Abstract
The light strand promoter of mammalian mitochondrial DNA gives rise to a primary transcript, but also to the RNA primer necessary for initiation of replication and 7S DNA synthesis as well as 7S RNA. Here we have studied the turnover of 7S DNA in isolated rat liver mitochondria and whether import of mitochondrial transcription factor A (mtTFA), which is necessary for transcription initiation, increases its rate of synthesis. 7S DNA was present as two species, probably due to two different sites of RNA–DNA transition. Time course and pulse–chase experiments showed that the half-life of this DNA is ∼45 min. Import of mtTFA, produced in vitro, into the mitochondrial matrix in stoichiometric amounts significantly increased the rate of 7S DNA formation. We conclude that isolated rat liver mitochondria faithfully synthesize and degrade 7S DNA and that increased matrix levels of mtTFA are sufficient to increase its rate of synthesis, strongly supporting the hypothesis that this process is transcription primed.
INTRODUCTION
In all vertebrates the mitochondrial DNA (mtDNA) molecule contains a non-coding region that harbors the promoters for transcription initiation and the origin of H-strand synthesis (OH), which is the leading strand during the orthodox, strand asynchronous mode of replication (for reviews see 1,2). In mammals transcription occurs from two adjacent promoters, designated the light strand promoter (LSP) and heavy strand promoter (HSP), depending on which strand serves as the template. In this region an arrested nascent H-strand (7S DNA) remains stably hybridized to the circular parental molecule, forming a triple-stranded structure characterized by a displaced H-strand (D-loop). The location of the LSP directly upstream of OH suggested a role for this promoter in initiation of H-strand synthesis. Indeed, precise mapping of RNA and DNA species in the D-loop region provided compelling evidence that RNA derived from the LSP serves as a primer for H-strand DNA replication and 7S DNA synthesis and, after polyadenylation, accumulates as 7S RNA (3,4). Comparison of the nucleotide sequences of human, mouse and rat mtDNA downstream of the LSP revealed three conserved sequence blocks (CSB I, II and III) where RNA–DNA transition events occur. The 3′-ends of the chimera are found at the ‘termination-associated sequences’ (TAS), which are located further upstream of the CSBs. Formation of this RNA–DNA chimera requires CSB II and does not occur on templates that contain other types of mitochondrial promoters. This chimera is extremely short lived and has a very high ‘turnover’. Altogether, analysis of vertebrate systems has provided overwhelming evidence that a transcription primed mechanism is used to initiate mtDNA replication at OH (4). Thus, linkage of transcription from the LSP to H-strand mtDNA replication suggests that mtDNA copy number may ultimately be controlled by the frequency of transcription initiation at the LSP, in concert with other proteins involved in processes at the CSBs and TASs, at least in orthodox replication mode.
In addition to mitochondrial RNA polymerase, mitochondrial transcription factor A (mtTFA; 5) is the primary protein component necessary for transcription initiation from the two mtDNA promoters (6). This protein specifically recognizes mtDNA sequences located 19–39 bp upstream of each transcription initiation site and binding to these upstream sequences is required for transcriptional activation. Cloning of the genes for human and mouse mtTFA revealed that it is a member of the HMG box class of DNA-binding proteins, since it contains two tandem HMG box domains and possesses DNA-binding properties that are characteristic of this class of proteins (7). The principal features of the HMG domain are its interaction with the minor groove of DNA and its capacity to distort DNA structure dramatically upon binding. Two additional regions, distinct from the HMG boxes, are present in human mtTFA. One is a 27 residue linker between the two HMG boxes and the other is a 25 residue C-terminal tail; both of these regions are rich in basic amino acid residues (8).
In vitro mtTFA is necessary and sufficient to stimulate transcription from both mammalian mtDNA promoters (9). We have shown that overexpression of mtTFA in HeLa cells as well as import into isolated rat liver mitochondria stimulates mitochondrial transcription from the HSP (10). Thus, we have concluded that a simple increase in intra-mitochondrial level of mtTFA is sufficient to increase transcription rate and that mtTFA is indeed a regulator of mitochondrial gene expression in vivo. Here we report that mtTFA, produced in vitro and imported into isolated rat liver mitochondria, is able to stimulate the synthesis of 7S DNA in this system in which transcription, replication and 7S DNA synthesis closely resemble the processes in vivo.
