Abstract
SPARC-like protein 1 (SPARCL1), a member of the family of secreted proteins which is acidic and rich in cysteine, is a potential tumor suppressor gene in most types of tumor. A systemic review and bioinformatics analysis was carried out to determine the associations between SPARCL1 and tumor progression and clinical factors. Downregulation of SPARCL1, thought to be regulated by epigenetic modifications including DNA methylation, serves important functions in tumor progression and development, with its regulatory functions on cell viability, migration, invasion, cell adhesion and drug resistance. Downregulation of SPARCL1 was markedly associated with a poor overall survival rate of patients with one of ≥7 solid tumors and predicted increased mortality in patients with one of ≥4 distinct tumor types. The present review indicated that SPARCL1 may be a therapeutic target for cancer treatment and a biomarker to determine prognosis.
Keywords: SPARC-like protein 1, tumor suppressor, cancer, progression, prognosis
1. SPARC-like protein 1 (SPARCL1) is a potential tumor suppressor gene
SPARCL1, a member of the family of secreted proteins that are acidic and rich in cysteine in the cellular matrix. Originally termed SC1, SPARCL1 was first cloned from the rat central nervous system and encodes an extracellular matrix glycoprotein, similar to osteonectin/basement membrane protein 40/secreted protein acidic and rich in cysteine (SPARC) (1). Subsequently, Schraml et al (2) and Girard et al (3) cloned the aforementioned gene from endothelial cells in non-small cell lung cancer and high endothelial venules in human tonsil lymphatic tissues, and termed it MAST9 and hevin, respectively. The mRNA of the gene is 3 kb in length and the theoretical molecular mass of the encoded protein, SPARCL1, is ~75 kDa. However, the protein expressed in vitro reveals molecular masses of ~75 and 150 kDa, suggesting that SPARCL1 protein may form a homodimer in vitro (4).
SPARC is a tumor suppressor gene in cancer, and it has been demonstrated to be involved in the regulation of tumor progression and drug resistance (5,6). SPARCL1 exhibits 62% identity with SPARC and the two proteins share three conservative structural domains (3), indicating functional similarity. SPARCL1 is localized on human chromosome 4, which contains a number of additional known tumor suppressor genes. Thus, SPARCL1 is considered to be a potential tumor suppressor gene and participates in tumor occurrence and development, by regulating tumor cell viability and differentiation (4). SPARCL1 may, additionally, be a potential oncogene and participates in tumor occurrence and development, by regulating tumor cell viability and affecting the production of tumor blood vessels (7).
In the present review, the mRNA expression of SPARCL1 in tumors was analyzed using Oncomine (www.oncomine.org/resource/login.html) (8). As presented in Fig. 1, of the ~20 different types of solid tumors included in the Oncomine database, SPARCL1 was downregulated >2-fold in the majority of tumors analyzed, with the exception of liver cancer, lymphoma and sarcoma, where SPARCL1 was upregulated. The downregulation of SPARCL1 was marked in bladder, breast, cervical, rectal, lung and ovarian cancer. Thus, it may be inferred that SPARCL1 is a tumor suppressor gene in cancer.
Figure 1.
