Abstract
Inverted repeats are important genetic elements for genome instability. In the current study we have investigated the role of inverted repeats in a DNA rearrangement reaction using a linear DNA substrate. We show that linear DNA substrates with terminal inverted repeats can efficiently transform Escherichia coli. The transformation products contain circular inverted dimers in which the DNA sequences between terminal inverted repeats are duplicated. In contrast to the recombination/rearrangement product of circular DNA substrates, which is exclusively one particular form of the inverted dimer, the rearrangement products of the linear DNA substrate consist of two isomeric forms of the inverted dimer. Escherichia coli mutants defective in RecBCD exhibit much reduced transformation efficiency, suggesting a role for RecBCD in the protection rather than destruction of these linear DNA substrates. These results suggest a model in which inverted repeats near the ends of a double-strand break can be processed by a helicase/exonuclease to form hairpin caps. Processing of hairpin capped DNA intermediates can then yield inverted duplications. Linear DNA substrates containing terminal inverted repeats can also be converted into inverted dimers in COS cells, suggesting conservation of this type of genome instability from bacteria to mammalian cells.
INTRODUCTION
Genome instability is part of the genetic program for cellular evolution and diversity. Repetitive DNA sequences are generally known to be the source of genome instability in both prokaryotes and eukaryotes. In contrast to the complex recombination pathways associated with direct repeats (1–5), DNA rearrangement involving inverted repeats has been less explored. Inverted repeats are often acknowledged to mediate genome instability through excision of the repeat-associated regions (6–9) and have recently been implicated in gene amplification processes (10–13). Gene amplification occurs during the development of many organisms (14), emergence of drug resistance (15) and progression of cancer (15–17). Intriguingly, amplified double-minute chromosomes (DMs) are often found as circular inverted dimers with two short unique sequences separating an otherwise perfect giant palindrome (18,19). Amplified genes identified to be in the form of circular inverted dimers include the DFR1 gene in Saccharomyces cerevisiae (20), the H-circles in Leishmania (10,21–24), the mdm2, myc and polyoma virus (Py) oncogenes in tumor cells (25–27) and the ADA, DHFR, AMPD, APRT and CAD genes in drug-resistant cell lines (26–30). Studies of the amplified H-circles from drug-resistant Leishmania have demonstrated that the unamplified H locus is bracketed by two pairs of inverted repeats (198 and 1241 bp, respectively), one at each end of the H locus (23,31). The possibility that these two pairs of inverted repeats may be crucial for forming the giant palindromic H-circles has been suggested (10,31). The most direct demonstration that inverted repeats can mediate formation of inverted dimers has been obtained in Escherichia coli plasmid systems (12,13,32) and in Tetrahymena (33). In this case, circular plasmid DNAs containing a pair of small inverted repeats are shown to lead to the formation of circular inverted dimers (12,13,32). However, the molecular mechanism(s) by which inverted repeats mediate genome instability is still unclear.
The abundance of inverted repeat sequences in mammalian cells and the stimulation of recombination by DNA double-strand breaks have led us to examine the role of inverted repeats in DNA rearrangement using a linear DNA substrate. We show in the current studies that linear DNA constructs containing terminal inverted repeats can increase the efficiency of transformation or transfection and produce circular inverted dimers in both E.coli and mammalian cells. Studies in E.coli have suggested a model in which the linear DNA substrate is first processed into dumbbell-like DNA with hairpins capping both ends. Subsequent replication of the dumbbell-like DNA leads to the formation of inverted dimers. These results suggest that inverted repeats may function as genetic elements for preventing degradation of damaged DNA and promoting DNA rearrangement.
MATERIALS AND METHODS
Enzymes, reagents and cells
Klenow polymerase and T4 DNA ligase were purchased from Gibco-BRL and New England Biolabs, respectively. Restriction enzymes were obtained from several commercial sources. The COS-7 cell line was obtained from the ATCC. Escherichia coli strains AB1157 (F– thr1 leuB6 thi1 lacY1 galK2 ara14 xyl5 mtl1 proA2 his4 argE3 rpsL31 tsx33 supE44 kdgK51) (Dr M. G. Marinus, University of Massachusetts Medical Center, MA), JC5519 (AB1157 recB21 recC22) (Dr A. J. Clark, University of California, Berkeley, CA), N3475 (AB1157 ruvC51 recG258) and N3476 (rvuC53 recG258) (Dr R. G. Lloyd, University of Nottingham, Nottingham, UK) were obtained from various laboratories.
Construction of pID-IP**
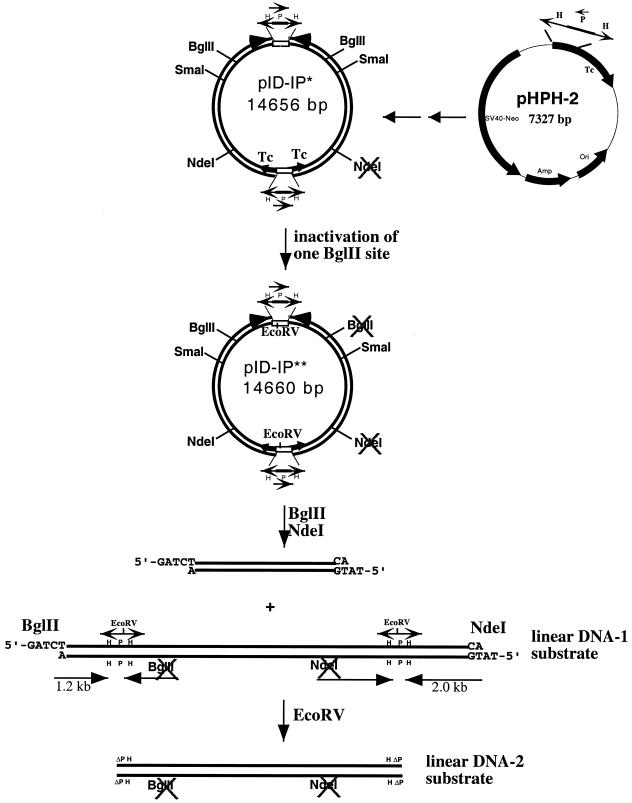
As shown in Figure 1, the fragment containing the SV40 origin and the neomycin resistance gene in pID-IP* (13) was obtained from BamHI digestion of pMAMneo (Clontech, CA). After blunting the ends with Klenow polymerase in the presence of dNTPs, the blunted fragment was cloned into the SspI site of pHPH (12) to generate pHPH-2 (13). Plasmid pHPH, which contains the HPH/tet (inverted repeats) cassette, was derived from pBR322. As reported previously, the HPH/tet cassette, which consists of a flipped Ptet promoter fragment including part of the coding region of the tet gene (the P fragment) flanked by inverted repeats (the two H fragments) can mediate efficient recA-independent recombination/rearrangement resulting in exclusive formation of a special inverted dimer (12). The HPH/tet cassette is basically a genetic switch controlling transcription of the functional tetracycline gene, depending on the orientation of the P fragment. The inverted dimer, pID-IP, was generated by transforming pHPH-2 (13) into E.coli DH5α (RecA–) followed by selection with tetracycline. This inverted dimer contains a functional tetracycline gene and therefore cells containing the inverted dimer can be readily selected for by resistance to tetracycline.
