Abstract
Speckle-type POZ domain protein (SPOP) has been acknowledged as a tumor suppressor gene in numerous types of cancer. However, SPOP expression and its prognostic role in human non-small cell lung cancer (NSCLC) remain unknown. The present study investigated SPOP expression in NSCLC and evaluated its prognostic significance in patients with NSCLC. The results demonstrated that SPOP expression was significantly downregulated in NSCLC tissues at the mRNA and protein level compared with normal lung tissues using reverse transcription-quantitative polymerase chain reaction, and western blot analysis. Immunohistochemical staining results also demonstrated that SPOP was expressed at a low level in 84.1% (132/157) of NSCLC samples and at a high level in 52.2% (12/23) of normal lung samples, whereby the difference was statistically significant (P<0.001). In addition, it was revealed that the level of SPOP was associated with histologic type (P=0.003), tumor differentiation (P=0.046), tumor size (P=0.0036), lymph node metastasis (P=0.041) and clinical stages (P=0.046). Furthermore, the overall survival of patients with high SPOP expression was significantly increased compared with that of patients with low SPOP expression (P=0.003). These results revealed that SPOP expression was downregulated in NSCLC tissues and associated with poor prognosis in patients with NSCLC, suggesting that SPOP is an independent prognostic marker candidate for NSCLC.
Keywords: speckle-type POZ protein, non-small cell lung cancer, prognosis
Introduction
Lung cancer (LC) remains a major cause of cancer-associated mortality worldwide (1). Despite the recent development of novel treatments, the survival rates of patients with lung cancer remain low, with a 5-year survival rate ≤15% (2). To improve the prognosis of patients, further research on lung cancer, particularly focusing on the underlying molecular mechanisms, is warranted.
There are two major types of lung cancer based on clinical classification: Non-small cell lung cancer (NSCLC) and small cell lung cancer, which account for ~80 and 20% of all lung cancer cases, respectively. NSCLC is a heterogeneous group of carcinomas derived from epithelial cells and they can be separated into three major histological subtypes: Adenocarcinoma (ADC), squamous cell carcinoma (SCC), and large cell carcinoma (3).
Considering the risk posed by NSCLC and relatively small progress in NSCLC treatment, efforts have been made to investigate specific molecular markers to understand the primary molecular mechanisms of this malignancy. Thus, this may benefit the diagnosis, therapy design and prognostic assessment of NSCLC.
The speckle-type POZ domain protein (SPOP) gene, which encodes the substrate-recognition component of a cullin3-based E3-ubiquitin ligase (Cul3) (4), is located at the 17q21 locus where a high allelic imbalance has been observed in primary tumors (5). SPOP is a 374-amino acid protein comprising an N-terminal meprin and TRAF-C homology (MATH) domain for recruiting substrate proteins, and a C-terminal poxvirus and zinc finger (POZ) domain (also known as a BTB domain) for interaction with Cul3 (4). It has been demonstrated that the MATH-BTB SPOP protein in mammals serves as an adapter of macroH2A in ubiquitination for regulating its deposition on the inactive X-chromosome (6). In addition, it is associated with the Daxx ubiquitination process in which Cul3-based ubiquitin ligase in the Hedgehog/Gli signaling pathway is involved for regulating transcriptional repression of proapoptotic proteins, including p53 (7). Ubiquitin modifications regulate various cellular processes and are involved in cancer pathogenesis to a certain extent (8). As an adaptor for Cul3-based ubiquitination, the SPOP gene may be mutated in certain malignancies (8–13). Based on previous analyses of genome-wide somatic mutation, frequent mutations of SPOP gene were observed in certain types of human cancer (9,10). Although SPOP protein is expressed in normal gastric, colonic and prostate epithelial cells, 30% of gastric cancer, 20% of colorectal cancer and 37% of prostate cancer do not express SPOP protein (14).