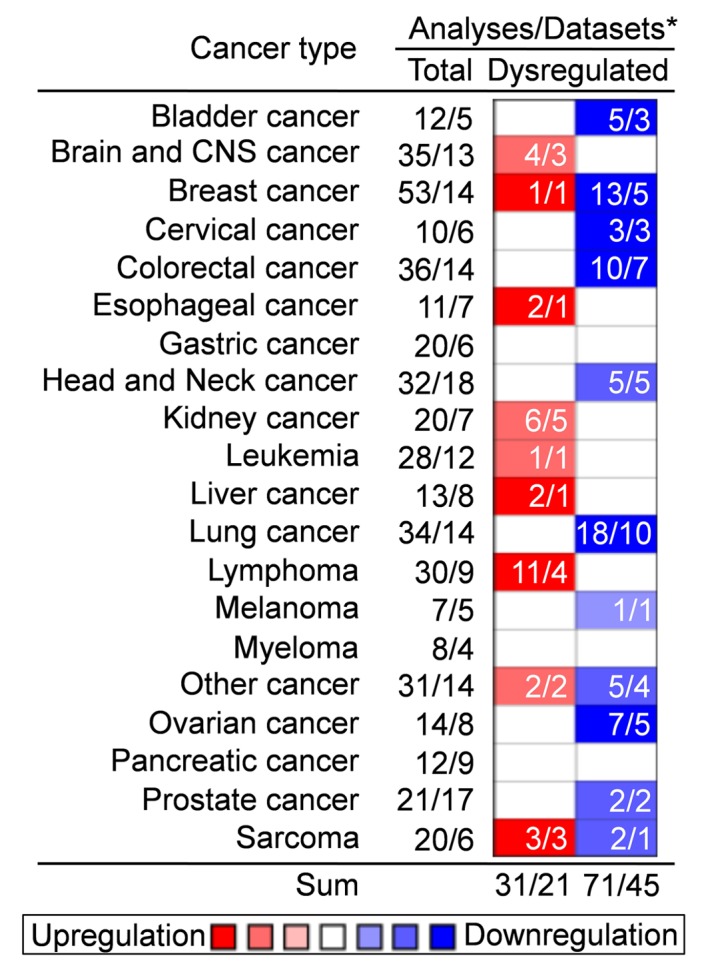
On the basis of the microarray data retrieved from Oncomine, SPARCL1 is differentially expressed in almost all tumors and a marked downregulation of SPARCL1 is observed in the majority of tumors. SPARCL1, SPARC-like protein 1; CNS, central nervous system; *total number of analyses in total number of datasets.
2. DNA methylation may be an important mechanism that contributes to the downregulation of SPARCL1
As presented in Table I, SPARCL1 is upregulated in liver cancer (7); however, SPARCL1 is markedly downregulated in prostate (9), lung (10), ovarian (11) and a number of other types of cancer. These results are consistent with Fig. 1. Downregulation of SPARCL1 in tumors may result from the epigenetic mechanisms, including DNA methylation, because SPARCL1 is not a classical tumor suppressor gene exhibiting a deletion or mutation. Isler et al (10) used microsatellite analysis, quantitative polymerase chain reaction and sequence analysis of all exons, including the intron-exon junctions and a portion of the putative promoter region, but did not identify a mutation or deletion that may be responsible for the downregulation of SPARCL1. This was indicative of other regulatory mechanisms resulting in the differential expression of SPARCL1 in tumors, including epigenetic modification. A previous study revealed that DNA methylation is the reason for the downregulation of SPARCL1 in pancreatic, ovarian and lung cancers (Table I), and demethylation of the gene partially reversed the abnormal expression in pancreatic cancer (12).
Table I.
Summary on the SPARCL1 correlated with tumor progression and development.
| Tumor type | SPARCL1 expression | Biological function | Molecular mechanism | Mechanism of gene expression |
|---|---|---|---|---|
| Prostate cancer | Downregulation (9) | Inhibition of cell migration and invasion (14) | Affects migration by regulating RhoC (19); inhibition of the assembly of focal adhesions (20) | – |
| Colorectal cancer | Downregulation (13,21) | Inhibition of cell proliferation, growth and invasion (13) | Affects tumor cell differentiation through EMT (13) | – |
| Hilar cholangiocarcinoma | Downregulation (15) | Inhibition of cell migration (15) | Inhibition of the expression of MMP-9, MMP-2, vimentin and fibronectin (15) | – |
| Gastric cancer | Downregulation (22,23) | – | Inactivation of its tumor suppressor functions (22) | Loss of heterozygosity (23) |
| Pancreatic cancer | Downregulation (12) | Inhibition of tumor cell proliferation and invasion (12) | Anti-invasive effects (12) | DNA methylation (12) |
| Ovarian cancer | Downregulation (11) | Associated with multiple drug resistance (18) | Interaction with drug resistance-related proteins including PTEN (18) | DNA methylation (24) |
| Lung cancer | Downregulation (10) | Decrease in luciferase activity (10) | – | DNA methylation (25) |
| Glioma | Downregulation (16) | Associated with cell cycle (16) | Anti-adhesion (16) | – |
| Liver cancer | Upregulation (7) | – | – | – |
| Uterine leiomyoma | Upregulation (26) | – | – | – |
SPARCL1, SPARC-like protein 1; MMP, matrix metalloproteinase; PTEN, phosphatase and tensin homolog.
