Abstract
DNA double-strand breaks (DSBs) are introduced in meiosis to initiate recombination and generate crossovers, the reciprocal exchanges of genetic material between parental chromosomes. Here we present high-resolution maps of meiotic DSBs in individual human genomes. Comparing DSB maps between individuals shows that along with DNA binding by PRDM9, additional factors may dictate the efficiency of DSB formation. We find evidence for both GC-biased gene conversion and mutagenesis around meiotic DSB hotspots, while frequent co-localization of DSB hotspots with chromosome rearrangement breakpoints implicates the aberrant repair of meiotic DSBs in genomic disorders. Furthermore, our data indicate that DSB frequency is a major determinant of crossover rate. These maps provide new insights into the regulation of meiotic recombination and the impact of meiotic recombination on genome function.
Introduction
Genetic variation in humans is shaped by homologous recombination. Meiotic recombination itself is required for correct chromosome segregation in gametes, however the initiation of recombination in humans remains poorly understood. Recombination events are tightly clustered in 1–2 kb wide hotspots whose position is primarily determined by the histone-lysine N-methyltransferase PRDM9 protein (1–6). PRDM9 is one of the most rapidly evolving genes in humans for which dozens of variants have been described (7–9) and allelic variants have been shown to specify different sets of hotspots (4). The PRDM9 protein binds DNA through a highly polymorphic tandem array of zinc fingers (ZnF) and is then thought to recruit the recombination initiation complex that includes meiotic recombination protein SPO11, the protein that introduces meiotic DSBs. These DSBs are subsequently repaired by homologous recombination to give rise to either genetic crossovers, where a reciprocal genetic exchange occurs between homologous chromosomes, or non-crossovers (10). Designation of a subset of events as crossovers is a tightly regulated process but the determinants of whether any particular DSB will become a crossover or not remain largely unknown (11).
Several approaches have been used to study recombination hotspots in humans. The most detailed maps of human recombination have been generated by computational inference of recombination rates from patterns of linkage disequilibrium (LD) in the human population (12–15). These maps do not, however, provide information about recombination rates in individuals. SNP genotyping in human pedigrees has been used to identify crossovers but the precision of crossover mapping is tens of Kb (16–20). Until recently, the study of meiotic recombination hotspots in individuals required sperm genotyping (21), a method that can define crossover sites with nucleotide resolution but that cannot be used for genome-wide analyses. Single cell sequencing techniques have facilitated the construction of genome-wide crossover maps from individual sperm and oocytes (22–25), however such approaches currently lack the resolution to perform fine scale analysis and hotspot detection. Furthermore, these approaches rely on the identification of crossovers which are just one of the possible outcomes of recombination. In mammals, only about 10% of DSBs are repaired as crossovers (26), therefore the vast majority of recombination events in meiosis remain unexplored.
We overcome these limitations by generating genome-wide maps of meiotic recombination initiation sites in individual human males. Our approach combines hotspot resolution that is comparable to sperm genotyping, with the gender, individual, and PRDM9-allele specificity that LD-based methods lack. In addition, we detect all sites where recombination can occur, independent of the subsequent repair pathway. Though PRDM9 dictates hotspot locations, we find that ~5% of hotspots are polymorphic between individuals with identical PRDM9 alleles. Sequence polymorphism at PRDM9 binding sites explains less than half of this variation. Through analysis of the DNA polymorphism spectrum at hotspots we identified distinct signatures of GC-biased gene conversion and of recombination mediated mutagenesis. We find evidence for a role of ectopic recombination in gross chromosomal rearrangements and identify 726 new potential rearrangement breakpoints. Finally, this first analysis of the recombination initiation landscape establishes that like crossovers, DSBs occur more frequently at subtelomeric regions. This suggests that initiation frequency is a major driver of crossover rate in human males.
Genome-wide DSB hotspot map in humans
To create a representative overview of the recombination initiation landscape in human males, we generated high-resolution maps of meiotic DSB hotspots from four unrelated individuals (Fig. 1, fig. S1, table S1). We performed chromatin immunoprecipitation followed by single-stranded DNA sequencing (SSDS) (27) to identify DNA fragments associated with DMC1 (meiotic recombination protein DMC1/LIM15 homolog), a specific marker of meiotic DSBs. The number of DMC1-associated ssDNA fragments provides an estimate of DSB frequency, although this estimate could be affected by the relative lifetime of ssDNA intermediates and by differences in DMC1 loading at individual hotspots. Two men in our sample set were homozygous for the most common PRDM9 allele (PRDM9A; individuals AA1 and AA2) and three were heterozygous for the PRDM9A allele and for the less frequent variants PRDM9B (AB1, AB2) and PRDM9C (AC). In total, we identified up to 38,946 DSB hotspots per individual (Fig. 1B, table S1). This number is substantially higher than the 15–20,000 hotspots identified in mouse (4), perhaps a reflection of the 2-fold higher recombination rate in humans (28). The SSDS signal was three to seven fold higher on the sex chromosomes than on the autosomes (fig. S1) and this may reflect continuous DSB formation (29) or extended DSB lifespan on the sex chromsomes (30, 31). Autosomal DSB hotspot strength varied by over three orders of magnitude (fig. S2) and ~100 hotspots (fig. S2, table S3, file S1) were stronger than the recombination hotspot with the highest known crossover rate (9, 32).
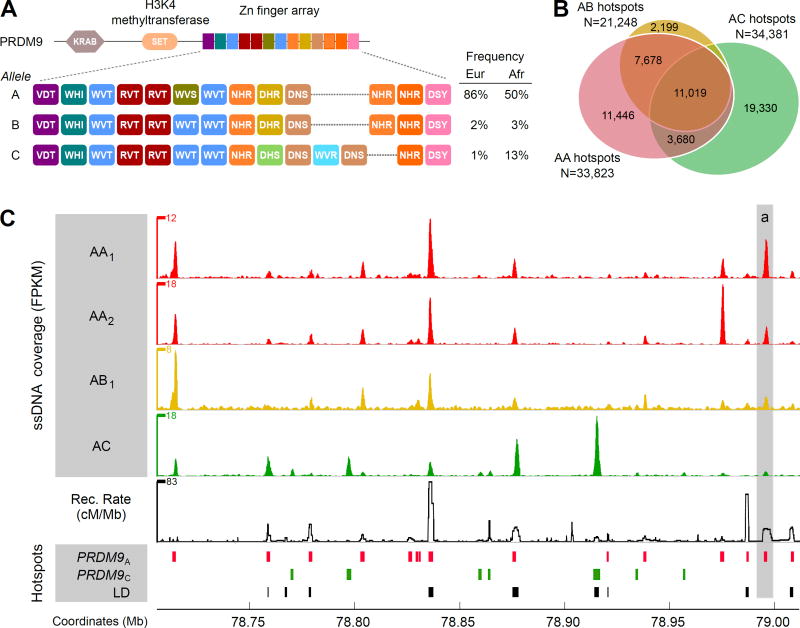
Figure 1. Genome-wide distribution of DSB hotspots in human individuals.