MATERIALS AND METHODS
Isolation of rat liver mitochondria
Adult male Wistar rats (200–300 g) were anesthetized with CO2 and killed by decapitation. The liver was rapidly removed, chilled in homogenization medium (0.32 M sucrose, 1 mM K-EDTA, 10 mM Tris–HCl, pH 7.4) and cut into small pieces. All further steps were carried out at 4°C using sterile solutions and glassware as described previously (11). Briefly, the minced tissue was resuspended in the same medium (4 ml/g liver) and homogenized in a Potter–Elvejhem homogenizer rotating at 750 r.p.m. using between two and four up-and-down strokes. The homogenate was spun at 1000 g for 5 min. Aliquots of 16 ml of the supernatant were placed in reaction tubes and centrifuged at 13 000 g for 2 min in a table centrifuge. The mitochondrial pellets were resuspended in 8 ml of homogenization buffer and pelleted again. This was repeated three times, each time reducing the buffer volume used for resuspension by half. The final mitochondrial pellet was resupended in incubation buffer (see below). The washes employed in this preparation scheme eliminate contaminating cytoplasmatic RNA. The mitochondrial protein concentration was determined by Waddell’s method (12).
Synthesis and analysis of DNA in isolated rat liver mitochondria
Mitochondrial protein (1 mg) was resuspended in 0.5 ml of incubation buffer (25 mM sucrose, 75 mM sorbitol, 100 mM KCl, 10 mM K2HPO4, 0.05 mM EDTA, 5 mM MgCl2, 1 mM ADP, 10 mM glutamate, 2.5 mM malate, 10 mM Tris–HCl, pH 7.4) also containing 1 mg/ml fatty acid-free bovine serum albumin (BSA), 50 µM each dATP, dCTP and dGTP and 20 µCi [α-32P]dTTP (800 Ci/mmol) (11). Incubation was carried out at 37°C for various times on a rotating wheel. For pulse–chase experiments, mitochondria were incubated with [α-32P]dTTP (final concentration 5 µM) for 30 min, followed by a chase with non-radiolabeled dTTP (5 mM) and further incubation for various times. After incubation, mitochondria were pelleted at 13 000 g for 1 min and washed twice with 10% glycerol, 10 mM Tris–HCl, pH 6.8, 0.15 mM MgCl2.
The pellet was then resuspended in 500 µl of pronase buffer (10 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA), lysed with 2% SDS in the presence of 100 µg pronase (autodigested to remove nucleases) and incubated for 15 min at 37°C. Total mitochondrial nucleic acids were extracted twice with an equal volume of a phenol/chloroform/isoamyl alcohol mixture (25:25:1) at room temperature. After ethanol precipitation and centrifugation, the pelleted nucleic acids were dissolved in 10 mM Tris–HCl, pH 7.4, 1 mM EDTA.
The labeled nucleic acids were denatured at 95°C for 15 min where indicated and analyzed by vertical 1% agarose slab gel electrophoresis (11). After the run, the gels were stained with ethidium bromide, photographed, dried and exposed to X-ray film.
Preparation of probes and Southern blot analysis
To prepare probes for Southern blot analysis, mtDNA extracted from rat liver mitochondria was used as template for PCR amplification. Primers for the D-loop region fragment were: sense primer, 5′-CCACCATCAACACCCAAAGCTG-3′ (nucleotides 15358–15380); antisense primer, 5′-TTTGGCATTGAAGTTTCAGGTG-3′ (nucleotides 16110–16088, EMBL/GenBank accession no. X14848) (13). The PCR reaction containing 0.5 µg rat mtDNA was heated for 1 min at 95°C. After 30 cycles of 90 s at 95°C, 90 s at 57°C and 90 s at 72°C, the temperature was cooled to 4°C. The PCR product was isolated from an agarose gel after staining with ethidium bromide and purified (Wizard purification kit; Promega). After purification, the PCR product was non-radioactively labeled (DIG-High-Prime; Boehringer Mannheim) and used to detect 7S DNA on Southern blots using an alkaline phosphatase-based system (CDP-Star; Boehringer Mannheim).
mtDNA extracted from isolated mitochondria was analyzed by Southern blot hybridization (14) after digestion with AccI and electrophoresis in a 1% vertical agarose slab gel. The unstained gel was transferred directly to a nylon membrane using a vacuum blotting system and immobilized. The PCR probe labeled to high specific activity was diluted in a volume of 4 ml of hybridization buffer (1% blocking reagent, 0.1 mg/ml ssDNA, 0.25% SDS, 0.2% PPi and 5× Denhardt’s solution). The membrane was hybridized at 68°C for 18 h in a hybridization oven. After hybridization, the membrane was washed for 2 × 15 min in 100 ml of washing solution 1 (2× SSC, 1% SDS) and for 2 × 10 min in 100 ml of washing solution 2 (0.2× SSC, 1% SDS) at 68°C and incubated for 30 min in blocking buffer (DIG-High-Prime). After blocking, the membrane was incubated for an additional 30 min with 20 ml of antibody solution and washed again several times. For detection, the membrane was equilibrated in 20 ml of 0.1 M Tris–HCl, 0.1 M NaCl, 50 mM MgCl2, pH 9.5. After equilibration, the membrane was incubated with CDP-Star and exposed to X-ray film for 15–20 min.