3. SPARCL1 contributes to tumor development and progression
There have been a limited number of studies on SPARCL1, but the gene has been identified to be markedly associated with tumor development and progression. SPARCL1 contributions to tumor cell viability (13), migration and invasion (12–16) and exhibits an anti-adhesive effect (12,16). In addition, SPARCL1 may be involved in the regulation of drug resistance in cancer. It has been identified that SPARCL1 is a recombinant gene in the extracellular matrix of osteosarcoma in children and is involved in the mechanism of multiple drug resistance (17). A previous study used comprehensive bioinformatics analysis to identify that the SPARCL1 gene was involved in the regulation of drug resistance in ovarian cancer (18).
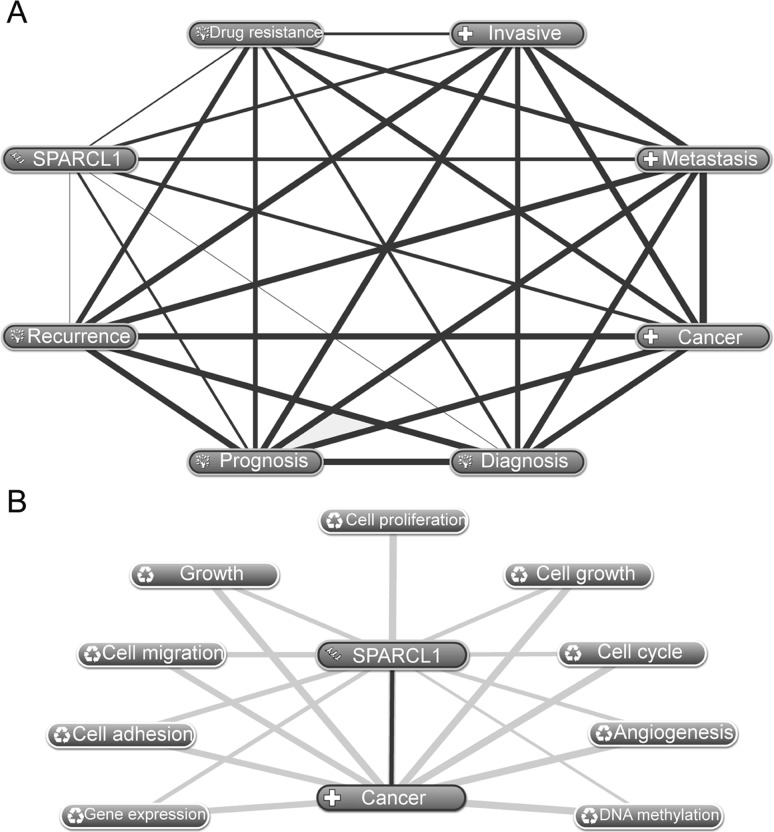
The association between SPARCL1 and tumor progression was investigated using Coremine Medical (http://www.coremine.com/medical). As presented in Fig. 2, using SPARCL1 and cancer as key words, SPARCL1 was identified to be associated with diagnosis, prognosis, recurrence, invasiveness, metastasis and drug resistance of cancer (Fig. 2A). In addition, the associations identified in the present review, between SPARCL1 and invasiveness, metastasis and drug resistance of cancer, were consistent with previous studies (Table I)(9–16,18–26). Furthermore, analysis indicated that SPARCL1 may participate in cancer development and progression, in 9 biological processes (P<0.001) including cell viability, cell cycle, migration and adhesion (Fig. 2B), which is consistent with previous studies (Table I). In addition, SPARCL1 and cancer were annotated with DNA methylation, supporting the hypothesis that DNA methylation may be an important mechanism which contributes to the downregulation of SPARCL1.
Figure 2.
Associations between SPARCL1 and tumors, as analyzed using Coremine Medical. (A) Associations between SPARCL1 and tumor progression and development. (B) Hypothetical pathways/biological processes in which SPARCL1 was involved. SPARCL1, SPARC-like protein 1.
4. Downregulation of SPARCL1 is associated with poor prognosis in cancer
Previous studies indicate that downregulation of SPARCL1 is markedly associated with poor prognosis and therefore the gene may be a prognostic marker in cancers. In prostate cancer, the downregulation of SPARCL1 has been markedly associated with biochemical recurrence, metastatic disease and poor overall survival (OS) time (19). Patients with stage II/III colorectal cancer who possessed increased p53 and decreased SPARCL1 expression levels exhibited ~50% decreased 3-year survival compared with controls (27). Furthermore, in gastric cancer, silenced expression of SPARCL1 predicted a poorer prognosis (23).