Not much is known regarding SPOP somatic mutations in lung cancer, and the prevalence of decreased SPOP expression has rarely been reported in lung cancer, in particular with respect to the influence of decreased SPOP expression in lung cancer. The present study aimed to examine the difference between in SPOP mRNA and protein levels in lung cancer tissues, and corresponding normal lung tissues derived from patients with NSCLC. In addition, the association between SPOP expression and clinicopathological features of patients with NSCLC was investigated. Potential candidate prognostic markers of NSCLC were identified by evaluating the role of SPOP in carcinogenesis, development and prognosis of NSCLC.
Materials and methods
Patients and tissue specimens
A total of 157 surgical specimens and 23 normal lung tissues were obtained from the Affiliated Hospital of Nantong University (Nantong, China) between January 2004 and January 2010, which included 115 adenocarcinoma cases and 42 squamous cell carcinoma cases. The carcinoma tissues and the adjacent normal tissues were snap frozen in liquid nitrogen and stored at −80°C. The patients involved in the present study did not receive any chemotherapy and/or radiation therapy prior to surgery. Written informed consent was obtained from patients or their legal guardians for the surgical procedures and permission to utilize resected tissue specimens for the current study. The present study was approved by the Ethics Committee of the Affiliated Hospital of Nantong University. All specimens had been confirmed through pathological diagnosis. The histological subtype was assessed using the current World Health Organization classification guidelines and the stage was classified in conformity with the 7th edition of the Tumor Node Metastasis classification of malignant tumors by two independent, experienced pathologists (15,16). The follow-up data of patients with lung cancer in the present study were available and complete. Overall survival, which was defined as the length of time between the surgery date and date of mortality or last follow-up, was used as a measure for prognosis. Postoperative follow-up was performed at the outpatient department of the Affiliated Hospital of Nantong University, and comprised clinical and laboratory examinations every quarter for the first 2 years, every 2 quarters during the 3rd to 5th years, and annually for an additional 5 years or until mortality.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue lysate using a TRIzol reagent extraction kit (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) followed by RT using the High Capacity cDNA Reverse Transcription kit (Qiagen, Inc., Valencia, CA, USA) in accordance with the manufacturer's protocol. The Roche Light Cycler 480 system (Roche Diagnostics, Burgess Hill, UK) was used for qPCR analysis. Primer pairs designed by Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA, USA) were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China), the sequences of which were as follows: SPOP forward, 5′-TGACCACCAGGTAGACAGCG-3′ and reverse, 5′-CCCGTTTCCCCCAAGTTA-3′. The GAPDH gene served as an internal control. The sequences of the primers for GAPDH were as follows: GAPDH forward, 5′-AACTTCCGTTGCTGCCAT-3′ and reverse, 5′-TTTCTTCCACAGGGCTTTG-3′. The PCR system (25 µl) comprised 1 µl of cDNA, 12.5 µl of 2X Fast EvaGreen™ qPCR Master mix (Biotium Inc., Freemont, CA, USA), 1 µl of primers (10 µM) and 10.5 µl of RNase/DNase-free water. The following thermocycling conditions were maintained: 96°C for 2 min; 40 cycles at 96°C for 15 sec; and 60°C for 1 min. Each sample was performed in triplicate, and the average value was computed. For comparative expression of SPOP, the 2−∆∆Cq method (17) was used as an indication of relative changes.
Western blot analysis
Lung cancer tissues from patients were homogenized in lysis buffer (50 mM Tris, pH 7.5, 5 mM EDTA, 1% SDS, 1% NP-40, 1% Triton X-100, 1% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride, 10 µg/ml leupeptin and 10 µg/ml aprotinin) and then centrifuged in a microcentrifuge at 12,800 × g at 4°C for 20 min. The supernatants were accumulated and protein concentrations were measured using a Bradford assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Approximately 20 µg protein was loaded into each well and samples were separated using 10% SDS-PAGE then electrophoretically transferred to a nitrocellulose membrane, which was blocked with 5% non-fat milk for 2 h at room temperature. Subsequently, the membrane was washed with TBS-Tween 20 three times, incubated with primary goat polyclonal antibodies directed against SPOP (1:500; cat. no. sc-66649; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and GAPDH (anti-rabbit, cat. no. sc-25778; 1:1,000; Santa Cruz Biotechnology, Inc.) at 4°C overnight, and incubated with horseradish peroxidase-conjugated goat-anti-rabbit secondary antibody (1:10,000; cat. no. sc-2007; Santa Cruz Biotechnology, Inc.) for 2 h at room temperature in the antibody buffer. The protein levels were normalized by GAPDH. The proteins were then detected using an Enhanced Chemiluminescence Detection system (Pierce; Thermo Fisher Scientific, Inc.). The band intensity was quantified using ImageJ (version 1.44p; National Institutes of Health, Bethesda, MD, USA).