Import of mtTFA into isolated rat liver mitochondria
A coupled in vitro transcription/translation reticulocyte lysate system (Promega) was used to produce mtTFA. It was incubated for 1 h at 37°C with T7 polymerase and pBS-KS-mtTFA or pBS-KS. Import of mtTFA into mitochondria was studied in 0.5 ml of the incubation reaction described above in the presence of 1 mg of mitochondrial protein and 10% rabbit reticulocyte lysate containing 35S-labeled mtTFA at 30°C. Some samples were treated with carbonyl cyanide m-chlorophenylhydrazone (CCCP) (2 µM), an uncoupler of the mitochondrial inner membrane potential during incubation. After an incubation period of 30 min, samples were divided into aliquots and further incubated on ice for 30 min. Where indicated, Triton X-100 (1%) or proteinase K (0.25 mg/ml) was added. Finally, mitochondria were pelleted, dissolved in SDS sample buffer and separated by SDS–PAGE. Gels were stained, destained, dried and exposed to X-ray films.
For stimulation of 7S DNA synthesis, 7% (v/v) reticulocyte lysate containing mtTFA or mock incubated lysate as a control was added to the DNA synthesis reaction described above.
Determination of mtTFA concentration and mtDNA copy number
In order to determine the stoichiometry between imported mtTFA and mtDNA in the in organello DNA synthesis system, aliquots of individual reticulocyte reactions were run on SDS–PAGE gels (n = 3). After drying the gels on filter paper, they were exposed to X-ray film, the localized mtTFA bands were excised and incorporation of [35S]methionine was measured by liquid scintillation counting (c.p.m.). A quench curve was prepared by spotting known amounts of [35S]methionine in the range 102–106 d.p.m. onto the back of a similar filter paper on which an empty PAGE gel had been dried. c.p.m. was then converted into d.p.m. and, from the specific activity of [35S]methionine (d.p.m./mol) and the number of methionine residues in the mtTFA protein, the molar concentration of mtTFA in the reticulocyte lysate was calculated. The specific activity of methionine was corrected for unlabeled methionine present in the lysate, which was determined to be 3 µM by Dr F. Lottspeich (MPI Martinsried, Germany) using an amino acid analyzer.
For the determination of mtDNA copy number in rat liver mitochondria, mtDNA was isolated from duplicate samples of mitochondrial preparations (1 mg protein each) isolated from five rat livers (14). mtDNA obtained from these 10 samples was blotted onto nitrocellulose in four 2-fold dilution steps, so that they represented DNA obtained from 0.1, 0.05, 0.025 and 0.0125 mg of mitochondrial protein, using a multiwell filtration apparatus. Known amounts of the plasmid pBR322-CO III, containing the gene for rat mitochondrial CO III, were subjected to the same extraction procedure in the presence of BSA and nominally 150 fmol were blotted onto the same blot in 12 2-fold dilution steps (75 fmol down to 18 amol). Rat liver mtDNA had been fragmented by digestion with XbaI and BamHI, while pBR322-CO III had been linearized with PstI prior to acid denaturation, neutralization and blotting. The blot was hybridized to a radioactively labeled CO III probe derived from pBR322-CO III, exposed and quantitated densitometrically. Using exclusively data which were in the linear range of the function densitometric units/blotted DNA, an average value for the concentration of mtDNA/mg mitochondrial protein was obtained.
In addition, the concentration of the DNA solutions extracted from rat liver mitochondria was determined by measurement of their optical density at 260 nm.
RESULTS
DNA synthesis in isolated mitochondria
It was shown previously that isolated rat liver mitochondria, when incubated in an appropriate medium, transcribe mtDNA in a way that closely resembles the process in vivo. RNA synthesis occurred efficiently for more than 3 h and a non-polyadenylated, small RNA, which is considered to be the primer for synthesis of the H-strand of mtDNA, could be detected, although at very low levels (15). In addition, faithful replication of mtDNA occurs under the same conditions (11).