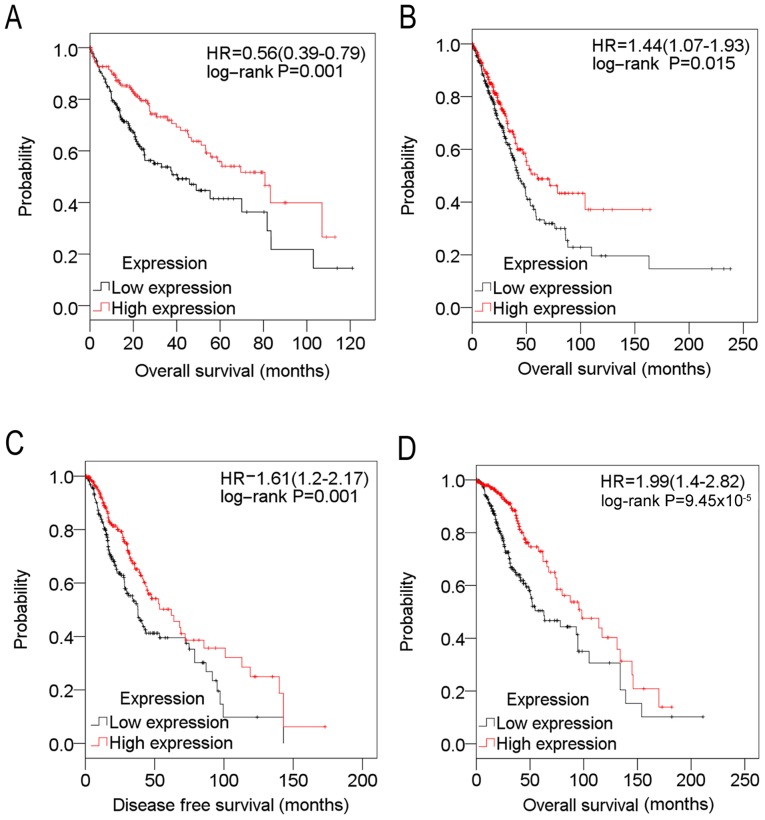
On the basis of The Cancer Genome Atlas (TCGA) (24) cohort data, the associations between SPARCL1 and cancer prognosis were analyzed. The expression value of SPARCL1 and the corresponding clinical data of each type of cancer in the TCGA cohort was retrieved from the cBioPortal database (cbioportal.org) (28). Expression values of SPARCL1 were divided into high and low expression using the median as the threshold value in a Kaplan-Meier estimator analysis, in accordance with a previous study (29). As presented in Table II and Fig. 3, downregulation of SPARCL1 was markedly associated with poor OS time in liver cancer (242 samples) and lung cancer (324 samples), and markedly associated with poorer disease-free survival and OS time in glioma (311 samples).
Table II.
Association between SPARCL1 expression and prognosis in glioma, lung cancer and liver cancer.
| DFS (median) | OS (median) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 95% confidence interval | 95% confidence interval | ||||||||
| Disease type | Group | Estimate | Standard error | Lower boundary | Upper boundary | Estimate | Standard error | Lower boundary | Upper boundary |
| Glioma | L | 37.900 | 3.983 | 30.093 | 45.707 | 62.900 | 12.971 | 37.476 | 88.324 |
| H | 62.000 | 10.173 | 42.061 | 81.939 | 98.200 | 19.627 | 59.731 | 136.669 | |
| Total | 43.500 | 4.524 | 34.633 | 52.367 | 87.400 | 10.232 | 67.346 | 107.454 | |
| Lung cancer | L | 42.500 | 3.920 | 34.818 | 50.182 | ||||
| H | 60.100 | 12.425 | 35.747 | 84.453 | |||||
| Total | 49.200 | 3.705 | 41.938 | 56.462 | |||||
| Liver cancer | L | 40.400 | 9.142 | 22.483 | 58.317 | ||||
| H | 80.700 | 10.853 | 59.428 | 101.972 | |||||
| Total | 55.600 | 8.087 | 39.749 | 71.451 | |||||
SPARCL1 expression and survival data in The Cancer Genome Atlas cohort were used for the analysis. Expression values of a gene were divided into high and low expression using the median as a cut-off. H, high expression; L, low expression; DFS, disease-free survival; OS, overall survival.
Figure 3.