Figure 1.
Construction of the linear DNA substrate containing two pairs of inverted repeats. pID-IP*, derived from pID-IP by inactivation one of its NdeI sites, was used to generate pID-IP**, by inactivation of a BglII site located on the same side of the inverted dimer as the inactivated NdeI site (see Materials and Methods for details). The linear DNA-1 substrate containing two pairs of inverted repeats is the larger of the two fragments generated by BglII and NdeI digestion of pID-IP**. The length of the repeated H fragment in the HPH cassette is 350 bp and the length of the P fragment is 651 bp. The linear DNA-2 substrate without HPH/tet cassettes (only ΔPH sequences) was generated by digesting linear DNA-1 with EcoRV.
pID-IP** was constructed from pID-IP* by destroying one of the two identical BglII sites in the inverted dimer. This was accomplished by partial digestion of pID-IP* with BglII, followed by gel purification of the full-length linear DNA. The linear DNA was then blunted by treatment with Klenow polymerase and dNTPs. Following cyclization with T4 DNA ligase, the DNA was used to transform E.coli DH5α. The resulting plasmid, pID-IP** (** indicates inactivation of two restriction sites), therefore contains a single NdeI site and a single BglII site.
Transformation of E.coli
The transformation of E.coli cells was performed by the standard calcium chloride procedure and a brief heat shock at 42°C for 30 s following mixing of DNA with competent cells. Transformation frequency was obtained by counting the colony number after plating the transformation mixture on LB (10 g/l Bacto-tryptone, 5 g/l Bacto-yeast extract and 10 g/l NaCl) plates supplemented with 100 µg/ml ampicillin. Plasmid DNAs isolated from transformants were analyzed by restriction enzyme digestion.
Denaturation of the linear DNA substrate
Denaturation of the linear DNA substrate was achieved by alkali denaturation (0.1 N NaOH), followed by rapid neutralization with an equal volume of 0.1 N HCl plus a 1/10 vol of 1 M Tris, pH 7.8. The neutralized DNA solution was briefly incubated at 37°C for 10 min and then kept on ice. As shown in a previous study (13), EcoRV can only digest double-stranded and not single-stranded DNA. EcoRV digestion was used to reduce residual supercoiled pID-IP** and linear double-stranded DNA substrates.
DNA transfection into COS cells
The linear DNA substrate used for transfection was generated by linearization of 50 µg of pID-IP** with BglII and NdeI, followed by phenol/CHCl3 extraction and ethanol precipitation. The larger linear DNA fragment (10.7 kb) was purified from agarose gels after electrophoresis in TAE buffer (Tris–acetate/EDTA). Aliquots of 5 µg of the gel-purified linear DNA substrate were electroporated (500 µF, 400 V) into COS-7 cells using a Gene Pulser apparatus from Bio-Rad (Hercules, CA). The transfected cells were resuspended in DMEM plus 10% FBS after placing on ice for 10 min.
DNA isolation from transfected COS cells
Twenty-four hours post-transfection the transfected COS-7 cells (in 100 mm Petri dishes) were washed once in DMEM and then lysed in 1 ml of a solution containing 1% SDS, 50 mM EDTA, 10 mM HEPES, pH 7.5, and 1 mg/ml proteinase K. The viscous lysate was passed through a 1 ml syringe plugged with glass wool to remove the cellular chromosome and then incubated at 65°C for 2 h. The episomal DNA in the filtrate was isolated by phenol/CHCl3 extraction and ethanol precipitation. The precipitated DNA was resuspended in TE (10 mM Tris, pH 8.0, and 1 mM EDTA).
Southern blotting analysis
DNA isolated from transfected COS-7 cells was digested with the indicated restriction enzymes and then analyzed by gel electrophoresis in TPE buffer (Tris–phosphate/EDTA). Alkaline DNA capillary transfer and Southern blotting were performed as described in the instruction manual for Zeta-Probe blotting membranes from Bio-Rad. Acid depurination was performed by soaking the gel in 0.25 N HCl for 20 min. DNA capillary transfer to Zeta-Probe membrane was performed with 0.4 N NaOH for 8–12 h. The probe was α-32P-labeled using a Random Primed Labeling Kit purchased from Boehringer Mannheim.
RESULTS
Construction of linear DNA substrates containing terminal inverted repeats
Our strategy to prepare the linear DNA substrate containing terminal inverted repeats is shown in Figure 1 (see Materials and Methods for details). We took advantage of the special head-to-head dimer (pID-IP) (inverted dimer with inverted P fragments) generated due to recombinational rearrangement of an HPH/tet cassette-containing plasmid (pHPH-2) (13). pID-IP* is derived from pID-IP by inactivation of one of its two identical NdeI sites (Fig. 1), as described in a previous study (13). In order to prepare the desired linear DNA substrates, we further eliminated one of the two identical BglII sites in pID-IP* to generate pID-IP** (** indicates inactivation of two restriction sites). The resulting plasmid, pID-IP**, was then digested with BglII and NdeI to generate two fragments. The larger DNA fragment is the linear DNA-1 substrate, which contains two pairs of HPH/tet cassettes, one located near each end of the linear DNA (Fig. 1). The linear DNA-2 substrate without HPH/tet cassettes (only ΔPH sequences) was generated by digesting linear DNA-1 with EcoRV. The linear DNA substrates (DNA-1 and DNA-2) were isolated from 0.8% agarose gels following electrophoresis and used subsequently to transform E.coli by the standard calcium chloride procedure or to transfect COS-7 cells by electroporation.
