.; Zharnikov M.; Terfort A. Adjustment of the bioresistivity by electron irradiation: self-assembled monolayers of oligo(ethyleneglycol)-terminated alkanethiols with embedded cleavable group. Phys. Chem. Chem. Phys. 2010, 12, 507–515. 10.1039/B915036F. [DOI] [PubMed] [Google Scholar]
- Christman K. L.; Schopf E.; Broyer R. M.; Li R. C.; Chen Y.; Maynard H. D. Positioning Multiple Proteins at the Nanoscale with Electron Beam Cross-Linked Functional Polymers. J. Am. Chem. Soc. 2009, 131, 521–527. 10.1021/ja804767j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau U. Y.; Saxer S. S.; Lee J.; Bat E.; Maynard H. D. Direct Write Protein Patterns for Multiplexed Cytokine Detection from Live Cells Using Electron Beam Lithography. ACS Nano 2016, 10, 723–729. 10.1021/acsnano.5b05781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconnet D.; Pasqui D.; Park S.; Eckert R.; Schift H.; Gobrecht J.; Barbucci R.; Textor M. A Novel Approach to produce Protein Nanopatterns by Combining Nanoimprint Lithography and Molecular Self-Assembly. Nano Lett. 2004, 4, 1909–1914. 10.1021/nl0489438. [DOI] [Google Scholar]
- Hoff J. D.; Cheng L.-J.; Meyhöfer E.; Guo L. J.; Hunt A. J. Nanoscale Protein Patterning by Imprint Lithography. Nano Lett. 2004, 4, 853–857. 10.1021/nl049758x. [DOI] [Google Scholar]
- Montague M.; Ducker R. E.; Chong K. S. L.; Manning R. J.; Rutten F. J. M.; Davies M. C.; Leggett G. J. Fabrication of Biomolecular Nanostructures by Scanning Near-Field Photolithography of Oligo(ethylene glycol) Terminated Self-Assembled Monolayers. Langmuir 2007, 23, 7328–7337. 10.1021/la070196h. [DOI] [PubMed] [Google Scholar]
- Reynolds N. P.; Tucker J. D.; Davison P. A.; Timney J. A.; Hunter C. N.; Leggett G. J. Site-Specific Immobilization and Micrometer and Nanometer Scale Photopatterning of Yellow Fluorescent Protein on Glass Surfaces. J. Am. Chem. Soc. 2009, 131, 896–897. 10.1021/ja8079252. [DOI] [PubMed] [Google Scholar]
- Ul-Haq E.; Patole S.; Moxey M.; Amstad E.; Vasilev C.; Hunter C. N.; Leggett G. J.; Spencer N. D.; Williams N. H. Photocatalytic Nanolithography of Self-Assembled Monolayers and Proteins. ACS Nano 2013, 7, 7610–7618. 10.1021/nn402063b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyer S. R.; García A. J.; Delamarche E. Facile Preparation of Complex Protein Architectures with Sub-100-nm Resolution on Surfaces. Angew. Chem., Int. Ed. 2007, 46, 6837–6840. 10.1002/anie.200700989. [DOI] [PubMed] [Google Scholar]
- Tizazu G.; el Zubir O.; Patole S.; McLaren A.; Vasilev C.; Mothersole D.; Adawi A.; Hunter C. N.; Lidzey D.; Lopez G.; Leggett G. Micrometer and Nanometer Scale Photopatterning of Proteins on Glass Surfaces by Photo-degradation of Films Formed from Oligo(Ethylene Glycol) Terminated Silanes. Biointerphases 2012, 7, 54. 10.1007/s13758-012-0054-6. [DOI] [PubMed] [Google Scholar]
- Alonso J. M.; Reichel A.; Piehler J.; del Campo A. Photopatterned Surfaces for Site-Specific and Functional Immobilization of Proteins. Langmuir 2008, 24, 448–457. 10.1021/la702696b. [DOI] [PubMed] [Google Scholar]
- del Campo A.; Arzt E. Fabrication Approaches for Generating Complex Micro- and Nanopatterns on Polymeric Surfaces. Chem. Rev. 2008, 108, 911–945. 10.1021/cr050018y. [DOI] [PubMed] [Google Scholar]