Kaplan-Meier estimator survival plots for SPARCL1 in (A) liver cancer, (B) lung cancer, (C) glioma and (D) glioma, on the basis of data retrieved from The Cancer Genome Atlas cohort. Expression values of SPARCL1 were divided into high (red line) and low (black line) expression using the median as the threshold value. SPARCL1, SPARC-like protein 1; HR, hazard ratio.
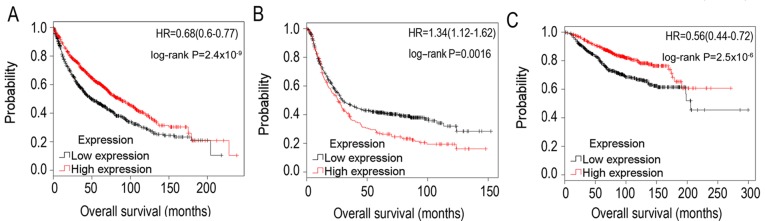
The association between SPARCL1 and OS time in lung cancer was additionally validated using Kaplan-Meier estimator analysis, which selected thousands of samples of ovarian, lung, breast and gastric cancer from microarrays deposited in the TCGA cohort and Gene Expression Omnibus profiles (30). Using the median expression as the threshold value, it was identified that the downregulation of SPARCL1 in lung cancer (3,021 samples) predicted decreased OS time (Fig. 4A), which was consistent with the results based on the TCGA cohort data (Table II and Fig. 3). Furthermore, downregulation of SPARCL1 predicted improved OS time in gastric cancer (1,223 samples) (Fig. 4B), although this result was in contrast with a previous study (23). In addition, in breast cancer (2,627 samples), downregulation of SPARCL1 predicted poorer OS time (Fig. 4C).
Figure 4.
Kaplan-Meier estimator survival plots for SPARCL1 in (A) lung cancer, (B) gastric cancer and (C) breast cancer. Expression values of SPARCL1 were divided into high (red line) and low (black line) expression using the median as the threshold value. SPARCL1, SPARC-like protein 1; HR, hazard ratio.
SPARCL1 was additionally associated with clinical features of a number of types of tumor. Downregulation of SPARCL1 was associated with increased mortality of patients with glioma, liver and lung cancer (P<0.05), and patients with cervical cancer (P=0.076; Table III). Furthermore, the downregulation rate of SPARCL1 increased considerably for surviving patients with cervical cancer and downregulation of the gene in ovarian cancer was markedly associated with a lower histological grade (P<0.05; Table IV).
Table III.
Association between SPARCL1 expression and vital status of patients with different tumors, in accordance with The Cancer Genome Atlas cohort.
| SPARCL1 expression | ||||
|---|---|---|---|---|
| Patient's vital status | No. of patients | Low (%) | High (%) | P-value |
| Glioma of lower grade | 528 | 0.001 | ||
| Deceased | 134 (25.4%) | 84 (62.7) | 50 (37.3) | |
| Alive | 394 (74.6%) | 180 (45.7) | 214 (54.3) | |
| Lung adenocarcinoma | 516 | 0.001 | ||
| Deceased | 187 (36.2%) | 111 (59.4) | 76 (40.6) | |
| Alive | 329 (63.8%) | 146 (44.4) | 183 (55.6) | |
| Hepatocellular carcinoma | 372 | 0.024 | ||
| Deceased | 130 (34.9%) | 75 (57.7) | 55 (42.3) | |
| Alive | 242 (65.1%) | 110 (45.5) | 132 (54.5) | |
| Cervical adenocarcinoma | 305 | 0.076 | ||
| Deceased | 73 (23.9%) | 43 (58.9) | 30 (41.1) | |
| Alive | 232 (76.1%) | 109 (47.0) | 123 (53.0) | |
| Subcutaneous melanoma | 470 | 0.42 | ||
| Deceased | 222 (47.2%) | 100 (45.0) | 122 (55.0) | |
| Alive | 248 (52.8%) | 135 (54.4) | 113 (45.6) | |
| Acute myeloid leukemia | 173 | 0.916 | ||
| Deceased | 114 (65.9%) | 57 (50.0) | 57 (50.0) | |
| Alive | 59 (34.1%) | 29 (49.2) | 30 (50.8) | |
| Lymphoma | 27 | 1 | ||
| Deceased | 6 (22.2%) | 3 (50.0) | 3 (50.0) | |
| Alive | 21 (77.8%) | 11 (52.4) | 10 (47.6) | |
| Prostate adenocarcinoma | 497 | 0.339 | ||
| Deceased | 10 (2.0%) | 7 (70.0) | 3 (30.0) | |
| Alive | 487 (98.0%) | 242 (49.7) | 245 (50.3) | |
| Sarcoma | 261 | 0.666 | ||
| Deceased | 99 (37.9%) | 51 (51.5) | 48 (48.5) | |
| Alive | 162 (62.1%) | 79 (48.8) | 83 (51.2) | |
| Esophageal carcinoma | 184 | 0.981 | ||
| Deceased | 77 (41.8%) | 38 (49.4) | 39 (50.6) | |
| Alive | 107 (58.2%) | 53 (49.5) | 54 (50.5) | |
Expression values of SPARCL1 were divided into high and low expression using the median as the threshold value. Cervical adenocarcinoma is defined as a cervical squamous cell carcinoma and endocervical adenocarcinoma. Lymphoma is defined as a lymphoid neoplasm diffuse large B-cell lymphoma. SPARCL1, SPARC-like protein 1.