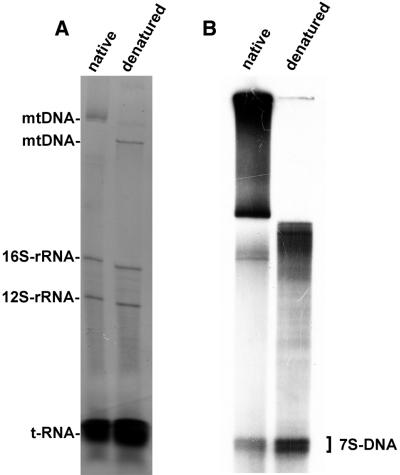
In order to study synthesis of 7S DNA, mitochondria were purified and incubated as described and the labeled nucleic acids were extracted and analyzed by electrophoresis. Gels were stained with ethidium bromide in order to document steady-state levels of mtDNA, 16S and 12S rRNAs and the tRNAs. 7S DNA and the non-polyadenylated 7S RNA precursor are present only in low amounts in rat mitochondria, precluding their detection by ethidium bromide staining (Fig. 1, left). An autoradiogram of the gel shows a multitude of species of high molecular weight DNA, relaxed circular molecules of mtDNA and catenanes (11) and two distinct, low molecular weight bands in native DNA. Denaturation by heating for 10 min at 95°C caused an increase in mobility of the high molecular weight DNA; the intensity of the two low molecular weight bands increased, suggesting that they are presumably additional 7S DNA molecules which had been melted from intact D-loop structures by this treatment (16). Digestion of mitochondrial DNA with DNase led to the disappearance of all labeled bands, while treatment with RNase A had no effect, neither on the quantity nor the mobility of the bands (data not shown). This shows that both low molecular weight bands are indeed DNA and that both are obviously not covalently linked to RNA.
Figure 1.
DNA synthesis in isolated rat liver mitochondria. (A) Ethidium bromide staining of total nucleic acids and (B) autoradiogram of labeled deoxyribonucleic acids isolated from rat liver mitochondria after incubation with [α-32P]dTTP followed by electrophoresis through a vertical 1% agarose slab gel. Nucleic acids were untreated (native) or heat denatured for 10 min at 95°C (denatured).
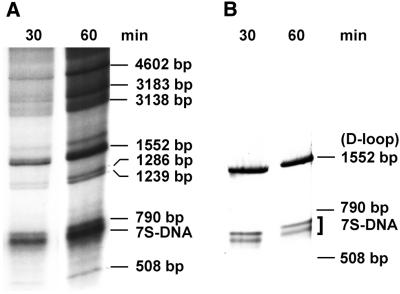
In order to identify these two bands as 7S DNA, newly synthesized labeled DNA was isolated and digested with AccI. The restriction pattern of the labeled products was that expected for rat mtDNA, leading to the appearance of eight bands (Fig. 2, left), and was identical to that obtained by ethidium bromide staining of purified rat mtDNA treated with the same enzyme (not shown). A Southern blot of isolated, restricted DNA was hybridized to a probe containing D-loop sequences (Fig. 2, right). As expected, this probe bound to the 1552 bp fragment containing the D-loop sequence, but also to the two low molecular weight bands, unequivocally identifying them as 7S DNA. These two molecules are present in rat liver mitochondria in approximately equal amounts, since both bands were of about the same intensity.
Figure 2.
Analysis of DNA synthesized in isolated rat liver mitochondria. (A) Autoradiogram of newly synthesized mtDNA after incubation with [α-32P]dTTP for 30 or 60 min. mtDNA was isolated after the indicated incubation times, digested with AccI at 55°C for 1 h and subjected to electrophoresis through a 1% agarose slab gel. (B) Hybridization of a Southern blot of a similar gel with a probe containing the D-loop sequence.