(A) Zn finger array structure of PRDM9 alleles in individuals from this study. Population allele frequencies are taken from (9). (B) The overlap between autosomal hotspots found in different individuals. AA hotspots are the hotspots found in either AA1 or AA2 individuals. (C) DSB hotspots from AA1, AA2, AB1 and AC individuals in a 300 Kb region on chromosome 17. The top four panels show normalized ssDNA coverage in fragments per Kb per million (FPKM), smoothed using a sliding window (window: 1 Kb; step: 0.1 Kb). The baseline of the y-axes is 0 FPKM. The fifth panel shows the recombination rate calculated from the population-averaged HapMap data (15). The lower panels show PRDM9A- and PRDM9C-defined DSB hotspots and LD-defined hotspots.
The PRDM9 protein defines most hotspot sites in mice (4). Consistent with this role, 89% of human DSB hotspots were found at the same locations in the two AA individuals (fig. S3). A similar proportion of hotspots in the AB individual overlapped AA1/AA2 hotspots (88%), suggesting that the PRDM9A-like PRDM9B allele does not specify a distinct set of hotspots (33) (fig. S3). Only 43% of DSB hotspots in the AC individual overlap PRDM9A-defined hotspots (Fig. 1B), therefore, the remaining 57% (19,330) are likely PRDM9C-defined. At the local level, many properties of hotspots, regardless of the PRDM9 allele, remain conserved between human and mouse (33) (fig. S4, S5). Common hotspot features include a purine-pyrimidine skew around hotspot centers and a local increase in GC content. Furthermore, as in mouse and consistent with the role of PRDM9 as a histone H3K4 trimethyltransferase (34), most human DSB hotspots coincide with H3K4me3 in testis (57%; fig. S6).
Consistent with the different DNA binding specificities of the PRDM9A and the PRDM9C alleles, we identified distinct consensus motifs enriched at the centers of PRDM9A and PRDM9C defined hotspots (fig. S7). Each motif matches the predicted PRDM9 binding site for the respective PRDM9 allele but not that of the other allele (fig. S7). Furthermore, the PRDM9A motif, but not the PRDM9C motif, is highly similar to a 13-mer motif previously found to be enriched at recombination hotspots (LD-hotspots) (13, 35) (fig. S7). This is consistent with the widespread predominance of the PRDM9A allele in human populations (84% in European, 50% in African populations (9)). In turn, the PRDM9C-motif is highly similar to a 17-mer motif, found at LD-defined recombination hotspots used in the African-American but not in the European population (20, 35). This agrees with the increased prevalence of the PRDM9C allele in Africans (13%) compared to Europeans (~1%) (9).
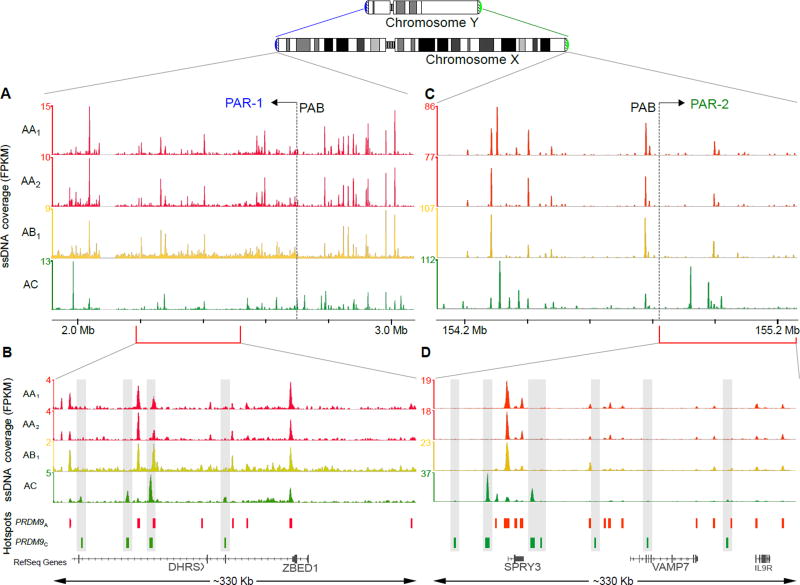
To ensure proper segregation of the sex chromosomes during male meiosis, a crossover must form in the short regions of sequence homology shared between the X and Y chromosomes (pseudoautosomal regions; PARs). In mouse there is a broad and extremely intense DSB signal adjacent to the pseudoautosomal boundary (PAB) Furthermore, this signal and several hotspots outside of the PAR but immediately adjacent to it appear to be formed independently of PRDM9 (4). Unlike in mouse, we did not observe a prominent DSB cluster near either human PAB (Fig. 2A,B) and PRDM9 allele specific hotspot formation was observed in both PAR regions (Fig. 2C,D). Thus, it is unlikely that PRDM9-independent DSB formation near the PAB is a conserved mechanism to ensure a mandatory crossover in the PAR. It is possible however, that PRDM9-independent DSB clusters are located in the very distal part of the human PARs close to the telomere. These regions are poorly assembled and replete with repetitive DNA, therefore they cannot be analyzed using high-throughput sequencing.
Figure 2. PRDM9-defined hotspots are found in the human PARs.
Overview of ~1 Mb regions on chromosome X overlapping the (A) PAR1 and (B) PAR2 pseudoautosomal boundaries (PABs). Normalized ssDNA coverage is shown as fragments per Kb per million (FPKM), smoothed using a sliding window (window: 1 Kb; step: 0.1 Kb). The baseline of the y-axes is 0 FPKM. Coverage is shown for the AA1, AA2, AB and AC individuals. Red and green bars depict PRDM9A-defined and PRDM9C-defined hotspots, respectively. Close up of a (C) 330 Kb region of the PAR1 and (D) the entire ~330 Kb PAR2 illustrating the presence of PRDM9C-defined hotspots (highlighted by grey bars) in both PARs.
Contribution of individual PRDM9 alleles to the LD-based recombination rate map
LD-based methods have provided the most comprehensive maps of human recombination to date (12–15). As these maps are intrinsically sex and population averaged they cannot distinguish between hotspots defined by different PRDM9 alleles. In contrast, our approach directly measures DSB frequency in a single male with a known combination of PRDM9 alleles. We thus explored the contribution of individual PRDM9 alleles to the LD map.