Transformation of the linear DNA substrates into E.coli
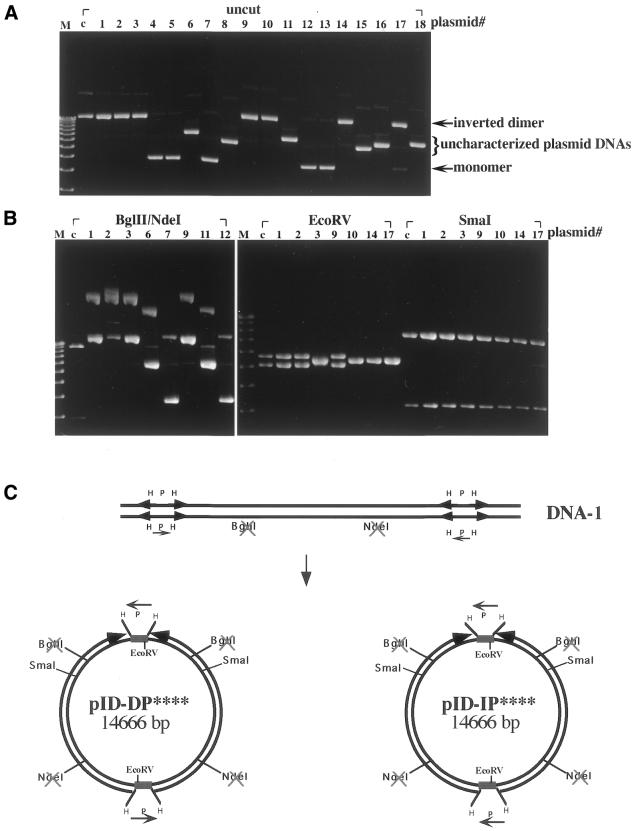
The linear DNA substrates with or without terminal inverted repeats were used to transform E.coli DH5α. The transformation frequencies (as measured by the number of Apr colonies) for the two linear DNA substrates, linear DNA-1 and linear DNA-2, were ∼0.5 and ∼0.001% of that of the supercoiled pID-IP** DNA, respectively (Table 1), suggesting that inverted repeats at each end of the linear DNA are responsible for the higher transformation frequency. When the plasmid DNAs isolated from transformants with linear DNA-1 were analyzed by restriction enzyme digestion, ∼42% was inverted dimers (Table 1 and discussion below). The rest of the plasmid DNA was monomeric DNA (29%) [see Fig. 4B(d), and Table 1] and uncharacterized plasmid DNAs with sizes in between those of the monomeric DNA and inverted dimers (29%) (Fig. 2A and Table 1). Selected inverted dimers (plasmid #s are marked at the top of each lane in Fig. 2) were further characterized by digestion with BglII and NdeI (Fig. 2B, left), EcoRV (Fig. 2B, middle) or SmaI (Fig. 2B, right). Surprisingly, two distinct populations of circular inverted dimers were identified, pID-DP**** (14%) and pID-IP**** (28%) (Table 1 and Fig. 2C; **** indicates inactivation of four restriction sites, two NdeI and two BglII sites, in the inverted dimer). These two circular inverted dimers differ in the relative orientation of the P fragments located within each HPH/tet cassette. In contrast to pID-IP****, in which the two P fragments are in the inverted orientation, pID-DP**** (inverted dimer with directly repeated P fragments) contains two P fragments in parallel orientation as direct repeats.
Table 1. Transformation of E.coli with the linear DNA substrate: DNA product distribution and effects of the recombination mutants.
| DNA sourcea | E.coli host | Relative transformation frequencyb,c (%) | No. of colonies analyzed | DNA products | ||||
| Inverted dimersd | Monomers | Otherse | ||||||
| |
|
|
|
pID-DP**** |
pID-IP**** |
pID-IP** |
|
|
| Supercoiled pID-IP** | DH5α (recA1) | 100b | 36 | 0 | 0 | 36 | 0 | 0 |
| Linear DNA-1 | DH5α | 0.5 ± 0.1b | 34 | 5 | 13 | 0 | 8 | 8 |
| Linear DNA-2 | DH5α | <0.001b | ND | ND | ND | ND | ND | ND |
| Denatured linear DNA-1 | DH5α | 3.1 ± 0.5b | 18 | 0 | 18 | 0 | 0 | 0 |
| Linear DNA-1 | AB1157 | 100c | 18 | 2 | 5 | 0 | 7 | 4 |
| Linear DNA-1 | N3475 (ruvC51 recG258) | 92 ± 6c | 18 | 2 | 5 | 0 | 8 | 3 |
| Linear DNA-1 | N3476 (ruvC53 recG258) | 93 ± 6c | ND | ND | ND | ND | ND | ND |
| Linear DNA-1 | JC5519 (recB21 recC22) | 15 ± 4c | 36 | 9 | 3 | 0 | 16 | 8 |
ND, not determined.
aLinear DNA-1 with two pairs of inverted repeats and linear DNA-2 without inverted repeats were isolated from 0.8% agarose gels following electophoresis.
bThe relative transformation frequency was defined as the number of Apr colonies from cells transformed with the linear DNA substrate over that with an equal amount of pID-IP** DNA. All determinations were averages of at least four separate experiments. The relative transformation frequency of the supercoiled pID-IP** DNA was taken as 100%.
cThe relative transformation frequency of the linear DNA substrate after normalization to supercoiled pID-IP** DNA in AB1157 was taken as 100%.
dThe two types of circular inverted dimers pID-DP**** and pID-IP**** were distinquished by digestion with EcoRV. **** indicates inactivation of four restriction sites, two NdeI and two BglII sites, on the inverted dimers determined by digestion with NdeI and BglII as shown in Figure 2B.
eThe sizes of these plasmids are heterogeneous and fall between those of monomers and circular inverted dimers.
Figure 4.