Metabolism of 7S DNA in isolated mitochondria
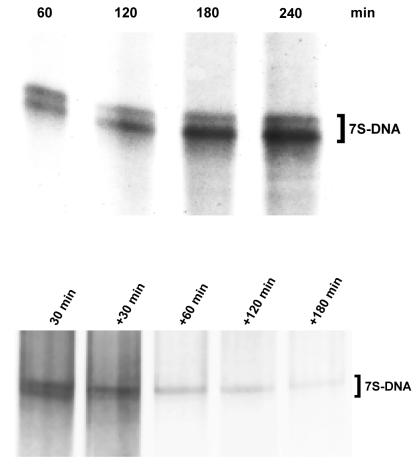
Incorporation of [α-32P]dTTP into 7S DNA was almost linear for 3 h, but did not increase further in the next hour (Fig. 3, upper). Three possible reasons could explain the cessation of label incorporation: (i) the capacity of the [α-32P]dTTP precursor pool is exhausted; (ii) because transport of nuclear encoded polymerase γ and other protein factors involved in 7S DNA synthesis is interrupted after the isolation procedure, mitochondria are unable to synthesize DNA for more than 3 h; (iii) the pools of 7S DNA and intra-mitochondrial [α-32P]dTTP have equilibrated. Since <10% of the [α-32P]dTTP pool had been incorporated after 3 h, the first explanation is very unlikely. In order to exclude the second possible explanation, mitochondria were pre-incubated for 2 h with unlabeled dTTP, then [α-32P]dTTP was added; the time course of labeling under these conditions was similar to the experiment depicted in Figure 3 (data not shown), showing that the rate of synthesis of 7S DNA was unchanged. Thus, we conclude that the cessation of labeling is due to equilibration between the intra-mitochondrial [α-32P]dTTP pool and 7S DNA after 3 h. This allowed us to perform pulse–chase experiments in order to estimate the half-life of 7S DNA in this system (Fig. 3, lower). Densitometric evaluation of band intensities allowed calculation of the half-life of 7S DNA. In the experiment shown, values for the upper band were 340 densitometric units after labeling and 235, 135, 120 and 49 units after 30, 60, 120 and 180 min of chase, respectively. Thus, its half-life was calculated to be ∼45 min. Interestingly, the lower 7S DNA band seems to turn over considerably faster, thus determination of its actual half-life was not feasible.
Figure 3.
Time course of 7S DNA synthesis and 7S DNA turnover in isolated rat liver mitochondria. (Upper) Autoradiogram showing incorporation of [α-32P]dTTP into 7S DNA after different times of incubation. DNA was isolated after the indicated incubation times and heat denatured for 10 min at 95°C before electrophoresis through a 1% agarose slab gel. (Lower) Autoradiogram showing incorporation of radioactivity into 7S DNA after 30 min pre-incubation with [α-32P]dTTP (pulse) followed by the indicated times of incubation with an excess of unlabeled dTTP (chase) (+ 30 to +180 min).
Import of mtTFA into isolated rat liver mitochondria
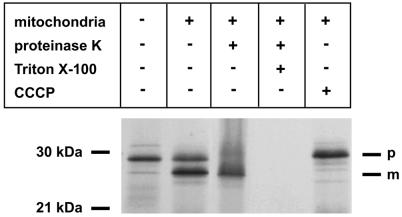
Human mtTFA was generated by in vitro translation in rabbit reticulocyte lysate and radiolabeled with [35S]methionine (Fig. 4). In the presence of mitochondria the 24 kDa mature protein (m) was generated from the 29 kDa mtTFA precursor (p). This was inhibited by the uncoupler CCCP, which was added during incubation. The 24 kDa protein was resistant to proteinase K treatment, while the 29 kDa precursor was accessible to digestion. After lysis of mitochondrial membranes by Triton X-100, both proteins were digested by proteinase K. This shows that the mtTFA precursor was imported into the mitochondrial matrix and processed to the mature form of the predicted molecular weight (Fig. 4).
Figure 4.
Import of mtTFA into isolated rat liver mitochondria. SDS–PAGE of mitochondrial protein from isolated rat liver mitochondria. p, precursor; m, mature protein. mtTFA was produced and labeled with [35S]methionine in rabbit reticulocyte lysate. After 1 h incubation with mitochondria, proteinase K and/or Triton X-100 plus proteinase K was added or the uncoupler CCCP was added during incubation, as indicated. In the first lane a 1/5 volume of programmed lysate was loaded.
Stoichiometry between newly imported mtTFA and mtDNA
The concentration of mtTFA in the lysate was 3 × 10–9 M, as determined from the incorporation of [35S]methionine into mtTFA (3580 ± 378 c.p.m./µl lysate, n = 3), the specific activity of [35S]methionine (1175 Ci/mmol), the number of methionine residues in mtTFA (six) and correction for quenching and unlabeled methionine. From this it can be calculated that ∼8 × 1011 molecules of mtTFA were present and could be imported into the matrix in the in organello transcription assay, which typically contained 35 µl of lysate.
By hybridization to a CO III probe of DNA extracted from known amounts of mitochondrial protein together with known amounts of the plasmid pBR322-CO III and by comparison of the densitometric values on an autoradiogram, we estimated that 1 mg of mitochondrial protein, which was used in the experiments, contains ∼6 × 10–13 mol or 3.6 × 1011 molecules of mtDNA. Using a value of 1.1 × 107 Da for the molecular mass of the 16.5 kb mtDNA molecule, this is equivalent to 6.6 µg mtDNA. From the optical density of the extracted DNA at 260 nm, a very similar value (5.9 ± 2.9 µg mtDNA/mg mitochondrial protein, n = 3) was obtained.