Immunohistochemistry analysis
Tissues were fixed in 10% formalin for 12 h at room temperature, embedded in paraffin for 4 h at 4°C, then cut into 5-µm thick serial sections. The tissue sections were deparaffinized in dimethylbenzene 15 min twice and rehydrated in graded ethanol solutions at room temperature. The slides were washed with running water and then washed in PBS to deactivate endogenous peroxidase 3 min three times at room temperature. With respect to the retrieval of antigen, the sections were immersed in buffer containing 0.01 M sodium citrate-hydrochloric acid (pH 6.0) and boiled by microwaving for 15 min. When the slides were naturally cooled to room temperature, they were subjected to washing twice in distilled water and three times in PBS. To block endogenous peroxidase, petroleum jelly was smeared on the tissue edge, and one drop of 3% H2O2 was added. After incubation at room temperature for 15 min, the slides were washed three times in PBS. The tissue sections went through the following steps: Incubation with goat polyclonal anti-human antibody directed against SPOP (1:100; no. sc-66649; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature; rinsing in 3% peroxidase quenching solution (Invitrogen; Thermo Fisher Scientific, Inc.) to block endogenous peroxidase 20 min at room temperature; incubation with a donkey anti-goat horseradish peroxidase-conjugated secondary antibody (1:5,000; cat. no. ab-6885; Abcam, Cambridge, UK) for 30 min at room temperature; and washing three times in PBS. The signal was developed with 3,3′-diaminobenzidine solution and all of the slides were counterstained with hematoxylin 5 min at room temperature. Normal lung tissues were processed simultaneously as negative controls with omission of the primary antibody. The specimens were analyzed by two consultant pathologists who were blinded to the patient data. The total SPOP immunoreactivity was determined by combining the percentage of positively stained tumor cells with the staining intensity. A total of five randomly selected high power fields were observed and scored based on the percentage of positive cells. Briefly, the percentage of positive cells were scored as 0 (0–5%), 1 (6–25%), 2 (26–50%, focal), 3 (51–75%) or 4 (>75%), and the intensity as 0, no staining; 1, weak staining; 2, moderate staining or 3, strong staining. The two scores were then added, and the expression level of SPOP was defined as follows: ‘−’ (negative, score of 0); ‘+’ (weakly positive, score of 1–3); ‘++’ (positive, score of 4–7); or ‘+++’ (strongly positive, score of 8–12). A total score of >3 was defined as strong SPOP expression, and a total score of ≤3 was defined as weak SPOP expression.