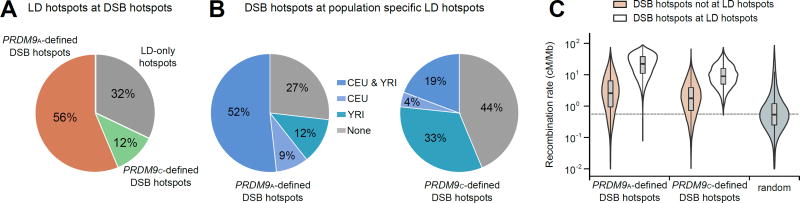
Overall, we found a good agreement between our individual-specific DSB maps and the LD map (Fig. 3A) with 68% of LD-hotspots coinciding with a DSB hotspot. Consistent with the high frequency of the PRDM9A allele across human populations, 56% of LD hotspots coincided with PRDM9A-defined hotspots (9) (Fig. 3A). The less frequent PRDM9C allele has also left a considerable footprint on patterns of linkage disequilibrium, as 12% of LD-hotspots overlapped PRDM9C-defined hotspots. These proportions are very close to the frequencies of these PRDM9 alleles in modern Africans (Fig. 1A) (9), suggesting that the PRDM9 allelic distribution in modern Africans is similar to that in ancestral human populations. Nevertheless, 32% of LD-hotspots do not overlap a DSB hotspot in any of our maps (Fig. 3A). These “LD-only” hotspots are likely the products of minor PRDM9 alleles, hotspots that vary between individuals and hotspots that are used more frequently in females than in males.
Figure 3. Comparison of LD-based recombination maps and DSB hotspots.
(A) The proportion of LD-hotspots that overlap PRDM9A- or PRDM9C-defined DSB hotspots. Most LD-hotspots are detected in the DSB hotspot maps. (B) The percentage of PRDM9A- (left) and PRDM9C-defined (right) hotspots overlapping LD-hotspots in the CEU (European) and YRI (African) populations. Most DSB hotspots are found in the LD maps. (C) Violin plots showing the population averaged recombination rate at DSB hotspots that overlap or do not overlap LD-hotspots defined in the CEU and YRI populations. 85% of PRDM9A-defined and 82% of PRDM9C-defined hotspots that do not overlap an LD-hotspot have a recombination rate above random expectation.
We next asked what proportion of DSB hotspots can be seen in LD data. Overall, 51.1% of DSB hotspots overlap population averaged LD-hotspots (15). Since there are substantial differences in PRDM9 allele frequencies between human populations we used LD-hotspots inferred from population-specific subsets of HapMap II data (19) (Utah residents with ancestry from northern and western Europe (CEU); Yoruba in Ibadan, Nigeria (YRI)) to assess the DSB overlap. In agreement with the higher prevalence of PRDM9C in African compared to European populations, the PRDM9C-defined DSB hotspots are better represented at YRI-specific hotspots (33% overlap; Fig. 3B) than at CEU-specific hotspots (4% overlap; Fig. 3B). Furthermore, at PRDM9C-defined DSB hotspots, the mean YRI-derived recombination rate is higher than the mean CEU-derived recombination rate (fig. S8). In contrast, PRDM9A-defined DSB hotspots are well represented in both CEU and YRI specific LD-hotspots with 52% overlapping hotspots in both populations (Fig. 3B). While the majority of DSB hotspots for each PRDM9 allele are found at LD-hotspots, 27% of PRDM9A-defined and 44% of PRDM9C-defined DSB hotspots are not found at an LD-hotspot in either population (Fig. 3B). We did find, however, that >80% of these DSB hotspots were located in a region with an elevated recombination rate (Fig. 3C, 1C(a)), suggesting that these hotspots have been active in human populations, but were simply below the detection thresholds used for LD-hotspot detection. Together, the DSB maps for different PRDM9 alleles clearly show that the population-averaged LD map is a combination of allele-specific maps.
Inter-individual variability in hotspot strength
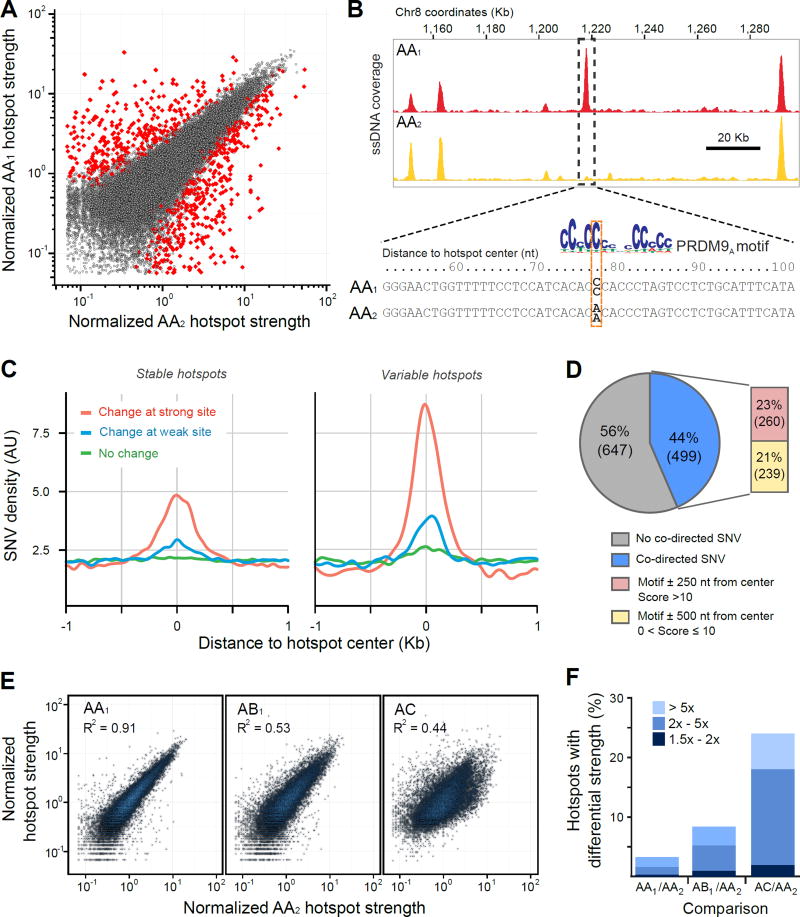
Although hotspot strength has been shown to vary between individuals sharing the same PRDM9 genotype (8, 9) these analyses were based on comparisons of crossover frequency at a limited number of strong hotspots and the extent of this variation genome-wide is not known. To explore variation in hotspot strength on a genome-wide scale, we compared our DSB maps between the AA1 and AA2 individuals (Fig. 4A; see Methods). Conservatively, we estimated that at least 3.2% (1,146) of hotspots varied in strength (1.25 to >30 fold; fig. S9), although the true proportion may be higher (fig. S9A).