In contrast to the experiments designed to show mtTFA import (Fig. 4), which were done at 30°C, almost all of the mtTFA protein synthesized in the lysate was imported into the matrix at 37°C, the conditions under which synthesis of 7S DNA was studied. Thus, it can be estimated that between two and three additional, newly imported mtTFA molecules per endogenous mtDNA molecule were available for binding.
Effect of mtTFA import on 7S DNA synthesis
Mitochondria were incubated in the presence of [α-32P]dTTP and reticulocyte lysate containing mtTFA for various times under identical conditions. Control incubations contained reticulocyte lysate which had been programmed with empty pBS vector alone. Ethidium bromide staining of the gels was used to confirm that the same amount of total nucleic acids had been extracted and loaded onto each lane. Incorporation of radioactivity into 7S DNA was significantly higher after import of mtTFA into the organelles (Fig. 5). This was only observed after incubation periods of <30 min, when equilibrium between [α-32P]dTTP and 7S DNA had not yet been reached (see above). Densitometric evaluation of band intensities from three independent observations showed that incorporation increased by 40–100% after 15 min incubation.
Figure 5.

Effect of imported mtTFA on 7S DNA synthesis in isolated rat liver mitochondria. Autoradiogram of DNA synthesized in isolated rat liver mitochondria and heat denatured for 10 min at 95°C before electrophoresis through a 1% agarose slab gel. Mitochondria were incubated with reticulocyte lysate pre-incubated with pBS-mtTFA (+) or empty pBS vector (–) for the indicated times.
DISCUSSION
In the present work we have obtained evidence that mtTFA, a mitochondrial DNA-binding protein of the HMG class, plays an important role in the synthesis of 7S DNA and, probably, replication initiation. This DNA molecule forms a three-stranded DNA structure located in the D-loop region of mtDNA of vertebrates; its function is largely unknown (1). Its high turnover rate, in contrast to the rate of synthesis of full-length mtDNA, rules out the possibility that it is simply an intermediate of leading strand synthesis during replication. The experiments described in this work show that isolated intact mitochondria from rat liver are able to support efficient synthesis of two species of low molecular weight DNA. Digestion experiments with DNase and hybridization experiments unequivocally confirmed that these two species are 7S DNA. Resistance to digestion with RNase demonstrated that the longer fragment is not the RNA–DNA chimera. Together with recent data on the preference of RNase MRP for cleavage of artificial products of the LSP, we propose that the difference in size is due to two sites of RNA/DNA transition (1,4,17), as was also previously shown in mouse cells (18,19).
In general, the initial steps in DNA replication involve regional duplex melting and primer synthesis and both processes can be accomplished efficiently by the transcription machinery, as exemplified by some well-studied prokaryotic systems, such as the bacterial ColE1 replicon and bacteriophage T7 (17). At the mtDNA OH a mechanism involving the action of a DNA primase is unlikely based on the length, structure and sequence content of the primer RNAs. Because the LSP transcripts extend beyond the OH region and the resulting polycistronic RNA encodes functional products, a processing activity was proposed to generate the mature primer RNA termini for DNA replication (20). Thus it was proposed that initiation of 7S DNA synthesis as well as replication is transcription primed. In order to test the hypothesis of transcription primed 7S DNA synthesis in a system more closely resembling the situation in vivo, we reasoned that increasing the transcription rate from the LSP should increase this process. The essential role of mtTFA in transcription initiation from the displacment loop region promoters of human mtDNA has been well documented in vitro (8). Mice with only one functioning mtTFA allele also confirm the important role of mtTFA in replication (21). Reduced mtDNA copy number and subtle biochemical defects have been detected in these heterozygous animals, while homozygous mutants died during embryogenesis. Southern analysis of these embryos failed to reveal mtDNA, unequivocally demonstrating that mtTFA is essential for the maintenance of mtDNA in vivo. The binding of mtTFA to different portions of the D-loop with different affinities is related to the multiple roles of this factor in mitochondrial transcription and replication. It has been proposed that variations in the level of mtTFA concentration within the mitochondrial matrix may regulate promoter selection such that at low concentrations L-strand transcription would predominate, which is linked to replication, whereas at higher mtTFA levels HSP transcription would also take place (22).