- Petersen S.; Alonso J. M.; Specht A.; Duodu P.; Goeldner M.; del Campo A. Phototriggering of cell adhesion by caged cyclic RGD peptides. Angew. Chem., Int. Ed. 2008, 47, 3192–3195. 10.1002/anie.200704857. [DOI] [PubMed] [Google Scholar]
- Stegmaier P.; Alonso J. M.; Campo A. d. Photoresponsive Surfaces with Two Independent Wavelength-Selective Functional Levels. Langmuir 2008, 24, 11872–11879. 10.1021/la802052u. [DOI] [PubMed] [Google Scholar]
- Erath J.; Cui J.; Schmid J.; Kappl M.; Campo A. d.; Fery A. Phototunable Surface Interactions. Langmuir 2013, 29, 12138–12144. 10.1021/la4021349. [DOI] [PubMed] [Google Scholar]
- Hoshi Y.; Xu Y.; Ober C. K. Photo-cleavable anti-fouling polymer brushes: A simple and versatile platform for multicomponent protein patterning. Polymer 2013, 54 (7), 1762–1767. 10.1016/j.polymer.2013.02.027. [DOI] [Google Scholar]
- Reuther C.; Tucker R.; Ionov L.; Diez S. Programmable Patterning of Protein Bioactivity by Visible Light. Nano Lett. 2014, 14, 4050–4057. 10.1021/nl501521q. [DOI] [PubMed] [Google Scholar]
- El Muslemany K. M.; Twite A. A.; ElSohly A. M.; Obermeyer A. C.; Mathies R. A.; Francis M. B. Photoactivated Bioconjugation Between ortho-Azidophenols and Anilines: A Facile Approach to Biomolecular Photopatterning. J. Am. Chem. Soc. 2014, 136, 12600–12606. 10.1021/ja503056x. [DOI] [PubMed] [Google Scholar]
- Weinrich D.; Köhn M.; Jonkheijm P.; Westerlind U.; Dehmelt L.; Engelkamp H.; Christianen P. C. M.; Kuhlmann J.; Maan J. C.; Nüsse D.; Schröder H.; Wacker R.; Voges E.; Breinbauer R.; Kunz H.; Niemeyer C. M.; Waldmann H. Preparation of Biomolecule Microstructures and Microarrays by Thiol–ene Photoimmobilization. ChemBioChem 2010, 11, 235–247. 10.1002/cbic.200900559. [DOI] [PubMed] [Google Scholar]
- Klán P.; Šolomek T.; Bochet C. G.; Blanc A.; Givens R.; Rubina M.; Popik V.; Kostikov A.; Wirz J. Photoremovable Protecting Groups in Chemistry and Biology: Reaction Mechanisms and Efficacy. Chem. Rev. 2013, 113, 119–191. 10.1021/cr300177k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor S. P.; Read J. L.; Pirrung M. C.; Stryer L.; Lu A. T.; Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science 1991, 251, 767–773. 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Sundberg S. A.; Barrett R. W.; Pirrung M.; Lu A. L.; Kiangsoontra B.; Holmes C. P. Spatially-Addressable Immobilization of Macromolecules on Solid Supports. J. Am. Chem. Soc. 1995, 117, 12050–12057. 10.1021/ja00154a003. [DOI] [Google Scholar]
- Pirrung M. C.; Wang L.; Montague-Smith M. P. 3′-Nitrophenylpropyloxycarbonyl (NPPOC) Protecting Groups for High-Fidelity Automated 5′ - 3′ Photochemical DNA Synthesis. Org. Lett. 2001, 3, 1105–1108. 10.1021/ol0069150. [DOI] [PubMed] [Google Scholar]
- Pirrung M. C.; Dore T. M.; Zhu Y.; Rana V. S. Sensitized two-photon photochemical deprotection. Chem. Commun. 2010, 46, 5313–5315. 10.1039/c0cc00782j. [DOI] [PubMed] [Google Scholar]
- Alang-Ahmad S. A.; Wong L. S.; ul-Haq E.; Hobbs J. K.; Leggett G. J.; Micklefield J. Micrometer- and Nanometer-Scale Photopatterning Using 2-Nitrophenylpropyloxycarbonyl-Protected Aminosiloxane Monolayers. J. Am. Chem. Soc. 2009, 131, 1513–1522. 10.1021/ja807612y. [DOI] [PubMed] [Google Scholar]