Table IV.
Association of SPARCL1 expression with neoplasm status and neoplasm histological grade in several cancers, in accordance with The Cancer Genome Atlas cohort.
| SPARCL1 expression | ||||
|---|---|---|---|---|
| Variables | No. of patients | Low (%) | High (%) | P-value |
| Glioma of lower grade | 442 | 0.036 | ||
| With tumor | 220 (49.8%) | 100 (45.5) | 120 (54.5) | |
| Tumor-free | 222 (50.2%) | 123 (55.4) | 99 (44.6) | |
| Cervical adenocarcinoma | 263 | 0.013 | ||
| With tumor | 76 (28.9%) | 47 (61.8) | 29 (38.2) | |
| Tumor-free | 187 (71.1%) | 84 (44.9) | 103 (50.2) | |
| Ovarian serous cystadenocarcinoma | 476 | 0.004 | ||
| Histological grade 2 | 56 (11.8%) | 18 (32.1) | 38 (67.9) | |
| Histological grade 3 | 420 (88.2%) | 220 (52.4) | 200 (47.6) | |
Expression values of SPARCL1 were divided into high and low expression using the median as the threshold value. Cervical adenocarcinoma is defined as a cervical squamous cell carcinoma and endocervical adenocarcinoma.
5. Conclusions
Previous studies on the association between SPARCL1 and tumor progression are relatively limited. One previous study suggested that SPARCL1 is an oncogene (7), but a number of contradictory studies have identified SPARCL1 as a potential tumor suppressor gene (4,12,13,15). A bioinformatic analysis, on the basis of the data retrieved from Oncomine and the TCGA cohort, was conducted to identify the associations between SPARCL1 and tumor progression. Oncomine included information concerning SPARCL1 expression in almost 20 solid tumors (Fig. 1) and this identified that downregulation of SPARCL1 is prevalent in the majority of tumors, suggesting that SPARCL1 is a tumor suppressor gene.
The present review revealed that the downregulation of the SPARCL1 was markedly associated with poor OS time of ≥7 solid tumors, which included prostate (19), colorectal (27), gastric (23), liver, lung, glioma and breast cancer (Table II; Figs. 3 and 4). In addition, decreased expression of SPARCL1 typically predicted increased mortality in glioma, lung, liver and cervical cancer (Table III). Therefore, SPARCL1 may be a universal prognostic marker of tumors in the clinic.
Previous studies have indicated that SPARCL1 is a tumor suppressor gene and is involved in tumor cell viability (13), migration and invasion (12–16) and cell adhesion (12,16), and is associated with drug resistance of tumors (17,18). Consistent with these studies, bioinformatics analysis in the present review revealed that SPARCL1 was associated with prognosis, invasiveness, metastasis, recurrence and drug resistance of cancer (Fig. 2A). It is hypothesized that SPARCL1 exhibits these aforementioned actions by interactions with a number of biological processes/signaling pathways including cell adhesion, cell viability, cell cycle and cell migration (Fig. 2B). These results indicate that SPARCL1 serves important functions in tumor progression.
The present review has elucidated the association between SAPRCL1 and cancer. SPARCL1 may be an important tumor suppressor gene in tumor progression and development, and it may be a therapeutic target for cancer treatment and a potential biomarker for prognosis.
Acknowledgements
The present review was supported by the National Natural Science Foundation of China (grant nos. 81302283, 81560424, 81660606 and 81460397), the China Postdoctoral Science Foundation (grant nos. 2014M552535XB and 2014M552291) and the Natural Science Foundation of Guangxi (grant nos. 2014GXNSFCA118010, 2015GXNSFBA139115, 2015GXNSFAA139151 and 2014GXNSFBA118155).