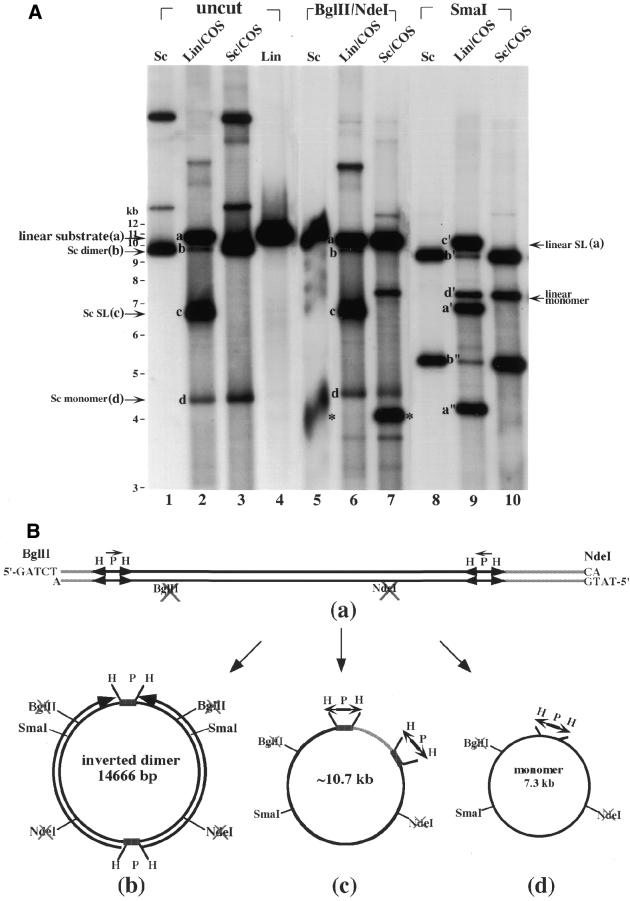
(A) Southern blotting analysis of episomal DNAs isolated from transfected COS cells. Episomal DNAs were isolated from transfected COS cells with either supercoiled (Sc) pID-IP** DNA (labeled Sc/COS at the top of the lanes) or the linear DNA substrate containing two pairs of inverted repeats (labeled Lin/COS at the top of the lanes) (see Materials and Methods). Untreated DNAs (labeled uncut at the top of the lanes) or DNAs digested with either BglII and NdeI or SmaI (see the respective labels at the top of the lanes) were analyzed by gel electrophoresis. Lanes 1, 5 and 8, supercoiled pID-IP** DNA controls (labeled Sc at the top of the lanes); lanes 2, 6 and 9, episomal DNAs isolated from COS cells transfected with the gel-purified linear DNA substrate (labeled Lin/COS at the top of the lanes); lanes 3, 7 and 10, episomal DNAs isolated from COS cells transfected with supercoiled pID-IP** (labeled Sc/COS at the top of the lanes); lane 4, the gel-purified linear DNA substrate derived from pID-IP** DNA restricted with BglII and NdeI (labeled Lin at the top of the lane). The bands marked * and a were generated from pID-IP** DNA digested with BglII and NdeI. The bands marked a–d correspond to the linear DNA substrate, supercoiled (Sc) inverted dimers (e.g. pID-IP**), supercoiled (Sc) self-ligated (SL) DNA substrate and supercoiled (Sc) monomer, respectively. (B) A schematic illustrating the formation of various DNA products in COS cells transfected with the linear DNA substrate. The linear DNA substrate with the two pairs of inverted repeats was transfected into COS cells. The proposed structures of various DNA products corresponding to bands a–d are shown schematically.
Figure 2.
Analysis of plasmid DNAs isolated from E.coli DH5α transformed with the linear DNA substrate. The linear DNA substrate containing terminal inverted repeats was used to transform E.coli DH5α. (A) Plasmid DNAs isolated from 18 different clones (see lanes labeled plasmid# 1–18 at the top of the lanes) were analyzed by electrophoresis in 0.8% agarose gel. (B) Plasmid DNAs isolated from various clones (plasmid# indicated at the top of the lanes) were digested with BglII and NdeI, EcoRV or SmaI. Lanes M, the 1 kb DNA ladder used as the marker; lanes c, pID-IP** DNA used as the control DNA. (C) A schematic illustrating the two isomeric forms of circular inverted dimers generated from linear DNA substrates.
Two isomeric forms of circular inverted dimers derived from linear DNA substrates via the dumbbell-like DNA intermediates
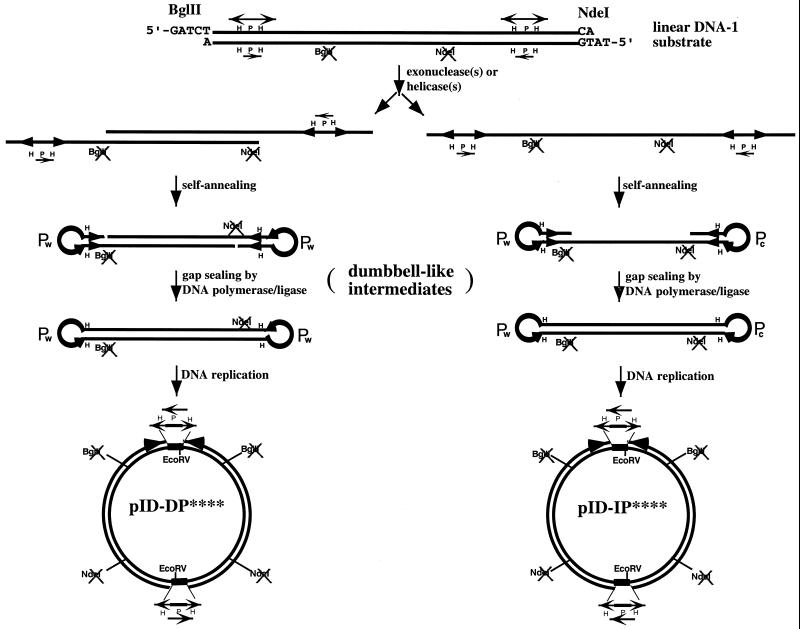
Previous studies in E.coli have demonstrated that dumbbell-like DNA can efficiently transform E.coli, producing circular inverted dimers as the exclusive transformation products (13). We have thus considered a possible mechanism by which circular inverted dimers can be generated from linear DNA substrates containing terminal inverted repeats via dumbbell-like DNA intermediates (Fig. 3). In this model the linear DNA substrate containing terminal inverted repeats is first processed by exonuclease (Fig. 3, left) or helicase/nuclease (Fig. 3, right) to expose the inverted repeats, including the HPH cassettes, in single-stranded form. The exposed single-stranded HPH cassettes can rapidly self-anneal to form terminal hairpin loops. Depending on the strand in which the hairpin loops are formed, two isomeric forms of dumbbell-like intermediates are produced. Subsequent replication of the dumbbell-like intermediates produces two isomeric forms of inverted dimers, pID-DP**** and pID-IP**** (Fig. 3). In this model no Holliday junction is formed during the entire process (note that extrusion of the inverted repeats into cruciforms followed by resolution of the Holliday junction could theoretically generate the proposed dumbbell-like intermediates).