Figure 4. Variation in hotspot strength between the AA1 and AA2 individuals.
(A) A comparison of PRDM9A-defined hotspot strength in the AA1 and AA2 individuals. Variable hotspots that satisfy our statistical criteria are shown in red. (B) Example of a variable hotspot between the two AA individuals where a homozygous SNV disrupts a PRDM9 motif near the hotspot center. The PRDM9 motif match is better in AA1 than in AA2 and the change in hotspot strength is co-directed with the change in motif score. (C) Motif-affecting SNVs are enriched around hotspot centers. The SNV density was smoothed using a gaussian kernel estimation and is shown for variants that change the motif score at a strong putative PRDM9 binding site (score > 10; red), at a weak putative PRDM9 binding site (0 < score ≤ 10; blue) or that do not change a putative binding site (green). Only changes > 1 bit are considered. Y-axis is expressed in arbitrary units (AU). Separate plots for stable (N=35,228; left) and for variable (N=1,146; right) hotspots from the AA1/AA2 comparison are shown. (D) The proportion of variable hotspots that contain a co-directed motif-affecting SNV near the hotspot centers. (E) PRDM9A-defined hotspot strength in the AA1, AB1 and AC individuals compared to the AA2 individual. Here, we used 15,051 PRDM9A-defined hotspots common to all four individuals. (F) Quantification of variable PRDM9A-defined hotspots between individuals. Bar colors indicate fold change.
Multiple factors may affect the DSB initiation frequency at a given hotspot. These include the DNA binding affinity of PRDM9, accessibility of the PRDM9 binding site and modulation of SPO11 recruitment and DNA cleavage. To determine how inter-individual sequence variation at PRDM9 binding sites affects DSB hotspot strength (Fig. 4B), we performed whole genome sequencing of the AA1, AA2 and AB individuals and identified 6.3 million sequence variants that differed from the reference genome (table S4). We then calculated the PRDM9 DNA-binding motif match scores associated with each variant. Motif-changing sequence variants were strongly enriched around the center of variable hotspots (Fig. 4C) and changes in motif score were positively correlated with changes in hotspot strength genome-wide (fig. S10). Furthermore, the enrichment of motif-score changing SNVs around hotspot centers is primarily driven by "co-directed" SNVs, where the change in motif score matches the direction of change in hotspot strength (fig. S11).
To estimate the proportion of variable hotspots likely explained by sequence variation at PRDM9 binding sites, we first evaluated the spatial distribution and strength of putative functional PRDM9 sites (sites found inside hotspots). We estimate that >70% of functional PRDM9 binding sites lie within 250 nt of hotspot centers while >92% lie within 500 nt (fig. S12A). 99% of hotspots contain a PRDM9 motif match with a score >10 within 250 nt of center (fig. S12B, C). However, analysis of PRDM9 motif loss through evolution suggests that even motifs with a score between 0 and 5 have been functionally active (fig. S12D). In total, 88% of variable hotspots differ in at least one sequence position between the genomes of AA1 and AA2 (table S5) providing an upper limit of proportion of variable hotspots caused by variation at PRDM9 binding sites. More realistically, between 23% (score > 10 within 250 nt from center) and 44% (score > 0 within 500 nt from center) of variable hotspots are likely explained by sequence differences at PRDM9 binding sites (Fig. 4D). Since sequence variation explains less than 44% of variable hotspots, other factors such as binding site accessibility must strongly affect DSB initiation frequency. The relatively minor impact of sequence variation at DNA binding sites is not unique to meiotic DSB hotspots; for transcription factors, sequence changes at putative DNA binding sites only explain a small fraction of differential transcription factor occupancy (36, 37).
Next, we asked if variable hotspots detected in the AA1/AA2 comparison were specific to these individuals or rather were polymorphic among humans. Individual-specific hotspots will not have left a footprint on historical recombination rates, and therefore we compared the LD-defined recombination rate between variable and stable hotspots. We observed little difference between the mean recombination rates (fig. S13), suggesting that few variable hotspots are specific to AA1 or AA2. In addition, over 60% of co-directed SNVs within 250 nt from center are commonly found (minor allele frequency >0.05) in both the CEU and YRI populations (fig. S14), suggesting that most variable hotspots driven by variation at the PRDM9 binding site are polymorphic in humans.
PRDM9 heterozygosity modulates hotspot strength
DSB hotspots can vary in strength between individuals homozygous for the PRDM9A allele. We next asked if a heterozygous combination of PRDM9 alleles contributes to hotspot strength variation at PRDM9A-defined hotspots. We compared hotspot strength in the AA1, AB and AC individuals to the strength in AA2 and found a much higher proportion of variable PRDM9A-defined hotspots in the AB and AC individuals (8.5%, 24.0%) compared to AA1/AA2 (3.5%; Fig. 4E, F, fig. S15). This difference is unlikely to be caused by sequence variation in PRDM9 binding sites, as the number of potential motif-disrupting sequence variants is similar in the AA and AB individuals (fig. S16; table S5). In the case of the PRDM9A/PRDM9B heterozygous individual, it is possible that the PRDM9B ZNF array has slightly different binding preferences compared to PRDM9A and therefore the apparent hotspot strength variation might be just a reflection of these changes in binding affinity. However, we do not observe any substantial changes in PRDM9B binding specificity (33) (fig. S3), and such a model cannot account for increased hotspot variation in the PRDM9A/PRDM9C individual.
One way to explain the increased hotspot variability in the AC individual is interference between neighboring hotspots. Indeed, correlated changes in crossover frequency at neighboring hotspots have been observed in humans (38). In Saccharomyces cerevisae there is also abundant evidence that the activation of a nearby hotspot can affect DSB frequency at other hotspots in its vicinity (39–42). Chromatin modifications following DSB formation could provide a mechanistic basis for such effects. For example, H2AX phosphorylation occurs rapidly following DSB formation and can span megabase-sized domains in mammals (43). This distance is clearly sufficient to affect nearby hotspots.
Alternatively, interactions between PRDM9 monomers, either direct or mediated by co-factors, may affect the DNA binding activity of PRDM9 and thus explain the increased hotspot strength variance in individuals heterozygous for PRDM9. Such cooperative interactions could modulate hotspot strength without changing binding specificity and may result in partial dominance (see (44) for discussion). In the case of PRDM9 binding we have clear partial dominance of one allele over the other (fig. S17) (4). Although we cannot clearly establish the mechanism of hotspot strength modulation, this effect is not restricted to humans. In a mouse hybrid derived from a 9Rx13R F1 cross we also observe increased variance in hotspot strength in heterozygous F1 animals relative to PRDM9 homozygotes (fig. S18). Taken together, these observations suggest that the presence of a second PRDM9 allele can influence DSB hotspot strength.