Statistical analysis
SPSS 21.0 (IBM Corp., Armonk, NY, USA) was utilized for statistical analysis, with the results presented as the mean ± standard deviation. One-way analysis of variance among groups or two-tailed Student's t-tests between groups was used to compare expression of SPOP protein and mRNA in tissues. Chi-squared or Fisher's exact tests were used to distinguish categorical variables. Survival curves were constructed using Kaplan-Meier method. Multivariate analysis was conducted using Cox proportional hazards method. P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulated expression of SPOP in human NSCLC tissue samples
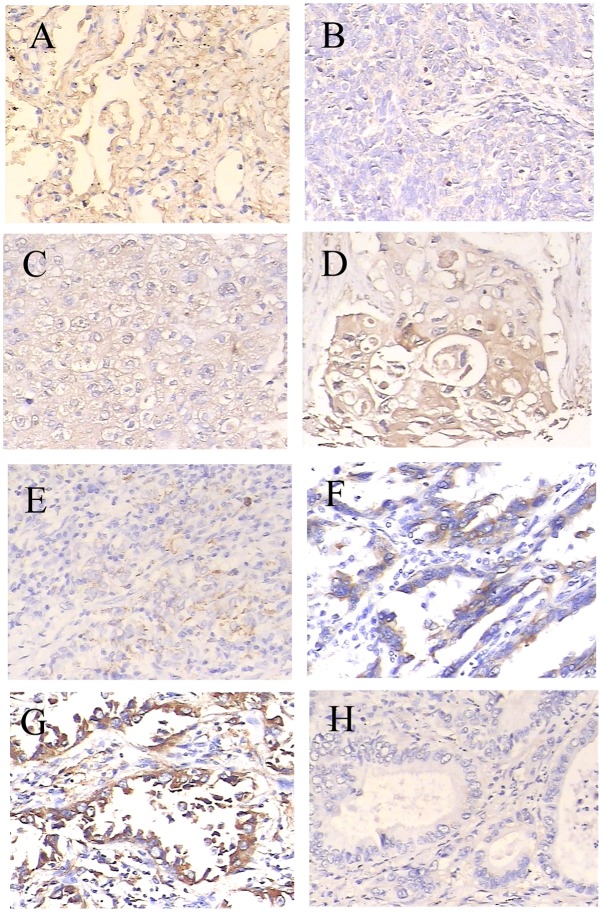
To investigate the role of SPOP in NSCLC, qPCR and western blot analysis was performed to detect the expression of SPOP in 12 pairs of newly collected NSCLC tissues, and adjacent normal lung tissues. It was demonstrated that SPOP mRNA was significantly downregulated in NSCLC tissues compared with normal lung tissues (P<0.05; Fig. 1A). Similar to mRNA concentration, the cellular concentration of SPOP protein in NSCLC tissues was significantly lower compared with that of normal lung tissues (P<0.05; Fig. 1B and C). For further verification, immunohistochemistry experiments were performed to detect the level of SPOP expression in 157 NSCLC tissues and 23 normal lung tissues. SPOP immunoreactivity was identified in the cytoplasm of representative NSCLC cells. Specifically, low expression level of SPOP protein was present in 84.1% (132/157) of lung cancer samples, while high expression level of SPOP was present in 52.2% (12/23) of normal lung samples (P<0.001; Table I and Fig. 2).
Figure 1.
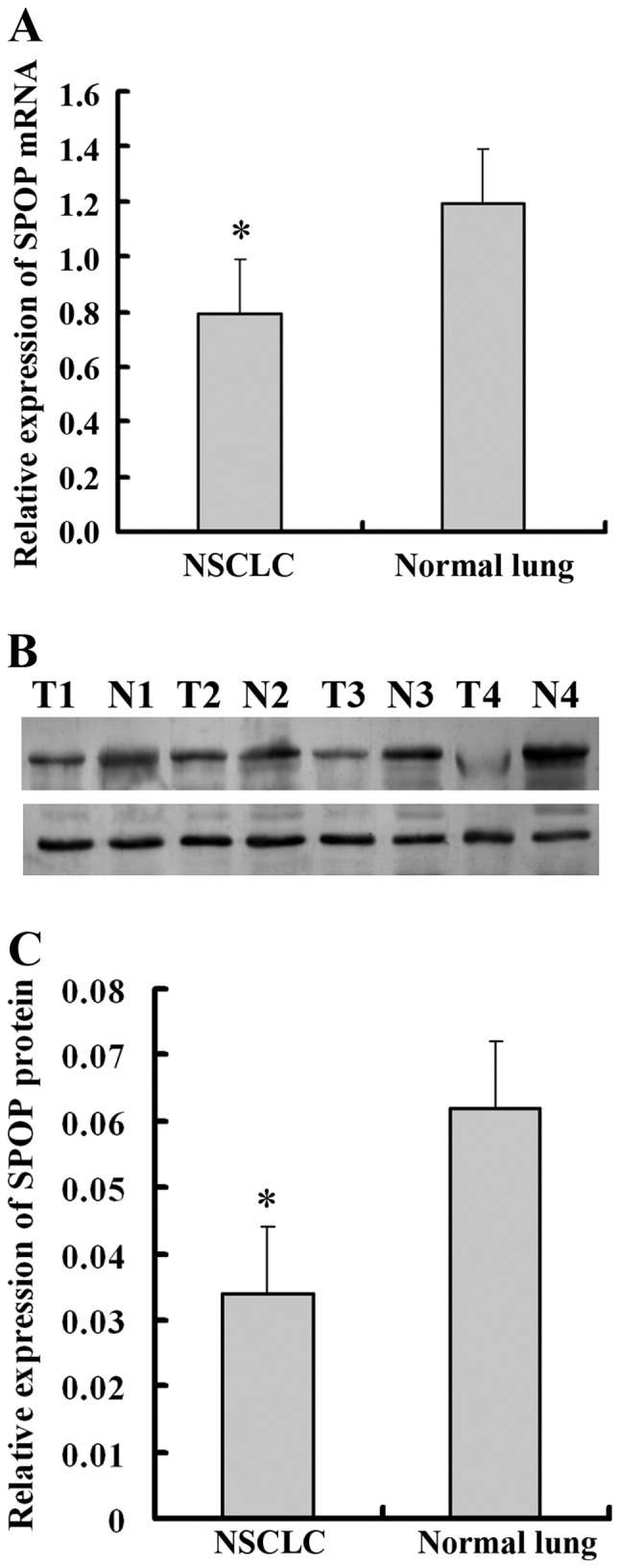
Expression of SPOP is downregulated in human NSCLC tissue samples. (A) Reverse transcription-quantitative polymerase chain reaction analysis revealed a significantly low level of SPOP mRNA in NSCLC. (B) Representative and (C) quantified western blot analysis, which revealed a significantly lower expression level of SPOP protein in 12 representative samples of NSCLC tumor tissues (T) compared with adjacent non-tumor tissues (N). GAPDH was used as a loading control. *P<0.05 compared with normal lung tissues. SPOP, speckle-type POZ domain protein; NSCLC, non-small cell lung cancer.