- Alang Ahmad S. A.; Wong L. S.; ul-Haq E.; Hobbs J. K.; Leggett G. J.; Micklefield J. Protein Micro- and Nanopatterning Using Aminosilanes with Protein-Resistant Photolabile Protecting Groups. J. Am. Chem. Soc. 2011, 133, 2749–2759. 10.1021/ja1103662. [DOI] [PubMed] [Google Scholar]
- Wöll D.; Smirnova J.; Galetskaya M.; Prykota T.; Bühler J.; Stengele K.-P.; Pfleiderer W.; Steiner U. E. Intramolecular Sensitization of Photocleavage of the Photolabile 2-(2-Nitrophenyl)propoxycarbonyl (NPPOC) Protecting Group: Photoproducts and Photokinetics of the Release of Nucleosides. Chem. - Eur. J. 2008, 14, 6490–6497. 10.1002/chem.200800613. [DOI] [PubMed] [Google Scholar]
- Kretschy N.; Holik A.-K.; Somoza V.; Stengele K.-P.; Somoza M. M. Next-Generation o-Nitrobenzyl Photolabile Groups for Light-Directed Chemistry and Microarray Synthesis. Angew. Chem., Int. Ed. 2015, 54, 8555–8559. 10.1002/anie.201502125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A.; Stengele K.-P.; Giegrich H.; Cornwell P.; Isham K. R.; Sachleben R. A.; Pfleiderer W.; Foote R. S. Photolabile protecting groups for nucleosides: Synthesis and photodeprotection rates. Tetrahedron 1997, 53, 4247–4264. 10.1016/S0040-4020(97)00154-3. [DOI] [Google Scholar]
- Sun S.; Leggett G. J. Matching the Resolution of Electron Beam Lithography by Scanning Near-Field Photolithography. Nano Lett. 2004, 4, 1381–1384. 10.1021/nl049540a. [DOI] [Google Scholar]
- Moxey M.; Johnson A.; El-Zubir O.; Cartron M.; Dinachali S. S.; Hunter C. N.; Saifullah M. S. M.; Chong K. S. L.; Leggett G. J. Fabrication of Self-Cleaning, Reusable Titania Templates for Nanometer and Micrometer Scale Protein Patterning. ACS Nano 2015, 9, 6262–6270. 10.1021/acsnano.5b01636. [DOI] [PubMed] [Google Scholar]
- Kremers G. J.; et al. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster Radius. Biochemistry 2006, 45, 6570–6580. 10.1021/bi0516273. [DOI] [PubMed] [Google Scholar]
- Betzig E.; Finn P. L.; Weiner J. S. Combined Shear Force and near-Field Scanning Optical Microscopy. Appl. Phys. Lett. 1992, 60, 2484–2486. 10.1063/1.106940. [DOI] [Google Scholar]
- Haq E. u.; Liu Z.; Zhang Y.; Ahmad S. A. A.; Wong L.-S.; Armes S. P.; Hobbs J. K.; Leggett G. J.; Micklefield J.; Roberts C. J.; Weaver J. M. R. Parallel Scanning Near-Field Photolithography: The Snomipede. Nano Lett. 2010, 10, 4375–4380. 10.1021/nl1018782. [DOI] [PubMed] [Google Scholar]
- Overney R.; Meyer E. Tribological investigations using friction force microscopy. MRS Bull. 1993, 18, 26–34. 10.1557/S0883769400047096. [DOI] [Google Scholar]
- Carpick R. W.; Salmeron M. Scratching the Surface: Fundamental Investigations of Tribology with Atomic Force Microscopy. Chem. Rev. 1997, 97, 1163–1194. 10.1021/cr960068q. [DOI] [PubMed] [Google Scholar]
- Mate C. M.Tribology on the Small Scale; Oxford University Press: Oxford, U.K., 2008. [Google Scholar]
- Brueck S. R. J. Optical and Interferometric Lithography - Nanotechnology Enablers. Proc. IEEE 2005, 93, 1704–1721. 10.1109/JPROC.2005.853538. [DOI] [Google Scholar]
- Lu C.; Lipson R. H. Interference lithography: a powerful tool for fabricating periodic structures. Laser Photon. Rev. 2010, 4, 568–580. 10.1002/lpor.200810061. [DOI] [Google Scholar]