The binding of mtTFA to CSB I probably underlies a further function of this factor. CSB I is universally conserved among vertebrates. Moreover, it contains the transition site between the most abundant RNA primer and the D-loop DNA. Therefore, it is likely that binding of mtTFA to CSB I might have rather a different role with respect to that hypothesized for the other regions of the D-loop. The most likely possibility is that the mtTFA–CSB I complex may be part of a recognition signal for the transition from L-RNA to H-DNA chains (20). Previous work has shown that the single-stranded DNA at CSB I may be associated with the RNA polymerase pause site. The existence of a transcription pause site in CSB I might have implications for the mechanism of replication primer formation. It could be hypothesized that formation of the 3′-end of the longest and presumably more active replication primer (7S RNA), which terminates in CSB I, might take place by transcriptional pausing and would not necessarily require the action of a nuclease.
We have shown previously that increasing matrix levels of mtTFA increases transcription rate from the HSP in isolated rat liver mitochondria (10). In the present work we now show that import of mtTFA into the mitochondrial matrix increases incorporation of radiolabel into 7S DNA at early time points, when the dTTP and 7S DNA pools were not equilibrated, confirming that synthesis of this DNA species, and presumably replication, is indeed transcription primed. It should also be noted that the human protein was used in a rat system. No significant changes in steady-state levels of 7S DNA nor full-length mtDNA was noticed within the observation period.
In this system it was also possible to determine the stoichiometry between mtDNA and newly imported mtTFA. The amount of mitochondria routinely used in the assay (1 mg protein) contains ∼3.6 × 1011 molecules of mtDNA. A value of 5.3 × 1010 rat liver mitochondria/mg protein was obtained previously by direct counting (23) and, using our own estimate of three mtDNA molecules per rat liver mitochondrion (24), a very similar value of 1.6 × 1011 molecules of mtDNA/mg protein was obtained. The number of mtTFA molecules, on the other hand, was determined to be ∼8 × 1011 molecules in the assay system. More than 90% of the protein was sedimented together with mitochondria and under the experimental conditions of the in organello mtDNA synthesis experiment almost all of it was imported. Thus, each copy of mtDNA had been challenged with two to three additional mtTFA molecules. As discussed above, in rat liver mitochondria the majority of mtDNA is occupied by mtTFA at the binding site near the LSP, but only a fraction is occupied at the HSP (25,26). The latter sites are probably those which are readily accessible to the newly imported molecules. Although our in organello system is difficult to compare to run-off experiments, in which only truncated promoter regions have been used lacking additional mtTFA-binding sites, it is striking that the relation between mtTFA levels and transcription rates of mitochondrial promoters is of the same order of magnitude in all systems. In run-off experiments in aqueous buffer solution up to 10-fold molar excesses of mtTFA proteins over naked template DNA stimulated transcription (8,27,28). Obviously, in the condensed mitochondrial matrix of rat liver mitochondria, as shown here, even more subtle increases in mtTFA over the endogenous pool will have a significant effect on transcription from the LSP.
In conclusion, isolated rat liver mitochondria faithfully synthesize and degrade 7S DNA. Increased matrix levels of mtTFA are sufficient to increase its rate of synthesis, strongly supporting the hypothesis that this process is transcription primed in mammalian mitochondria.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr F. Lottspeich (Martinsried, Germany) for measuring the concentration of methionine in the reticulocyte lysate. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Wi 889/3-2 to R.J.W.), by the DAAD (Acciones Integradas to R.J.W., S.G. and J.M.) and by the Spanish Direccion de Ensenanza Superior e Investigacion (PB97-1019 to J.M.).