The mutagenic effect of meiotic recombination
Meiotic recombination influences genome evolution through the shuffling of parental alleles, and broad scale recombination rates are positively correlated with genetic diversity (45). At finer scales, the recombination rate has also been found to positively correlate with genetic diversity in humans (46, 47). However, the use of polymorphisms themselves to infer LD-defined recombination rates may confound such analyses (15). In order to better understand how meiotic recombination influences genome diversity at a local scale, we explored the patterns of DNA variation around DSB hotspots.
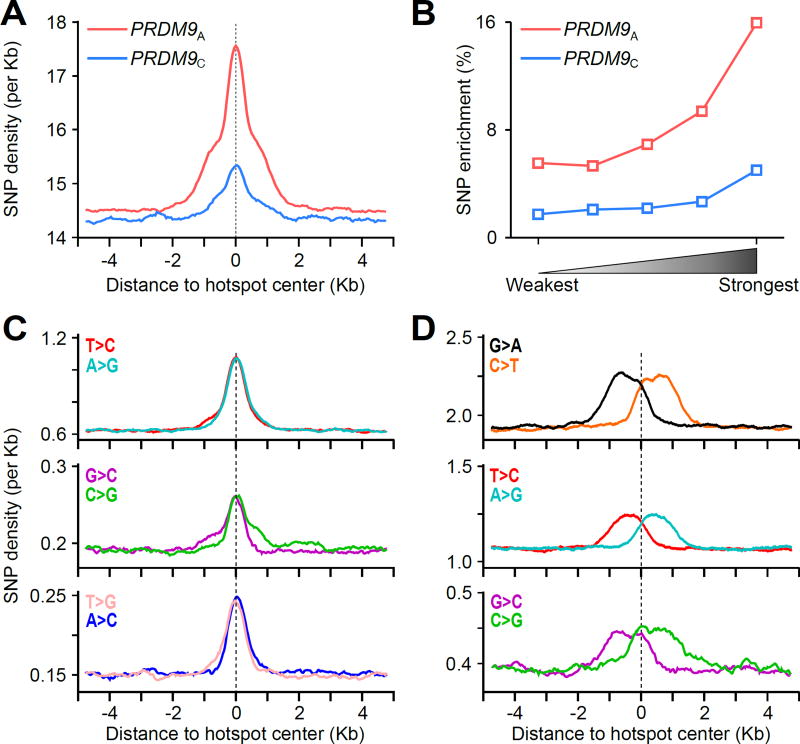
Our initial analyses revealed a local increase in single nucleotide polymorphism (SNP) density in the ~3 Kb region around both PRDM9A- and PRDM9C-defined hotspot centers (Fig. 5A). This local increase in SNP density is likely a direct consequence of meiotic DSBs as the magnitude of enrichment was positively correlated with hotspot strength (Fig. 5B). In addition, SNP enrichment reflects historical hotspot usage, as enrichment at PRDM9A-defined hotspots was three times greater than at PRDM9C-defined hotspots. To account for the effects of selection and population history, we investigated the distribution of SNPs with different derived allele frequencies (DAFs) around DSB hotspots. In general, common SNPs represent relatively old mutations that have become established in the population as a result of selection or genetic drift. Rare variants are less likely to be influenced by selection therefore they will more accurately reflect the spectrum of mutagenesis events (48). Parsing by DAF revealed two distinct spatial profiles of SNP enrichment, each of which was correlated with hotspot strength (fig. S19, S20); a signal in the central ± 0.5 Kb derived primarily from common variants, and a broader signal extending to the ± 1.5Kb shoulders of hotspots, derived from rare variants.
Figure 5. Signatures of increased genetic diversity at DSB hotspots.
(A) A local increase in SNP density is observed at both PRDM9A- (red) and PRDM9C-defined (blue) hotspots. All SNPs from the 1,000 genomes project were used. (B) The magnitude of SNP enrichment at hotspots is positively correlated with hotspot strength. SNP enrichment is calculated as the SNP density in the central ± 1.5 Kb relative to the mean SNP density in the region from 4 to 5 Kb from the hotspot center. (C) Common AT>GC and GC>CG variants are enriched in the ± 0.5 Kb region around the hotspot center. (D) Rare variants are enriched in a region ± 1.5 Kb around the hotspot center. Only variants enriched at hotspots are shown. Variants exhibit rotational symmetry around the hotspot center.
Among common variants (DAF >0.01), AT>GC, AT>CG and GC>CG variants were enriched in the narrow central ±0.5 Kb region of hotspots (Fig. 5C, fig. S21). This polymorphism spectrum is indicative of GC-biased gene conversion (gBGC) (49) while the 1 Kb extent of this signature closely approximates recent measurements of gBGC at mouse hotspots (50). Consistent with gBGC, we observe fixation of and enrichment for GC nucleotides at the hotspot center (fig. S22). The polymorphism spectrum of enriched rare variants (DAF < 0.01) was quite different from that observed for common variants. Among rare variants C>T (G>A) transitions, T>C (A>G) transitions, and to a lesser extent C>G (G>C) transversions were enriched in the broad ±1.5 Kb region around the hotspot centers (Fig. 5D; fig. S21). Like DNA resection, the rare polymorphism spectrum exhibited 180° rotational symmetry around the hotspot center (Fig. 5D; fig. S21, S23). Together with the strength-dependence of enrichment (fig. S19) this symmetry suggests that these variants arose directly from DSB repair processes. A more detailed analysis of the tri-nucleotide context of SNPs shows that a majority of rare C>T and G>A variants occurred at ancestral CpG dinucleotides (fig. S24A, B). Nevertheless, the polarity of these variants around DSBs makes it unlikely that cytosine deamination, a major mutagenic mechanism affecting methylated CpG dinucleotides (51), was the mechanism of their formation (fig. S24C). Exactly which mechanism drives this diversity remains unclear, however error prone DNA synthesis by trans-lesion polymerases may have a role in meiotic DSB repair (52). Together, our data show unequivocally that meiotic DSB repair processes increase local genetic diversity by both gBGC and by mutagenesis.
DSB frequency is a major determinant of crossover rate
In meiosis, a DSB can be repaired as either a crossover or as a non-crossover (Fig. 6A)(26). Since the proportion of DSBs resolved as crossovers might vary from hotspot to hotspot (11), the frequency of crossing over need not necessarily reflect the DSB formation rate. We thus asked whether the crossover landscape is largely shaped by variation in crossover/non-crossover resolution or if it is mostly determined by the DSB frequency.