Table I.
Summary of immunohistochemical results in NSCLC and normal lung tissues.
| SPOP expression | ||||
|---|---|---|---|---|
| Group | N | Low (n) | High (n) | P-value |
| Tumor | 157 | 132 | 25 | <0.001 |
| Normal | 23 | 11 | 12 | |
NSCLC, non-small cell lung cancer; SPOP, speckle-type POZ domain protein.
Figure 2.
Expression of SPOP in lung cancer specimens of different tumor differentiation grades was determined by immunohistochemical experiments. SPOP immunoreactivity shows homogeneous brown-yellow staining in the cytoplasm of tumor cells along with hematoxylin (blue) counterstain. Samples include (A) normal lung tissue, (B) poorly differentiated squamous cell carcinoma, (C) moderately differentiated squamous cell carcinoma, (D) well differentiated squamous cell carcinoma, (E) poorly differentiated adenocarcinoma, (F) moderately differentiated adenocarcinoma, (G) well differentiated adenocarcinoma and (H) negative control. Original magnification, ×200. With poorer tumor differentiation, the SPOP expression in NSCLC was significantly decreased (P<0.05 compared with normal lung). SPOP, speckle-type POZ domain protein; NSCLC, non-small cell lung cancer.
Association between SPOP protein expression and clinicopathological variables in human NSCLC tissues
The association between the SPOP protein expression and clinicopathological variables in patients with NSCLC was analyzed. For statistical analysis of the SPOP expression, the NSCLC tissue specimens were separated into a low expression group and a high expression group according to the final scores of staining. According to Table II, the level of SPOP was positively associated with histologic type (P=0.003), tumor differentiation (P=0.046), tumor size (P=0.0036), lymph node metastasis (P=0.041) and clinical stages (P=0.046) in patients with NSCLC. However, no significant association was identified between SPOP expression and other clinical factors.