References
- 1.Shadel G.S. and Clayton,D.A. (1997) Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem., 66, 409–435. [DOI] [PubMed] [Google Scholar]
- 2.Holt I.J., Lorimer,H.E. and Jacobs,H.T. (2000) Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell, 100, 515–524. [DOI] [PubMed] [Google Scholar]
- 3.Kang D.C., Miyako,K., Kai,Y., Irie,T. and Takeshige,K. (1997) In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J. Biol. Chem., 272, 15275–15279. [DOI] [PubMed] [Google Scholar]
- 4.Lee D.Y. and Clayton,D.A. (1998) Initiation of mitochondrial DNA replication by transcription and R-loop processing. J. Biol. Chem., 273, 30614–30621. [DOI] [PubMed] [Google Scholar]
- 5.Larsson N.G., Barsh,G.S. and Clayton,D.A. (1997) Structure and chromosomal localization of the mouse mitochondrial transcription factor A gene (Tfam). Mamm. Genome, 8, 139–140. [DOI] [PubMed] [Google Scholar]
- 6.Fisher R.P. and Clayton,D.A. (1988) Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol., 8, 3496–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parisi M.A. and Clayton,D.A. (1991) Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science, 252, 965–969. [DOI] [PubMed] [Google Scholar]
- 8.Dairaghi D.J., Shadel,G.S. and Clayton,D.A. (1995) Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol., 249, 11–28. [DOI] [PubMed] [Google Scholar]
- 9.Fisher R.P. and Clayton,D.A. (1985) A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J. Biol. Chem., 260, 11330–11338. [PubMed] [Google Scholar]
- 10.Montoya J., Perez,M.A., Garstka,H.L. and Wiesner,R.J. (1997) Regulation of mitochondrial transcription by mitochondrial transcription factor A. Mol. Cell. Biochem., 174, 227–230. [PubMed] [Google Scholar]
- 11.Enriquez J.A., Ramos,J., Perez,M.A., Lopez,P.M. and Montoya,J. (1994) Highly efficient DNA synthesis in isolated mitochondria from rat liver. Nucleic Acids Res., 22, 1861–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waddell W.J. (1956) A simple ultraviolet spectrophotometric method for the determination of protein. J. Lab. Clin. Med., 48, 311–314. [PubMed] [Google Scholar]
- 13.Gadaleta G., Pepe,G., De,C.G., Quagliariello,C., Sbisa,E. and Saccone,C. (1989) The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J. Mol. Evol., 28, 497–516. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Enriquez J.A., Lopez,P.M. and Montoya,J. (1991) Saturation of the processing of newly synthesized rRNA in isolated brain mitochondria. FEBS Lett., 280, 32–36. [DOI] [PubMed] [Google Scholar]
- 16.Ojala D. and Attardi,G. (1978) Precise localization of the origin of replication in a physical map of HeLa cell mitochondrial DNA and isolation of a small fragment that contains it. J. Mol. Biol., 122, 301–319. [DOI] [PubMed] [Google Scholar]
- 17.Clayton D.A. (1982) Replication of animal mitochondrial DNA. Cell, 28, 693–705. [DOI] [PubMed] [Google Scholar]
- 18.Gillum A.M. and Clayton,D.A. (1979) Mechanism of mitochondrial DNA replication in mouse L-cells: RNA priming during the initiation of heavy-strand synthesis. J. Mol. Biol., 135, 353–368. [DOI] [PubMed] [Google Scholar]
- 19.Gillum A.M. and Clayton,D.A. (1978) Displacement-loop replication initiation sequence in animal mitochondrial DNA exists as a family of discrete lengths. Proc. Natl Acad. Sci. USA, 75, 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee D.Y. and Clayton,D.A. (1997) RNase mitochondrial RNA processing correctly cleaves a novel R loop at the mitochondrial DNA leading-strand origin of replication. Genes Dev., 11, 582–592. [DOI] [PubMed] [Google Scholar]
- 21.Larsson N.G., Wang,J., Wilhelmsson,H., Oldfors,A., Rustin,P., Lewandoski,M., Barsh,G.S. and Clayton,D.A. (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genet., 18, 231–236. [DOI] [PubMed] [Google Scholar]
- 22.Dairaghi D.J., Shadel,G.S. and Clayton,D.A. (1995) Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim. Biophys. Acta, 1271, 127–134. [DOI] [PubMed] [Google Scholar]
- 23.Pak Y.K. and Weiner,H. (1990) Import of chemically synthesized signal peptides into rat liver mitochondria. J. Biol. Chem., 265, 14298–14307. [PubMed] [Google Scholar]
- 24.Wiesner R.J., Ruegg,J.C. and Morano,I. (1992) Counting target molecules by exponential polymerase chain reaction: copy number of mitochondrial DNA in rat tissues. Biochem. Biophys. Res. Commun., 183, 553–559. [DOI] [PubMed] [Google Scholar]
- 25.Ghivizzani S.C., Madsen,C.S., Nelen,M.R., Ammini,C.V. and Hauswirth,W.W. (1994) In organello footprint analysis of human mitochondrial DNA: human mitochondrial transcription factor A interactions at the origin of replication. Mol. Cell. Biol., 14, 7717–7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantatore P., Daddabbo,L., Fracasso,F. and Gadaleta,M.N. (1995) Identification by in organello footprinting of protein contact sites and of single-stranded DNA sequences in the regulatory region of rat mitochondrial DNA. Protein binding sites and single-stranded DNA regions in isolated rat liver mitochondria. J. Biol. Chem., 270, 25020–25027. [DOI] [PubMed] [Google Scholar]
- 27.Parisi M.A., Xu,B. and Clayton,D.A. (1993) A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol., 13, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antoshechkin I. and Bogenhagen,D.F. (1995) Distinct roles for two purified factors in transcription of Xenopus mitochondrial DNA. Mol. Cell. Biol., 15, 7032–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]