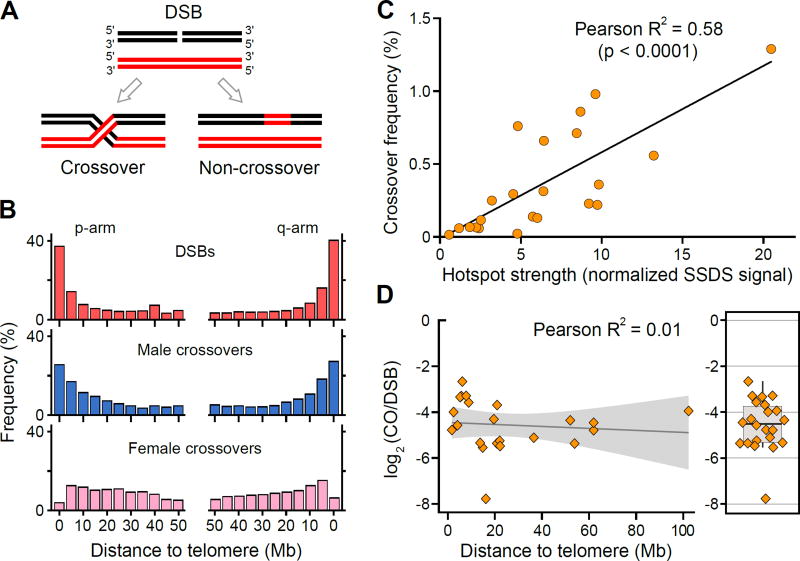
Figure 6. DSB frequency is correlated with the crossover rate.
(A) Meiotic DSBs can be repaired as either a crossover or a non crossover. (B) The distribution of PRDM9A-defined DSBs from the AA2 individual (red) and broad scale distribution of male (blue) and female (pink) derived crossovers from (18). (C) The average DSB frequency from the two AA individuals is correlated with crossover frequency at 22 PRDM9A-defined hotspots (P < 10−4, two tailed t-test). Crossover frequency is taken from (9) and (32) (D) The CO:DSB ratio is not strongly dependent on the distance to the telomere. The CO:DSB ratio for each of the hotspots analyzed in (C) is plotted against the absolute distance of the hotspot to the closest telomere. The shaded region represents the standard error for the linear fit line. The boxplot (right) illustrates that the CO:DSB ratio is highly variable among individual hotspots.
It has been well established that crossovers in human males are enriched in subtelomeric regions (16, 18, 19, 22, 23, 25, 53) (Fig. 6B, fig. S25). Here we found that DSB hotspots were also stronger and more densely spaced in the distal parts of chromosomes (Fig. 6B, fig. S1, S26). This enrichment spans approximately 10 Mb, independent of chromosome size (fig. S27). Quantitavely, at the megabase scale, the SSDS signal was strongly correlated to the male (Pearson R2 = 0.96, n = 14, p < 0.0001) but not to the female crossover frequency (18) (R2 = −0.07, n = 14, p = 0.36) (Fig. 6B). Furthermore, at PRDM9A-defined hotspots, we also found a positive correlation between the SSDS signal and the LD-defined recombination rates (Spearman R2 = 0.2) (33) despite the influence of female recombination and hotspot erosion on these rate estimates. The relationship between crossover frequency and SSDS signal remains strong at the level of individual hotspots. We find that the mean SSDS signal in AA individuals is strongly correlated with the mean crossover frequency determined by sperm genotyping (Pearson R2 = 0.58; p < 0.0001; Fig. 6C, table S3) (9, 32) although the CO:DSB ratio varies among individual hotspots. The preferential resolution of DSBs as COs near telomeres may also contribute to the observed CO distribution and we therefore analyzed the relationship between CO:DSB ratio and proximity to telomeres. Based on the limited set of available hotspots we found no strong evidence that the CO:DSB ratio depends on the distance to the telomere (Fig. 6D), although non-linear fitting suggests that the CO:DSB ratio may be higher near the telomere (fig. S28A). This increase could be driven by stronger CO hotspots (fig. S28B) which are themselves known to be enriched near telomeres, therefore more data will be required to establish if the CO:NCO ratio is elevated in telomere adjacent regions. Taken together, though biased CO/NCO resolution may contribute, our per-hotspot and genome-wide observations collectively indicate that crossover frequency is largely determined by the rate of DSB formation.
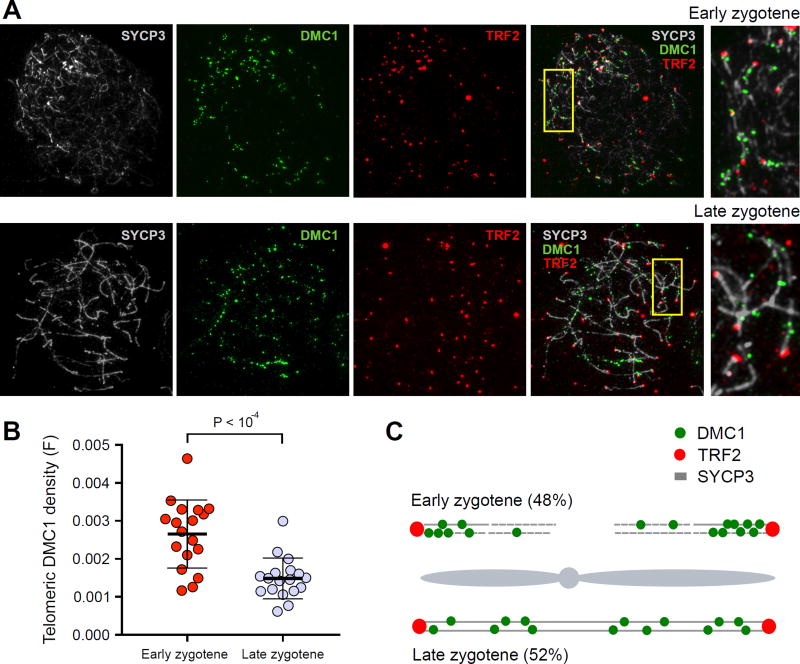
The SSDS signal is a reflection of DSB frequency, but it may be influenced by the rate at which DSBs are repaired. To obtain an independent estimate of DSB frequency we performed immunostaining for DMC1, a marker for meiotic DSBs, and quantified the distribution of DMC1 foci with respect to telomeres. We compared the density of DMC1 foci in the telomere proximal area to foci density in interstitial regions and found that early in meiosis, when DMC1 foci are first detected (early zygotene), there is a ~1.8-fold excess of DMC1 foci in the telomere proximal region (fig. S29A). Later in zygotene, though the number of DMC1 foci remains similar (early zygotene: 185 ± 37 (SD); late zygotene: 166 ± 41 (SD); P = 0.27, two-tailed Mann Whitney), the proportion of telomere-proximal DMC1 foci decreases (Fig. 7A, B, fig. S29, S30). Though we cannot rule out that some sub-telomeric DSBs have extended lifespan, our consistent observation of a decrease in DMC1 foci density near telomeres using different approaches, indicates that most of these DSBs do not persist as meiosis progresses. Taken together, our observations suggest that increased DSB formation close to telomeres in early zygotene shapes the global DSB distribution in our genome-wide maps (Fig. 7C, fig. S31).