Figure 3.
Two isomeric forms of circular inverted dimers derived from the linear DNA substrate. The linear DNA substrate containing terminal inverted repeats is converted to the dumbbell-like DNA intermediate via processing by exonuclease or helicase/nuclease. Two types of circular inverted dimers can be generated depending on specificity of the exonuclease/helicase. As shown to the left, a double-stranded 5′→3′ or 3′→5′ exonuclease can expose the two HPH/tet cassettes as protruding single-stranded DNA on opposite DNA strands. Formation of terminal hairpin loops at the HPH sites converts the DNA into a dumbbell-like intermediate with identical terminal loops (P elements at the terminal loops have the same sequence). Subsequent DNA replication converts the dumbbell-like DNA into the circular inverted dimer, pID-DP****. To the right, the same or a different exonuclease/helicase (such as RecBCD) can convert the linear DNA substrate into single-stranded DNA. This can be achieved by either a single exonuclease or helicase working processively from one end. Annealing of the inverted repeats on the single-stranded DNA results in formation of dumbbell-like DNA with complementary terminal loops. Subsequent DNA replication converts the dumbbell-like DNA into the circular inverted dimer, pID-IP****.
Formation of inverted dimers is unaffected in E.coli recG ruvC mutants but is reduced in an E.coli recBC mutant
To test whether formation of circular inverted dimers from linear DNA substrates containing terminal inverted repeats involves recombination intermediates containing Holliday junctions, E.coli mutants defective in RuvABC/RusA Holliday junction resolving systems [E.coli N3475 (ruvC51 recG258) and N3476 (ruvC53 recG258)] were used (34). The relative transformation frequencies of the linear DNA-1 substrate in these mutants and wild-type cells (E.coli AB1157) (35) were nearly the same (Table 1) after normalization with supercoiled pID-IP**. In addition, the distribution of the two isomeric forms of circular inverted dimers appeared to be similar (Table 1). These results suggest that Holliday junctions are probably not involved in formation of the inverted dimers. The known function of RecBCD in processing linear DNA had also led us to test a possible role of RecBCD in processing of the linear DNA substrate. The transformation frequency of the linear DNA-1 substrate in E.coli JC5519 (recB21 recC22) (36) was only 15% of that in E.coli AB1157 (Table 1) after normalization to supercoiled pID-IP**. In addition, the distribution of the two isomeric forms of circular inverted dimers was also altered, 25% pID-DP**** and 8% pID-IP**** (Table 1). These results suggest that RecBCD (nuclease/helicase) is the major enzyme responsible for transformability of the linear DNA-1 substrate.
Analysis of DNA products isolated from COS cells transfected with the linear DNA-1 substrate
The structure of the inverted dimer obtained in E.coli strikingly resembles that of the extrachromosomal gene amplification products in eukaryotic cells (25–30). To determine whether the transfected linear DNA-1 substrate can also be converted into an inverted dimer in COS cells, DNA isolated from transfected COS cells was analyzed by Southern blotting using 32P-labeled pID-IP** DNA as the probe. As shown in Figure 4A, four major DNA products were detected from transfected cells (see bands a–d in Fig. 4A, lane 2). Band a was identified to be the residual linear DNA-1 substrate [see Fig. 4B(a), for the structure] based on restriction enzyme analysis. Band a was converted into bands a′ and a″ by digestion with SmaI (see lane 9 in Fig. 4A). Band b co-migrated with supercoiled pID-IP** (Fig. 4A, compare lanes 1 and 2). However, in contrast to supercoiled pID-IP** which was digested by BglII and NdeI into two bands (Fig. 4A, compare lanes 1 and 5 with 3 and 7), band b (and the other bands) was resistant to BglII and NdeI digestion (Fig. 4A, compare lanes 2 and 6). Furthermore, band b was converted into bands b′ and b″ by digestion with SmaI (see Fig. 4A, lane 9). Based on these results and results from additional restriction enzyme digestions (data not shown), band b was identified as an inverted dimer with the structure shown in Figure 4B(b). Band c was identified as the self-ligated (cyclization) product of the linear substrate [see Fig. 4B(c) for the structure]. Band c was converted into band c′ (unit length linear) by digestion with SmaI. Band d was identified as the monomeric DNA circle [see Fig. 4B(d) for the structure] generated due to a possible recombinational event between two directly repeated H segments each located at one of the two HPH/tet cassettes on the linear DNA substrate. Band d was converted into band d′ (unit length linear) by digestion with SmaI.
When supercoiled pID-IP** DNA (Fig. 4A, lane 1; supercoiled pID-IP** DNA had the same mobility as band b, the inverted dimer) was used as a control to transfect COS cells, the major DNA product was pID-IP** itself (Fig. 4A, compare lanes 1, 3, 5, 7 and 10). A small amount (<10%) of supercoiled monomeric DNA (band d in Fig. 4A, lane 3) was also produced. The supercoiled monomeric DNA (band d in lanes 2 and 3) obtained from transfection of COS cells with either linear DNA-1 or supercoiled pID-IP** could be generated through the same pathway (described in the previous paragraph), since the DNA could not be re-cut by BglII and NdeI (see band d in Fig. 4A, lanes 6 and 7) and was converted to linear form by SmaI (see band d′ in Fig. 4A, lanes 9 and 10).
Although the major DNA product following transfection of the linear DNA-1 substrate was the self-ligated DNA (band c), the inverted dimer (band b) was clearly detectable and represented several percent of the total DNA products. Resistance of the inverted dimer (band b) to digestion by BglII and NdeI supported the conclusion that it was generated in COS cells and was not derived from supercoiled pID-IP** DNA which might have contaminated the linear DNA-1 substrate during gel isolation.
In view of the two isomeric inverted dimers (pID-DP**** and pID-IP****) in E.coli transformed with the linear DNA-1 substrate, we also analyzed circular inverted dimers (band b in Fig. 4A) formed in COS cells transfected with the linear DNA-1 substrate, by transforming gel-isolated band b into E.coli DH5α. Again, both forms of the inverted dimers were observed, pID-IP**** (30%) and pID-DP**** (40%) (Table 2).