Table II.
Association between the clinicopathological characteristics and the expression level of SPOP in lung cancer tissues.
| SPOP expression | ||||
|---|---|---|---|---|
| Characteristic | N | Low | High | P-value |
| Sex | 0.266 | |||
| Male | 94 | 82 | 12 | |
| Female | 63 | 50 | 13 | |
| Age | 0.266 | |||
| ≤60 | 94 | 50 | 13 | |
| >60 | 63 | 82 | 12 | |
| Histologic type | 0.003a | |||
| Adenocarcinoma | 115 | 91 | 24 | |
| Squamous cell carcinoma | 42 | 41 | 1 | |
| Tumor differentiation | 0.046a | |||
| Well | 29 | 21 | 8 | |
| Moderate | 100 | 84 | 16 | |
| Poor | 28 | 27 | 1 | |
| Tumor size | 0.0036a | |||
| T1 | 83 | 69 | 14 | |
| T2 | 62 | 56 | 6 | |
| T3 | 7 | 4 | 3 | |
| T4 | 5 | 2 | 3 | |
| Lymph node metastasis | 0.041a | |||
| Absent | 104 | 83 | 21 | |
| Present | 53 | 49 | 4 | |
| Lung cancer stages | 0.046a | |||
| 1 | 29 | 21 | 8 | |
| 2 | 100 | 84 | 16 | |
| 3 | 28 | 27 | 1 | |
Statistically significant. SPOP, speckle-type POZ domain protein.
Prognostic significance of SPOP expression level in human NSCLC samples
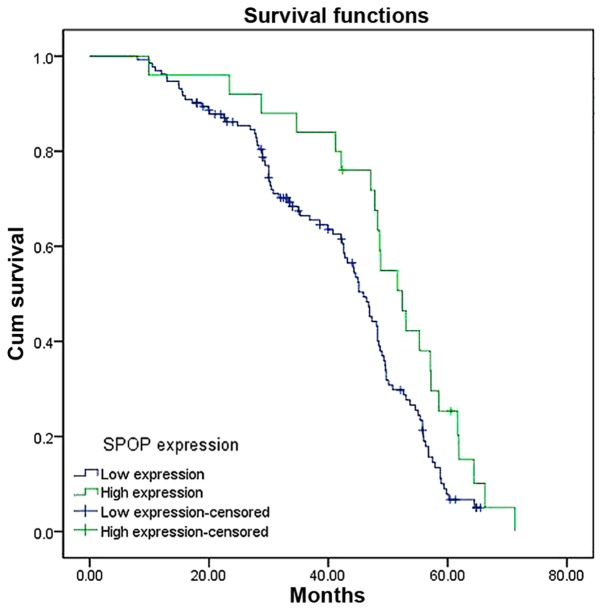
To determine the prognostic significance of SPOP in NSCLC, the association between expression level of SPOP and clinical outcome was analyzed. At the end of clinical follow-up, survival information of all patients was available. Log-rank test analysis indicated that the overall survival time of patients with high SPOP expression level was significantly increased compared with that of patients having low SPOP expression level (P=0.003; Fig. 3). Based on univariate Cox regression analyses, tumor size, clinical stages and SPOP expression level were significantly associated with overall survival. In addition, a multivariate Cox regression analysis validated clinical stages and SPOP expression level for predicting the overall survival of NSCLC patients (Table III).
Figure 3.
Kaplan-Meier curves for overall survival in patients with NSCLC with low SPOP expression level and high SPOP expression level (P=0.003). SPOP, speckle-type POZ domain protein; NSCLC, non-small cell lung cancer; cum, cumulative.
Table III.