References
- 1.Johnston IG, Paladino T, Gurd JW, Brown IR. Molecular cloning of SC1: A putative brain extracellular matrix glycoprotein showing partial similarity to osteonectin/BM40/SPARC. Neuron. 1990;4:165–176. doi: 10.1016/0896-6273(90)90452-L. [DOI] [PubMed] [Google Scholar]
- 2.Schraml P, Shipman R, Stulz P, Ludwig CU. cDNA subtraction library construction using a magnet-assisted subtraction technique (MAST) Trends Genet. 1993;9:70–71. doi: 10.1016/0168-9525(93)90216-5. [DOI] [PubMed] [Google Scholar]
- 3.Girard JP, Springer TA. Cloning from purified high endothelial venule cells of hevin, a close relative of the antiadhesive extracellular matrix protein SPARC. Immunity. 1995;2:113–123. doi: 10.1016/1074-7613(95)90083-7. [DOI] [PubMed] [Google Scholar]
- 4.Claeskens A, Ongenae N, Neefs JM, Cheyns P, Kaijen P, Cools M, Kutoh E. Hevin is down-regulated in many cancers and is a negative regulator of cell growth and proliferation. Br J Cancer. 2000;82:1123–1130. doi: 10.1054/bjoc.1999.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Socha MJ, Said N, Dai Y, Kwong J, Ramalingam P, Trieu V, Desai N, Mok SC, Motamed K. Aberrant promoter methylation of SPARC in ovarian cancer. Neoplasia. 2009;11:126–135. doi: 10.1593/neo.81146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tai IT, Dai M, Owen DA, Chen LB. Genome-wide expression analysis of therapy-resistant tumors reveals SPARC as a novel target for cancer therapy. J Clin Invest. 2005;115:1492–1502. doi: 10.1172/JCI23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau CP, Poon RT, Cheung ST, Yu WC, Fan ST. SPARC and Hevin expression correlate with tumour angiogenesis in hepatocellular carcinoma. J Pathol. 2006;210:459–468. doi: 10.1002/path.2068. [DOI] [PubMed] [Google Scholar]
- 8.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlomm T, Luebke AM, Sültmann H, Hellwinkel OJ, Sauer U, Poustka A, David KA, Chun FK, Haese A, Graefen M, et al. Extraction and processing of high quality RNA from impalpable and macroscopically invisible prostate cancer for microarray gene expression analysis. Int J Oncol. 2005;27:713–720. [PubMed] [Google Scholar]
- 10.Isler SG, Ludwig CU, Chiquet-Ehrismann R, Schenk S. Evidence for transcriptional repression of SPARC-like 1, a gene downregulated in human lung tumors. Int J Oncol. 2004;25:1073–1079. [PubMed] [Google Scholar]
- 11.Biade S, Marinucci M, Schick J, Roberts D, Workman G, Sage EH, O'Dwyer PJ, Livolsi VA, Johnson SW. Gene expression profiling of human ovarian tumours. Br J Cancer. 2006;95:1092–1100. doi: 10.1038/sj.bjc.6603346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esposito I, Kayed H, Keleg S, Giese T, Sage EH, Schirmacher P, Friess H, Kleeff J. Tumor-suppressor function of SPARC-like protein 1/hevin in pancreatic cancer. Neoplasia. 2007;9:8–17. doi: 10.1593/neo.06646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu H, Zhang H, Ge W, Liu X, Loera S, Chu P, Chen H, Peng J, Zhou L, Yu S, et al. Secreted protein acidic and rich in cysteines-like 1 suppresses aggressiveness and predicts better survival in colorectal cancers. Clin Cancer Res. 2012;18:5438–5448. doi: 10.1158/1078-0432.CCR-12-0124. [DOI] [PubMed] [Google Scholar]
- 14.Xiang Y, Qiu Q, Jiang M, Jin R, Lehmann BD, Strand DW, Jovanovic B, DeGraff DJ, Zheng Y, Yousif DA, et al. SPARCL1 suppresses metastasis in prostate cancer. Mol Oncol. 2013;7:1019–1030. doi: 10.1016/j.molonc.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Y, Chen Y, Ma J, Yu X, Yu G, Li Z. SPARCL1 is a novel predictor of tumor recurrence and survival in hilar cholangiocarcinoma. Tumour Biol. 2016;37:4159–4167. doi: 10.1007/s13277-015-4206-4. [DOI] [PubMed] [Google Scholar]