Table 2. Transformation of E.coli with episomal or gel-isolated DNAs from transfected COS-7 cells: DNA product distribution and DNA topology effects of the transfecting DNA.
| DNA used for transfection | E.coli host | No. of Tcr colonies analyzed | DNA products | ||||
| Inverted dimersa | Monomers | Othersb | |||||
| |
|
|
pID-DP**** |
pID-IP**** |
pID-IP** |
|
|
| Supercoiled pID-IP**c | DH5α | 36 | 0 | 0 | 35d | 1 | 0 |
| Linear DNA-1e | DH5α | 10 | 3 | 6 | 0 | 1 | 0 |
| Denatured linear DNA-1f | DH5α | 16 | 0 | 15 | 0 | 1 | 0 |
aThe two types of circular inverted dimers pID-DP**** and pID-IP**** were distinquished by digestion with EcoRV. **** indicates inactivation of four restriction sites, two NdeI and two BglII sites, on the inverted dimers determined by digestion with NdeI and BglII as shown in Figure 2B.
bThe sizes of these plasmids are heterogeneous and fall between those of monomers and circular inverted dimers as shown in Figure 2A and Table 1.
cSupercoiled pID-IP** DNA was used to transfect COS-7 cells. Episomal DNAs from transfected COS-7 cells were used to transform E.coli DH5α and Tcr colonies were analyzed.
dThe circular inverted dimer was identified as pID-IP** based on sensitivity to NdeI and BglII.
eAs in footnote c except that the linear DNA-1 substrate containing the terminal inverted repeats was used to transfect COS-7 cells and transformation was with gel-isolated band b in Figure 4, lane 2.
fAs in footnote c except that the linear DNA substrate was alkali-denatured prior to transfection (see Materials and Methods).
In order to test that the formation of two types of circular inverted dimers (pID-DP**** and pID-IP****) is due to the presence of two types of intermediates (dumbbell-like DNAs) as proposed in Figure 3, one type of dumbbell-like DNA (Fig. 3, right) generated by alkaline denaturation and rapid renaturation (see Materials and Methods) was used to transfect COS cells. As predicted in Figure 3, only one type of inverted dimer (pID-IP****) was isolated (Table 2) after transforming E.coli DH5α with the episomal DNA. The structure of the dumbbell-like DNA was confirmed by restriction enzyme analysis as in previous studies (13; data not shown).
DISCUSSION
Our studies have clearly demonstrated that linear DNAs containing terminal inverted repeats can transform E.coli efficiently. Interestingly, the transformation products contain the DNA as an inverted dimer. Previous studies have demonstrated that circular inverted dimers can be generated by head-to-head joining of linearized plasmid DNAs in both yeast (37) and human cells (38). The inverted dimers (14.7 kb) recovered from transfected COS cells or transformed E.coli DH5α in this study could not have been generated by head-to-head joining of the linear DNA substrates. Head-to-head joining would have produced a perfect palindromic circle of larger size (∼21.4 kb). We did not observe this kind of larger DNA following either transfection of COS cells or transformation of E.coli DH5α with the linear DNA-1 substrate.
Inverted dimers could theoretically be produced by intermolecular end-to-end joining through annealing of the exposed HPH cassettes. When two exposed single-stranded HPH cassettes initially anneal, only the H sequences on the two strands can actually form base pairs; the two P sequences on opposite strands will not be able to form base pairs since P itself is not an inverted repeat. Thus in the initial formation of an inverted dimer plasmid, there would have to be two HPH annealing events to form a circle and two bubbles the size of P sequences 180° from each other on the plasmid. First, if such a structure is replicated directly, then direct and inverted dimers with the direct P orientation by double strand annealing or direct and inverted dimers with the inverted P orientation through single strand annealing (SSA) would be generated. Secondly, the mismatch or bubble might be a target for the mismatch repair system. When a bubble is being repaired, which strand is excised and which strand is used as the template for repair synthesis will determine the ultimate orientation of P and thus generate the mix of direct and inverted dimers with either direct P or inverted P sequences. Clearly, only inverted dimers (pID-DP**** and pID-IP****) have been detected in both linear DNA-1 transformed E.coli and linear DNA-1 transfected COS cells and only one type of inverted dimer (pID-IP****) has been observed with denatured linear DNA-1 (Table 2 and Fig. 3, right). Therefore, the possibility of intermolecular end-to-end joining seems very unlikely.
The presence of two isomeric forms of circular inverted dimers could in principle be generated by homologous recombination between the two homologous halves of the inverted dimers subsequent to their formation. However, when supercoiled pID-IP** or dumbbell-like DNA was used to transfect COS cells, only one type of inverted dimer (pID-IP** or pID-IP****) was isolated (Table 2). These results therefore support the notion that the formation of two types of circular inverted dimers is due to the presence of two types of intermediates (dumbbell-like DNAs), as proposed in Figure 3, rather than subsequent homologous recombination between the duplicated regions within the circular inverted dimer.
Consistent with the dumbbell model (Fig. 3) proposed for the formation of two isomeric inverted dimers, mutants (recG ruvC) defective in Holliday junction resolution systems (RuvABC and RecG/RusA) (39–41) had no detectable effect on the frequency of formation of the inverted dimers. In contrast, an E.coli mutant (E.coli JC5519) defective in RecBCD nuclease/helicase (36,42,43) significantly reduced the frequency of transformation and changed the distribution of the two types of circular inverted dimers (Table 1). These results suggest that a RecBCD-like helicase/nuclease is the major enzyme responsible for the transformability of the linear DNA-1 substrate.
The increase in the relative distribution of pID-DP**** over pID-IP**** in recBC mutants is interesting and can be explained by the proposed model. In the model, the formation of pID-DP**** involves end processing on both strands by exonucleases. RecBCD is probably just one of the exonucleases capable of processing the ends of the linear DNA substrate. In the absence of RecBCD, other exonucleases can still process the ends leading to formation of pID-DP****. However, formation of pID-IP**** may require the helicase/nuclease activity of RecBCD (44,45) to generate the single-stranded DNA template (the intermediate shown in Fig. 3, right). In this case, RecBCD may be the major DNA helicase/nuclease capable of processing the linear DNA substrate into its single-stranded form. In the recBC mutants (defective in both nuclease and helicase activities) the pID-IP**** pathway is more severely inhibited due to lack of RecBCD activity.