Figure 7. Increased frequency of DSB formation near telomeres.
(A) Early and late zygotene spermatocytes stained with SYCP3 (which detects axial elements) in grey, DMC1 (a marker of DSBs) in green, and TRF2 (a marker of telomeres) in red. DMC1 foci are clustered near telomeres at early zygotene. (B) Telomere-proximal DMC1 density is about 2-fold higher in early compared to late zygotene cells. For each cell, we manually traced all chromosome axes that unambiguously initiated at a TRF2 focus. We defined telomere-proximal regions as the 1μm of axis adjacent to the TRF2 focus (Fig. S30A). For each cell, the telomeric density of DMC1 foci (F) was calculated as ((Σ DMC1 foci in telomere proximal 1μm regions) / (total DMC1 foci) / (total TRF2 foci)). We do not count DMC1 foci adjacent to TRF2 foci that could not be unambiguously attributed to a particular chromosomal axis. This is particularly punitive for early cells as the DMC1 density appears highest in regions where TRF2 foci are clustered and individual axes are difficult to distinguish. Error bars indicate mean ± SD. P-values are calculated using a Mann-Whitney U-test. (C) Cartoon showing the distribution of DMC1 foci in early and late zygotene spermatocytes. 48% of zygotene cells are at the early stage where they show significantly higher frequency of DSB formation near telomeres. At late zygotene, DSBs are more evenly distributed. The combined signal from these two populations may result in the telomeric bias we observe in our genome-wide maps.
We finally explored other aspects of DSB formation that may contribute to increased hotspot strength in subtelomeric regions. The PRDM9-dependent H3K4me3 ChIP-Seq peak intensity is correlated with hotspot strength in mice (4, 54), therefore we examined the H3K4me3 ChIP-Seq signal at human hotspots. We found that despite increased hotspot strength in distal regions (fig. S26B, S27, S32A) the H3K4me3 signal at hotspots did not increase (fig. S32B). Furthermore, the correlation between hotspot strength and H3K4me3 signal decreased in distal relative to interstitial regions (fig. S32C). This suggests that in subtelomeric regions, PRDM9 may define DSBs independent of its methyltransferase activity or that in these regions, other factors such as the chromatin environment and/or proximity to the nuclear envelope, may modulate hotspot strength. Given the biased distribution of crossovers in males but not in females, such modulation may be acting uniquely in the male germ line.
Meiotic recombination drives genome instability
Meiotic recombination has been implicated as a potential source of gross structural variants (SVs) (55) therefore we examined the association between meiotic DSBs and SVs. We found that SVs generated by homology-based mechanisms (non-allelic homologous recombination (NAHR) or shrinking or expansion of variable number tandem repeats (VNTR)) (55) were enriched at PRDM9A-defined hotspots (table S6). Structural rearrangements derived from unequal crossovers are known to cause genomic diseases (56), therefore we asked whether disease-causing SV breakpoints occur at DSB hotspots. We found that 14 out of 27 disease-associated breakpoints that have been mapped to < 1.5 Kb coincided with a PRDM9A-defined hotspot (table S7). These hotspot-associated breakpoints include those responsible for X-linked ichthyosis (fig. S33A), Charcot-Marie-Tooth disease, Hunter and Potocki-Lupski/Smith-Magenis syndromes (fig. S33B) among others (table S7). No disease-causing SV breakpoints coincided with PRDM9C-defined DSB hotspots. This implies that individuals homozygous for the PRDM9C allele are not at risk for these and highlights the utility of PRDM9 genotyping for future studies of genomic disorders. Most of these disease-associated breakpoints occur within directly paralagous (DP-LCRs) or inverted paralagous low copy repeats (IP-LCRs), genomic regions which are susceptible to NAHR-mediated recombination events (57, 58). The 726 PRDM9A-defined hotspots that occur at DP/IP-LCR regions (file S1) represent targets for future research into human genetic disease.
Conclusions
We have generated comprehensive maps of meiotic recombination initiation in human individuals. Our comparison of recombination initiation maps between individuals sharing PRDM9 alleles clearly demonstrates that the recombination initiation frequency varies between individuals on a hotspot level. We can explain less than half of the variation in hotspot intensity by sequence changes at PRDM9 binding sites. This suggests that the chromatin environment or other factors mediate the rate of recombination initiation. Our maps allowed us to deduce genome-wide sets of human hotspots defined by different alleles of the PRDM9 protein. A comparison of DSB hotspot maps with LD-based map suggests that an LD-based recombination map is an apparent superposition of allele-specific maps and indicates that a significant proportion of LD-defined hotspots are defined by minor PRDM9 alleles. Unlike previous methods that rely on the detection of only crossovers, we mapped hotspots by directly identifying the sites of early DSB repair intermediates. Our observations indicate that the DSB frequency itself is largely shaping the crossover distribution.
The high resolution of DSB hotspot mapping allowed us to carefully evaluate the impact of meiotic recombination on genome evolution. We have found clear evidence for GC-biased gene conversion and recombination-associated mutagenesis at sites of DSB hotspots. SVs associated with genomic disease are clearly associated with DSB hotspots defined by the PRDM9A, but not by the PRDM9C allele suggesting that the PRDM9 genotype should be considered in assessing predisposition to genomic disorders. Taken together, our data open a broad window into future studies of human recombination, meiosis and genome evolution and provide a rich data source for future research into human genomic disease.
Methods Summary
We performed SSDS (27) using antibodies against DMC1 (Santa Cruz; C-20, sc 8973), to identify meiotic DSB hotspots in testis tissue from five individual human males. The genotype at the PRDM9 locus was established in each of these individuals using the primers and method described in (9). SSDS samples were compared to a matched control and DSB hotspots were defined using MACS 2.0.10 (59). Whole genome sequencing libraries were prepared for three individuals according to an established protocol (Illumina). GATK best practices (60) were followed to identify variants using the GATK Genome Analysis Toolkit v2.3.9 (61). Chromatin immunoprecipitation using antibodies against H3K4me3 (Abcam; ab8580) followed by high throughput sequencing was performed to identify sites of trimethylated H3K4 in one individual. Peaks in H3K4me3 data were called using SICER v1.1 (62). All high throughput sequencing was performed on an Illumina HiSeq 2500. Spermatocyte spreads were prepared for immunofluorescence microscopy using the method described in (63). Detailed methods are available in Supplementary Materials.