Univariate and multivariate analyses of overall survival of patients with NCSLC.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Sex (male vs. female) | 0.948 | 0.660–1.362 | 0.772 | 0.968 | 0.643–1.456 | 0.875 |
| Age (≤60 vs. >60) | 1.159 | 0.810–1.658 | 0.419 | 1.216 | 0.996–2.681 | 0.303 |
| Histologic type (adenocarcinoma vs. squamous cell carcinoma) | 0.855 | 0.477–1.529 | 0.597 | 1.099 | 0.574–2.105 | 0.776 |
| Tumor differentiation (well vs. moderate vs. poor) | 0.862 | 0.462–1.548 | 0.829 | 1.289 | 0.656–1.857 | 0.721 |
| Tumor size (T1 vs. T2 vs. T3 vs. T4) | 1.976 | 0.425–4.088 | 0.046a | 0.802 | 0.117–4.740 | 0.379 |
| Lymph node metastasis (absent vs. present) | 1.261 | 0.441–1.192 | 0.365 | 1.543 | 0.619–3.848 | 0.352 |
| Lung cancer stages (1 vs. 2 vs. 3) | 1.344 | 1.072–2.466 | 0.032a | 1.815 | 1.347–2.802 | 0.041a |
| SPOP expression (low vs. high) | 0.652 | 0.306–0.861 | 0.039a | 0.634 | 0.422–0.712 | 0.045a |
Statistically significant. HR, hazard ratio; CI, confidence interval; SPOP, speckle-type POZ domain protein.
Discussion
Although SPOP has been acknowledged as a tumor suppressor gene in numerous types of cancer (9,18), little has been reported regarding SPOP expression and the prognostic implications thereof in lung cancer. In the current study, it was demonstrated that, compared with normal lung tissues, the expression of SPOP was significantly downregulated at the mRNA and protein level in NSCLC tissues. Furthermore, low expression level of SPOP was observed to be significantly associated with short postsurgical survival. Notably, the data in the present study demonstrated that SPOP is expressed in normal lung tissue, providing a basis for further research on the potential role of SPOP in lung development.
SPOP is an adaptor protein facilitating the degradation of several substrates that are essential to cellular development and physiology (18,19). Specifically, it is an E3 ubiquitin ligase adaptor protein that has been frequently mutated in prostate and endometrial cancer, which cluster in the MATH domain (11,20). All cancer-associated SPOP mutations presumably affect substrate binding in the ubiquitination process. Ubiquitination regulates essential cellular processes and is associated with tumor progression (21). Ubiquitin is activated by the E1 activating enzyme, provisionally carried by the E2 conjugating enzyme, and then transferred to its specific substrates by the E3 ubiquitin ligase (18), which confers substrate specificity to ubiquitin ligation. Of the E3 ligase family, the Cullin-RING E3 ubiquitin ligase is the most notable (22). In addition, SPOP is a unique substrate-binding adaptor protein that renders the E3 ubiquitin ligase complex specific, and gains increasing attention due to its far-reaching effects in cellular physiology and in pathological conditions (18). It has been demonstrated that the deregulation of the ubiquitin system results in the development of numerous types of tumors, and alterations in the ubiquitin system occur during the initiation and progression of cancer (23). Thus, various approaches have been made to target the ubiquitin system in cancer therapy, in which E3 ubiquitin ligases are considered as the most important components as they bind directly to target proteins (23,24). As an E3 ubiquitin ligase adaptor protein, SPOP serves a tumor suppressor role in numerous types of cancer (18).
In the present study, the downregulated expression of SPOP in NSCLC tissues suggests that SPOP is mutated in NSCLC and may contribute to cancer development. The level of SPOP expression was positively associated with tumor differentiation and clinical stages in the patients with NSCLC, suggesting that SPOP mutations are be incapable of serving as a tumor suppressor gene and were early events in NSCLC tumorigenesis. Furthermore, the low expression level of SPOP was associated with poor postsurgical survival. Combined with univariate and multivariate analyses on overall survival, there are sufficient reasons to believe that SPOP may serve as a novel independent candidate of prognostic marker for NSCLC.
In summary, SPOP expression in NSCLC tissues and its prognostic significance was confirmed, which will support further research on NSCLC development. It was also proposed that it may be a significant indicator for unfavorable progression in patients with NSCLC. However, further studies are required to expound the molecular mechanisms underlying the regulatory role of SPOP in NSCLC pathogenesis for the development of novel cancer therapeutics.