Similar to the dumbbell model, two pathways for inverted repeat-mediated linear giant rDNA palindrome formation in Tetrahymena thermophila have been proposed (46). Following chromosome breakage, the single copy of excised rDNA is converted to a giant palindrome by inverted repeat-mediated recombinational rearrangement at cruciform structures (46). This pathway of palindrome formation has been shown in T.thermophila (46) and Saccharomyces (11). Alternatively, it was proposed that linear palindromic DNA can be formed by SSA of the exposed single-stranded inverted repeats (46). Recent studies have provided supporting results for formation of long palindromic DNAs by inverted repeat-mediated single-strand (intra-strand) annealing in Streptomyces (47). Moreover, our results further support the pathway of intra-strand annealing of inverted repeats for inverted dimer (palindrome) formation in E.coli (see discussions above).
Our model can also be used to explain the formation of circular inverted dimers (e.g. H-circles and certain DMs) during gene amplification processes (10,19,21,23,27,30,31,48–52). Excision (or release) of the amplicon either as a double- or single-stranded DNA fragment produces a linear DNA substrate capable of forming a dumbbell-like DNA intermediate which is subsequently converted into an inverted dimer. The importance of inverted repeats in generating the giant inverted repeats (i.e. inverted dimers) has been demonstrated in the case of the H-circles in drug-resistant Leishmania (10,31). Whether inverted repeats are important genetic elements in gene amplification or other DNA sequence rearrangement remains to be determined.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Drs M. G. Marinus, A. J. Clark and R. G. Lloyd for providing bacterial strains. We also thank Dr Leroy F. Liu for helpful discussions. This work was supported by an NHRI Taiwan grant.
References
- 1.Bi X. and Liu,L.F. (1994) recA-independent and recA-dependent intramolecular plasmid recombination. Differential homology requirement and distance effect. J. Mol. Biol., 235, 414–423. [DOI] [PubMed] [Google Scholar]
- 2.Bi X. and Liu,L.F. (1996) A replicational model for DNA recombination between direct repeats. J. Mol. Biol., 256, 849–858. [DOI] [PubMed] [Google Scholar]
- 3.Bierne H., Vilette,D., Ehrlich,S.D. and Michel,B. (1997) Isolation of a dnaE mutation which enhances RecA-independent homologous recombination in the Escherichia coli chromosome. Mol. Microbiol., 24, 1225–1234. [DOI] [PubMed] [Google Scholar]
- 4.Klein H.L. (1995) Genetic control of intrachromosomal recombination. Bioessays, 17, 147–159. [DOI] [PubMed] [Google Scholar]
- 5.Lovett S.T., Gluckman,T.J., Simon,P.J., Sutera,V.A.,Jr and Drapkin,P.T. (1994) Recombination between repeats in Escherichia coli by a recA-independent, proximity-sensitive mechanism. Mol. Gen. Genet., 245, 294–300. [DOI] [PubMed] [Google Scholar]
- 6.Collins J., Volckaert,G. and Nevers,P. (1982) Precise and nearly-precise excision of the symmetrical inverted repeats of Tn5; common features of recA-independent deletion events in Escherichia coli. Gene, 19, 139–146. [DOI] [PubMed] [Google Scholar]
- 7.Leach D.R. and Stahl,F.W. (1983) Viability of lambda phages carrying a perfect palindrome in the absence of recombination nucleases. Nature, 305, 448–451. [DOI] [PubMed] [Google Scholar]
- 8.Henderson S.T. and Petes,T.D. (1993) Instability of a plasmid-borne inverted repeat in Saccharomyces cerevisiae. Genetics, 134, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordenin D.A., Lobachev,K.S., Degtyareva,N.P., Malkova,A.L., Perkins,E. and Resnick,M.A. (1993) Inverted DNA repeats: a source of eukaryotic genomic instability. Mol. Cell. Biol., 13, 5315–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olmo A., Arrebola,R., Bernier,V., Gonzalez-Pacanowska,D. and Ruiz-Perez,L.M. (1995) Co-existence of circular and multiple linear amplicons in methotrexate-resistant Leishmania. Nucleic Acids Res., 23, 2856–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler D.K., Yasuda,L.E. and Yao,M.C. (1996) Induction of large DNA palindrome formation in yeast: implications for gene amplification and genome stability in eukaryotes. Cell, 87, 1115–1122. [DOI] [PubMed] [Google Scholar]
- 12.Bi X. and Liu,L.F. (1996) DNA rearrangement mediated by inverted repeats. Proc. Natl Acad. Sci. USA, 93, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin C.T., Lyu,Y.L. and Liu,L.F. (1997) A cruciform-dumbbell model for inverted dimer formation mediated by inverted repeats. Nucleic Acids Res., 25, 3009–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kafatos F.C., Mitsialis,S.A., Spoerel,N., Mariani,B., Lingappa,J.R. and Delidakis,C. (1985) Studies on the developmentally regulated expression and amplification of insect chorion genes. Cold Spring Harbor Symp. Quant. Biol., 50, 537–547. [DOI] [PubMed] [Google Scholar]
- 15.Stark G.R., Debatisse,M., Giulotto,E. and Wahl,G.M. (1989) Recent progress in understanding mechanisms of mammalian DNA amplification. Cell, 57, 901–908. [DOI] [PubMed] [Google Scholar]
- 16.Bishop J.M. (1991) Molecular themes in oncogenesis. Cell, 64, 235–248. [DOI] [PubMed] [Google Scholar]
- 17.Brison O. (1993) Gene amplification and tumor progression. Biochim. Biophys. Acta, 1155, 25–41. [DOI] [PubMed] [Google Scholar]
- 18.Hahn P.J. (1993) Molecular biology of double-minute chromosomes. Bioessays, 15, 477–484. [DOI] [PubMed] [Google Scholar]
- 19.Fried M., Feo,S. and Heard,E. (1991) The role of inverted duplication in the generation of gene amplification in mammalian cells. Biochim. Biophys. Acta, 1090, 143–155. [DOI] [PubMed] [Google Scholar]
- 20.Huang T. and Campbell,J.L. (1995) Amplification of a circular episome carrying an inverted repeat of the DFR1 locus and adjacent autonomously replicating sequence element of Saccharomyces cerevisiae. J. Biol. Chem., 270, 9607–9614. [DOI] [PubMed] [Google Scholar]
- 21.Ellenberger T.E. and Beverley,S.M. (1989) Multiple drug resistance and conservative amplification of the H region in Leishmania major. J. Biol. Chem., 264, 15094–15103. [PubMed] [Google Scholar]
- 22.Hightower R.C., Ruiz-Perez,L.M., Wong,M.L. and Santi,D.V. (1988) Extrachromosomal elements in the lower eukaryote Leishmania. J. Biol. Chem., 263, 16970–16976. [PubMed] [Google Scholar]