Supplementary Material
Acknowledgments
We would like to thank Harold Smith and the NIDDK Genomics Core for sequencing assistance. We thank Michael Lichten and Peggy Hsieh for critical feedback on the manuscript and also extend our gratitude to Professor Sir Alec Jeffreys for providing details of human crossover rates. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD. (http://biowulf.nih.gov). This research was supported by NIH grant 1R01GM084104-01A1 from NIGMS (G.V.P.), March of Dimes grant # 1-FY13-506 (G.V.P.) and by the NIDDK Intramural Research Program (R.D.C.O.). The sequencing data reported in this paper are archived at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) as accession number GSE59836.
Footnotes
Materials and Methods
Figs. S1-S33
Tables S1-S7
References (64–103): [Note: These numbers refer to references cited only within the Supplementary Materials]
File S1
References and Notes
- 1.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 6.de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563–599. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 7.Oliver PL, et al. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS genetics. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg IL, et al. Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. Proc Natl Acad Sci U S A. 2011;108:12378–12383. doi: 10.1073/pnas.1109531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg IL, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42:859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007;15:565–577. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- 12.McVean GA, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 13.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 14.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The 1000 Genomes Project Consortium, A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong A, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 17.Kong A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 18.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 19.Khil PP, Camerini-Otero RD. Genetic crossovers are predicted accurately by the computed human recombination map. PLoS Genet. 2010;6:e1000831. doi: 10.1371/journal.pgen.1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinch AG, et al. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnheim N, Calabrese P, Nordborg M. Hot and cold spots of recombination in the human genome: the reason we should find them and how this can be achieved. Am J Hum Genet. 2003;73:5–16. doi: 10.1086/376419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu S, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou Y, et al. Genome analyses of single human oocytes. Cell. 2013;155:1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 25.Kirkness EF, et al. Sequencing of isolated sperm cells for direct haplotyping of a human genome. Genome Res. 2013;23:826–832. doi: 10.1101/gr.144600.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cole F, Keeney S, Jasin M. Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol Cell. 2010;39:700–710. doi: 10.1016/j.molcel.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 2012;22:957–965. doi: 10.1101/gr.130583.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen-Seaman MI, et al. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004;14:528–538. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thacker D, Mohibullah N, Zhu X, Keeney S. Homologue engagement controls meiotic DNA break number and distribution. Nature. 2014;510:241–246. doi: 10.1038/nature13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver-Bonet M, Turek PJ, Sun F, Ko E, Martin RH. Temporal progression of recombination in human males. Mol Hum Reprod. 2005;11:517–522. doi: 10.1093/molehr/gah193. [DOI] [PubMed] [Google Scholar]
- 31.Moens PB, et al. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma. 1997;106:207–215. doi: 10.1007/s004120050241. [DOI] [PubMed] [Google Scholar]
- 32.Webb AJ, Berg IL, Jeffreys A. Sperm cross-over activity in regions of the human genome showing extreme breakdown of marker association. Proc Natl Acad Sci U S A. 2008;105:10471–10476. doi: 10.1073/pnas.0804933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Materials and methods are available as supporting material on Science Online.
- 34.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 35.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 36.Reddy TE, et al. Effects of sequence variation on differential allelic transcription factor occupancy and gene expression. Genome Res. 2012;22:860–869. doi: 10.1101/gr.131201.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tiemann-Boege I, Calabrese P, Cochran DM, Sokol R, Arnheim N. High-resolution recombination patterns in a region of human chromosome 21 measured by sperm typing. PLoS Genet. 2006;2:e70. doi: 10.1371/journal.pgen.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu L, Kleckner N. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 1995;14:5115–5128. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu TC, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocco V, Nicolas A. Sensing of DNA non-homology lowers the initiation of meiotic recombination in yeast. Genes Cells. 1996;1:645–661. doi: 10.1046/j.1365-2443.1996.00256.x. [DOI] [PubMed] [Google Scholar]
- 42.Fan QQ, Xu F, White MA, Petes TD. Competition between adjacent meiotic recombination hotspots in the yeast Saccharomyces cerevisiae. Genetics. 1997;145:661–670. doi: 10.1093/genetics/145.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iacovoni JS, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bost B, Veitia RA. Dominance and interloci interactions in transcriptional activation cascades: models explaining compensatory mutations and inheritance patterns. Bioessays. 2014;36:84–92. doi: 10.1002/bies.201300109. [DOI] [PubMed] [Google Scholar]
- 45.Nachman MW. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 2001;17:481–485. doi: 10.1016/s0168-9525(01)02409-x. [DOI] [PubMed] [Google Scholar]
- 46.Spencer CC, et al. The influence of recombination on human genetic diversity. PLoS Genet. 2006;2:e148. doi: 10.1371/journal.pgen.0020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Auton A, et al. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimura M, Ota T. The age of a neutral mutant persisting in a finite population. Genetics. 1973;75:199–212. doi: 10.1093/genetics/75.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 50.Clément Y, Arndt PF. Meiotic Recombination Strongly Influences GC-Content Evolution in Short Regions in the Mouse Genome. Mol Biol Evol. 2013 doi: 10.1093/molbev/mst154. [DOI] [PubMed] [Google Scholar]
- 51.Ehrlich M, Wang RY. 5-Methylcytosine in eukaryotic DNA. Science. 1981;212:1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- 52.Arbel-Eden A, et al. Trans-lesion DNA Polymerases may be Involved in Yeast Meiosis. G3 (Bethesda) 2013 doi: 10.1534/g3.113.005603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barlow AL, Hultén MA. Crossing over analysis at pachytene in man. Eur J Hum Genet. 1998;6:350–358. doi: 10.1038/sj.ejhg.5200200. [DOI] [PubMed] [Google Scholar]
- 54.Smagulova F, et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mills RE, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dittwald P, et al. NAHR-mediated copy-number variants in a clinical population: mechanistic insights into both genomic disorders and Mendelizing traits. Genome Research. 2013 doi: 10.1101/gr.152454.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dittwald P, et al. Inverted low-copy repeats and genome instability--a genome-wide analysis. Hum Mutat. 2013;34:210–220. doi: 10.1002/humu.22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van der Auwera G, et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Vol. 43. Wiley Online Library, Current Protocols in Bioinformatics; 2013. pp. 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Page J, et al. Inactivation or non-reactivation: what accounts better for the silence of sex chromosomes during mammalian male meiosis? Chromosoma. 2012;121:307–326. doi: 10.1007/s00412-012-0364-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.