Acknowledgements
This study was supported by the Clinical Medicine Center of Suzhou (grant no. Szzx201502) and the Jiangsu Provincial Key Medical Discipline (Laboratory)(grant no. ZDXKB2016007).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Zhang T, Zhang DM, Zhao D, Hou XM, Liu XJ, Ling XL, Ma SC. The prognostic value of osteopontin expression in non-small cell lung cancer: A meta-analysis. J Mol Histol. 2014;45:533–540. doi: 10.1007/s10735-014-9574-3. [DOI] [PubMed] [Google Scholar]
- 3.Esposito L, Conti D, Ailavajhala R, Khalil N, Giordano A. Lung cancer: Are we up to the challenge? Curr Genomics. 2010;11:513–518. doi: 10.2174/138920210793175903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagai Y, Kojima T, Muro Y, Hachiya T, Nishizawa Y, Wakabayashi T, Hagiwara M. Identification of a novel nuclear speckle-type protein, SPOP. FEBS Lett. 1997;418:23–26. doi: 10.1016/S0014-5793(97)01340-9. [DOI] [PubMed] [Google Scholar]
- 5.De Marchis L, Cropp C, Sheng ZM, Bargo S, Callahan R. Candidate target genes for loss of heterozygosity on human chromosome 17q21. Br J Cancer. 2004;90:2384–2389. doi: 10.1038/sj.bjc.6601848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernández-Muñoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M. Stable X chromosome inactivation involves the PRC1 polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA. 2005;102:7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon JE, La M, Oh KH, Oh YM, Kim GR, Seol JH, Baek SH, Chiba T, Tanaka K, Bang OS, et al. BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J Biol Chem. 2006;281:12664–12672. doi: 10.1074/jbc.M600204200. [DOI] [PubMed] [Google Scholar]
- 8.Ande SR, Chen J, Maddika S. The ubiquitin pathway: An emerging drug target in cancer therapy. Eur J Pharmacol. 2009;625:199–205. doi: 10.1016/j.ejphar.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 9.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 10.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le GM, O'Hara AJ, Rudd ML, Urick ME, Hansen NF, O'Neil NJ, Price JC, Zhang S, England BM, Godwin AK, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44:1310–1315. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Ao J, Fu J, Lee DF, Xu J, Lonard D, O'Malley BW. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30:4350–4364. doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim MS, Je EM, Oh JE, Yoo NJ, Lee SH. Mutational and expressional analyses of SPOP, a candidate tumor suppressor gene, in prostate, gastric and colorectal cancers. APMIS. 2013;121:626–633. doi: 10.1111/apm.12030. [DOI] [PubMed] [Google Scholar]
- 15.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JH, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, et al. The 2015 World Health Organization classification of lung tumors: Impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 16.Edge SB, Compton CC. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Mani RS. The emerging role of speckle-type POZ protein (SPOP) in cancer development. Drug Discov Today. 2014;19:1498–1502. doi: 10.1016/j.drudis.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding D, Song T, Jun W, Tan Z, Fang J. Decreased expression of the SPOP gene is associated with poor prognosis in glioma. Int J Oncol. 2015;46:333–341. doi: 10.3892/ijo.2014.2729. [DOI] [PubMed] [Google Scholar]
- 20.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varshavsky A. Discovery of the biology of the ubiquitin system. JAMA. 2014;311:1969–1970. doi: 10.1001/jama.2014.5549. [DOI] [PubMed] [Google Scholar]
- 22.Pei XH, Bai F, Li Z, Smith MD, Whitewolf G, Jin R, Xiong Y. Cytoplasmic CUL9/PARC ubiquitin ligase is a tumor suppressor and promotes p53-dependent apoptosis. Cancer Res. 2011;71:2969–2977. doi: 10.1158/0008-5472.CAN-10-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 24.Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell. 2010;143:686–693. doi: 10.1016/j.cell.2010.11.016. [DOI] [PubMed] [Google Scholar]