- 23.Ouellette M. and Borst,P. (1991) Drug resistance and P-glycoprotein gene amplification in the protozoan parasite Leishmania. Res. Microbiol., 142, 737–746. [DOI] [PubMed] [Google Scholar]
- 24.White T.C., Fase-Fowler,F., van Luenen,H., Calafat,J. and Borst,P. (1988) The H circles of Leishmania tarentolae are a unique amplifiable system of oligomeric DNAs associated with drug resistance. J. Biol. Chem., 263, 16977–16983. [PubMed] [Google Scholar]
- 25.Fakharzadeh S.S., Rosenblum-Vos,L., Murphy,M., Hoffman,E.K. and George,D.L. (1993) Structure and organization of amplified DNA on double minutes containing the mdm2 oncogene. Genomics, 15, 283–290. [DOI] [PubMed] [Google Scholar]
- 26.Ford M., Davies,B., Griffiths,M., Wilson,J. and Fried,M. (1985) Isolation of a gene enhancer within an amplified inverted duplication after “expression selection”. Proc. Natl Acad. Sci. USA, 82, 3370–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ford M. and Fried,M. (1986) Large inverted duplications are associated with gene amplification. Cell, 45, 425–430. [DOI] [PubMed] [Google Scholar]
- 28.Ma C., Looney,J.E., Leu,T.H. and Hamlin,J.L. (1988) Organization and genesis of dihydrofolate reductase amplicons in the genome of a methotrexate-resistant Chinese hamster ovary cell line. Mol. Cell. Biol., 8, 2316–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nalbantoglu J. and Meuth,M. (1986) DNA amplification–deletion in a spontaneous mutation of the hamster aprt locus: structure and sequence of the novel joint. Nucleic Acids Res., 14, 8361–8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nonet G.H., Carroll,S.M., DeRose,M.L. and Wahl,G.M. (1993) Molecular dissection of an extrachromosomal amplicon reveals a circular structure consisting of an imperfect inverted duplication. Genomics, 15, 543–558. [DOI] [PubMed] [Google Scholar]
- 31.Ouellette M., Hettema,E., Wust,D., Fase-Fowler,F. and Borst,P. (1991) Direct and inverted DNA repeats associated with P-glycoprotein gene amplification in drug resistant Leishmania. EMBO J., 10, 1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyu Y.L., Lin,C.T. and Liu,L.F. (1999) Inversion/dimerization of plasmids mediated by inverted repeats. J. Mol. Biol., 285, 1485–1501. [DOI] [PubMed] [Google Scholar]
- 33.Yasuda L.F. and Yao,M.C. (1991) Short inverted repeats at a free end signal large palindromic DNA formation in Tetrahymena. Cell, 67, 505–516. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd R.G. (1991) Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J. Bacteriol ., 173, 5414–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bachmann B.J. (1972) Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev., 36, 525–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen A. and Clark,A.J. (1986) Synthesis of linear plasmid multimers in Escherichia coli K-12. J. Bacteriol ., 167, 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunes S., Botstein,D. and Fox,M.S. (1990) Synapsis-mediated fusion of free DNA ends forms inverted dimer plasmids in yeast. Genetics, 124, 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derbyshire M.K., Epstein,L.H., Young,C.S., Munz,P.L. and Fishel,R. (1994) Nonhomologous recombination in human cells. Mol. Cell. Biol., 14, 156–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitby M.C., Vincent,S.D. and Lloyd,R.G. (1994) Branch migration of Holliday junctions: identification of RecG protein as a junction specific DNA helicase. EMBO J., 13, 5220–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinagawa H. and Iwasaki,H. (1996) Processing the holliday junction in homologous recombination. Trends Biochem. Sci., 21, 107–111. [PubMed] [Google Scholar]
- 41.Shah R., Cosstick,R. and West,S.C. (1997) The RuvC protein dimer resolves Holliday junctions by a dual incision mechanism that involves base-specific contacts. EMBO J., 16, 1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kowalczykowski S.C., Dixon,D.A., Eggleston,A.K., Lauder,S.D. and Rehrauer,W.M. (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev., 58, 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaudhury A.M. and Smith,G.R. (1984) A new class of Escherichia coli recBC mutants: implications for the role of RecBC enzyme in homologous recombination. Proc. Natl Acad. Sci. USA, 81, 7850–7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson D.G. and Kowalczykowski,S.C. (1998) SSB protein controls RecBCD enzyme nuclease activity during unwinding: a new role for looped intermediates. J. Mol. Biol., 282, 275–285. [DOI] [PubMed] [Google Scholar]
- 45.Yu M., Souaya,J. and Julin,D.A. (1998) The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc. Natl Acad. Sci. USA, 95, 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butler D.K., Yasuda,L.E. and Yao,M.C. (1995) An intramolecular recombination mechanism for the formation of the rRNA gene palindrome of Tetrahymena thermophila. Mol. Cell. Biol., 15, 7117–7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin Z. and Cohen,S.N. (2000) Long palindromes formed in Streptomyces by nonrecombinational intra-strand annealing. Genes Dev., 14, 1789–1796. [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen S., Hassin,D., Karby,S. and Lavi,S. (1994) Hairpin structures are the primary amplification products: a novel mechanism for generation of inverted repeats during gene amplification. Mol. Cell. Biol., 14, 7782–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heard E., Williams,S.V., Sheer,D. and Fried,M. (1991) Gene amplification accompanied by the loss of a chromosome containing the native allele and the appearance of the amplified DNA at a new chromosomal location. Proc. Natl Acad. Sci. USA, 88, 8242–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hyrien O., Debatisse,M., Buttin,G. and de Saint Vincent,B.R. (1988) The multicopy appearance of a large inverted duplication and the sequence at the inversion joint suggest a new model for gene amplification. EMBO J., 7, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma C., Martin,S., Trask,B. and Hamlin,J.L. (1993) Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes Dev., 7, 605–620. [DOI] [PubMed] [Google Scholar]
- 52.Passananti C., Davies,B., Ford,M. and Fried,M. (1987) Structure of an inverted duplication formed as a first step in a gene amplification event: implications for a model of gene amplification. EMBO J., 6, 1697–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]