- 16.Turtoi A, Musmeci D, Naccarato AG, Scatena C, Ortenzi V, Kiss R, Murtas D, Patsos G, Mazzucchelli G, De Pauw E, et al. Sparc-like protein 1 is a new marker of human glioma progression. J Proteome Res. 2012;11:5011–5021. doi: 10.1021/pr3005698. [DOI] [PubMed] [Google Scholar]
- 17.Mintz MB, Sowers R, Brown KM, Hilmer SC, Mazza B, Huvos AG, Meyers PA, Lafleur B, McDonough WS, Henry MM, et al. An expression signature classifies chemotherapy-resistant pediatric osteosarcoma. Cancer Res. 2005;65:1748–1754. doi: 10.1158/0008-5472.CAN-04-2463. [DOI] [PubMed] [Google Scholar]
- 18.Yin F, Liu X, Li D, Wang Q, Zhang W, Li L. Bioinformatic analysis of chemokine (C-C motif) ligand 21 and SPARC-like protein 1 revealing their associations with drug resistance in ovarian cancer. Int J Oncol. 2013;42:1305–1316. doi: 10.3892/ijo.2013.1819. [DOI] [PubMed] [Google Scholar]
- 19.Hurley PJ, Marchionni L, Simons BW, Ross AE, Peskoe SB, Miller RM, Erho N, Vergara IA, Ghadessi M, Huang Z, et al. Secreted protein, acidic and rich in cysteine-like 1 (SPARCL1) is down regulated in aggressive prostate cancers and is prognostic for poor clinical outcome. Proc Natl Acad Sci USA. 2012;109:14977–14982. doi: 10.1073/pnas.1203525109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurley PJ, Hughes RM, Simons BW, Huang J, Miller RM, Shinder B, Haffner MC, Esopi D, Kimura Y, Jabbari J, et al. Androgen-Regulated SPARCL1 in the Tumor Microenvironment Inhibits Metastatic Progression. Cancer Res. 2015;75:4322–4334. doi: 10.1158/0008-5472.CAN-15-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Widegren E, Wang DW, Sun XF. SPARCL1: A potential molecule associated with tumor diagnosis, progression and prognosis of colorectal cancer. Tumour Biol. 2011;32:1225–1231. doi: 10.1007/s13277-011-0226-x. [DOI] [PubMed] [Google Scholar]
- 22.Jakharia A, Borkakoty B, Singh S. Expression of SPARC like protein 1 (SPARCL1), extracellular matrix-associated protein is down regulated in gastric adenocarcinoma. J Gastrointest Oncol. 2016;7:278–283. doi: 10.3978/j.issn.2078-6891.2015.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li P, Qian J, Yu G, Chen Y, Liu K, Li J, Wang J. Down-regulated SPARCL1 is associated with clinical significance in human gastric cancer. J Surg Oncol. 2012;105:31–37. doi: 10.1002/jso.22025. [DOI] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research Network: Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato T, Arai E, Kohno T, Takahashi Y, Miyata S, Tsuta K, Watanabe S, Soejima K, Betsuyaku T, Kanai Y. Epigenetic clustering of lung adenocarcinomas based on DNA methylation profiles in adjacent lung tissue: Its correlation with smoking history and chronic obstructive pulmonary disease. Int J Cancer. 2014;135:319–334. doi: 10.1002/ijc.28684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mencalha AL, Levinsphul A, Deterling LC, Pizzatti L, Abdelhay E. SPARC-like1 mRNA is overexpressed in human uterine leiomyoma. Mol Med Rep. 2008;1:571–574. [PubMed] [Google Scholar]
- 27.Yu SJ, Yu JK, Ge WT, Hu HG, Yuan Y, Zheng S. SPARCL1, Shp2, MSH2, E-cadherin, p53, ADCY-2 and MAPK are prognosis-related in colorectal cancer. World J Gastroenterol. 2011;17:2028–2036. doi: 10.3748/wjg.v17.i15.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hedditch EL, Gao B, Russell AJ, Lu Y, Emmanuel C, Beesley J, Johnatty SE, Chen X, Harnett P, George J, et al. ABCA transporter gene expression and poor outcome in epithelial ovarian cancer. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju149. pii: dju149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]